Abstract
Soil-transmitted nematodes (STNs) infect over a billion people and place several billion more at risk of infection. Hookworm disease is the most significant of these STNs, with over 500 million people infected. Hookworm infection can result in debilitating and sometimes fatal iron-deficiency anemia, which is particularly devastating in children and pregnant women. Currently, hookworms and other soil-transmitted helminths (STHs) are controlled by administration of a single dose of a benzimidazole to targeted populations in endemic areas. While effective, people are quickly re-infected, necessitating frequent treatment. Widespread exposure to anthelmintic drugs can place significant selective pressure on parasitic nematodes to generate resistance, which has severely compromised benzimidazole anthelmintics for control of livestock nematodes in many areas of the world. Here we report, to our knowledge, the first naturally occurring multidrug-resistant strain of the canine hookworm Ancylostoma caninum. We reveal that this isolate is resistant to fenbendazole at the clinical dosage of 50 mg/kg for 3 days. Our data shows that this strain harbors a fixed, single base pair mutation at amino acid 167 of the β-tubulin isotype 1 gene, and by using CRISPR/Cas9 we demonstrate that introduction of this mutation into the corresponding amino acid in the orthologous β-tubulin gene of Caenorhabditis elegans confers a similar level of resistance to thiabendazole. We also show that the isolate is resistant to the macrocyclic lactone anthelmintic ivermectin. Understanding the mechanism of anthelmintic resistance is important for rational design of control strategies to maintain the usefulness of current drugs, and to monitor the emergence of resistance. The isolate we describe represents the first multidrug-resistant strain of A. caninum reported, and our data reveal a resistance marker that can emerge naturally in response to heavy anthelminthic treatment.
Keywords: Anthelmintic resistance, Hookworm, Benzimidazole, Ivermectin, Ancylostoma caninum
1. Introduction
Parasitic nematodes continue to place a tremendous burden on the health and economies of developed and developing countries worldwide. An estimated 1.45 billion people are infected with at least one soil-transmitted nematode (STN) species, and 4 billion people are at risk of infection (de Silva et al., 2003; Pullan et al., 2014). The STNs, which include Ascaris lumbricoides, Trichuris trichiura, and the hookworms Necator americanus, Ancylostoma duodenale and Ancylostoma ceylanicum, cause widespread morbidity, accounting for the loss of approximately 5.2 million disability adjusted life years (DALYs), a burden exceeding that of malaria (IHME, 2013; Hotez et al., 2014; Pullan et al., 2014). Of these infections, hookworms are the most significant. More than 400 million people are infected, and of the approximately 4.98 million years lost to disability (YLD) from STN infections, hookworm accounts for 65% (IHME, 2013; Pullan et al., 2014). Heavy infection results in debilitating and sometimes fatal iron-deficiency anemia, caused by blood lost to feeding adults in the intestine. Hookworm infection is a leading cause of anemia globally (Kassebaum et al., 2014), and is especially devastating in growing children, pregnant women, and the elderly (Bundy et al., 1995; Dreyfuss et al., 2000; Bethony et al., 2002).
Currently, hookworms and other STNs are controlled by treatment with anthelmintic drugs that kill the mature worms in the host. Efforts are underway to control morbidity by mass drug administration (MDA) using the anthelmintics albendazole (ABZ) and mebendazole (MBZ), members of the benzimidazole (BZ) class of drugs, with the goal of eliminating STHs as a public health problem by 2020 (WHO, 2012a, b). While relatively effective, people in endemic regions are rapidly re-infected, necessitating treatment on an annual or biannual schedule. Heavy exposure to BZ anthelmintics has selected populations of livestock parasitic nematodes that are no longer susceptible to the drugs, rendering this class of anthelmintics useless in many areas of the world (Kaplan, 2004; Wolstenholme et al., 2004). While no cases of BZ resistance in hookworms have been confirmed, the rapidity with which anthelmintic resistance (AR) developed to these drugs suggests that increasing the selective pressure on human helminths could quickly generate resistant worm populations as well. As MDA programs are scaled up, detecting emerging AR will become critical to avoid losing the most effective treatments for hookworms (Albonico et al., 2004a).
Developing methods for detecting emerging resistance requires an understanding of the underlying resistance mechanisms. BZ anthelmintics work by binding to β-tubulin and inhibiting microtubule polymerization (Lacey, 1988, 1990). BZ resistance in trichostrongyle nematodes is associated primarily with single nucleotide polymorphisms (SNPs) in the β-tubulin isotype 1 gene (Kwa et al., 1994; Silvestre and Cabaret, 2002; Wolstenholme et al., 2004). While the mechanism of BZ resistance is known in livestock nematodes, the mechanism in hookworms appears to be different, as known mutations do not appear to correlate with worm populations that show apparent resistance or increased allele frequencies (Albonico et al., 2004b; Schwenkenbecher et al., 2007; Diawara et al., 2013a). The absence of a confirmed resistant hookworm population makes investigation of the genetic basis of BZ resistance problematic. Here we report the isolation and characterization of a BZ-resistant strain of the canine hookworm Ancylostoma caninum. The strain is also phenotypically resistant to ivermectin (IVM), and represents, to our knowledge, both the first BZ, as well as the first multidrug-resistant hookworm strain reported.
2. Materials and methods
2.1. Ethics statement
All experiments were conducted in strict accordance with the recommendations of the National Institute of Health (USA) Guide for the Care and Use of Laboratory Animals. The George Washington University (USA) Institutional Animal Care and Use Committee approved the protocols (numbers A313, A270) used in this study.
2.2. Nomenclature
The molecular phylogeny of tubulin genes is currently in flux, and there are several designations for β-tubulin genes in the literature (Demeler et al., 2013; Saunders et al., 2013). We use the nomenclature of Saunders et al. (2013), in which the hookworm β-tubulin isotype 1 gene is referred to as tbb-iso-1, and the isotype 2 gene as tbb-iso-2.
2.3. Parasites
The wild-type laboratory strain WMD (US National Parasite Collection No. 106970) was isolated from a rescued Labrador retriever in 2013 and maintained in the Hawdon laboratory by continuous passage. The exact geographical origin of the strain is unclear, but is thought to be western Maryland or West Virginia, USA. The resistant KGR strain was isolated in 2016 from a retired racing greyhound originally from Florida, USA that had a history of monthly heartworm preventative treatment. The dog presented to a veterinary clinic with a hookworm infection that was refractory to multiple treatments with fenbendazole (Panacur). Infective L3s (iL3s), reared from eggs collected from the feces of the infected dog, were used to infect a hookworm-naïve beagle. All strains were maintained in beagles as described previously (Schad and Page, 1982; Krepp et al., 2011).
2.4. Isolation of hookworm L1
Hookworm eggs were isolated from infected dog feces by a modified salt floatation protocol. Briefly, feces homogenized in water were strained through a tea strainer and then through a Mini-Sieve Microsieve (Bel-Art SP Scienceware, USA) using a #25 and #45 mesh insert. The sieved feces solution was centrifuged at 500 g for 3 min and washed with water twice, then resuspended in 15 ml of saturated NaCl solution. The saturated salt solution was centrifuged at 300 g for 5–10 min, and the top 5 ml of the supernatant collected and washed five times in BU buffer (50 mM Na2HPO4, 22 mM KH2PO4, 70 mM NaCl, pH 6.8) (Hawdon and Schad, 1991) before counting the eggs.
To isolate L1s, eggs were washed by centrifugation in BU buffer containing antibiotics (100 U/mL of penicillin, 100 μg/mL of kanamycin, and 100 μg/mL of ampicillin), then incubated for 16–18 h at 27°C to allow the eggs to hatch. The hatched L1s were centrifuged and washed in BU buffer prior to counting.
2.5. Larval development assay
We used a modified larval development assay (LDA) (Kotze et al., 2005). A culture of Escherichia coli strain OP50 was grown overnight at 37°C in lysogeny broth (LB), and the concentration determined using the “cell cultures” program on a NanoDrop ND-1000 (ThermoFisher Scientific, USA). For each drug concentration and control, triplicate wells of 3.12 × 106 E. coli cells and 100 L1s in 250 μL of BU buffer were added to wells of a 24 well tissue culture plate. Stocks of thiabendazole (TBZ) (10 mg/ml) and IVM (20 mg/ml) (Sigma-Aldrich, St. Louis, MO, USA) were prepared in dimethyl sulfoxide (DMSO), and then diluted to a concentration twice the desired in-well concentration with BU buffer. An equal volume (250 μL) of the diluted drug was added to the appropriate well. Controls without drug contained the same DMSO concentration as the highest DMSO concentration in the drug wells. The plates were incubated at 27°C for 7 days, after which 100 μL of Lugol’s iodine was added to each well, and the number of each larval stage (L1, L2, L3) was counted. The percentage developing across all replicates was determined using the formula L3total / (L1total + L2total + L3total) x 100. To calculate the half maximal inhibitory concentration (IC50), the average percentage of larvae developing to the L3 stage was plotted against the log of the drug concentration, and the curve analyzed with the sigmoidal dose-response (variable slope) function in GraphPad Prism (ver. 6, GraphPad Software San Diego, CA, USA) to generate the IC50 and the Hill slope value, from which the IC95 was calculated (https://www.graphpad.com/quickcalcs/Ecanything1/). Resistance ratios (RRs) at the IC50 and IC95 concentrations were calculated by dividing the KGR IC50 and IC95 values by the corresponding WMD values.
2.6. cDNA synthesis and sequencing of β-tubulin genes
Total RNA was isolated from KGR and WMD iL3s using Trizol (ThermoFisher), DNase treated, and purified using the Purelink RNA kit (Ambion) according to the manufacturer’s instructions. Pooled cDNA was synthesized using a Verso cDNA synthesis kit (Thermo Scientific). For PCR amplification of β-tubulin isotypes 1 and 2, we designed primers to amplify the entire coding sequence using the isotype 1 (ANCCAN_08153) and isotype 2 (ANCCAN_23405) sequences in the draft A. caninum genome assembly (PRJNA72585) available at WormBase ParaSite (Howe et al., 2017). The primers used to amplify tbb-iso-1 were SHH282 (5’-atgcgtgagatcgtgcatgt-3’) and SHH286 (5’- ctactcctcggggtaagcct-3’), and those for tbb-iso-2 were SHH287 (5’- tgcgtgagatcgttcatgttc-3’) and SHH291 (5’- ttactcttccgcgtagg-3’). All PCRs were performed using high fidelity Phusion Hotstart MasterMix (New England Biolabs). PCR products of approximately 1.3 kb of both isotypes 1 and 2 from KGR and WMD were gel purified using Wizard® SV Gel and PCR Clean-Up (Promega), and Sanger sequenced by MCLAB (South San Francisco, CA, USA) using several primers to cover the whole cDNA. The primers used for sequencing tbb-iso-1 were SHH282 (5’-atgcgtgagatcgtgcatgt-3’), SHH283 (5’-gcaaagaggcggaagg-3’), SHH284 (5’-caacagagaacgaagacatg-3’), SHH285 (5’-tttgctccactttcggc-3’), and SHH286 (5’- ctactcctcggggtaagcct-3’). The primers used to sequence tbb-iso-2 were SHH287 (5’- tgcgtgagatcgttcatgttc-3’), SHH288 (5’-gtacgcaaggaagctg-3’), SHH289 (5’-taggcgacggtacaac-3’), SHH290 (5’-gccaggatttgcaccac-3’), SHH291 (5’- ttactcttccgcgtagg-3’), SHH346 (5’-aaagttgtcagggcggaaca-3’), and SHH347 (5’-agatggctgccacgtttgtt-3’). Sequences of isotypes 1 and 2 from WMD and KGR strains were submitted to GenBank with the accession numbers MH253567 through MH253570.
2.7. Allele frequency determination
The frequency of tbb-iso-1 alleles was determined as described previously (Germer et al., 2000). Genomic DNA (gDNA) was isolated from WMD and KGR iL3s using the Wizard Genomic DNA Purification Kit (Promega). Ten nanograms of gDNA were used in separate reactions to amplify the sensitive or resistant allele. To target the SNP in codon 167 of the tbbiso-1 gene, two forward primers were designed such that the 3’ terminal base matched the second base of the wildtype or mutant codon 167. The forward primers were combined with a common reverse primer that binds in an intron in separate PCRs. The sequences of primers were: sensitive (wildtype) forward primer, 5’-gataggatcatgtcctcgtt-3’; resistant (mutant) forward primer, 5’-gataggatcatgtcctcgta-3’; and common reverse primer, 5’-gctggcgccttcgccttttc-3’ (Schwenkenbecher et al., 2007). Genomic DNA was used as template for quantitative PCR using Brilliant II SYBR Green QPCR Master mix (Agilent Technology) on a CFX96 detection system (BioRad) following the manufacturer’s suggested conditions. Allele frequencies were calculated using the formula
where ΔCt = (Ct of allele1-specific PCR) - (Ct of allele2-specific PCR). Ct values were the mean of at least three technical replicates.
2.8. Re-creation of TBZ resistance in C. elegans using CRISPR/Cas9
The F167Y mutation has been associated with BZ resistance in trichostrongyles (Silvestre and Cabaret, 2002), but has never been demonstrated conclusively to confer resistance. To determine if this mutation conferred TBZ resistance, we used homology-directed CRISPR/Cas9 genome editing (Paix et al., 2015) to recapitulate the identical mutation in the orthologous ben-1 gene (C54C6.2) of C. elegans. In this method, ribonucleotide protein (RNP) complexes formed by mixing Cas9 protein, CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), together with a single stranded oligonucleotide (ssODN) that acts as the repair template, were microinjected into the hermaphrodite gonad. The repair template was designed with flanking homologous regions 41 bp upstream and 40 bp downstream of the targeted region. In the repair template, the targeted TTC codon that codes for phenylalanine was changed to TAC, which codes for tyrosine. The PAM site adjacent to the TTC codon was also converted from CGG to CAG to prevent subsequent edits. We employed a co-CRISPR strategy using Celdpy-10 as a marker for edits (Arribere et al., 2014). Together with targeting Cel-ben-1, a crRNA targeting the Cel-dpy-10 gene was introduced to act as a visible marker in the F1 generation. Mutation in the Cel-dpy-10 gene generates a dominant roller phenotype in heterozygous worms, and a dumpy phenotype in homozygous worms. Selection of rollers leads to a higher proportion of the offspring containing the targeted (ben-1) mutation. Wild-type F2 progeny from these rollers were screened for the ben-1 mutation by sequencing the Cel-ben-1 locus.
All CRISPR/Cas9 components were purchased from Dharmacon (Lafayette, CO, USA). The injection mix contained 30 pM Cas9 protein, 320 pM tracrRNA, 160 pM of dpy-10 and ben-1 crRNAs, 38 pM ssODN, and 200 mM KCl. The injection mix was incubated at 37°C for 15 min prior to injection. One or both gonadal arms of 15 young adult C. elegans were injected and subsequently maintained at 20°C. After 72 h, the plates were screened for rollers in the F1 generation. A total of 25 rollers and 10 dumpy worms were observed in the F1 generation. Individual rollers were separated from the wild type animals and allowed to lay eggs on 10 cm nematode growth media (NGM) plates containing 50 μg/ml of TBZ in DMSO. Surviving F2 rollers (180) were cloned to individual 3.5 mm plates containing TBZ, and after F3 progeny were detected on the plate, the F2 parents (96) were frozen in the worm lysis buffer (30 mM Tris pH 8.0, 8 mM EDTA, 100 mM NaCl, 0.7% NP40, and 0.7% Tween 20) with 20 mg/ml of proteinase K. The frozen F2s were lysed at 65°C for 1 h followed by 95°C for 15 min. Five microliters of the lysate were used for a PCR containing OneTaq DNA polymerase (NEB), forward primer Fwd-ACGCGCTGTTCTCGTCG, and reverse primer Rev- CTCGGCGACCGTAAGTGC. The primers amplify a region 800 bp around the targeted mutation. The PCR products were sequenced by Sanger sequencing, and the resulting sequences aligned using CLUSTALW (Thompson et al., 1994).
2.9. Caenorhabditis elegans egg hatch assay
The effect of drug on egg hatch of wild type and edited C. elegans was determined. Stock drug solutions in DMSO were added to NGM to obtain the desired concentrations as follows: TBZ (100 mg/ml of stock) plates containing 0, 10, 15, 20, 30, 40, 60, 80 μg/ml (50–400 μM) and IVM (10 μg/ml) plates containing 0, 0.8, 1, 1.5, 2, 2.5, 3 ng/ml (0.9–3.4 nM). Control plates (without drug) contained the highest concentration of DMSO used for each drug. For the egg hatch assay, a synchronous population of hermaphrodites from wild type and two edited ben-1 lines were bleached to obtain eggs (Stiernagle, 2006). The eggs were plated onto the drug plates, and the hatch frequency determined 24 – 30 h later by counting the number of hatched and unhatched eggs. Larval development assays were conducted similarly, except that the total number of adults and larvae on the plate was counted after 48–53 h. Assays were performed on four different replicate plates, each containing more than 100 eggs. The percentage of eggs that hatched or developed was normalized to the control and plotted against the log10 of the drug concentration. IC50 and IC95 concentrations were determined as described above.
2.10. Caenorhabditis elegans larval development assay
The effect of the drug on the development of wild-type and the edited C. elegans was also determined. Drug stock solutions in DMSO were added to NGM to obtain the desired concentrations as follows: TBZ (100 mg/ml of stock) plates containing 0, 10, 15, 20, 30, 40, 60, μg/ml and IVM (10 μg/ml of stock) plates containing 0, 0.8, 1, 1.5, 2, 2.5, 3, ng/ml. Control plates (without drug) contained the highest concentration of DMSO used for each drug. For the larval development assay, a synchronous population of hermaphrodites from wild-type as well as the two CRISPR edited ben-1 lines were bleached to obtain eggs. The eggs were aseptically transferred to a 15 ml falcon tube in 7 ml of M9 buffer and incubated overnight at 20°C to obtain starved L1 animals (Stiernagle, 2006). The following day, starved L1s were plated on the drug plates. After 48–53 h, the total number of adults and the total number of larvae on the plate were counted. Assays were performed on four replicate plates, each containing at least 100 larvae. The percentage of larvae that developed and became adults was normalized to the control as per the egg hatch assay above and plotting against the log10 of the drug concentration. IC50 and IC95 concentrations were determined as described above.
3. Results
3.1. Isolation and characterization of anthelmintic resistance in KGR strain
The attending veterinarian at the George Washington University animal facility encountered a dog in her clinic in April 2016 with a persistent A. caninum infection. The dog was formerly a racing greyhound from Florida, and had a history of treatment with a monthly heartworm preventative containing a macrocyclic lactone. The veterinarian treated the dog with three separate, 3-day courses of fenbendazole (50 mg/kg), 3–5 weeks apart. Aware of our interest in AR, the veterinarian obtained feces prior to the final treatment. Infective L3s raised from these feces were used to establish the resistant strain, which we named KGR, in a naïve puppy.
To characterize the BZ resistance phenotype of KGR, we first determined the IC50 and IC95 of our wild-type, sensitive strain WMD to TBZ in a LDA. As shown in Table 1, TBZ prevents development of WMD larvae at low concentrations, with an IC50 of 9.1 ng/ml and an IC95 of 12.5 ng/ml. However, the WMD IC50/IC95 concentrations of TBZ had no effect on KGR larval development (Fig. 1). Next, we determined the dose response curves of KGR and WMD to TBZ (Fig. 2A) and used them to calculate the IC50/IC95 and the RR at each concentration. As shown in Table 1, the IC50 of KGR was 18 ng/ml, and the IC95 was 32.3 ng/ml, with IC50 and IC95 RRs of 1.98 and 2.58, respectively.
Table 1.
Half maximal inhibitory concentration (IC50) and IC95 of Ancylostoma caninum WMD and KGR strains to thiabendazole and ivermectin.
| Thiabendazole | Ivermectin | |||||
|---|---|---|---|---|---|---|
| WMD | KGR | RR | WMD | KGR | RR | |
| IC50 (μg/ml) | 0.0091 | 0.018 | 1.98 | 3.34×10−6 | 0.0049 | 1467 |
| IC95 (μg/ml) | 0.0125 | 0.0323 | 2.58 | 22.4×10−6 | 0.0300 | 1339 |
| SE (n) | 0.05 (57) | 0.08 (180) | 0.27 (63) | 0.47 (86) | ||
WMD, susceptible strain; KGR, resistant strain; RR, resistance ratio. SE, standard error of the logIC50 as reported by Graphpad Prism 7. n, average number of larvae per replicate. The IC95 was calculated from the IC50 using the Graphpad calculator (www.graphpad.com/quickcalcs/Ecanything1/).
Fig. 1.
Ancylostoma caninum KGR strain is resistant to thiabendazole. (A) Larvae were incubated at the IC50 concentration of the susceptible strain WMD. (B) Larvae were incubated at the IC95 concentration of WMD. First stage hookworm larvae were incubated in drug or DMSO vehicle alone (control) for 7 days, and the number of larvae that developed to L3 counted. Experiments were performed in triplicate (average number per replicate of 98 for A, and 111 for B). Data were analyzed by one-way ANOVA with multiple comparisons in GraphPad Prism.
Fig. 2.
Dose response curves of Ancylostoma caninum susceptible strain WMD and resistant strain KGR to thiabendazole (TBZ) and ivermectin (IVM). L1s of strains KGR and WMD were incubated with increasing concentrations of drug and the number of larvae that developed to L3s determined after 6–7 days. ((A) WMD n=57, KGR n = 180; (B) WMD n = 63, KGR n= 86). Curves were analyzed with the sigmoidal dose-response (variable slope) function in GraphPad Prism to generate the IC50 and the Hill coefficient (nH) values (TBZ: WMD strain = −7.447, KGR strain = −3.795; IVM: WMD strain = −1.547, KGR strain = −1.627), from which the IC95 was determined.
Considering the history of anthelmintic exposure, we tested whether KGR was resistant to IVM. Using the LDA assay, we constructed a dose-response curve and determined the IC50 and IC95 values and the corresponding RRs for IVM in WMD and KGR larvae. As shown in Fig. 2B and Table 1, KGR strain larvae were resistant to IVM compared with the wild-type WMD larvae, with IC50 and IC95 RRs of 1467 and 1339, respectively. We also tested KGR for resistance to pyrantel pamoate, but found no difference in susceptibility from the wild type WMD strain (not shown). These data indicate that the KGR strain of A. caninum is resistant to members of two of the major classes of anthelmintics.
3.2. Sequencing of β-tubulin genes
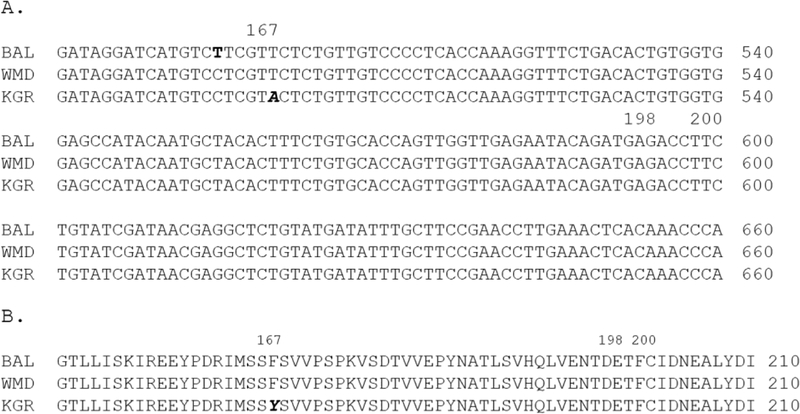
Unlike the C. elegans tub-1 ortholog, ben-1, trichostrongyle nematodes typically have two closely related β-tubulin isotypes, referred to as isotype 1 (tbb-iso-1) and isotype 2 (tbb-iso-2). BZ resistance is most frequently associated with three SNPs in isotype 1 (F200Y, E198L, F167Y) and, rarely, similar mutations in, or complete loss of, isotype 2 (Kwa et al., 1993a,b, 1994; Beech et al., 1994; Elard and Humbert, 1999; Silvestre and Cabaret, 2002; Ghisi et al., 2007; Kotze et al., 2012; Hahnel et al., 2018). To determine if any of these mutations were present in KGR-resistant worms, we amplified and sequenced the tbb-iso-1 and tbb-iso-2 genes from WMD and KGR cDNA and compared them to the A. caninum reference genome (Baltimore strain, BioProject PRJNA72585) sequences. In the tbb-iso-1 genes, we found 12 synonymous mutations, and a single non-synonymous SNP that changed TTC (Phe) to TAC (Tyr) in codon 167 of the KGR strain (Fig. 3). This SNP has been associated with BZ resistance in several trichostrongyles, but not hookworms (Silvestre and Cabaret, 2002; Albonico et al., 2004b; Schwenkenbecher et al., 2007; Vercruysse et al., 2011). No mutations were found at amino acid 198 or 200 of tbb-iso-1 or at any of the corresponding positions in the isotype 2 gene of either strain (not shown).
Fig. 3.
Sequences of β-tubulin isotype 1 cDNAs from three Ancylostoma caninum strains. (A) DNA sequences. The non-synonymous mutation in codon 167 associated with benzimidazole resistance is in bold italics. A synonymous mutation in codon 165 of the BAL sequence is in bold. (B) Protein translation of the relevant region of the β-tubulin isotype 1sequence. The F167Y mutation is in bold italics. BAL, Baltimore strain reference genome (Bioproject PRJNA72585); WMD, thiabendazole (TBZ) -susceptible strain; KGR, TBZ-resistant strain.
3.3. Frequency of the TBZ resistance allele
Although we confirmed the SNP in codon 167 by sequencing the tbb-iso-1 gene amplified from pooled KGR cDNA, the fraction of worms carrying this mutation in the resistant KGR or the sensitive WMD strains of A. caninum was unknown. To determine the resistant allele frequency in each population, we used a sensitive quantitative PCR approach to measure the relative ratio of the sensitive allele verses the resistant (mutant) allele (Germer et al., 2000; Schwenkenbecher et al., 2007; Kotze et al., 2012). This method uses specific primers to amplify the different alleles by quantitative PCR, using equal amounts of pooled gDNA from individual samples as templates. We determined the frequency of the sensitive and resistant alleles in the wild type WMD strain, and in the first four passages of the resistant KGR strain. An example of the quantitative PCR result is shown in Supplementary Fig. S1. As shown in Table 2, the WMD population had a sensitive allele (TTC) frequency in each generation of greater than 99%. In the BZ-resistant KGR worms, however, the frequency of the resistant allele (TAC) was greater than 99% in L3s from each of the first four passages, which were performed without drug selection. This indicates that the resistant allele frequency is fixed at 100% (i.e. homozygous) in the KGR strain.
Table 2.
β-tubulin isotype 1 allele frequency at the 167 loci of susceptible and resistant Ancylostoma caninum as determined by allele-specific quantitative PCR (qPCR).
| gDNA pool | qPCR target | Ct Mean | Ct SD | ΔCt = Ct-sen − Ct-res | Sensitive allele frequency (%) | Resistant allele frequency (%) |
|---|---|---|---|---|---|---|
| WMD (1st) | Susceptible | 24.02 | 0.067 | −14.92 | 99.996 | 0.003 |
| Resistant | 38.94 | 0.000 | ||||
| KGR (1st) | Susceptible | 35.15 | 0.359 | 11.50 | 0.034 | 99.965 |
| Resistant | 23.65 | 0.057 | ||||
| WMD (2nd) | Susceptible | 24.04 | 0.085 | −14.33 | 99.995 | 0.004 |
| Resistant | 38.37 | 0.049 | ||||
| KGR (2nd) | Susceptible | 35.26 | 0.442 | 10.93 | 0.051 | 99.948 |
| Resistant | 24.33 | 0.145 | ||||
| WMD (3rd) | Susceptible | 22.33 | 0.089 | −13.12 | 99.988 | 0.005 |
| Resistant | 35.45 | 0.651 | ||||
| KGR (3rd) | Susceptible | 29.01 | 0.151 | 7.55 | 0.528 | 99.471 |
| Resistant | 21.45 | 0.105 | ||||
| WMD (4th) | Susceptible | 25.04 | 0.035 | −7.79 | 99.549 | 0.450 |
| Resistant | 32.83 | 0.000 | ||||
| KGR (4th) | Susceptible | 29.14 | 1.430 | 7.19 | 0.678 | 99.321 |
| Resistant | 21.95 | 1.420 |
WMD, susceptible strain; KGR, resistant strain.
WMD (1st), pooled F1 L3; KGR (1st), pooled F1 L3; WMD (2nd), pooled F2 L3; KGR (2nd), pooled F2 L3; WMD (3rd), pooled F3 L3; KGR (3rd), pooled F3 L3; WMD (4th), pooled F4 L3; KGR (4th), pooled F4 L3. Allele frequencies were calculated using the formula Freq = 1/(2ΔCt + 1). Cycle threshold (Ct) values are the mean of four technical replicates. ΔCT S.E.M.: WMD ± 1.62; KGR ± 1.11.
3.4. Re-creation of TBZ resistance in C. elegans using CRISPR/Cas9
To confirm that the F167Y mutation was responsible for the TBZ-resistant phenotype in KGR worms, we sought to recreate the same mutation in the orthologous β-tubulin gene in C. elegans using CRISPR/Cas9. We used C. elegans as a surrogate for this experiment because CRISPR technology has yet to be established in hookworms. Twenty-eight mutations confer resistance to BZs in C. elegans, all of which map to the ben-1 gene (Driscoll, 1989). To identify amino acid 167 in Ce-ben-1, we aligned the amino acid sequence with Aca-TBB-iso-1 (Fig. 4A). Our strategy was to use homology directed CRISPR/Cas9 genome editing (Paix et al., 2015) to change the second nucleotide of the ben-1 codon 167 from a thymine to an adenine, thereby changing the amino acid from a phenylalanine to a tyrosine and mimicking the mutation in the KGR tbb-iso-1 gene. We also designed the crRNA to introduce a synonymous mutation in the PAM targeting site to prevent subsequent cutting of the integrated repair template ( Fig. 4B). The gonads of 15 adult C. elegans hermaphrodites were injected with the crRNA, the tracRNA, the ssODN, and Cas9 protein, together with a dpy-10 co-injection marker, and the F1s were screened for roller worms. After selection on TBZ plates, 96 F2s were lysed and sequenced to identify mutations. A total of 22 of the 96 sequences were edited at the Cel-ben-1 locus (Fig. 4C). We selected two different sequence-confirmed lines to test for their response to TBZ. An egg hatch assay confirmed that the CRISPR-induced edit of ben-1 codon 167 conferred TBZ resistance on C. elegans (Fig. 5A). IC50 values obtained in egg hatch assay revealed that both ben-1 edited lines were resistant to TBZ compared with wild-type worms. The IC50 value for wild-type is 13.78 μg/ml, and 26.59 μg/ml and 26.74 μg/ml for the ben-1 mutant lines, resulting in a RR of approximately 2 for both (Table 3).
Fig. 4.
An introduced F167Y mutation imparts thiabendazole resistance on wild type Caenorhabditis elegans. CRISPR/Cas9 was used to introduce the T to A mutation in the corresponding codon of the C. elegans ben-1 gene. (A) Alignment of predicted protein sequences in the region surrounding amino acid 167 in Ancylostoma caninum TBB-iso-1 and Caenorhabditis elegans BEN-1 proteins. Amino acid 167 is in bold, and asterisks indicate identical amino acids. (B) Diagram of the CRISPR/Cas9 strategy. The black arrow indicates the protospacer adjacent motif (PAM) site, and the white arrow indicates the antisense CRISPR RNA. The donor template (ssODN) is shown, with the changes in codon 167 and the PAM site indicated in bold italics. (C) Sequences of the CRISPR/Cas9 targeted region of eight independent lines of C. elegans. The first sequence represents the wild type DNA sequence, and the second sequence is the expected mutation in codon 167 and the targeted PAM site. Codon 167 and the PAM site are overlined. Bases that differ from the wild type are shown in bold, and asterisks indicate bases identical to the wild type ben-1 sequence.
Fig. 5.
Dose response curves of Caenorhabditis elegans wild type (Wt) and ben-1 F167Y edited lines to thiabendazole (TBZ) and ivermectin (IVM). (A) Effect of TBZ on egg hatch of C. elegans wild type and edited lines (n=221). (B) Effect of TBZ on larval development of C. elegans wild type and edited lines (n=332). (C) Effect of IVM on larval development of C. elegans wild type and edited lines (n=106).
Table 3.
Effect of changing amino acid 167 of the ben-1 gene from phenylalanine to tyrosine on resistance to thiabendazole and ivermectin in two lines of Caenorhabditis elegans.
| TBZ (μg/ml) | IVM (ng/ml) | |||||
|---|---|---|---|---|---|---|
| IC50 | SE (n) | RR | IC50 | SE (n) | RR | |
| N2 | 13.8 | 0.017 (332) | -- | 2.07 | 0.5 (106) | -- |
| Line 1 | 27.6 | 0.02 (287) | 2.0 | 1.61 | 0.06 (102) | 0.78 |
| Line 2 | 26.7 | 0.02 (251) | 1.9 | 1.43 | 0.06 (110) | 0.69 |
IC50, half maximal inhibitory concentration; RR, resistance ratio; TBZ, thiabendazole; IVM, ivermectin. SE, standard error of the logIC50 as reported by Graphpad Prism 7. n, average number of larvae per replicate.
We next asked if the F167Y mutation conferred resistance to IVM on the F167Y edited C. elegans lines. We were unable to use an egg hatch assay for IVM, as eggs failed to hatch on any of the tested concentrations. Therefore, we performed LDAs on wild-type and edited worms using the two drugs. The dose response curve indicated that the IC50 of mutant worms to TBZ in the LDA is almost double that of wild-type worms (Fig. 5B). However, there was no increase in the IC50 values of IVM in edited worms compared with wild-type, suggesting that resistance to IVM in KGR is not mediated by the F167Y mutation (Fig. 5C).
4. Discussion
The specter of anthelmintic resistance threatens to render the current treatment for hookworms and other STNs ineffective if current mass deworming practices are accelerated (Geary, 2012; Vercruysse et al., 2011, 2012; Furtado et al., 2016b). There is a critical need for new treatments, as well as for tools to detect emerging resistance alleles before large-scale phenotypic resistance develops. In order to build such tools, underlying mechanisms of resistance must be determined. While resistance has yet to be confirmed in any of the STNs, resistance to all the major anthelmintics has appeared in parasitic nematodes of livestock, and resistant strains of several important species are available (Gilleard, 2013; Gilleard and Redman, 2016; Kotze and Prichard, 2016). These strains have led to a deeper understanding of the mechanism of resistance against BZ anthelmintics and have provided tools to investigate resistance mechanisms of other anthelmintics including the macrocyclic lactone IVM. Until now, no naturally occurring, confirmed BZ- or ML-resistant strain of hookworm has been identified (Kopp et al., 2007), and no resistant laboratory strains have been reported. Here we report the first description of a multidrug-resistant strain of the canine hookworm A. caninum. This strain, named Aca-KGR, contains a fixed mutation associated with BZ resistance in multiple parasitic nematodes of livestock. The strain shows a two-fold increase in the IC50 concentration of the BZ anthelmintic TBZ in a larval development assay. We confirmed that this mutation confers TBZ resistance by using CRISPR/Cas9 to replicate the SNP in the homologous ben-1 gene of C. elegans, and demonstrated a similar doubling of the RR between edited and wild-type C. elegans nematodes. Furthermore, the KGR strain also exhibits profound resistance to the ML anthelmintic IVM, with an IC50 RR of 1467. This strain, isolated from a retired racing greyhound, has likely had extensive exposure to several anthelmintics that selected for mutations conferring resistance to both classes of drug.
It is generally accepted that BZ resistance in veterinary parasites is caused by SNPs found in one of three codons of the β-tubulin isotype 1 gene (Schwenkenbecher et al., 2007). SNPs in codons 200 and 167 cause the substitution of tyrosine for phenylalanine (TTC→TAC), while a SNP in codon 198 substitutes alanine for glutamate (GAG→GCG) (Kwa et al., 1994; Silvestre and Cabaret, 2002; Ghisi et al., 2007; Rufener et al., 2009). Although the 200 and 167 SNPs are more commonly associated with resistance, all three of these SNPs have been found to confer resistance independently (Kwa et al., 1994; Silvestre and Cabaret, 2002; Ghisi et al., 2007; Vercruysse et al., 2011). These amino acid substitutions cause conformational changes in the β-tubulin protein, which inhibits the binding of the BZs (Lacey and Prichard, 1986). However, direct evidence that any of these mutations confer resistance has been lacking. Previously, Kwa et al. (1995) indirectly showed that the F200Y mutation conferred BZ resistance by demonstrating that H. contortus isotype-1 susceptible alleles (containing phenylalanine at amino acid 200) expressed in trans increased susceptibility of resistant C. elegans ben-1 mutants to BZs, but resistant alleles with the tyrosine at amino acid 200 did not. Using CRISPR/Cas9 to edit the C. elegans ben-1 gene, we directly demonstrated that the F167Y mutation also confers resistance. The IC50 RR in the two C. elegans ben-1 edited lines we tested were similar to that of the KGR strain of A. caninum, suggesting that this mutation accounts for the resistant phenotype displayed by KGR worms. Similarly, Hahnel et al. (2018) used CRISPR/Cas9 to introduce the F200Y mutation in the Ce-ben-1 and showed that it conferred BZ resistance. Recently, however, experiments using near isogenic lines (NILs) of C. elegans suggest that additional mutations may contribute to BZ resistance (Zamanian et al., 2018). We therefore cannot rule out other mutations contributing to the BZ resistance. However, given the similar RRs in Aca-KGR worms and C. elegans with the same mutation introduced by gene editing, it is likely that the F167Y mutation accounts for a large part of the resistance seen in Aca-KGR. Furthermore, our results indicate that the F167Y mutation can become fixed in naturally occurring populations of A. caninum under current drug pressure, and thereby impart significant resistance to BZ anthelmintics.
The KGR strain of A. caninum is the first confirmed report of BZ resistance in any hookworm species. Treatment failures of BZs, especially MBZ, against human hookworms were reported (Albonico et al., 2003; Flohr et al., 2007). However, efforts to identify any of the three SNPs associated with BZ resistance in these populations have been unsuccessful (Albonico et al., 2004b; Schwenkenbecher et al., 2007; Diawara et al., 2013a,b). Similarly, low to nonexistent frequencies of BZ resistance alleles have been found in naturally occurring populations of A. caninum. The SNP in amino acid 200 was detected at low (0.8%) frequency, and the amino acid 167 or 198 SNPs were not detected, in hookworms from dogs in Brazil, none of which were phenotypically resistant to BZ (Furtado et al., 2014, 2016a; Furtado and Rabelo, 2015). A survey of A. caninum recovered from dogs in Georgia also failed to detect any of the resistance alleles at significant levels (Schwenkenbecher and Kaplan, 2009). This is probably due to low drug pressure or large effective population sizes. Aca-KGR was isolated from a former racing greyhound that was likely exposed to repeated doses of anthelmintics. Housing conditions and treatment regimens associated with racetracks may exert extreme selective pressure on small effective populations of hookworms, resulting in selection for, and rapid fixation of, resistance alleles.
While the mechanism of resistance to BZ anthelmintics is well known, resistance to ML anthelmintics, including IVM and moxidectin (MOX), is currently unknown. MLs are glutamate-gated chloride channel (GluCl) agonists that irreversibly open the channel, causing hyperpolarization of the cell and flaccid paralysis of the parasite (Cully et al., 1994). The channels are composed of five subunits of multiple types. Simultaneous mutation of three subunit genes confers IVM resistance in C. elegans (Dent et al., 2000), however no convincing association between mutations in these or other candidate genes and ML resistance has been demonstrated (Laing et al., 2017). Changes in members of the P-glycoprotein (Pgp)/multidrug resistance-associated protein (MRP) subfamily of ATP binding cassette (ABC) transporters have been implicated in ML resistance (Lespine et al., 2012; Ardelli, 2013). These proteins are involved with uptake and efflux of xenobiotics including MLs. Changes in Pgp/MRPs could result in decreased uptake or increased efflux of the drug, resulting in reduced access to the target, and consequently resistance. Pgps have been implicated in resistance to IVM and other MLs in H. contortus (Blackhall et al., 1998; Xu et al., 1998; Kerboeuf et al., 2003; Prichard and Roulet, 2007; Lespine et al., 2012), Teladorsagia circumcincta (Dicker et al., 2011), and Onchocerca volvulus (Ardelli, 2013; Ardelli and Prichard, 2007; Bourguinat et al., 2008; Lespine et al., 2012). Transporters have been implicated in mediating multidrug resistance in several parasitic nematodes, including a triple resistant strain of H. contortus (Williamson et al., 2011) and in laboratory-generated multidrug-resistant C. elegans (James and Davey, 2009; Yan et al., 2012). Overexpression of transporters may affect BZ resistance (Beugnet et al., 1997), and ML may select on β-tubulin (Eng et al., 2006; Bourguinat et al., 2007), including codons 167 or 200 in H. contortus (Mottier and Prichard, 2008). Co-selection of BZ-resistant alleles might explain the presence of both BZ and IVM resistance in our KGR strain.
Recently, genetic crosses and genome resequencing experiments have identified a single major IVM resistance locus common to two geographically and genetically different strains of H. contortus (Rezansoff et al., 2016; Doyle et al., 2018. A major locus for ivermectin resistance in a parasitic nematode. bioRxiv, https://www.biorxiv.org/content/10.1101/298901v2). Interestingly, none of the candidate IVM resistance genes maps to this locus, suggesting yet another potential mechanism of IVM resistance. Our newly isolated multidrug-resistant A. caninum provides another tool to investigate and clarify the mechanisms of IVM and BZ resistance in a parasitic nematode.
In summary, we have identified a naturally occurring, multidrug-resistant strain of the canine hookworm A. caninum. The strain, Aca-KGR, has a fixed SNP in amino acid codon 167 of the β-tubulin isotype 1 gene that results in a change from a phenylalanine to a tyrosine, a mutation long associated with BZ resistance in trichostrongyles. We confirmed its role in BZ resistance by using CRISPR/Cas9 to introduce the same SNP in the corresponding amino acid at codon 167 of the C. elegans tbb-iso1 ortholog, ben-1, which conferred a similar level of BZ resistance on the edited nematodes. Furthermore, the KGR strain also shows profound resistance to the ML anthelmintic IVM. This strain represents the first multidrug-resistant strain of A. caninum reported, and indicates that under the proper conditions, resistance to commonly used anthelmintics can develop in natural populations of hookworms, a potential concern for current and future mass deworming campaigns.
Supplementary Material
Supplementary Fig. S1. Representative real time quantitative PCR amplification curves of the kinetic PCRs for allele frequency determination in Ancylostoma caninum. (A) Susceptible strain WMD reaction. (B) Resistant strain (F1) KGR reaction. DNA (10 ng) isolated from pooled L3s from each strain was amplified with primers designed to detect sensitive alleles (blue) or resistant alleles (red). Threshold cycle (CT) values from the curves were used to calculated allele frequency as described in Section 2.7of the main text. RFU, relative fluorescence units.
Highlights.
Ancylostoma caninum strain KGR is phenotypically resistant to thiabendazole
Aca-KGR has a fixed, non-synonymous single nucleotide polymorphism in β-tubulin isotype 1 codon 167
CRISPR/Cas9 editing of the orthologous Caenorhabditis elegans gene confers resistance
Aca-KGR is also phenotypically resistant to ivermectin
Acknowledgements
The authors would like to thank Melissa Keaney and Tamara Outler for parasite maintenance, and Kayleen Gloor for veterinary contributions. This work was supported by the National Institutes of Health, USA, grant 1R21AI115012. The sponsors had no role in the study design, collection, analysis, or interpretation of data, writing the manuscript, or the decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: Nucleotide sequence data reported in this paper are available in the GenBank™, EMBL and DDBJ databases under the accession numbers MH253567 - MH253570.
References
- Albonico M, Bickle Q, Ramsan M, Montresor A, Savioli L, Taylor M, 2003. Efficacy of mebendazole and levamisole alone or in combination against intestinal nematode infections after repeated targeted mebendazole treatment in Zanzibar. Bull World Health Organ 81, 343–352. [PMC free article] [PubMed] [Google Scholar]
- Albonico M, Engels D, Savioli L, 2004a. Monitoring drug efficacy and early detection of drug resistance in human soil-transmitted nematodes: a pressing public health agenda for helminth control. Inter J Parasitol 34, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Albonico M, Wright V, Bickle Q, 2004b. Molecular analysis of the ß-tubilin gene of human hookworms as a basis for possible benzimidazole resistance on Pemba Island. Mol Biochem Parasitol 134, 1–4. [DOI] [PubMed] [Google Scholar]
- Ardelli BF, 2013. Transport proteins of the ABC systems superfamily and their role in drug action and resistance in nematodes. Parasitol Int 62, 639–646. [DOI] [PubMed] [Google Scholar]
- Ardelli BF, Prichard RK, 2007. Reduced genetic variation of an Onchocerca volvulus ABC transporter gene following treatment with ivermectin. Trans R Soc Trop Med Hyg 101, 1223–1232. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ, 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech RN, Prichard RK, Scott ME, 1994. Genetic variability of the beta-tubulin genes in benzimidazole-susceptible and -resistant strains of Haemonchus contortus. Genetics 138, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethony J, Chen J, Lin S, Xiao S, Zhan B, Li S, Xue H, Xing F, Humphries D, Yan W, Chen G, Foster V, Hawdon JM, Hotez PJ, 2002. Emerging patterns of hookworm infection: influence of aging on the intensity of Necator infection in Hainan Province, People’s Republic of China. Clin Infect Dis 35, 1336–1344. [DOI] [PubMed] [Google Scholar]
- Beugnet F, Gauthey M, Kerboeuf D, 1997. Partial in vitro reversal of benzimidazole resistance by the free-living stages of Haemonchus contortus with verapamil. Vet Rec 141, 575–576. [DOI] [PubMed] [Google Scholar]
- Blackhall WJ, Liu HY, Xu M, Prichard RK, Beech RN, 1998. Selection at a P-glycoprotein gene in ivermectin- and moxidectin-selected strains of Haemonchus contortus. Mol Biochem Parasitol 95, 193–201. [DOI] [PubMed] [Google Scholar]
- Bourguinat C, Ardelli BF, Pion SD, Kamgno J, Gardon J, Duke BO, Boussinesq M, Prichard RK, 2008. P-glycoprotein-like protein, a possible genetic marker for ivermectin resistance selection in Onchocerca volvulus. Mol Biochem Parasitol 158, 101–111. [DOI] [PubMed] [Google Scholar]
- Bourguinat C, Pion SD, Kamgno J, Gardon J, Duke BO, Boussinesq M, Prichard RK, 2007. Genetic selection of low fertile Onchocerca volvulus by ivermectin treatment. PLoS Negl Trop Dis 1, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy DAP, Chan MS, Savioli L, 1995. Hookworm infection in pregnancy. Trans. Royal Soc. Trop. Med. Hyg 89, 521–522. [DOI] [PubMed] [Google Scholar]
- Cully DF, Vassilatis DK, Liu KK, Paress PS, Van der Ploeg LH, Schaeffer JM, Arena JP, 1994. Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans. Nature 371, 707–711. [DOI] [PubMed] [Google Scholar]
- de Silva NR, Brooker S, Hotez PJ, Montresor A, Engels D, Savioli L, 2003. Soil-transmitted helminth infections: updating the global picture. Trends Parasitol 12, 547–551. [DOI] [PubMed] [Google Scholar]
- Demeler J, Krüger N, Krücken J, von der Heyden VC, Ramünke S, Küttler U, Miltsch S, López Cepeda M, Knox M, Vercruysse J, Geldhof P, Harder A, von Samson-Himmelstjerna G, 2013. Phylogenetic characterization of β-tubulins and development of pyrosequencing assays for benzimidazole resistance in cattle nematodes. PLOS ONE 8, e70212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent JA, Smith MM, Vassilatis DK, Avery L, 2000. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc Natl Acad Sci U S A 97, 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diawara A, Halpenny CM, Churcher TS, Mwandawiro C, Kihara J, Kaplan RM, Streit TG, Idaghdour Y, Scott ME, Basanez MG, Prichard RK, 2013a. Association between response to albendazole treatment and beta-tubulin genotype frequencies in soil-transmitted helminths. PLoS Negl Trop Dis 7, e2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diawara A, Schwenkenbecher JM, Kaplan RM, Prichard RK, 2013b. Molecular and biological diagnostic tests for monitoring benzimidazole resistance in human soil-transmitted helminths. Am J Trop Med Hyg 88, 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicker AJ, Nisbet AJ, Skuce PJ, 2011. Gene expression changes in a P-glycoprotein (Tcipgp-9) putatively associated with ivermectin resistance in Teladorsagia circumcincta. Int J Parasitol 41, 935–942. [DOI] [PubMed] [Google Scholar]
- Dreyfuss ML, Stoltzfus RJ, Shrestha JB, Pradhan EK, LeClerq SC, Khatry SK, Shrestha SR, Katz J, Albonico M, West KP Jr., 2000. Hookworms, malaria and vitamin A deficiency contribute to anemia and iron deficiency among pregnant women in the plains of Nepal. J Nutr 130, 2527–2536. [DOI] [PubMed] [Google Scholar]
- Elard L, Humbert JF, 1999. Importance of the mutation of amino acid 200 of the isotype 1 beta-tubulin gene in the benzimidazole resistance of the small-ruminant parasite Teladorsagia circumcincta. Parasitol Res 85, 452–456. [DOI] [PubMed] [Google Scholar]
- Eng JK, Blackhall WJ, Osei-Atweneboana MY, Bourguinat C, Galazzo D, Beech RN, Unnasch TR, Awadzi K, Lubega GW, Prichard RK, 2006. Ivermectin selection on β-tubulin: evidence in Onchocerca volvulus and Haemonchus contortus. Mol Biochem Parasitol 150, 229–235. [DOI] [PubMed] [Google Scholar]
- Flohr C, Tuyen LN, Lewis S, Minh TT, Campbell J, Britton J, Williams H, Hien TT, Farrar J, Quinnell RJ, 2007. Low efficacy of mebendazole against hookworm in Vietnam: two randomized controlled trials. Am J Trop Med Hyg 76, 732–736. [PubMed] [Google Scholar]
- Furtado LF, Bello AC, dos Santos HA, Carvalho MR, Rabelo EM, 2014. First identification of the F200Y SNP in the beta-tubulin gene linked to benzimidazole resistance in Ancylostoma caninum. Vet Parasitol 206, 313–316. [DOI] [PubMed] [Google Scholar]
- Furtado LF, Rabelo EM, 2015. Molecular analysis of the F167Y SNP in the beta-tubulin gene by screening genotypes of two Ancylostoma caninum populations. Vet Parasitol 210, 114–117. [DOI] [PubMed] [Google Scholar]
- Furtado LFV, Alves WP, Moreira TB, Costa Junior LM, Miranda RRC, Rabelo EML, 2016a. Standardization and application of the tetraprimer ARMS-PCR technique for screening of the E198A SNP in the beta-tubulin gene of hookworm populations in Brazil. Vet Parasitol 224, 65–67. [DOI] [PubMed] [Google Scholar]
- Furtado LFV, de Paiva Bello ACP, Rabelo EML, 2016b. Benzimidazole resistance in helminths: From problem to diagnosis. Acta Trop 162, 95–102. [DOI] [PubMed] [Google Scholar]
- Geary TG, 2012. Are new anthelmintics needed to eliminate human helminthiases? Curr Opin Infect Dis 25, 709–717. [DOI] [PubMed] [Google Scholar]
- Germer S, Holland MJ, Higuchi R, 2000. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res 10, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisi M, Kaminsky R, Maser P, 2007. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet Parasitol 144, 313–320. [DOI] [PubMed] [Google Scholar]
- Gilleard JS, 2013. Haemonchus contortus as a paradigm and model to study anthelmintic drug resistance. Parasitology 140, 1506–1522. [DOI] [PubMed] [Google Scholar]
- Gilleard JS, Redman E, 2016. Genetic Diversity and Population Structure of Haemonchus contortus. Adv Parasitol 93, 31–68. [DOI] [PubMed] [Google Scholar]
- Hahnel SR, Zdraljevic S, Rodriguez BC, Zhao Y, McGrath PT, Andersen EC, 2018. Extreme allelic heterogeneity at a Caenorhabditis elegans beta-tubulin locus explains natural resistance to benzimidazoles. PLOS Pathog 14, e1007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawdon JM, Schad GA, 1991. Long-term storage of hookworm infective larvae in buffered saline solution maintains larval responsiveness to host signals. J. Helm. Soc. Wash 58, 140–142. [Google Scholar]
- Hotez PJ, Alvarado M, Basanez MG, Bolliger I, Bourne R, Boussinesq M, Brooker SJ, Brown AS, Buckle G, Budke CM, Carabin H, Coffeng LE, Fevre EM, Furst T, Halasa YA, Jasrasaria R, Johns NE, Keiser J, King CH, Lozano R, Murdoch ME, O’Hanlon S, Pion SD, Pullan RL, Ramaiah KD, Roberts T, Shepard DS, Smith JL, Stolk WA, Undurraga EA, Utzinger J, Wang M, Murray CJ, Naghavi M, 2014. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS Negl Trop Dis 8, e2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M, 2017. WormBase ParaSite - a comprehensive resource for helminth genomics. Mol Biochem Parasitol 215, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IHME, 2013. The Global Burden of Disease: Generating Evidence, Guiding Policy Institute for Health Metrics and Evaluation, Seattle, WA. [Google Scholar]
- James CE, Davey MW, 2009. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int J Parasitol 39, 213–220. [DOI] [PubMed] [Google Scholar]
- Kaplan RM, 2004. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol 20, 477–481. [DOI] [PubMed] [Google Scholar]
- Kassebaum NJ, Jasrasaria R, Naghavi M, Wulf SK, Johns N, Lozano R, Regan M, Weatherall D, Chou DP, Eisele TP, Flaxman SR, Pullan RL, Brooker SJ, Murray CJ, 2014. A systematic analysis of global anemia burden from 1990 to 2010. Blood 123, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerboeuf D, Blackhall W, Kaminsky R, von Samson-Himmelstjerna G, 2003. P-glycoprotein in helminths: function and perspectives for anthelmintic treatment and reversal of resistance. Inter J Antimicrob Agents 22, 332–346. [DOI] [PubMed] [Google Scholar]
- Kopp SR, Kotze AC, McCarthy JS, Coleman GT, 2007. High-level pyrantel resistance in the hookworm Ancylostoma caninum. Vet Parasitol 143, 299–304. [DOI] [PubMed] [Google Scholar]
- Kotze AC, Coleman GT, Mai A, McCarthy JS, 2005. Field evaluation of anthelmintic drug sensitivity using in vitro egg hatch and larval motility assays with Necator americanus recovered from human clinical isolates. Inter J Parasitol 35, 445–453. [DOI] [PubMed] [Google Scholar]
- Kotze AC, Cowling K, Bagnall NH, Hines BM, Ruffell AP, Hunt PW, Coleman GT, 2012. Relative level of thiabendazole resistance associated with the E198A and F200Y SNPs in larvae of a multi-drug resistant isolate of Haemonchus contortus. Int J Parasitol Drugs Drug Resist 2, 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotze AC, Prichard RK, 2016. Anthelmintic Resistance in Haemonchus contortus: History, Mechanisms and Diagnosis. Adv Parasitol 93, 397–428. [DOI] [PubMed] [Google Scholar]
- Krepp J, Gelmedin V, Hawdon JM, 2011. Characterisation of hookworm heat shock factor binding protein (HSB-1) during heat shock and larval activation. Int J Parasitol 41, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa MS, Kooyman FN, Boersema JH, Roos MH, 1993a. Effect of selection for benzimidazole resistance in Haemonchus contortus on beta-tubulin isotype 1 and isotype 2 genes. Biochem Biophys Res Comm 191, 413–419. [DOI] [PubMed] [Google Scholar]
- Kwa MS, Veenstra JG, Roos MH, 1994. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol Biochem Parasitol 63, 299–303. [DOI] [PubMed] [Google Scholar]
- Kwa MS, Veenstra JG, Van Dijk M, Roos MH, 1995. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J Mol Biol 246, 500–510. [DOI] [PubMed] [Google Scholar]
- Kwa MSG, Veenstra JG, Roos MH, 1993b. Molecular characterization of B-tubulin genes present in benzimidazole-resistant populations of Haemonchus contortus. Mol Biochem Parasitol 60, 133–144. [DOI] [PubMed] [Google Scholar]
- Lacey E, 1988. The role of the cytoskeletal protein, tubulin, in the mode of action and mechanism of drug resistance to benzimidazoles. Inter J Parasitol 18, 885–936. [DOI] [PubMed] [Google Scholar]
- Lacey E, 1990. Mode of action of benzimidazoles. Parasit. Today 6, 112–115. [DOI] [PubMed] [Google Scholar]
- Lacey E, Prichard RK, 1986. Interactions of benzimidazoles (BZ) with tubulin from BZ-sensitive and BZ-resistant isolates of Haemonchus contortus. Mol Biochem Parasitol 19, 171–181. [DOI] [PubMed] [Google Scholar]
- Laing R, Gillan V, Devaney E, 2017. Ivermectin - Old Drug, New Tricks? Trends Parasitol 33, 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespine A, Menez C, Bourguinat C, Prichard RK, 2012. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: Prospects for reversing transport-dependent anthelmintic resistance. Int J Parasitol Drugs Drug Resist 2, 58–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier M, Prichard RK, 2008. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenetics and genomics 18, 129–140. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G, 2015. High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard RK, Roulet A, 2007. ABC transporters and beta-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology 134, 1123–1132. [DOI] [PubMed] [Google Scholar]
- Pullan RL, Smith JL, Jasrasaria R, Brooker SJ, 2014. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezansoff AM, Laing R, Gilleard JS, 2016. Evidence from two independent backcross experiments supports genetic linkage of microsatellite Hcms8a20, but not other candidate loci, to a major ivermectin resistance locus in Haemonchus contortus. Int J Parasitol 46, 653–661. [DOI] [PubMed] [Google Scholar]
- Rufener L, Kaminsky R, Maser P, 2009. In vitro selection of Haemonchus contortus for benzimidazole resistance reveals a mutation at amino acid 198 of beta-tubulin. Mol Biochem Parasitol 168, 120–122. [DOI] [PubMed] [Google Scholar]
- Saunders GI, Wasmuth JD, Beech R, Laing R, Hunt M, Naghra H, Cotton JA, Berriman M, Britton C, Gilleard JS, 2013. Characterization and comparative analysis of the complete Haemonchus contortus beta-tubulin gene family and implications for benzimidazole resistance in strongylid nematodes. Int J Parasitol 43, 465–475. [DOI] [PubMed] [Google Scholar]
- Schad GA, Page MR, 1982. Ancylostoma caninum : adult worm removal, corticosteroid treatment, and resumed development of arrested larvae in dogs. Exp. Parasitol 54, 303–309. [DOI] [PubMed] [Google Scholar]
- Schwenkenbecher JM, Albonico M, Bickle Q, Kaplan RM, 2007. Characterization of beta-tubulin genes in hookworms and investigation of resistance-associated mutations using real-time PCR. Mol Biochem Parasitol 156, 167–174. [DOI] [PubMed] [Google Scholar]
- Schwenkenbecher JM, Kaplan RM, 2009. Real-time PCR assays for monitoring benzimidazole resistance associated mutations in Ancylostoma caninum. Exp Parasitol 122, 6–10. [DOI] [PubMed] [Google Scholar]
- Silvestre A, Cabaret J, 2002. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol Biochem Parasitol 120, 297–300. [DOI] [PubMed] [Google Scholar]
- Stiernagle T, 2006. Maintenance of C. elegans, in: Community, T.C.e.R. (Ed.), WormBook, pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ, 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J, Albonico M, Behnke JM, Kotze AC, Prichard RK, McCarthy JS, Montresor A, Levecke B, 2011. Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? Int J Parasitol Drugs Drug Resist 1, 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J, Levecke B, Prichard R, 2012. Human soil-transmitted helminths: implications of mass drug administration. Curr Opin Infect Dis 25, 703–708. [DOI] [PubMed] [Google Scholar]
- WHO, 2012a. Accelerating work to overcome the global impact of neglected tropical diseases - a roadmap for implementation World Health Organization, Geneva. [Google Scholar]
- WHO, 2012b. Soil-transmitted helminthiases: Eliminating Soil-transmitted helminthiases as a public health problem in children: progress report 2001–2010 and strategic plan 2011–2020 World Health Organization, Geneva. [Google Scholar]
- Williamson SM, Storey B, Howell S, Harper KM, Kaplan RM, Wolstenholme AJ, 2011. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol Biochem Parasitol 180, 99–105. [DOI] [PubMed] [Google Scholar]
- Wolstenholme AJ, Fairweather I, Prichard R, von Samson-Himmelstjerna G, Sangster NC, 2004. Drug resistance in veterinary helminths. Trends Parasitol 20, 469–476. [DOI] [PubMed] [Google Scholar]
- Xu M, Molento M, Blackhall W, Ribeiro P, Beech R, Prichard R, 1998. Ivermectin resistance in nematodes may be caused by alteration of P-glycoprotein homolog. Mol Biochem Parasitol 91, 327–335. [DOI] [PubMed] [Google Scholar]
- Yan R, Urdaneta-Marquez L, Keller K, James CE, Davey MW, Prichard RK, 2012. The role of several ABC transporter genes in ivermectin resistance in Caenorhabditis elegans. Vet Parasitol 190, 519–529. [DOI] [PubMed] [Google Scholar]
- Zamanian M, Cook DE, Zdraljevic S, Brady SC, Lee D, Lee J, Andersen EC, 2018. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl Trop Dis 12, e0006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Representative real time quantitative PCR amplification curves of the kinetic PCRs for allele frequency determination in Ancylostoma caninum. (A) Susceptible strain WMD reaction. (B) Resistant strain (F1) KGR reaction. DNA (10 ng) isolated from pooled L3s from each strain was amplified with primers designed to detect sensitive alleles (blue) or resistant alleles (red). Threshold cycle (CT) values from the curves were used to calculated allele frequency as described in Section 2.7of the main text. RFU, relative fluorescence units.