Abstract
Context-dependent signaling, as a ubiquitous phenomenon in nature, is a dynamic molecular process at nano- and microscales, but how to mimic its essence using non-covalent synthesis in cellular environment has yet to be developed. Here we show a dynamic continuum of non-covalent filaments formed by instructed-assembly (iA) of a supramolecular phosphoglycopeptide (sPGP) as context-dependent signals for controlling death and morphogenesis of cells. Specifically, while enzymes (i.e., ectophosphatases) on cancer cells catalyze the formation of the filaments of the sPGP to result in cell death, damping the enzyme activity induces 3D cell spheroids. Similarly, relying on the ratio of stromal and cancer cells in a co-culture to modulate the expression of the ectophosphatase, the iA process enables cell spheroids. The spheroids act as a mimic of tumor microenvironment for drug screening. As the first demonstration of iA as multifunctional processes according to local enzyme activity for controlling the behavior of same cells, this work illustrates context-dependent biological functions of non-covalent synthesis in cellular environment.
Keywords: self-assembly, context-dependent, enzyme, cell spheroid, extracellular matrix
Graphical Abstract
Bio-functional phosphoglycopeptide formed by non-covalent synthesis to control cell fate and morphogenesis in a context-dependent manner.
This work reports the use of instructed-assembly[1] as context-dependent, active, and multifunctional signals for controlling cell behavior. Being ubiquitous in nature, context-dependent signaling[2] plays major roles in many important cellular functions. For example, TNFR1, being activated by membrane-bound TNFα, induces cell death, but, interacting with secreted TNF-α, it leads prosurvival NF-κB signaling.[3] With advances on the molecular details of other TNF family ligands and receptors, now it is suggested that all of the death receptors exhibit such a context-dependent feature.[4] That is, the biochemical and biophysical nature of the interactions between these receptors and their extracellular ligands (or intracellular signaling partners) determines cell fate. In fact, many signaling pathways are context-dependent, such as TGF-β,[5] IKK-β,[6] Notch,[7] and Wnt-β-catenin[8] pathways, just to name a few. Despite the large diversity and daunting complexity of these context-dependent processes, they share several key common features: spatiotemporal regulated phosphorylation/dephosphorylation;[9] higher-order organizations;[10] adaptive conformational transitions (or protein dynamics). Particularly, the last two features have emerged as new insights in cell signaling processes.[10–11] In addition to advancing knowledge, these new insights of cell biology also provide guiding principles for developing synthetic molecules or materials that mimic the unique features of biological processes. For example, the understanding of self-organization of tubulins[12] has led to the development of active matters.[13] However, how to use non-covalent synthesis for mimicking the essence of context-dependent signaling remains less developed,[14] especially in live cells.[15] Our recent study on the dynamic continuum (i.e., dynamics of all higher-order assemblies formed by system components which offer synergistic functions) of supramolecular phosphoglycopeptides (sPGP)[16] implies that instructed-assembly (iA) likely would act as context-dependent signals in cellular environment.
Instructed-assembly refers to forming ordered superstructures of molecules as the consequence of a reaction, especially enzymatic reactions.[1] Like self-assembly, iA relies on non-covalent interactions to form large structures from small, simple building blocks; unlike self-assembly, iA includes a process away from equilibrium (ΔG ≠ 0), that is, the enzymatic reaction(ΔG < 0), and is inherently irreversible. In addition, the resulting structures of iA highly depend on the path, that is, the history of molecular transformation.[1, 17] More importantly, the intermediate status of instructed-assembly, which couples with reaction-diffusion processes,[18] plays key roles for determining the functions of resulted assemblies. Although this concept appears obvious, it remains a challenge to use non-covalent synthesis to form bioactive assemblies that exhibit multiple functions in a context-dependent manner.
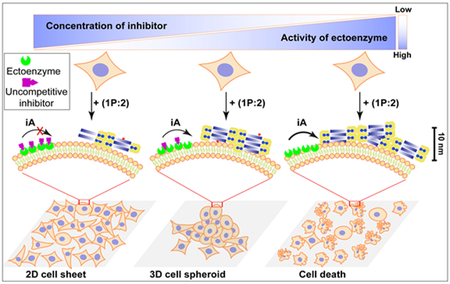
Here we show a dynamic continuum[16, 19] of non-covalent filaments formed by iA[1] of a sPGP as context-dependent signaling for controlling cell death and morphogenesis. As shown in Figure 1, while the enzyme (i.e., tissue non-specific alkaline phosphatase (ALPL)[20]) on cancer cells (Saos-2[21]) catalyze the formation of the filaments of the sPGP to result in cell death, damping the enzyme activity with a synthetic inhibitor of the enzyme induces 3D cell spheroids. Controlling the ratio of stromal (HS-5[22]) and cancer (Saos-2) cells in a co-culture also modulates the expression of ALPL, which regulates the iA process to enable cell spheroids. Acting as a mimic of tumor microenvironment, the spheroids are useful for drug screening. Demonstrating iA as the multifaceted signal according to local enzyme activity for controlling cell behavior, this work, for the first time, illustrates the promises of non-covalent synthesis for context-dependent and active biological functions in cellular environment.
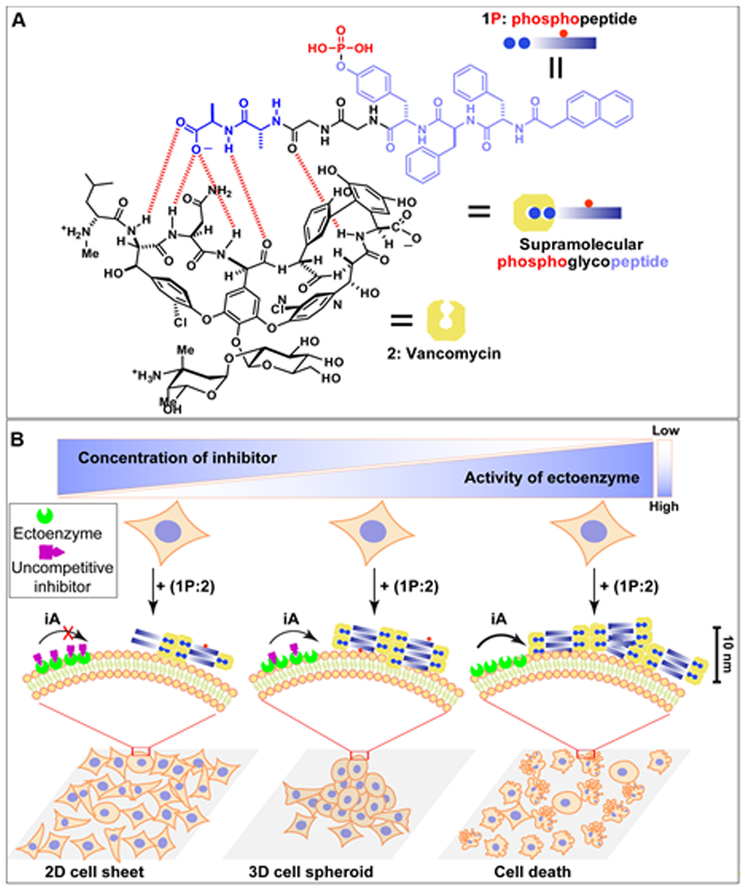
Figure 1. iA as context-dependent signals for cell death and cell morphogenesis.
(A) Molecular structures of NapFFpYGGaa (1P, Nap: naphthylacetic acid; F: L-phenylalanine; pY: L-phosphotyrosine; G: glycine; a: D-alanine), vancomycin (2), and their non-covalent complex 1P:2. (B) Illustration of the context-dependent dynamic continuum of molecular assemblies for controlling cell fate. By controlling the activity of the ectophosphatase (e.g., ALPL) on cancer cells via a corresponding enzyme inhibitor, dynamic continuum of the assemblies of supramolecular phosphoglycopeptides (sPGP) resulting in a 2D cell sheet, 3D cell spheroids, or cell death.
Using our recently developed sPGP (i.e., 1P:2, Figure 1A) as the building blocks,[16] we tested the response of Saos-2 cells to 1P:2. We chose Saos-2 cells and an uncompetitive inhibitor (DQB)[23] to inhibit the activity of the ALPL anchored on the plasma membrane of Saos-2 cell. Live-dead assay (Figure 2A and B) shows red fluorescence after 48 h incubation with 1P:2, indicating that most of the cells are dead. Co-incubating DQB (at a low concentration, 5 μM) with 1P:2 transformed the 2D cell sheet of Saos-2 to 3D cell spheroids, which exhibit green fluorescence, indicating that the cells are alive. Increasing the concentration of DQB results in a well-grown 2D cell sheet of Saos-2 (Figure S1). While incubating 1P:2 (500 μM) induces about 50.0% death of Saos-2 cells, adding DQB (20 μM) rescues the cells (about 90% viability). Either a pan-caspase inhibitor (zVAD-fmk)[24] or a PARP inhibitor (PJ34)[25] recuses cells from exposure to 1P:2 (Figure 2C), indicating that 1P:2 induces the apoptosis of Saos-2 cells. Co-incubation of Saos-2 cells with 1:2 (at the same concentration as of 1P:2) hardly affect the cell viability, indicating that directly using 1:2 is unable to deposit enough 1:2 on cell surface to kill the cells. This result agrees with our previous observations.[26] These results together suggest that iA, which generates the dynamic continuum of non-covalent assemblies (containing 1P:2 and 1:2), controls cell death and morphogenesis depending on the activity of ALPL, that is, in a context-dependent manner.
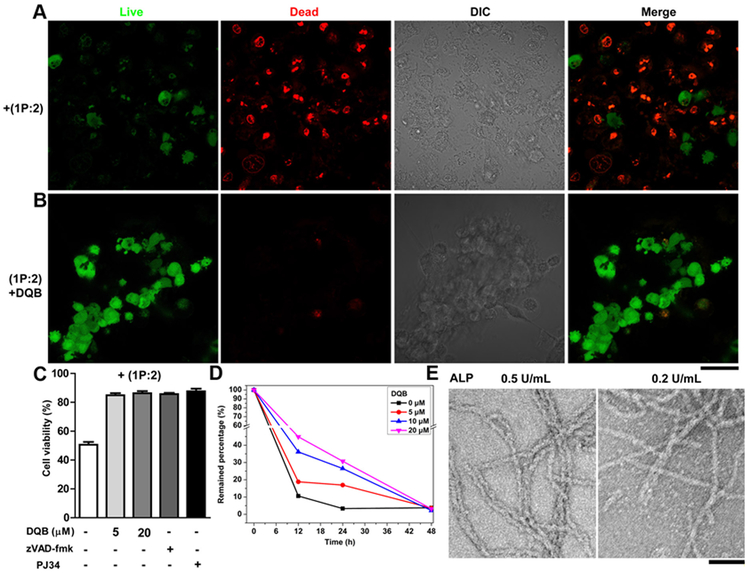
Figure 2. iA of sPGP controls cell fate depending on the activity of enzyme.
Fluorescent images of live-dead assay of Saos-2 cell lines treated with (A) 1P (500 μM) plus vancomycin (2, 500 μM) and (B) 1P, 2, and DQB (5 μM) for 48 h. Green indicates the live cells and red the dead cells. (C) Viability of Saos-2 cells cultured with (1P:2) plus different concentration of DQB, zVAD-fmk (50 μM), or PJ34 (1 μM). (D) Saos-2 cells catalyzed conversion of 1P:2 within 48 h. (E) TEM images of 1P:2 (300 μM) after treated by a phosphatase (ALP, 24 h) at 0.5 U/mL or 0.2 U/mL. Scale bar is 50 nm (in B and E).
To gain more insights into the transformation of 1P:2 to 1:2 on the cells that resulted in different cellular phenotypes, we investigated the enzymatic dephosphorylation rate of 1P:2 without or with the addition of DQB at different concentrations in the presence of Saos-2 cells. Liquid chromatography mass spectrometry (LC-MS) results show that 1P:2 undergoes ALPL catalyzed dephosphorylation at a much faster rate in absence of DQB (with t1/2 = 6.70 h), while the addition of 5, 10, and 20 μM DQB increases the t1/2 to 7.41, 9.37, and 10.9 h, respectively. After 24 h, the percentages of remaining 1P:2 are 3.3%, 16.9%, 26.5%, and 30.7% when the DQB concentrations are 0, 5, 10 and 20 μM, respectively. During this process, the activity of ALPL dictates the conversation of 1P:2 to 1:2, which further controls the cell behaviors. Moreover, co-incubating 1:2 with Saos-2 cells hardly changes the cell morphology and cell viability (Figure S4 and 5). These results suggest that the process of iA and the dynamic continuum of the assemblies rather than the final structures determine the morphogenesis or death of the Saos-2 cells. Transmission electron microscopy (TEM) images show that 1P:2 exhibits different morphologies upon treatment of alkali phosphatase (ALP) at different concentrations: high concentration of ALP results in longer and uniform nanofibers with diameter of 8±2 nm (Figure 2E), while low concentration of ALP generates the mixture of nanofibers (8±2 nm) and nanoparticles (about 2 nm). These results imply that the morphology of 1P:2 controls the morphogenesis and fate of cells, which is useful for designing adaptive materials for controlling cell behaviors.
To directly visualize the distribution of nanostructures formed by 1P:2, we used NBD-1P, a fluorescent analogue of 1P,[16] and 2 to co-incubate with Saos-2 cells. NBD is used because it acts as a sensitive fluorophore for reporting molecular self-assembly in cellular environment.[27] Being incubated with NBD-1P, Saos-2 cells only exhibit weak fluorescence on cell surface (Figure S7). The cells show bright fluorescence after being treated with NBD-1P:2 (Figure 3A), suggesting the non-covalent interactions between NBD-1P and 2 plays key role for generating the nanostructures on cell surface. Unlike the continuous fluorescent nanostructures on cell surface formed by NBD-1P:2 (Video clip S1), fluorescent dots appeared on cell surface when co-incubating NBD-1P:2 with DQB (Figure 3B and C, Video clip S2), Since DQB inhibits ALPL, the dephosphorylation rate of NBD-1P depends on the concentration of DQB. Thus, this result indicates that the rate of dephosphorylation is a key control for iA to act as context-dependent signals. This result agrees with the TEM image that 1P:2 forms nanoparticles and nanofibers at low concentration of ALP, while exhibits uniform nanofibers at higher concentration of ALP.
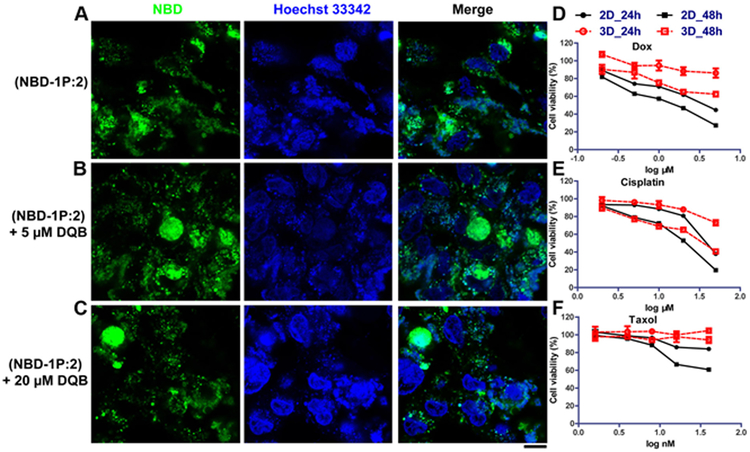
Figure 3. Extracellular iA of sPGP generates a biomimetic 3D model for drug screening.
Confocal images of Saos-2 cell lines treated with (A) NBD-1P plus 2 (300 μM) and (B) NBD-1P, 2 and DQB (5 μM) and (C) NBD-1P, 2 and DQB (20 μM) for 48 h. Scar bar is 10 μm. Cytotoxicity of three representative chemotherapy drugs (D) doxorubicin, (E) cisplatin, and (F) taxol on the 3D cell spheroids and the 2D cell culture.
Recapitulating the complex features of cell processes in tissues, three dimensional (3D) cell models have served as a toxicity and activity assay of lead compounds in drug discovery for decades.[28] Among several osteosarcoma models, hanging drop cultures is the common strategy to induce 3D cancer cell spheroids for drug screening. But it is inefficient and requires long term culture for spheroid formation.[29] Being able to induce cell spheroids rapidly, this dynamic sPGP assemblies should provide an alternative strategy for generating 3D cell models to assessing the efficacy of chemotherapy drug against osteosarcoma cells in vitro.[30] So we next evaluated the responses of Saos-2 cells, in 3D versus 2D, to different drugs. As shown in Figure 3, compared with 2D culture, Saos-2 cells in 3D culture show more resistance to anticancer drugs, such as doxorubicin (Dox), cisplatin, and taxol. Specifically, the IC50 of Dox against Saos-2 cells increases more than 3 times in 3D culture (> 6 μM, 48 h) than 2D culture (1.4 μM) after 48 h incubation. For cisplatin, the IC50 increases by 12 μM in 3D culture (33.3 μM, 48 h). For the Saos-2 cell treated by taxol (40 nM, 48 h), the cell viability increases from 60.84 % to 94.25% when the cells are cultured from as a 2D sheet to the 3D spheroids. Moreover, co-incubating DQB or vancomycin with chemotherapy drugs hardly influence their cytotoxicity against Saos-2 cells, which preclude the possibility of synergistic effect between DQB (or vancomycin) and the drugs.[31] These observations agree with the results obtained by using other 3D osteosarcoma models[30] and in vivo tumor models.[32]
Although different strategies have been developed to produce tumor spheroids of a single cell type, it remains a challenge to mimic tumor microenvironment that consists more than one cell types. As a hallmark of cancer, heterogeneity is an essential property of tumor in vivo. Thus, it is important to develop a 3D co-culture of human tumor cells with different types of stromal cells and other cell types.[28] We next investigated whether iA of sPGP could induce spheroid formation in the co-culture consisting of normal stromal cells (HS-5) and cancer cells (Saos-2). Being treated with 1P:2 for 48 h, the 2D cell sheet of the co-cultured cells transforms to 3D cell spheroids, while the cells without any treatment or treated with one component (1P or 2) remain a 2D cell sheet (Figure S8). Co-incubation of DQB at higher concentration (10 μM, Figure 4B) abolishes the ability of 1P:2 to induce tumor spheroids, agreeing with that controlling activity of ALPL results in different cell phenotypes. Live-dead assay indicates that almost all the cells are live in the cell spheroid (Figure 4C and video clip S3). To visualize the distribution of normal cells and cancer cells within the 3D spheroids, we first labelled Saos-2 cells with a membrane dye[33] (green fluorescence) and HS-5 cells with nucleus dye (red fluorescence), and then co-cultured with 1P:2. As shown in Figure 4D (Video clip S4), Saos-2 cells and HS-5 cells interact with each other inside the cell spheroids. Western blot analysis (Figure 4E) indicates that the expression level of ALPL in co-culture system rises with increasing the percentage of Saos-2 cells in the co-culture of the cells. The ratio of HS-5 and Saos-2 cells in the co-culture controls the activity of ALPL, which further influences the process of iA of 1P:2 for the morphogenesis of the co-cultured cells. We also evaluated the differential drug response of the co-culture cells in 2D versus 3D. Both Dox and cisplatin exhibits dosage dependent cell toxicity in 2D cell sheets within the tested concentrations, while showing little toxicity in 3D cell spheroids. For example, 5 μM of Dox or 50 nM of cisplatin kills about half of cells in 2D cell sheets, but hardly causes cell death in the 3D cell spheroids.
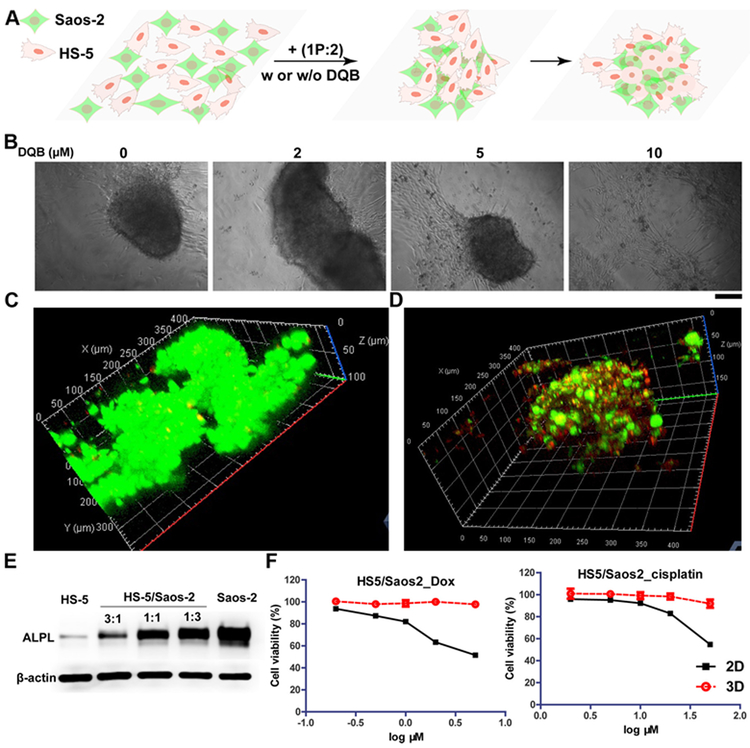
Figure 4. iA of sPGP induces formation of spheroids of heterotypic cells.
(A) The illustration of the formation of 3D cell spheroids made of HS-5 and Saos-2 cells from a 2D cell sheet upon the addition of 1P:2. (B) Microscope images of the co-culture of HS-5 and Saos-2 (ratio is 1:1) cells at the density of 3×104 treated with mixture of 1P:2 (300 μM) with different concentration of DQB for 48 h. Scar bar is 150 μm. (C) CLSM images of live-dead assay of the co-culture of HS-5 and Saos-2 cells treated with mixture of 1P:2 (300 μM) for 48 h. (D) CLSM images of the co-culture of HS-5 and Saos-2 (ratio is 1:1) cells at the density of 3×104 treated with mixture of 1P:2 (300 μM) for 48 h. HS-5 cell lines were treated with Hoechst 33342 (red) for 10 minutes prior to co-cultur with the Saos-2 cell that were treated with membrane probe (green) for 1 h. (E) Expression levels of ALPL on HS-5, Saos-2, and the mixture of HS-5 and Saos-2 at different ratios. The total cell numbers in each group are same. (F) Cytotoxicity of two representative chemotherapy drugs (doxorubicin and cisplatin) on co-cultured 3D spheroids and 2D cell culture for 24 h.
In summary, this work describes that the iA of sPGP serves as a context-dependent signal for controlling the death a and morphogenesis of homotypic or heterotypic cells. Our work, for the first time, employs the process of non-covalent synthesis to mimic the signal transduction in a context-dependent manner, which advances the application of supramolecular chemistry in cell milieu.[34] This work shows that the degree and formation rate of assemblies, as the consequence of context-dependent iA, controls cell behaviors. The processes of iA of sPGP at the cellular level, not only serve as a dynamic molecular process at nano- and microscales,[18] but also highlight the important role of iA for biological function. The use of iA for cell morphogenesis is significant because it provides a practical way to generate cell spheroids, which play increasingly important roles in biology and medicine. Although this work utilizes vancomycin non-covalent peptide complex as the self-assembler, it should be plausible to use simpler peptide motifs (e.g., dipeptides[35]) for achieving iA-based context-dependent signaling. The concept demonstrated in this work may find applications in neural tissues or wounds, which use transient and context-dependent signals for functions.[36]
Supplementary Material
Acknowledgements
This work is partially supported by NIH (CA142746), NSF (DMR-1420382), and W. M. Keck Foundation. ZF is supported by NIH (F99CA234746).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].He H, Xu B, Bull. Chem. Soc. Jpn 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moon RT, Kohn AD, De Ferrari GV, Kaykas A, Nat. Rev. Genet. 2004, 5, 691–701. [DOI] [PubMed] [Google Scholar]
- [3].Grell M, Douni E, Wajant H, Löhden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Cell 1995, 83, 793–802. [DOI] [PubMed] [Google Scholar]
- [4].Nair P, Lu M, Petersen S, Ashkenazi A, Methods Enzymol. 2014, 544, 99–128. [DOI] [PubMed] [Google Scholar]
- [5].Yan X, Xiong X, Chen Y-G, Acta Biochim. Biophys. Sin 2018, 50, 37–50. [DOI] [PubMed] [Google Scholar]
- [6].Page A, Navarro M, Suarez-Cabrera C, Bravo A, Ramirez A, Genes 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nowell CS, Radtke F, Nat. Rev. Cancer 2017, 17, 145–159. [DOI] [PubMed] [Google Scholar]
- [8].Angers S, Moon RT, Nat. Rev. Mol. Cell Biol 2009, 10, 468–477. [DOI] [PubMed] [Google Scholar]
- [9].Day EK, Sosale NG, Lazzara MJ, Curr. Opin. Biotechnol. 2016, 40, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu H, Cell 2013, 153, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Miskei M, Gregus A, Sharma R, Duro N, Zsolyomi F, Fuxreiter M, FEBS Lett. 2017, 591, 2682–2695. [DOI] [PubMed] [Google Scholar]
- [12] a).Borisy G, Taylor E, J. Cell Biol 1967, 34, 535–548; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mitchison T, Kirschner M, nature 1984, 312, 237; [DOI] [PubMed] [Google Scholar]; c) Ndlec F, Surrey T, Maggs AC, Leibler S, Nature 1997, 389, 305. [DOI] [PubMed] [Google Scholar]
- [13] a).Sanchez T, Chen DT, DeCamp SJ, Heymann M, Dogic Z, Nature 2012, 491, 431; [DOI] [PMC free article] [PubMed] [Google Scholar]; b)Kawamura R, Kakugo A, Shikinaka K, Osada Y, Gong JP, Biomacromolecules 2008, 9, 2277–2282. [DOI] [PubMed] [Google Scholar]
- [14] a).Yang ZM, Xu KM, Guo ZF, Guo ZH, Xu B, Adv. Mater 2007, 17, 3152–3156; [Google Scholar]; b) Kuang Y, Shi J, Li J, Yuan D, Alberti KA, Xu Q, Xu B, Angew. Chem., Int. Ed 2014, 53, 8104–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15] a).Feng Z, Zhang T, Wang H, Xu B, Chem. Soc. Rev 2017, 46, 6470–6479; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liang G, Ren H, Rao J, Nat. Chem 2010, 2, 54–60; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tanaka A, Fukuoka Y, Morimoto Y, Honjo T, Koda D, Goto M, Maruyama T, J. Am. Chem. Soc 2015, 137, 770–775; [DOI] [PubMed] [Google Scholar]; d) Zhan J, Cai Y, He S, Wang L, Yang Z, Angew. Chem., Int. Ed 2018, 57, 1813–1816; [DOI] [PubMed] [Google Scholar]; e) Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV, Nat. Nanotechnol 2009, 4, 19. [DOI] [PubMed] [Google Scholar]
- [16].Wang H, Shi J, Feng Z, Zhou R, Wang S, Rodal AA, Xu B, Angew. Chem., Int. Ed 2017, 56, 16297–16301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a).Mattia E, Otto S, Nat. Nanotechnol 2015, 10, 111; [DOI] [PubMed] [Google Scholar]; b) Korevaar PA, George SJ, Markvoort AJ, Smulders MM, Hilbers PA, Schenning AP, De Greef TF, Meijer E, Nature 2012, 481, 492. [DOI] [PubMed] [Google Scholar]
- [18].Epstein IR, Xu B, Nat. Nanotechnol. 2016, 11, 312–319. [DOI] [PubMed] [Google Scholar]
- [19].Wu H, Fuxreiter M, Cell 2016, 165, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Millán JL, Mammalian alkaline phosphatases: from biology to applications in medicine and biotechnology, John Wiley & Sons, 2006. [Google Scholar]
- [21].PAUTKE C, SCHIEKER M, TISCHER T, KOLK A, NETH P, MUTSCHLER W, MILZ S, Anticancer Res. 2004, 24, 3743–3748. [PubMed] [Google Scholar]
- [22].Roecklein BA, Torok-Storb B, Blood 1995, 85, 997–1005. [PubMed] [Google Scholar]
- [23].Dahl R, Sergienko EA, Su Y, Mostofi YS, Yang L, Simao AM, Narisawa S, Brown B, Mangravita-Novo A, Vicchiarelli M, J. Med. Chem 2009, 52, 6919–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huijun Z, MacFARLANE M, NICHOLSON DW, COHEN GM, Biochem. J 1996, 315, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Abdelkarim GE, Gertz K, Harms C, Katchanov J, Dirnagl U, Szabo C, Endres M, Int. J. Mol. Med 2001, 7, 255–260. [PubMed] [Google Scholar]
- [26].Zhou J, Du X, Yamagata N, Xu B, J. Am. Chem. Soc 2016, 138, 3813–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gao Y, Shi J, Yuan D, Xu B, Nat. Commun 2012, 3, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Weeber F, Ooft SN, Dijkstra KK, Voest EE, Cell Chem. Biol 2017, 24, 1092–1100. [DOI] [PubMed] [Google Scholar]
- [29].Fang Y, Eglen RM, SLAS DISCOVERY: Advancing Life Sciences R&D 2017, 22, 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rimann M, Laternser S, Gvozdenovic A, Muff R, Fuchs B, Kelm JM, Graf-Hausner U, J. Biotechnol 2014, 189, 129–135. [DOI] [PubMed] [Google Scholar]
- [31].Lee MJ, Albert SY, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB, Cell 2012, 149, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sodek KL, Ringuette MJ, Brown TJ, Int. J. Cancer 2009, 124, 2060–2070. [DOI] [PubMed] [Google Scholar]
- [33].Wang H, Feng Z, Del Signore SJ, Rodal AA, Xu B, J. Am. Chem. Soc. 2018, 140, 3505–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang H, Feng Z, Xu B, Chem. Soc. Rev 2017, 46, 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Reches M, Gazit E, Science 2003, 300, 625–627. [DOI] [PubMed] [Google Scholar]
- [36] a).Schultz W, Trends Neurosci. 2007, 30, 203–210; [DOI] [PubMed] [Google Scholar]; b) Edlund T, Jessell TM, Cell 1999, 96, 211–224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.