SUMMARY
The ability to distinguish harmful and beneficial microbes is critical for the survival of an organism. Here we show that bloating of the intestinal lumen of Caenorhabditis elegans caused by microbial colonization elicits a microbial aversion behavior. Bloating of the intestinal lumen also activates a broad innate immune response, even in the absence of bacterial pathogens or live bacteria. Neuroendocrine pathway genes are upregulated by intestinal bloating and are required for microbial aversion behavior. We propose that microbial colonization and bloating of the intestine may be perceived as a danger signal that activates an immune fight-and-flight response. These results reveal how inputs from the intestine can aid in the recognition of a broad range of microbes and modulate host behavior via neuroendocrine signaling.
Graphical Abstract
eTOC Blurb
Singh and Aballay describe a mechanism by which a fight-and-flight response against pathogenic microbes is activated. They show that in C. elegans microbial colonization induces bloating of the intestinal lumen, which enhances the expression of innate immune genes and neuroendocrine pathway genes required for the elicitation of a microbial aversion behavior.
INTRODUCTION
Organisms live in complex environments replete with potential dangers. The ability to distinguish potential dangers from useful resources is critical for the survival of an organism. Discrimination of microbial pathogens from commensal organisms by the host is one such example (Blander and Sander, 2012). To effectively counteract microbial pathogens, metazoans have evolved mechanisms to recognize them and deploy counter attacks at molecular and behavioral levels. Potential pathogenic microorganisms are detected by receptors that recognize microbial-associated molecular patterns (MAMPs) (Stuart et al., 2013). In addition, the recognition of endogenous signals, or damage-associated molecular patterns (DAMPs), that result from the cellular damage caused by infections seems to play an essential role in the activation of innate immune responses (Matzinger, 2002). A reliance on specific cellular receptors that are capable of sensing specific microbial patterns limits the range of pathogens that a given host may be able to detect. In contrast, host physiological mechanisms that are capable of sensing alterations caused by infections provide a broad response to microbial pathogens.
Caenorhabditis elegans is a free-living nematode that forages on decomposing organic matter rich in bacteria (Schulenburg and Felix, 2017). Behaviors of C. elegans as diverse as feeding, locomotion, and aerotaxis are affected by exposure to different bacterial species (Bretscher et al., 2011; Sawin et al., 2000; Schulenburg and Félix, 2017). In addition, several pathogens that are present in the natural habitat of C. elegans can impair growth and induce stress (Samuel et al., 2016). Pathogenic bacteria infect and kill C. elegans and elicit a protective avoidance behavior, which is a crucial defense response employed by the animal (Martin et al., 2017; Meisel and Kim, 2014; Pujol et al., 2001; Styer et al., 2008). Bacterial metabolites, such as serrawettin W2 from Serratia marcescens, have been implicated in elicitation of the pathogen avoidance behavior (Pradel et al., 2007). Similarly, the secondary metabolites phenazine-1-carboxamide and pyochelin from Pseudomonas aeruginosa have been shown to activate neuroendocrine signaling, potentially leading to elicitation of the avoidance behavior (Meisel et al., 2014).
In this study, we determined that different bacterial pathogens elicit different patterns of avoidance in C. elegans. We found that while certain pathogens elicited a rapid avoidance behavior that could be observed within 5 minutes of exposure, other pathogens induced a late avoidance behavior that was only evident after 12 hours of exposure. This late avoidance behavior was induced by bacterial colonization and bloating of the intestinal lumen of C. elegans, which also led to activation of the expression of innate immune genes. The increased aversion required a neuroendocrine signaling that was activated by bloating of the intestinal lumen. We propose that the bloating induced fight-and-flight response (referred to as BIFF response) is a general mechanism used by C. elegans to activate immune pathways and elicit microbial aversion.
RESULTS
P. aeruginosa Colonization of the Intestine Induces Pathogen Avoidance
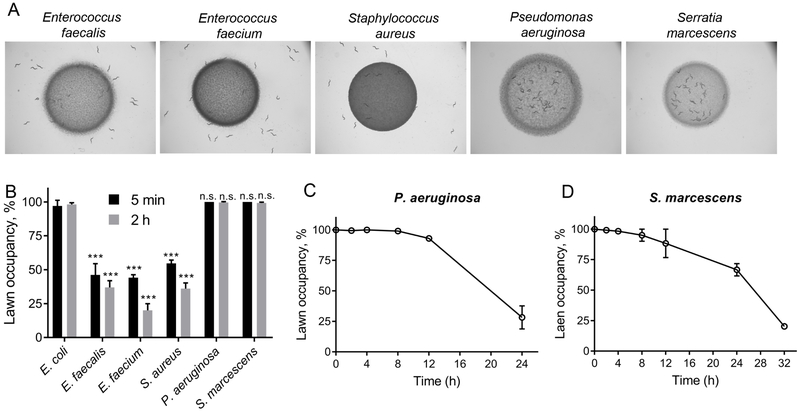
Because behavioral pathogen avoidance is a crucial defense response employed by C. elegans and significantly improves the survival of animals (Martin et al., 2017; Meisel and Kim, 2014; Reddy et al., 2009; Styer et al., 2008), we focused on elucidating the mechanism by which this behavior is elicited. We exposed the animals to lawns of different bacterial pathogens, including Enterococcus faecalis, Enterococcus faecium, Staphylococcus aureus, P. aeruginosa, and S. marcescens. While the animals were initially attracted towards the lawns of the Gram-negative pathogens P. aeruginosa and S. marcescens, they avoided the lawns of the Gram-positive pathogens E. faecalis, E. faecium, and S. aureus (Figures 1A and 1B). Colonization of the intestine by these pathogens is only observed after several hours of exposure (Garsin et al., 2001; Irazoqui et al., 2010; Yuen and Ausubel, 2018). Thus, the rapid avoidance phenotype that is observed after 5 minutes of pathogen exposure seems to be independent of microbial colonization of the intestine. The animals continued to avoid the lawns of E. faecalis, E. faecium, and S. aureus after longer durations of incubation (Figure S1A), indicating that C. elegans actively avoided these bacteria throughout the course of the infection. In contrast to its rapid avoidance of the studied Gram-positive pathogens, C. elegans only began to avoid the lawns of P. aeruginosa and S. marcescens after 12 hours of incubation (Figures 1C and 1D), as shown in previous studies (Pujol et al., 2001; Sun et al., 2011). These results suggest that while pathogen avoidance directed towards E. faecalis, E. faecium, and S. aureus is rapidity elicited, avoidance of P. aeruginosa and S. marcescens develops after a period of interaction between the host and these pathogens.
Figure 1. Microbial Pathogens Elicit Different Patterns of Avoidance in C. elegans.
(A) Distribution of wild-type N2 C. elegans on lawns of different pathogenic bacteria. Representative images are shown at 2 hours after transfer of C. elegans to the bacterial lawns.
(B) Occupancy of different bacterial lawns by N2 animals at 5 minutes and 2 hours after transfer to the lawns. p-values for E. faecalis, E. faecium, S. aureus, P. aeruginosa, and S. marcescens are relative to E. coli for the corresponding time points. ***p < 0.001 via the t-test. n.s., non-significant.
(C)-(D) Occupancy of P. aeruginosa (C) and Serratia marcescens (D) lawns by C. elegans over time.
See also Figure S1.
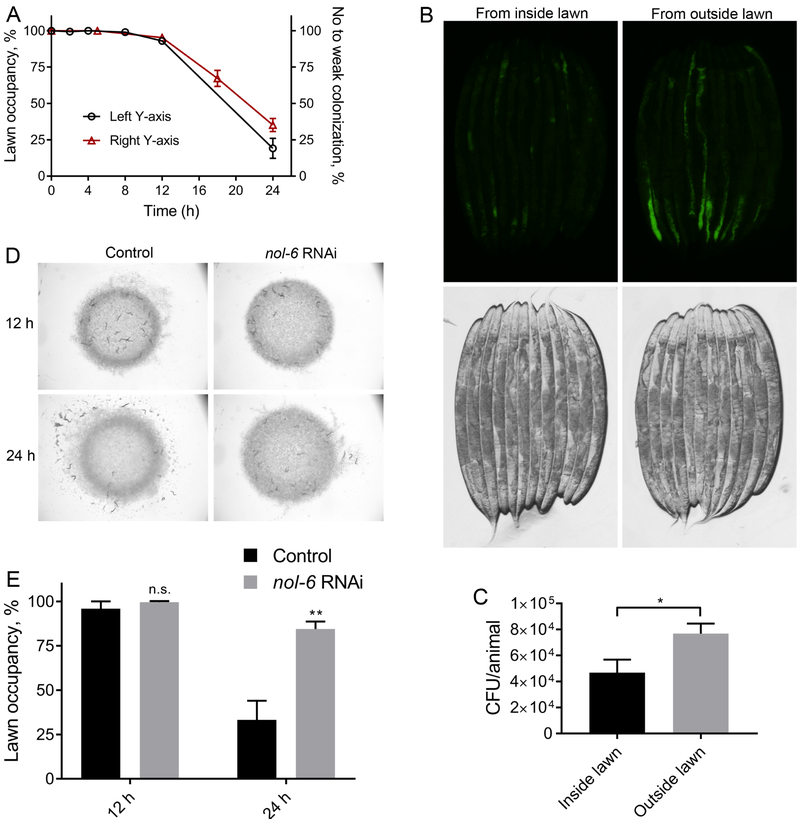
To gain insight into the host mechanism(s) involved in the elicitation of the late avoidance behavior exhibited by C. elegans, we focused on the effects of P. aeruginosa on the animals. We studied the rate of colonization of the C. elegans intestine by P. aeruginosa. As shown in Figure 2A, the kinetics of colonization of the C. elegans intestine by P. aeruginosa was similar to the kinetics of avoidance of the pathogen. To study the relationship between colonization and avoidance behavior, we analyzed the bacterial colonization of animals present inside and outside of the lawn after 18 hours of incubation on P. aeruginosa expressing green fluorescent protein (GFP). We found that the animals that had left the lawn showed higher levels of colonization than those that remained on the lawn at the same time point (Figures 2B and 2C), suggesting that colonization of the intestine may induce the avoidance behavior. We reasoned that if the colonization of the intestine indeed induces the avoidance behavior, then animals that resist colonization should exhibit diminished avoidance behavior. To test this hypothesis, we studied the avoidance behavior of animals after knockdown of the gene nol-6. Previous studies have shown that RNA interference (RNAi)-mediated knockdown of nol-6, a nucleolar RNA-associated protein, prevents colonization of the intestine by microbial pathogens, including P. aeruginosa (Fuhrman et al., 2009). Consistent with the idea that colonization results in the elicitation of avoidance behavior, inhibition of nol-6 blocked the pathogen-avoidance phenotype of P. aeruginosa (Figures 2D and 2E) but not the rapid avoidance of Gram-positive pathogens (Figure S1B). Taken together, these results suggest that the intestinal colonization by P. aeruginosa elicits the observed avoidance behavior.
Figure 2. P. aeruginosa Colonization of the Intestine Induces Pathogen Avoidance.
(A) Percent occupancy of N2 animals on P. aeruginosa lawn over time (left y-axis) and percent of N2 animals with no or weak colonization of P. aeruginosa-GFP over time (right y-axis). Animals showing colonization in a small fraction of the intestine were considered weakly colonized.
(B) Representative photomicrographs of animals that were picked either from the inside or outside of P. aeruginosa-GFP lawn after 18 hours of incubation on the lawns.
(C) Colony forming units per animal of animals that were picked either from the inside or outside of P. aeruginosa-GFP lawn after 18 hours of incubation on the lawns. *p < 0.05 via the t-test.
(D) Representative photomicrographs of the distribution of control as well as nol-6 RNAi animals after 12 hours and 24 hours of incubation on P. aeruginosa lawns.
(E) Percent occupancy of the control as well as nol-6 RNAi animals after 12 hours and 24 hours of incubation on P. aeruginosa lawns. **p < 0.01 via the t-test. n.s., non-significant.
See also Figure S1.
Bloating of the Intestinal Lumen Enhances the Microbial Aversion Behavior
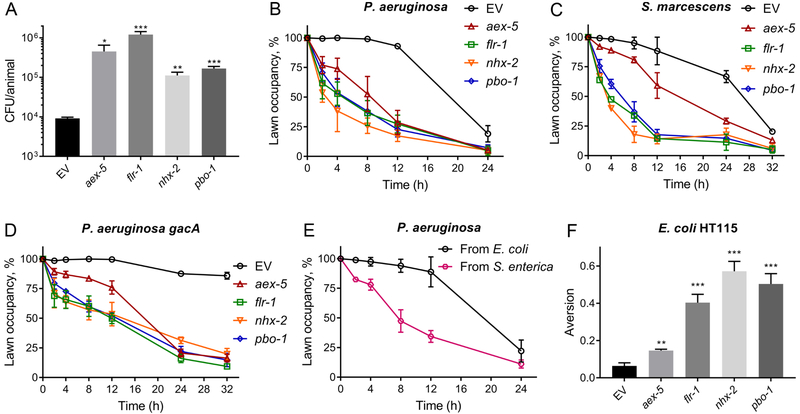
Next, we tested whether increased colonization of the intestine could enhance the avoidance behavior. We studied animals that are defective in the defecation motor program (DMP), and are prone to bacterial colonization (Shapira and Tan, 2008). The genes aex-5, flr-1, nhx-2, and pbo-1 were selected for the current study because their inhibition is known to lead to defects in the DMP, which results in bloating of the intestinal lumen by the accumulation of bacteria (Branicky and Hekimi, 2006; Pfeiffer et al., 2008; Thomas, 1990). As expected, defects in the DMP led to enhanced accumulation of P. aeruginosa (Figures 3A and S2A). Animals with bloated lumens showed enhanced avoidance of P. aeruginosa lawns, and within two hours of the transfer, the animals were observed to avoid the P. aeruginosa lawns (Figure 3B). Additional mutants that lead to mild defects in the DMP also showed enhanced P. aeruginosa accumulation (Figure S2B and S2C) and exhibited enhanced avoidance behavior (Figure S2D). Animals with bloated lumens also showed enhanced avoidance of the pathogen S. marcescens (Figure 3C).
Figure 3. Microbial Colonization of the Intestine Leads to Enhanced Microbial Aversion Behavior.
(A) Colony forming units per animal for N2 animals grown on control as well as aex-5, flr-1, nhx-2, and pbo-1 RNAi followed by exposure to P. aeruginosa-GFP for 5 hours at 25°C. EV, empty vector RNAi control. ***p < 0.001, **p < 0.01, and *p < 0.05 via the t-test.
(B)-(D) Percent occupancy of P. aeruginosa (B), Serratia marcescens (C), and P. aeruginosa gacA mutant (D) lawns over time by N2 animals grown on control as well as aex-5, flr-1, nhx-2, and pbo-1 RNAi. EV, empty vector RNAi control.
(E) Percent occupancy of P. aeruginosa lawns over time by N2 animals grown on S. enterica for 48 hours post L4 larval stage. The control animals were grown on E. coli.
(F) Aversion to E. coli HT115 upon knockdown of aex-5, flr-1, nhx-2, and pbo-1 by RNAi. ***p < 0.001 and **p < 0.01 via the t-test.
See also Figures S2 and S3.
Animals that are defective in feeding manifest pharyngeal anomalies (Avery, 1993; Straud et al., 2013), which lead to enhanced bacterial colonization (Portal-celhay et al., 2012). Thus, we studied several feeding defective mutants to explore the relationship between bacterial colonization and avoidance behavior further. As shown in Figures S2E and S2F, six out of eight tested mutants exhibited enhanced accumulation of P. aeruginosa. Consistent with the idea that bacterial colonization elicits an avoidance behavior, all of the animals that had enhanced bacterial accumulation also showed enhanced avoidance of P. aeruginosa (Figure S2G). On the other hand, eat-5(ad1402) and eat-7(ad450) animals did not exhibit enhanced P. aeruginosa accumulation or enhanced avoidance behavior (Figures S2E-S2G). Taken together, these results suggested that animals with bloated lumens show enhanced avoidance of the pathogen P. aeruginosa.
Non-virulent strains of P. aeruginosa with mutations in the virulence factor gacA do not cause infection and fail to colonize the intestine of C. elegans (Tan et al., 1999). As shown previously (Hao et al., 2018), the gacA mutant of P. aeruginosa does not elicit an avoidance behavior in C. elegans (Figure S3). P. aeruginosa gacA mutants are also deficient in the production of the secondary metabolites phenazines and pyochelin, which have been suggested to be important for elicitation of the avoidance behavior in C. elegans (Meisel et al., 2014). Thus, the inability of P. aeruginosa gacA mutants to elicit an avoidance behavior in C. elegans could be because of either their inability to colonize the intestine or a lack of production of secondary metabolites or virulence factors that are important for eliciting the avoidance behavior. We found that animals with bloated lumens started to avoid the lawns of P. aeruginosa gacA within only two hours of transfer, and approximately 75% of the animals were outside the lawns by 24 hours of incubation (Figure 3D). In contrast, control animals showed no avoidance even at 32 hours after transfer to P. aeruginosa gacA lawns (Figure 3D). These results suggest that intestinal colonization and bloating of the lumen play an important role in elicitation of the pathogen avoidance behavior.
Next, we tested whether colonization and bloating of the C. elegans intestinal lumen by a given pathogen could elicit avoidance to another pathogen. We transferred larval L4 animals to lawns of Salmonella enterica for 48 hours. We used S. enterica because infection by this pathogen leads to persistent colonization and bloating of the intestinal lumen, but it does not elicit an avoidance behavior (Fuhrman et al., 2009; Tenor and Aballay, 2008). Wild-type animals grown on S. enterica showed increased avoidance of P. aeruginosa compared with control animals grown on E. coli (Figure 3E). Finally, we tested the aversive behavior of animals with bloated intestinal lumens towards the non-pathogenic bacteria E. coli. Animals with bloated lumens showed an increased aversion to E. coli (Figure 3F). It is not known whether there is an attractant on the S. enterica lawn (Fuhrman et al., 2009; Tenor and Aballay, 2008) that may override the bloating-induced aversion. Currently, S. enterica appears to be the exception and not the rule.
Bloating of the Intestinal Lumen Activates a Broad Innate Immune Response
C. elegans is capable of mounting a distinct transcriptional response against different bacterial pathogens (Wong et al., 2007). However, diverse bacterial pathogens, including E. faecalis, E. faecium, S. aureus, S. marcescens, and Photorhabdus luminescens, also activate the expression of the same innate immune genes (Irazoqui et al., 2010; Wong et al., 2007; Yuen and Ausubel, 2018). The aforementioned bacterial pathogens all cause colonization and bloating of the C. elegans intestinal lumen. Thus, we reasoned that animals might sense bloating of the intestinal lumen as a potential pathogenic attack and trigger a defense response not only by eliciting pathogen avoidance but also by activating innate immune genes. In such a case, bloating of the intestine by defects in the DMP would trigger the activation of innate immune genes in the absence of pathogens.
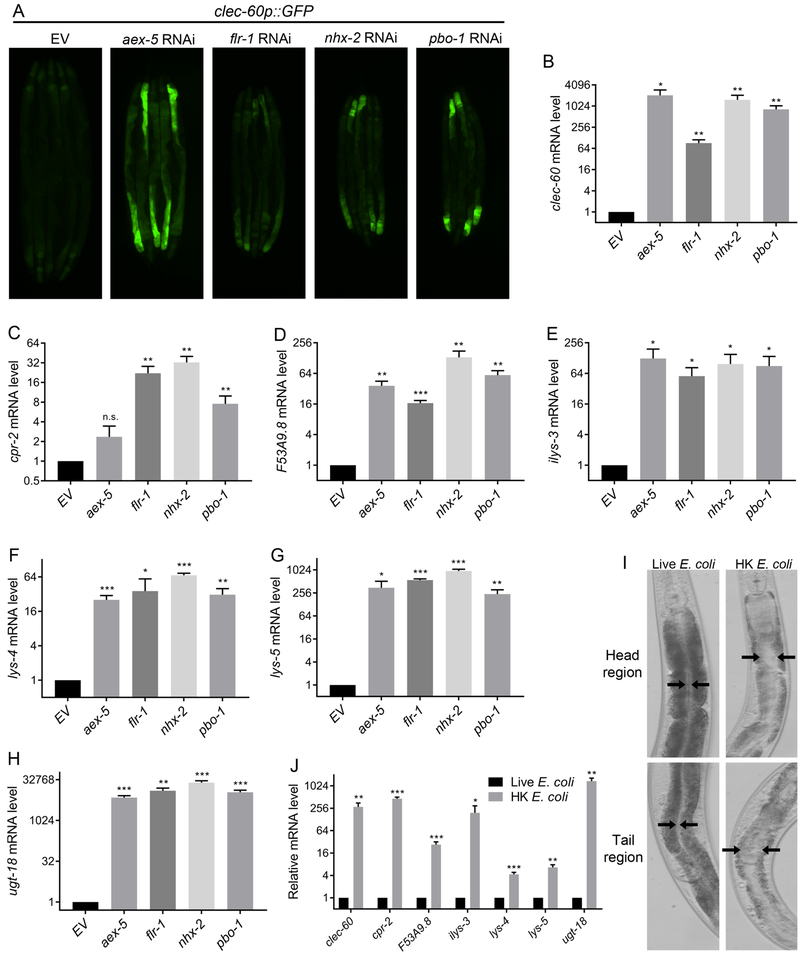
To address whether bloating of the C. elegans intestinal lumen on E. coli can engage transcriptional responses that are only observed upon exposure to pathogens, we examined the expression of the genes clec-60, cpr-2, F53A9.8, ilys-3, lys-4, lys-5, and ugt-18, which are induced by two or more of the bacterial pathogens capable of colonizing the intestine of the animals (Irazoqui et al., 2010; Wong et al., 2007; Yuen and Ausubel, 2018). A reporter strain expressing GFP under the promoter of clec-60 was induced in the DMP-defective animals on E. coli (Figure 4A). The endogenous mRNA levels of clec-60 were also induced in the DMP-defective animals (Figure 4B). Similarly, the immune genes cpr-2, F53A9.8, ilys-3, lys-4, lys-5, and ugt-18 showed significantly increased levels in the DMP-defective animals compared with the control animals (Figures 4C-4H). Several of these immune genes have been shown to be important for the survival of C. elegans on pathogenic bacteria (Gravato-Nobre et al., 2016; Irazoqui et al., 2010; Luhachack et al., 2012). Therefore, the upregulation of these genes will likely improve the survival of C. elegans on pathogenic bacteria. However, investigations of the survival of animals with defects in the DMP on pathogens would not be informative because these animals are known to accumulate pathogens, resulting in an enhanced susceptibility to infection. We reasoned that the animals with bloated intestinal lumens, which exhibit an upregulated immune response, might show increased survival on E. coli since the latter can be mildly pathogenic, especially as the animals age (Garigan et al., 2002; Podshivalova et al., 2017). Indeed, animals with defects in the DMP showed a significantly enhanced lifespan compared with control animals grown on E. coli (Figure S4).
Figure 4. Intestinal Lumen Bloating Activates Innate Immune Genes.
(A) Representative photomicrographs of clec-60p::GFP animals grown on RNAi control bacteria as well as bacteria for RNAi against aex-5, flr-1, nhx-2, and pbo-1. EV, empty vector RNAi control.
(B)-(H) Quantitative reverse transcription-PCR (qRT-PCR) for the immune genes clec-60 (B), cpr-2 (C), F53A9.8 (D), ilys-3 (E), lys-4 (F), lys-5 (G), and ugt-18 (H) in N2 animals grown on control as well as aex-5, flr-1, nhx-2, and pbo-1 RNAi. EV, empty vector RNAi control. ***p < 0.001, **p < 0.01, and *p < 0.05 via the t-test. n.s., non-significant.
(I) Representative photomicrographs of N2 animals grown on live E. coli until the young adult stage, followed by incubation on HK E. coli for 24 hours at 20°C. The control animals were maintained on live E. coli. Representative photomicrographs of the head and tail regions are shown. Arrows point to the border of the intestinal lumen.
(J) Gene expression levels for the immune genes in N2 animals grown on live as well as HK E. coli. The animals were grown on live E. coli until the young adult stage before incubation on HK E. coli for 24 hours at 20°C. ***p < 0.001, **p < 0.01, and *p < 0.05 via the t-test.
See also Figures S4 and S5.
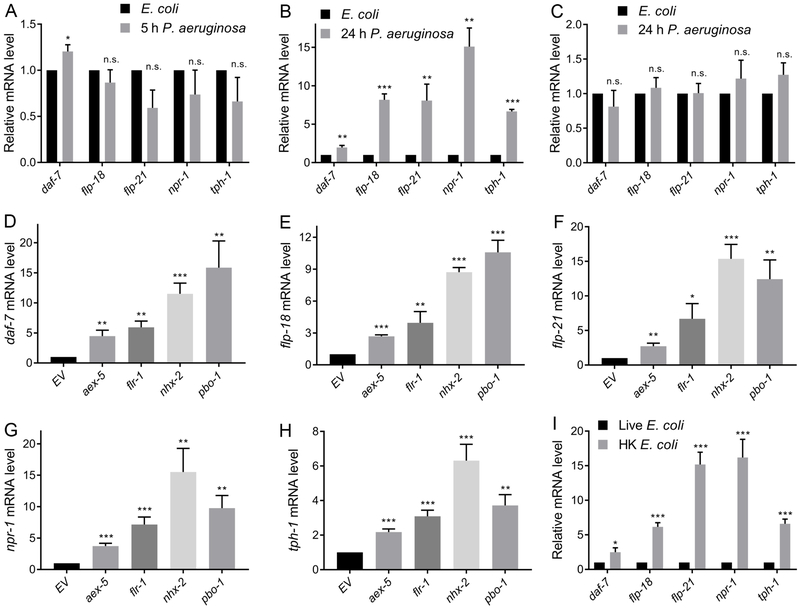
Next, we sought to address whether bloating of the intestinal lumen in the absence of RNAi for DMP genes or bacterial infection up-regulated the expression of immune genes. We found that animals that were fed heat-killed E. coli (HK E. coli) exhibited bloated intestinal lumens (Figure 4I). The pharyngeal pumping rate and the length of the DMP cycle of animals that were fed HK E. coli was comparable to that of animals fed live bacteria (Figure S5A and Figure S5B), suggesting that the viscous lawn of HK E. coli may mechanically block the intestine. As shown in Figure 4J, animals fed on HK E. coli had significantly higher levels of mRNAs for innate immune genes compared with control animals grown on live E. coli. Taken together, these results suggest that bloating of the intestinal lumen may be perceived as a danger signal that induces a fight-and-flight response.
Intestinal Lumen Bloating Enhances the Expression of Neuroendocrine Signaling Pathway Genes
Neurosensory inputs are necessary for pathogen avoidance behavior (Cao et al., 2017; Chang et al., 2011; Pradel et al., 2007; Reddy et al., 2009; Styer et al., 2008) ; however, how animals integrate information to modulate neuroendocrine signaling pathways remains largely unknown. To test whether colonization and bloating of the intestinal lumen may modulate neuroendocrine signaling, we studied the gene expression of different components of neuroendocrine pathways that might influence the avoidance behavior, including the NPR-1/GPCR pathway (npr-1, flp-18, flp-21) (Styer et al., 2008), the DAF-7/TGF-β pathway (Meisel et al., 2014), and the serotonin biosynthesis pathway (Shivers et al., 2009). We found that the expression of genes of the aforementioned pathways did not change after 5 hours of exposure to P. aeruginosa (Figure 5A). However, all the studied genes were up-regulated after 24 hours of P. aeruginosa exposure (Figure 5B). To test whether the increased expression observed after 24 hours of P. aeruginosa infection was related to the colonization of the intestine rather than a longer exposure alone, we took advantage of nol-6 RNAi animals. These animals, which resist intestinal colonization up to 24 hours after P. aeruginosa infection (Fuhrman et al., 2009), did not develop an aversion to the pathogen (Figures 2C and 2D). Figure 5C shows no changes in gene expression after 24 hours of P. aeruginosa infection in nol-6 RNAi animals in comparison to control animals grown on E. coli. These results suggested that colonization of the intestine by P. aeruginosa is required to induce neuroendocrine pathways linked to the pathogen avoidance behavior.
Figure 5. Intestinal Lumen Bloating Enhances the Expression of Neuroendocrine Signaling Pathways.
(A)-(B) Gene expression analysis of N2 animals grown on E. coli HT115 until the young adult stage, followed by incubation on P. aeruginosa for 5 hours (A) or 24 hours (B). ***p < 0.001, **p < 0.01, and *p < 0.05 via the t-test. n.s., non-significant.
(C) Gene expression analysis of N2 animals grown on nol-6 RNAi bacteria until the young adult stage, followed by incubation on P. aeruginosa for 24 hours. n.s., non-significant.
(D)-(H) Relative mRNA levels of daf-7 (D), flp-18 (E), flp-21 (F), npr-1 (G), and tph-1 (H) in animals grown on RNAi control bacteria as well as bacteria expressing RNAi against aex-5, flr-1, nhx-2, and pbo-1. ***p < 0.001, **p < 0.01, and *p < 0.05 via the t-test. n.s., non-significant.
(I) Gene expression analysis of N2 animals grown on live E. coli until the young adult stage, followed by incubation on HK E. coli for 24 hours at 20°C. The control animals were maintained on live E. coli. ***p < 0.001, and *p < 0.05 via the t-test.
The activation of neuroendocrine pathway genes by P. aeruginosa colonization could be a consequence of either an increased load of the pathogen or bloating of the intestinal lumen. To distinguish these possibilities, we studied the changes in gene expression in the DMP-defective animals in the presence of E. coli. We reasoned that if the P. aeruginosa load is required for the induction of gene expression, then no changes would be expected in animals with bloated intestinal lumens on E. coli. However, if bloating of the intestinal lumen rather than the pathogen load induces gene expression, then the changes should be observed in the animals with bloated intestines, even in the absence of P. aeruginosa. The animals with bloated intestinal lumens on E. coli had significant induction of the neuroendocrine pathway genes compared with the control animals (Figures 5D-5H). Animals fed HK E. coli, which causes luminal bloating, also exhibited significantly higher levels of mRNAs for the neuroendocrine pathway genes compared with control animals grown on live E. coli (Figure 5I). These results indicate that bloating of the intestinal lumen triggers a ‘flight’ response in the animals through the induction of different neuroendocrine pathways that are required for pathogen avoidance.
The NPR-1 Neuroendocrine Pathway is Required for Pathogen Avoidance Induced by Intestinal Bloating
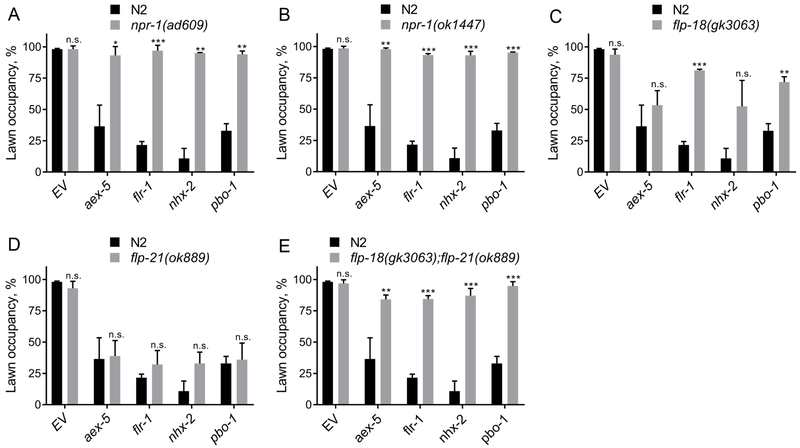
Next, we assessed the neuroendocrine signaling pathway(s) that is required for the pathogen aversive behavior induced by bacterial colonization and bloating of the intestinal lumen. We inactivated genes required for the DMP in the loss-of-function mutants daf-7, npr-1, and tph-1, which are deficient in DAF-7, NPR-1, and serotonin signaling, respectively. While the tph-1(mg280) animals did not show any significant differences in avoidance compared to the wild-type N2 animals, daf-7(e1372) animals showed a partial, but significant, suppression of the enhanced P. aeruginosa avoidance behavior (Figure S6). In contrast, npr-1(ad609) animals showed complete suppression of the enhanced P. aeruginosa avoidance behavior of animals with bloated intestinal lumen (Figure 6A). This suppression is not due to lack of bacterial colonization as the inhibition of DMP genes in npr-1(ad609) animals indeed led to enhanced bacterial colonization (Figure S7A and S7B). Another allele of npr-1, npr-1(ok1447) also led to complete suppression of the enhanced P. aeruginosa avoidance behavior of the DMP-defective animals (Figure 6B).
Figure 6. The NPR-1 Neural Pathway is Required for Pathogen Avoidance Induced by Intestinal Lumen Bloating.
(A)-(E) Percent lawn occupancy after 12 hours of incubation on P. aeruginosa of npr-1(ad609) (A), npr-1(ok1447) (B), flp-18(gk3063) (C), flp-21(ok889) (D), andflp-18(gk3063);flp-21(ok889) (E) animals compared to control wild-type N2 animals. ***p < 0.001, **p < 0.01, and *p < 0.05 via the t-test. n.s., non-significant.
See also Figures S6 and S7.
The FMRF-like peptide FLP-18 is expressed in several neurons and FLP-21 is expressed in both neurons and intestine (Rogers et al., 2003). These peptides play important roles in behaviors associated with NPR-1, such as behavioral quiescence and social feeding (Choi et al., 2013; Rogers et al., 2003). However, their role in pathogen avoidance behavior has not been studied to date. As shown in Figures 6C and 6D, while loss of function mutants offlp-18 showed partial suppression of the enhanced pathogen avoidance of the DMP-defective animals, flp-21 mutants fully retained the enhanced ability to avoid P. aeruginosa. However, flp-18;flp-21 double mutant animals showed complete suppression of the enhanced avoidance of P. aeruginosa (Figure 6E), suggesting that they might act redundantly. This suppression of the avoidance behavior was not due to lack of bacterial colonization (Figure S7C and S7D). Taken together, these results suggest that DAF-7 and NPR-1 and its ligands play a role in elicitation of the pathogen avoidance behavior upon microbial colonization and bloating of C. elegans intestinal lumen.
DISCUSSION
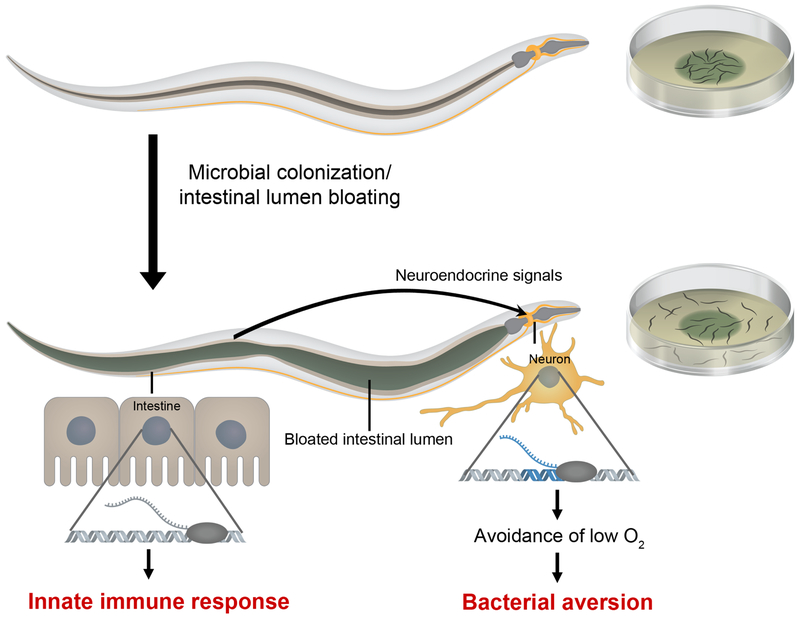
Despite being a simple organism, C. elegans has evolved complex mechanisms to recognize pathogens as such and to respond to their presence by eliciting avoidance behavioral responses and innate immune pathways. Here we show that intestinal lumen bloating induced by microbial colonization activates both immune pathways with microbicidal function and a microbial aversion behavior. We demonstrate that bloating activates different neuroendocrine signaling pathways and that the increased aversion behavior requires the DAF-7/TGF-β and NPR-1 pathways. We propose a model wherein bloating of the intestinal lumen caused by microbial colonization may be perceived as a danger signal that activates an immune fight-and-flight response or BIFF response (Figure 7). The results suggest that inputs from the intestine can aid in the recognition of a broad range of microbes and modulate host behavior via neuroendocrine signaling.
Figure 7. Model for the Microbial Colonization-Induced Immune Fight-and-Flight Response.
Bloating of the intestinal lumen caused by microbial colonization activates a fight-and-flight response that results in the activation of innate immunity and a bacterial avoidance behavior. Bloating results in the up-regulation of genes encoding the neuropeptide receptor NPR-1, its ligands, and TGF-β/DAF-7, which may promote bacterial avoidance by eliciting avoidance to low oxygen.
Bloating of the intestine leads to reduced levels of intestinal fat, which may result in a nutritional deficit in C. elegans (Sheng et al., 2015). Because poor nutrition may lead to food avoidance (Shtonda and Avery, 2006), it is possible that the avoidance behavior is elicited by reduced levels of intestinal fat in bloated animals. However, eat-5(ad1402) and eat-7(ad450) animals do not exhibit intestinal bloating or pathogen avoidance behavior, despite having starvation phenotypes (Avery, 1993; Raizen et al., 2006), indicating that the nutritional status of the intestine is unlikely to mediate the bloating-induced avoidance behavior. Another possible mechanism by which intestinal bloating may elicit the avoidance behavior is through the mechanosensation of changes in the intestine and/or hypodermis. While the exact nature of the signaling mechanism that communicates the intestine with the nervous system is not understood, our data indicate that bloating induced by bacterial colonization might act as a ‘danger signal’ that elicits the pathogen avoidance behavior.
In support of the idea that the physiological alterations caused by bloating of the intestinal lumen result in the induction of innate immune genes, earlier studies have shown that C. elegans mounts innate immune responses when core cellular activities are disrupted (Dunbar et al., 2012; McEwan et al., 2012; Melo and Ruvkun, 2012). RNAi and toxin-mediated disruption of core cellular activities, including translation, respiration, and protein turnover, induce the expression of detoxification and innate immune effectors (Melo and Ruvkun, 2012). Similarly, inhibition of C. elegans translation by P. aeruginosa exotoxin A triggers an innate immune response (Dunbar et al., 2012; McEwan et al., 2012). Together with these earlier findings, our study highlights the ability of C. elegans to sense physiological changes induced by pathogen infection or toxins and, in turn, induce an innate immune response. With such capabilities, the animals would be equipped to recognize a broader range of pathogens and toxins and mount defense responses.
One of our most surprising findings is the activation of neuroendocrine pathway gene expression by intestinal lumen bloating. This activation was only observed post-colonization of the intestine by pathogens and not at early time points of exposure, suggesting that the modulation of neuroendocrine signaling by pathogen exposure was triggered by bloating and not by simple exposure to the pathogen. Activation of the neuroendocrine signaling in animals with bloated intestinal lumens on live or heat-killed E. coli further supports the conclusion that neuroendocrine signaling was modulated by bloating of the intestinal lumen and not by pathogen exposure. While the nervous system is known to control the physiology and behavior of animals by modulating the functionality of other tissues (Durieux et al., 2011; Prahlad et al., 2008; Reddy et al., 2009; Styer et al., 2008; Sun et al., 2011), the modulation of neural functions by non-neuronal cells to control physiology and behaviors remains understudied. Recent studies have shown that the intestine of C. elegans can modulate the nervous system (Chikka et al., 2016; Lee and Mylonakis, 2017), but the role of intestinal lumen bloating in this regulation has not been previously explored.
Intestinal lumen bloating was found to activate the expression of neuroendocrine pathway genes, including those that encode NPR-1/GPCR, DAF-7/TGF-β, and TPH-1, which is required for serotonin signaling. While the serotonin pathway was not found to be required for the elicitation of pathogen avoidance, it has been shown to be important for aversion of non-pathogenic E. coli when core cellular activities are disrupted (Melo and Ruvkun, 2012). Inhibition of the DAF-7/TGF-β pathway partially suppressed the enhanced pathogen avoidance behavior of animals with bloated intestinal lumens, while inhibition of NPR-1 caused complete suppression. Both of these pathways are important for the control of an aerotaxis behavior exhibited by C. elegans that is likely to impact microbial aversion (Meisel and Kim, 2014; Reddy et al., 2009, 2011; Styer et al., 2008). Inhibition of the NPR-1 and DAF-7/TGF-β pathways elicit avoidance of high oxygen, while higher activity of these pathways induces avoidance of low oxygen (De Bono and Bargmann, 1998; Chang et al., 2006; Meisel et al., 2014; Rogers et al., 2003). Therefore, the higher expression of npr-1 and daf-7 induced by bloating may elicit C. elegans avoidance of low oxygen, resulting in the avoidance of bacterial lawns, as they have low oxygen due to microbial metabolism (Figure 7). Consistent with this idea, the avoidance of E. coli lawns of DMP defective animals is not as strong as that of P. aeruginosa lawns (Figure 3), which have lower oxygen levels than E. coli lawns (Reddy et al., 2011).
Our study suggests that bloating of the intestine may be a general mechanism involved in the recognition of a broad range of microbes and modulates host behavior via neuroendocrine signaling. In the future, it will be important to understand how intestinal bloating modulates neuroendocrine signaling. NPR-1 is closely related to the mammalian neuropeptide Y (NPY) receptors that are involved in the control of a diverse set of behavioral processes, including appetite, circadian rhythm, and anxiety (Heilig, 2004; Vona-Davis and McFadden, 2007; Yulyaningsih et al., 2011). Therefore, the identity and mode of action of the cues that are used by the intestine to activate the NPR-1 pathway may provide broad insights into the modulation of NPY receptor-related behaviors such as appetite and anxiety.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alejandro Aballay (aballay@ohsu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains
The following bacterial strains were used: Escherichia coli OP50, E. coli HT115(DE3), Pseudomonas aeruginosa PA14, P. aeruginosa PA14 gacA mutant, P. aeruginosa PA14-GFP, Serratia marcescens Db11, Salmonella enterica serovar Typhimurium 1344, Enterococcus faecalis OG1RF, Enterococcus faecium E007, and Staphylococcus aureus NCTC8325. The E. coli OP50, E. coli HT115(DE3), P. aeruginosa PA14 strains, S. marcescens, and S. enterica cultures were grown in Luria-Bertani (LB) broth at 37°C. The E. faecalis and E. faecium cultures were grown in brain heart infusion (BHI) broth at 37°C. The S. aureus cultures were grown in tryptic soy (TS) broth at 37°C.
C. elegans strains and growth conditions
C. elegans hermaphrodites were maintained on E. coli OP50 at 20 °C unless otherwise indicated. Bristol N2 was used as the wild-type control unless otherwise indicated. Strains DA609 npr-1(ad609), RB1330 npr-1(ok1447), VC2016 flp-18(gk3063), RB982 flp-21(ok889), CB1372 daf-7(e1372), MT15434 tph-1(mg280), DA597 phm-2(ad597), DA465 eat-2(ad465), DA819 eat-4(ad819), DA1402 eat-5(ad1402), DA521 eat-7(ad450), RB2247 eat-17(ok3041), DA820 eat-18(ad820), MT1083 egl-8(n488), VC2117 rab-6.2(ok2254), CB156 unc-25(e156), and CB307 unc-47(e307) were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). Strain AU185 agIs26[myo-2p::mCherry,clec-60p::gfp] was provided by Javier E. Irazoqui (University of Massachusetts Medical School, Worcester MA 01605). The flp-18(gk3063);flp-21(ok889) double mutant was obtained by standard genetic crosses.
METHOD DETAILS
RNA interference (RNAi).
RNAi was used to generate loss-of-function RNAi phenotypes by feeding nematodes E. coli strain HT115(DE3) expressing double-stranded RNA (dsRNA) homologous to a target gene (Fraser et al., 2000; Timmons and Fire, 1998). RNAi was carried out as described previously (Singh and Aballay, 2017). Briefly, E. coli with the appropriate vectors were grown in LB broth containing ampicillin (100 μg/mL) and tetracycline (12.5 μg/mL) at 37°C overnight and plated onto NGM plates containing 100 μg/mL ampicillin and 3 mM isopropyl β-D-thiogalactoside (IPTG) (RNAi plates). RNAi-expressing bacteria were allowed to grow overnight at 37°C. Gravid adults were transferred to RNAi-expressing bacterial lawns and allowed to lay eggs for 2 hours. The gravid adults were removed, and the eggs were allowed to develop at 20°C to young adults for subsequent assays. unc-22 RNAi was included as a positive control to account for the RNAi efficiency. All RNAi clones except pbo-1 were from the Ahringer RNAi library.
Construction of the pbo-1 RNAi clone
The 591-base-pair full-length cDNA of pbo-1 was amplified using the forward primer 5’-GCTCCCGGGATGGGCCAAAATTCGTCTCA-3’ and the reverse primer 5’-AGGTCTAGATGTCAAAAAACGGAAGGACATC-3’. The fragment was cloned into the SmaI and XbaI sites of pL4440 (Open Biosystems) and transformed into E. coli HT115(DE3) cells.
Avoidance assay on pathogens
The bacterial lawns were prepared by inoculating individual bacterial colonies into 2 mL of the corresponding broth mentioned above and growing them for 8-10 hours on a shaker at 37°C. Then, 20 μL of the culture was plated onto the center of a 3.5-cm plate and incubated at 37°C for 12-16 hours except for S. aureus which was incubated for 6 hours. For P. aeruginosa PA14 strains, S. marcescens and S. enterica modified NGM (3.5% instead of 2.5% peptone) plates were used. For E. faecalis and E. faecium, BHI agar plates were used. For S. aureus, TS agar plates were used. Thirty synchronized young gravid adult hermaphroditic animals grown on E. coli HT115(DE3) containing control vector or an RNAi clone targeting a gene were transferred outside the bacterial lawns, and the numbers of animals on and off the lawns were counted at the indicated times for each experiment. Three 3.5-cm plates were used per trial in every experiment. Experiments were performed at 25°C. The percent occupancy was calculated as (Nonlawn/Ntotal)×100. At least three independent experiments were performed.
Avoidance assay on E. coli
Saturated overnight cultures of the E. coli HT115 RNAi bacteria were concentrated 30–40 times in LB. A 50-μL aliquot was plated onto the center of a 3.5-cm RNAi plate and incubated at 37°C overnight. The following day, the plates were cooled at room temperature for 1 hour, and then gravid adults were transferred to bacterial lawns and allowed to lay eggs for 2 hours. The gravid adults were removed, and the eggs were allowed to develop at 20°C. After 72 hours of incubation at 20°C, the numbers of animals on and off the lawns were counted to calculate aversion as Nofflawn/Ntotal. Three independent experiments were performed.
P. aeruginosa-GFP colonization assay
Bacterial lawns were prepared by inoculating individual bacterial colonies into 2 mL of LB with 50 μg/mL kanamycin and growing them for 8-10 hours on a shaker at 37°C. For the colonization assays, bacterial lawns of P. aeruginosa-GFP were prepared by spreading 20 μL of the culture on the complete surface of 3.5-cm-diameter modified NGM agar plates (0.35% instead of 0.25% peptone). For P. aeruginosa-GFP colonization assays on partial lawns, bacterial lawns were prepared by placing 20 μL of the culture at the center of 3.5-cm-diameter modified NGM agar plates. The plates were incubated at 37°C for 12-16 hours and then cooled to room temperature for at least 1 h before seeding with young gravid adult hermaphroditic animals. The assays were performed at 25°C. At the indicated times for each experiment, the animals were transferred from P. aeruginosa-GFP plates to fresh E. coli OP50 plates and visualized within 5 minutes under a fluorescence microscope.
Quantification of Intestinal Bacterial Loads
P. aeruginosa-GFP lawns were prepared as described above. For quantification of colony forming units (CFU) at 5 hours of exposure, bacterial lawns of P. aeruginosa-GFP were prepared by spreading 20 μL of the culture on the complete surface of 3.5-cm-diameter modified NGM agar plates (0.35% instead of 0.25% peptone). For quantification of CFU on partial lawns, bacterial lawns were prepared by placing 20 μL of the culture at the center of 3.5-cm-diameter modified NGM agar plates. The plates were incubated at 37°C for 12-16 hours and then cooled to room temperature for at least 1 hour before seeding with young gravid adult hermaphroditic animals. The assays were performed at 25°C. At the indicated times for each experiment, the animals were transferred from P. aeruginosa-GFP plates to the center of fresh E. coli plates for 10 min to eliminate P. aeruginosa-GFP stuck to their body. Animals were transferred to the center of a new E. coli plate for 10 min to further eliminate external P. aeruginosa-GFP.
Animals were transferred to fresh E. coli plates a third time for 10 min. Afterward, ten animals/condition were transferred into 50 μL of PBS plus 0.01% Triton X-100 and ground. Serial dilutions of the lysates (101, 102, 103, 104) were seeded onto LB plates containing 50 μg/mL of kanamycin to select for P. aeruginosa-GFP cells and grown overnight at 37 °C. Single colonies were counted the next day and represented as the number of bacterial cells or CFU per animal. Three independent experiments were performed for each condition.
Fluorescence imaging
Fluorescence imaging was carried out as described previously (Singh and Aballay, 2017). Briefly, animals were anesthetized using an M9 salt solution containing 30 mM sodium azide and mounted onto 2% agar pads. The animals were then visualized using a Leica M165 FC fluorescence stereomicroscope.
S. enterica colonization followed by P. aeruginosa avoidance
Wild-type N2 L4 animals grown on E. coli HT115 were transferred to the lawns of S. enterica, followed by incubation at 25°C. Control animals on E. coli HT115 were also incubated at 25°C. After 48 hours of incubation, S. enterica-colonized animals, along with the animals grown on E. coli HT115, were transferred to P. aeruginosa lawns, and the numbers of animals on and off the lawns at the indicated times were counted. The experiments were performed at 25°C. The percent occupancy was calculated as (Non lawn/Ntotal)×100. Three independent experiments were performed.
Cultivation of C. elegans on heat-killed E. coli OP50
A single colony of E. coli OP50 was inoculated in 100 mL of LB broth in a 500 mL conical flask and incubated at 37°C at 225 rpm shaking for 24 hours. Bacteria were then concentrated 10 to 20 times and heat-killed at 100°C for 1 hour. Bacterial death was confirmed by failure to grow on LB plates at 37°C overnight. The heat-killed bacteria were seeded on NGM plates containing 50 μg/mL of kanamycin and 100 μg/mL of streptomycin. Young adult wild-type N2 animals grown on E. coli HT115 RNAi control plates were washed with M9 medium and incubated at room temperature for 1 hour with M9 medium containing 50 μg/mL of kanamycin to remove live bacteria from their intestinal lumen. The animals were then washed with M9 medium and transferred to NGM plates containing heat-killed E. coli OP50 and incubated at 20°C for 24 hours.
Pharyngeal pumping assay
Wild-type N2 animals grown on heat-killed E. coli for 24 hours at 20°C were used for pharyngeal pumping assay with animals grown on live E. coli as controls. The number of contractions of the terminal bulb was counted over 1 minute. A contraction was defined as the backward movement of the grinder in the terminal bulb of the pharynx. The pumping rates for 20 nematodes were recorded for each condition.
Defecation assay
Wild-type N2 animals grown on heat-killed E. coli for 24 hours at 20°C were used for defecation assay with animals grown on live E. coli as controls. The DMP cycle length was scored by assessing the time between expulsions (which are preceded by posterior and anterior body wall muscle contraction, and the contraction of enteric muscles in a normal regular pattern) (Thomas, 1990). Ten cycles each were measured for six different animals per condition.
RNA isolation and quantitative reverse transcription-PCR (qRT-PCR)
Animals were synchronized by egg laying. Approximately 35 N2 gravid adult animals were transferred to 10-cm RNAi plates seeded with E. coli HT115 expressing the appropriate vectors and allowed to lay eggs for 4 hours. The gravid adults were then removed, and the eggs were allowed to develop at 20°C. For the RNAi against the DMP genes, the animals were grown until they were one-day-old adults at 20°C. For the P. aeruginosa infection assays, the animals were first grown on 10-cm RNAi plates seeded with E. coli HT115 expressing either the empty vector RNAi control or nol-6 RNAi until the young adult stage. Subsequently, the animals were collected, washed with M9 buffer, and transferred to 10-cm modified NGM plates (0.35% instead of 0.25% peptone) seeded with 300 μL of P. aeruginosa culture grown overnight. The P. aeruginosa plates were prepared by spreading 300 μL of the P. aeruginosa culture on the surface of the modified NGM plates, followed by an overnight incubation at 37°C. After transfer of the animals, the P. aeruginosa plates were incubated at 25°C for the indicated time points. Animals on the control E. coli plates were also incubated at 25°C. After the desired treatment (RNAi against the DMP genes, growth on heat-killed E. coli for 24 hours or P. aeruginosa infection), the animals were collected, washed with M9 buffer, and frozen in TRIzol reagent (Life Technologies, Carlsbad, CA). Total RNA was extracted using the RNeasy Plus Universal Kit (Qiagen, Netherlands). Residual genomic DNA was removed using TURBO DNase (Life Technologies, Carlsbad, CA). A total of 6 μg of total RNA was reverse-transcribed with random primers using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA).
qRT-PCR was conducted using the Applied Biosystems One-Step Real-time PCR protocol using SYBR Green fluorescence (Applied Biosystems) on an Applied Biosystems 7900HT real-time PCR machine in 96-well-plate format. Twenty-five-microliter reactions were analyzed as outlined by the manufacturer (Applied Biosystems). The relative fold-changes of the transcripts were calculated using the comparative CT(2−ΔΔCT) method and normalized to pan-actin (act-1, −3, −4). The cycle thresholds of the amplification were determined using StepOnePlus software (Applied Biosystems). All samples were run in triplicate. The primer sequences are available upon request.
C. elegans longevity assays
Lifespan assays were performed on RNAi plates containing E. coli HT115(DE3) with the appropriate vector in the presence of 50 μg/mL of 5-fluorodeoxyuridine (FUdR). Animals were synchronized on RNAi plates without FUdR and incubated at 20°C. At the L4 larval stage, the animals were transferred onto the corresponding RNAi plates containing 50 μg/mL of FUdR. Animals were scored on a daily basis as alive, dead, or gone. Animals that failed to display touch-provoked movement were scored as dead. Experimental groups contained 60 to100 animals. Young adult animals were considered as day 0 for the lifespan analysis. The assays were performed at 20°C. Three independent experiments were performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed with Prism 7 (GraphPad). All error bars represent the standard deviation (SD). The two-sample t test was used when needed, and the data were judged to be statistically significant when p < 0.05. In the figures, asterisks (*) denote statistical significance as follows: *, p < 0.05, **, p < 0.001, ***, p < 0.0001, as compared with the appropriate controls. The Kaplan-Meier method was used to calculate the survival fractions, and statistical significance between survival curves was determined using the log-rank test. For Figure S6, two-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test was used. All experiments were performed in triplicate.
Supplementary Material
Highlights.
P. aeruginosa colonization elicits immune genes and pathogen aversion in C. elegans
Bloating of the intestinal lumen by microbial colonization upregulates immune genes
Intestinal bloating upregulates neuroendocrine pathways, inducing a flight response
NPR-1 signaling is required for a flight response that elicits pathogen aversion
ACKNOWLEDGMENTS
This work was supported by NIH grants GM0709077 and AI117911 (to A.A.). Some strains used in this study were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Javier E. Irazoqui (University of Massachusetts Medical School, Worcester MA 01605) for providing the clec-60p::GFP strain and Meta Kuehn (Duke University Medical Center, Durham, NC) for providing the P. aeruginosa gacA mutant. We are also grateful for the critiques and comments of the reviewers that improved our manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
The Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.devcel.2019.02.001
REFERENCES
- Avery L (1993). The Genetics of Feeding in Caenorhabditis elegans. Genetics 917, 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM, and Sander LE (2012). Beyond pattern recognition: Five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol 12, 215–225. [DOI] [PubMed] [Google Scholar]
- De Bono M, and Bargmann CI (1998). Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94, 679–689. [DOI] [PubMed] [Google Scholar]
- Branicky R, and Hekimi S (2006). What keeps C. elegans regular: the genetics of defecation. Trends Genet. 22, 571–579. [DOI] [PubMed] [Google Scholar]
- Bretscher AJ, Kodama-Namba E, Busch KE, Murphy RJ, Soltesz Z, Laurent P, and de Bono M (2011). Temperature, oxygen, and salt-sensing neurons in C. elegans are carbon dioxide sensors that control avoidance behavior. Neuron 69, 1099–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Kajino-Sakamoto R, Doss A, and Aballay A (2017). Distinct Roles of Sensory Neurons in Mediating Pathogen Avoidance and Neuropeptide-Dependent Immune Regulation. Cell Rep. 21, 1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AJ, Chronis N, Karow DS, Marietta MA, and Bargmann CI (2006). A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 4, e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Paek J, and Kim DH (2011). Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature 480, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikka MR, Anbalagan C, Dvorak K, Dombeck K, and Prahlad V (2016). The Mitochondria-Regulated Immune Pathway Activated in the C. elegans Intestine Is Neuroprotective. Cell Rep. 16, 2399–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, and Kaplan JM (2013). Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C.elegans. Neuron 78, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar TL, Yan Z, Balla KM, Smelkinson MG, and Troemel ER (2012). C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, and Dillin A (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, and Ahringer J (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Fuhrman LE, Goel AK, Smith J, Shianna KV, and Aballay A (2009). Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLoS Genet. 5, e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Abringet J, and Kenyon C (2002). Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, and Ausubel FM (2001). A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci 98, 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato-Nobre MJ, Vaz F, Filipe S, Chalmers R, and Hodgkin J (2016). The Invertebrate Lysozyme Effector ILYS-3 Is Systemically Activated in Response to Danger Signals and Confers Antimicrobial Protection in C. elegans. PLoS Pathog. 12, e1005826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Yang W, Ren J, Hall Q, Zhang Y, and Kaplan JM (2018). Thioredoxin shapes the C. elegans sensory response to Pseudomonas produced nitric oxide. Elife 7, e36833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M (2004). The NPY system in stress, anxiety and depression. Neuropeptides 38, 213–224. [DOI] [PubMed] [Google Scholar]
- Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, and Ausubel FM (2010). Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6, e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, and Mylonakis E (2017). An Intestine-Derived Neuropeptide Controls Avoidance Behavior in Caenorhabditis elegans. Cell Rep. 20, 2501–2512. [DOI] [PubMed] [Google Scholar]
- Luhachack LG, Visvikis O, Wollenberg AC, Lacy-Hulbert A, Stuart LM, and Irazoqui JE (2012). EGL-9 controls C. elegans host defense specificity through prolyl hydroxylation-dependent and -independent HIF-1 pathways. PLoS Pathog. 8, e1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Singh J, and Aballay A (2017). Natural Genetic Variation in the Caenorhabditis elegans Response to Pseudomonas aeruginosa. G3 (Bethesda) 7, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P (2002). The danger model: A renewed sense of self. Science 296, 301–305. [DOI] [PubMed] [Google Scholar]
- McEwan DL, Kirienko NV, and Ausubel FM (2012). Host translational inhibition by Pseudomonas aeruginosa exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11, 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JD, and Kim DH (2014). Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol. 35, 465–470. [DOI] [PubMed] [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, and Kim DH (2014). Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, and Ruvkun G (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J, Johnson D, and Nehrke K (2008). Oscillatory Transepithelial H+Flux Regulates a Rhythmic Behavior in C. elegans. Curr. Biol 18, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podshivalova K, Kerr RA, and Kenyon C (2017). How a Mutation that Slows Aging Can Also Disproportionately Extend End-of-Life Decrepitude. Cell Rep. 19, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal-celhay C, Bradley ER, and Blaser MJ (2012). Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, and Ewbank JJ (2007). Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 104, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, and Morimoto RI (2008). Regulation of the Cellular Heat Shock by Thermosensory Neurons. Science 320, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, and Ewbank JJ (2001). A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11, 809–821. [DOI] [PubMed] [Google Scholar]
- Raizen DM, Cullison KM, Pack AI, and Sundaram MV (2006). A Novel Gain-of-Function Mutant of the Cyclic GMP-Dependent Protein Kinase egl-4 Affects Multiple Physiological Processes in Caenorhabditis elegans. Genetics 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, and Kim DH (2009). A Polymorphism in npr-1 Is a Behavioral Determinant of Pathogen Susceptibility in C. elegans. Science 323, 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Hunter RC, Bhatla N, Newman DK, and Kim DH (2011). Caenorhabditis elegans NPR-1-mediated behaviors are suppressed in the presence of mucoid bacteria. Proc. Natl. Acad. Sci 108, 12887–12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, and de Bono M (2003). Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat. Neurosci 6, 1178–1185. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Rowedder H, Braendle C, Félix M-A, and Ruvkun G (2016). Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci 113, E3941–E3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, and Horvitz HR (2000). C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, and Félix MA (2017). The natural biotic environment of Caenorhabditis elegans. Genetics 206, 55–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, and Tan M (2008). Genetic Analysis of Caenorhabditis elegans Innate Immunity. In Methods in Molecular Biology, pp. 429–442. [DOI] [PubMed] [Google Scholar]
- Sheng M, Hosseinzadeh A, Muralidharan SV, Gaur R, Selstam E, and Tuck S (2015). Aberrant fat metabolism in Caenorhabditis elegans mutants with defects in the defecation motor program. PLoS One 10, e0124515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers RP, Kooistra T, Chu SW, Pagano DJ, and Kim DH (2009). Tissue-Specific Activities of an Immune Signaling Module Regulate Physiological Responses to Pathogenic and Nutritional Bacteria in C. elegans. Cell Host Microbe 6, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, and Avery L (2006). Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol 209, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, and Aballay A (2017). Endoplasmic reticulum stress caused by lipoprotein accumulation suppresses immunity against bacterial pathogens and contributes to immunosenescence. MBio 8, e00778–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straud S, Lee I, Song B, Avery L, and You Y-J (2013). The Jaw of the Worm: GTPase-activating Protein EAT-17 Regulates Grinder Formation in Caenorhabditis elegans. Genetics 195, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Paquette N, and Boyer L (2013). Effector-triggered versus pattern-triggered immunity: How animals sense pathogens. Nat. Rev. Immunol 13, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, and Aballay A (2008). Innate Immunity in Caenorhabditis elegans Is Regulated by Neurons Expressing NPR-1/GPCR. Science 322, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Singh V, Kajino-Sakamoto R, and Aballay A (2011). Neuronal GPCR Controls Innate Immunity by Regulating Noncanonical Unfolded Protein Response Genes. Science 332, 729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, and Ausubel FM (1999). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A 96, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenor JL, and Aballay A (2008). A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 9, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH (1990). Genetic analysis of defecation in Caenorhabditis elegans. Genetics 124, 855–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, and Fire A (1998). Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Vona-Davis LC, and McFadden DW (2007). NPY Family of Hormones: Clinical Relevance and Potential Use in Gastrointestinal Disease. Curr. Top. Med. Chem 7, 1710–1720. [DOI] [PubMed] [Google Scholar]
- Wong D, Bazopoulou D, Pujol N, Tavernarakis N, and Ewbank JJ (2007). Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 8, R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen GJ, and Ausubel FM (2018). Both live and dead Enterococci activate Caenorhabditis elegans host defense via immune and stress pathways. Virulence 9, 683–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yulyaningsih E, Zhang L, Herzog H, and Sainsbury A (2011). NPY receptors as potential targets for anti-obesity drug development. Br. J. Pharmacol 163, 1170–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.