Abstract
Krüppel-like factor 8 (KLF8) plays many important roles in various diseases, especially cancer. Previous studies have shown that KLF8 is regulated by ubiquitylation. The molecular mechanism underlying this posttranslational modification of KLF8, however, has not been investigated. Reported here is our identification of the neural precursor cell expressed, developmentally down-regulated 4 (NEDD4) as the E3 ubiquitin ligase for this modification. By co-immunoprecipitation and ubiquitylation assays, we determined that KLF8 interacts with NEDD4 and is ubiquitylated by NEDD4. By site-directed mutagenesis and pharmacological inhibition of MEK, we found that the ubiquitylation of KLF8 by NEDD4 depends upon the phosphorylation of KLF8 at serine 48 by ERK. Cycloheximide chase analysis, target gene promoter reporter assay and fluorescent staining indicated that NEDD4 plays a critical role in promoting the stability and transcriptional activity of KLF8 in the nucleus. Taken together, this work identified NEDD4 as a novel E3 ubiquitin ligase for KLF8 that provides insights into targeting the KLF8-NEDD4 axis to treat various types of cancer associated with overexpression of both proteins.
Keywords: KLF8, NEDD4, interaction, ubiquitylation
Introduction
Krüppel-like factor 8 (KLF8) is a member of the Krüppel-like transcription factor family and acts as a transcriptional activator or repressor to regulate the expression of many target genes [1-9]. Through these target genes, KLF8 plays a critical role in multiple types of cancer where KLF8 is aberrantly upregulated [1-4,6-18] by promoting tumor formation and metastasis. In addition to cancer, other diseases such as Alzheimer’s [19] and adipogenesis [20] have also been linked to KLF8. Our recent studies have demonstrated multiple types of posttranslational modification (PTM) on KLF8 protein including sumoylation, acetylation, ubiquitylation and phosphorylation [8,12,21,22]. While the E3 SUMO ligases and the acetyltransferase responsible for the corresponding PTMs on KLF8 have been discovered [8,21], the E3 ubiquitin ligase responsible for the ubiquitylation of KLF8 that is closely correlated with the latest finding of ERK2 phosphorylation at the serine 48 site [22] remains unknown.
Neural Precursor Cell Expressed, Developmentally Down-Regulated 4 (NEDD4, also known as Nedd4-1) belongs to the HECT E3 ubiquitin ligase family. NEDD4 plays important roles in multiple cellular functions [23] by interacting with and ubiquitylating many proteins including epithelial sodium channel (ENaC) [24,25], multiple endocytic or vesicle sorting proteins [26-32] and autophagy regulating proteins [33,34] as well as the tumor suppressors pTEN [35-37] and LATS1 [27,29] and the proto-oncogene EGFR [29,38]. The WW domain-mediated substrate protein binding [39] and subsequent ubiquitylation alter the substrate protein’s stability, subcellular transportation, translocation or compartmentalization and thus cellular function [25,29,33,34,38,40,41]. NEDD4 loss or gain of function has been linked to pathogenesis of various diseases, including Liddle Syndrome [24] and cancer [36-38,42]. Like KLF8, NEDD4 has been found to be aberrantly overexpressed in various cancer types and is highly correlated with the malignant progression and poor patient survival [37,38,42].
In this report, we identified NEDD4 as the E3 ubiquitin ligase that interacts with and ubiquitylates KLF8 in the nucleus. This PTM depends upon the phosphorylation at the serine 48 site by ERK2 and is critical for the stability and transcriptional activity of KLF8.
Materials and methods
Antibodies and reagents
The anti-HA-probe F-7 antibody (sc-7392), anti-HA-probe Y-11 antibody (sc-805), anti-c-Myc 9E10 antibody (sc-40), anti-ubiquitin P4D1 antibody (sc-8017), anti-β-actin (sc-47778), anti-pERK E-4 antibody (sc-7383), anti-ERK C-16 antibody (sc-93), Protein A-Agarose (sc-2001) and Protein G PLUS-Agarose (sc-2002) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Secondary antibodies were HRP-conjugated donkey anti-mouse (715-035-150) and donkey anti-rabbit IgG (711-035-152) from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). Fluorescent dye-conjugated secondary antibody Alexa Fluor Plus 488 (A-11034) and 594 (A-11032) were purchased from Invitrogen (Carlsbad, CA, USA). Cycloheximide (239764) and MEK inhibitor PD98059 (51-3000) were purchased from Calbiochem (San Diego, CA, USA). The proteasome inhibitor MG132 was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Plasmid construction, cell culture and transfection
The mammalian expression plasmids pHAN, pKH3, pKH3-KLF8, pKH3-KLF8 S48A, pKH3-KLF8-S48D, pcDNA3-HA-NEDD4 and promoter reporter plasmid pGL3b-Cyclin D1-p were previously described [29,43,44]. To construct pHAN-NEDD4 plasmid, we double digested pcDNA3-HA-NEDD4 plasmid with Bam HI and Eco RI and cloned into pHAN vector between the same sites. NIH3T3 and HEK293 cells were cultured in DMEM with 10% calf serum and 10% fetal bovine serum, respectively at 37°C and 5% CO2. In the experiments for protein-protein interaction and protein ubiquitylation analyses, MG132 (20 µM) was included in the media for 12 hours prior to cell lysis. Transfections of the DNA plasmid were performed using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions.
Co-immunoprecipitation (co-IP), IP and immunoblotting (IB)
These assays were done as previously described [34]. Cells were rinsed once with pre-cold phosphate-buffered saline (PBS) and lysed in pre-cold NP-40 lysis buffer (1% NP-40, 137 mM NaCl, 10% glycerol, 20 mM Tris, pH8, 1 mM Na3VO4, 1 mM PMSF, 20 mg/ml leupeptin, and 0.06 trypsin inhibitor unit [TIU]/ml aprotinin). Whole cell lysates were centrifuged at 14,000 rpm for 15 min at 4°C to remove the insoluble fraction. In co-IP or IP experiments, the primary antibody was added to the lysates, incubated at 4°C for 30 min, then 25 μl of the Protein G PLUS-Agarose beads was added, and incubated with rotation overnight. The immunoprecipitates were washed three times with lysis buffer. When the whole cell lysates or immunoprecipitated proteins were denatured in SDS-PAGE sample buffer and boiled at 100°C for 5-10 min, these proteins were resolved by 8%-12% SDS-PAGE. The proteins were transferred from the gel to nytrocellulose membranes, western blotted and quantified using Image Lab 6.0 (Bio-Rad, Hercules, CA).
Ubiquitylation assay
This assay was carried out as previously described [45]. Specifically, HEK293 cells were grown to ~70% density in 6-well plates, transfected with pHAN-NEDD4 and pKH3-KLF8 or its mutants and incubated for 48 hours. The cells were then treated with the proteasome inhibitor MG132 (20 µM) for 12 hours. Whole cell lysates were prepared for immunoprecipitation using an anti-HA antibody. The ubiquitin adducts was examined by western blotting using an anti-ubiquitin antibody. The protein interaction in the same samples was verified by co-IP with an anti-HA antibody followed by western blotting with anti-HA and anti-Myc antibodies.
Cycloheximide chase assay
Cycloheximide chase assay was performed essentially as previously described [12,22]. The HA-KLF8 transiently overexpressed along or co-overexpressed with NEDD4 in HEK293 cells. Thirty-six hours after transfection, the cells were treated with 50 μg/ml of cycloheximide. At various time points post-treatment, whole cell lysates were prepared and analyzed by anti-HA western blotting.
Promoter reporter assay
The cyclin D1 gene promoter luciferase reporter assays were performed as described previously [9,44]. NIH3T3 cells were grown to ~70% density in 12-well plates. The promoter reporter plasmid pGL3b-Cyclin D1-p (0.2 μg) and Renilla luciferase reporter vector pRLSV40 (4 ng) were co-transfected into NIH3T3 cells, alongside pKH3 vector, pKH3-KLF8 or pKH3-KLF8 mutants (0.2 μg), with or without pHAN-NEDD4 (1.0 μg).
Immunofluorescence staining
The procedure for immunofluorescence staining was described previously [33,46]. The cells were rinsed with PBS twice, fixed with 3.7% paraformaldehyde at room temperature for 20 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 1% bovine seral albumin (BSA) for 30 min. The cells were then incubated with the primary antibody anti-HA (Y-11) and/or anti-Myc (9E10) antibodies (1:200) overnight at 4°C, followed by a fluorescent dye-conjugated secondary antibody for 1 hour at room temperature. Cells were mounted with SlowFade® Gold Antifade Reagent with DAPI (Invitrogen), and imaged using a Zeiss 710 microscope.
Statistical analysis
The three experiments at minimum were completed for each analysis. Data are presented as mean ± standard deviation. Statistical difference was analyzed using Student’s t test. P value less than 0.05 was considered statistically significant.
Results
KLF8 interacts with and is ubiquitylated by NEDD4
As shown in Figure 1A, NEDD4 contains four WW domains that mediate its interaction with substrate proteins. On the substrate side, various motifs can interact with the NEDD4 WW domains. Examples of such motifs are proline-rich motifs such as PPXY, PPLP, PPR, and phospho-serine motif pSP or phospho-threonine motif pTP [47,48]. Interestingly, our previous study demonstrated a pS48P site in KLF8 that is phosphorylated by ERK2 and is associated with ubiquitylation of KLF8 [22]. Thus, we speculated that NEDD4 may directly bind to KLF8 at the pS48P site via its WW domain. To test this notion, we co-transfected the HEK293 cells with Myc-tagged NEDD4 and HA-tagged KLF8 and performed co-immunoprecipitation (co-IP) assay. As shown in Figure 1B, the Myc-NEDD4 protein was co-IPed together with the HA-KLF8 protein using an antibody against HA peptide only when co-expressed with HA-KLF8, but not with the HA-vector alone. This result indicates that KLF8 binds to NEDD4 and raises the possibility that KLF8 could be ubiquitylated by NEDD4.
Figure 1.
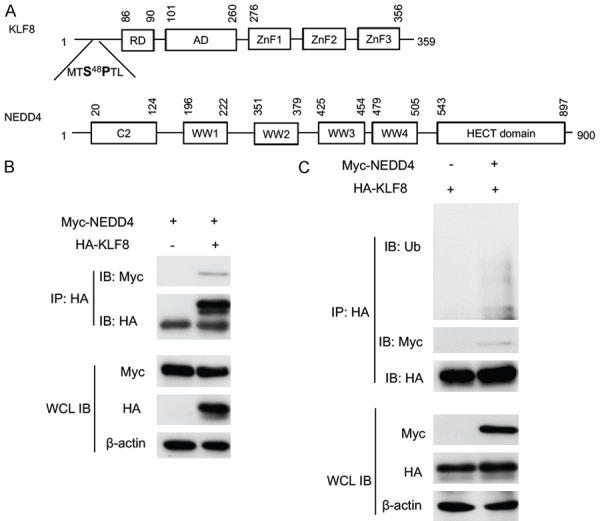
KLF8 interacts with and is ubiquitylated by NEDD4. A. The domain structure of KLF8 and NEDD4. Amino acid positions are numbered on the shoulders of the individual domains. Potential interacting S48P motif of KLF8 and WW domain of NEDD4 are highlighted. B. NEDD4 is co-IPed with KLF8. Myc-tagged NEDD4 was co-transfected with HA-tagged KLF8 in HEK293 cells. Whole cell lysates (WCL) were prepared 48 h later for IP with an anti-HA antibody for the KLF8 and co-IPed NEDD4 determined by subsequent immunoblotting (IB) with an anti-HA and anti-Myc antibody, respectively. C. KLF8 is ubiquitylated by NEDD4. NEDD4 was co-transfected with HA-KLF8 in HEK293 cells. KLF8 was IPed with an anti-HA antibody and ubiquitylated KLF8 was detected by immunoblotting the precipitates with an anti-ubiquitin antibody. Expression of the KLF8 and NEDD4 in the WCL was confirmed by IB with an anti-actin antibody serving as a loading control.
To test if KLF8 is a ubiquitylation substrate of NEDD4, we performed in vivo ubiquitylation assay. The HA-tagged KLF8 was expressed alone or co-expressed with the Myc-NEDD4 in HEK293 cells. Ubiquitylation of KLF8 was determined by anti-HA IP followed by immunoblot with an anti-ubiquitin antibody. The KLF8 protein was heavily ubiquitylated when co-expressed with NEDD4, whereas the ubiquitylation of KLF8 was barely detectable in the absence of NEDD4 co-expression (Figure 1C). This result clearly indicates that KLF8 is targeted by NEDD4 for ubiquitylation in the cell.
S48 phosphorylation by ERK is required for KLF8 interaction with and ubiquitylation by NEDD4
To determine if the pS48P site of KLF8 is responsible for its interaction with NEDD4, we compared the NEDD4-binding capacity between the wild-type (WT) KLF8 and mutant KLF8 at the S48 site that either prevents the S48 from being phosphorylated (S48A) or mimics the phosphorylated S48 (S48D). Co-expression followed by co-IP and immunoblot analysis showed that the S48A mutant lost its ability to bind NEDD4, whereas the S48D mutant interaction with NEDD4 was strongly enhanced when compared to the WT KLF8 (Figure 2A).
Figure 2.
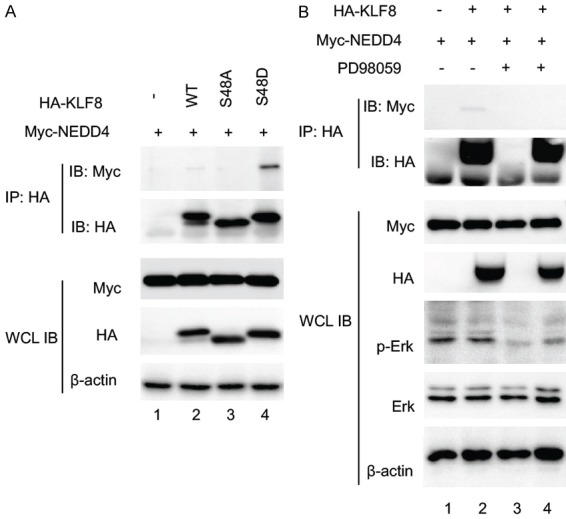
S48 phosphorylation by ERK is required for KLF8 to interact with NEDD4. A. The Serine 48 residue of KLF8 is responsible for the binding of NEDD4 and KLF8. HA-KLF8, wild-type or mutants indicated, was co-expressed with Myc-NEDD4 in HEK293 cells for 48 hours. Co-IP and IB were carried out similarly as described in Figure 1. B. ERK activity is required for the binding of KLF8 to NEDD4. Similarly treated HEK293 cells overexpressing Myc-NEDD4 and HA-KLF8 were treated with the MEK inhibitor PD98059 (50 mM) or DMSO for 30 min prior to cell lysate preparation for the similar co-IP assay. Effective inhibition of ERK activity was verified by IB, the WCL with an antibody against active ERK (p-Erk) and an antibody against total ERK (Erk). Expression of the KLF8 and NEDD4 was confirmed by immunoblotting the WCL with b-actin serving as a loading control.
To further examine the requirement of S48 phosphorylation for the interaction between KLF8 and NEDD4, we inhibited the ERK activity in the HEK293 cells co-expressing Myc-NEDD4 and HA-KLF8 by treating the cells with the MEK inhibitor PD98059 prior to the co-IP/immunoblot analysis (Figure 2B). The interaction was abolished when the ERK activity was inhibited (compare lane 4 to lane 2). These results demonstrate that KLF8 interaction with NEDD4 depends upon the phosphorylation of the S48 site by ERK. Similar to the NEDD4-binding capacity, the level of ubiquitylation was significantly reduced in the S48A while enhanced in the S48D mutant (Figure 3).
Figure 3.
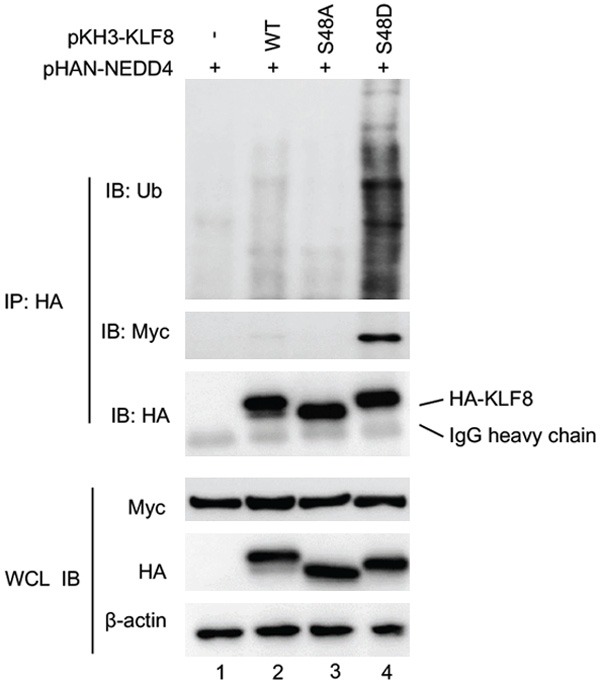
S48 phosphorylation is required for KLF8 ubiquitylation by NEDD4. Myc-tagged NEDD4 was co-expressed with HA-KLF8 or its mutants S48A and S48D in HEK293 cells for 48 hours followed by 12-hour treatment with MG132. Whole cell lysates were prepared for IP with anti-HA followed by IB with anti-ubiquitin. Interaction between KLF8 and NEDD4 was examined by co-IP similarly. Expression of the KLF8 and NEDD4 was confirmed by immunoblotting the WCL with b-actin serving as a loading control.
Taken together, these results strongly suggest that KLF8 is a substrate of the NEDD4 E3 ubiquitin ligase and its ubiquitylation is regulated by the ERK-mediated phosphorylation of S48.
NEDD4 co-expression enhances the stability of KLF8
We next sought to see if the steady-state level of KLF8 protein is regulated by NEDD4. HEK293 cells were transfected with HA-KLF8 alone or together with Myc-NEDD4 and followed by the cycloheximide chasing assay. The result showed a dramatic increase in the level of KLF8 in the presence of NEDD4 (Figure 4), suggesting that NEDD4-mediated ubiquitylation of KLF8 does not shorten the overall life span of KLF8 but rather prolongs it.
Figure 4.
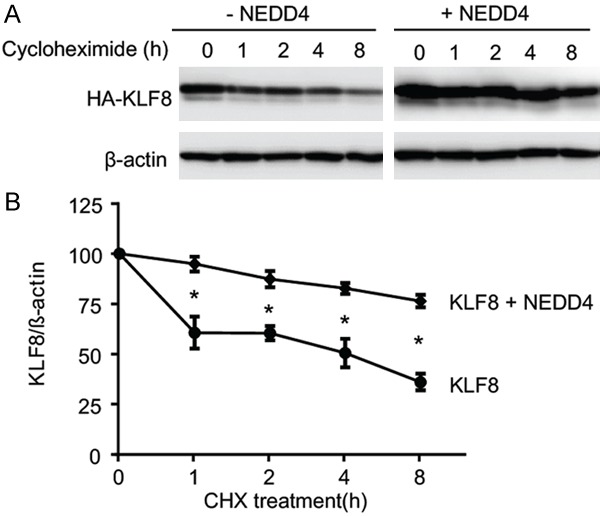
NEDD4 co-expression enhances the stability of KLF8. A. The half-life of KLF8 protein is prolonged by NEDD4. HA-KLF8 was co-expressed with an empty vector (-NEDD4) or Myc-NEDD4 (+NEDD4) in HEK293 cells for 48 hours. The cells were treated with 50 mg/ml cycloheximide for various times indicated. Whole cell lysates were prepared at each of the time points for IB with an anti-HA antibody with the b-actin loading control. B. The chemiluminescence intensity of KLF8 protein in the blots was quantified using Image Lab 3.0 (Bio-Rad, Hercules, CA) as we previously reported [12] and graphed in relative to the zero point of time. *P<0.005.
NEDD4 co-expression enhances the transcriptional activity of KLF8 in the pS48 dependent manner
We then tested the effect of NEDD4 on the molecular function of KLF8 as a transcription factor using the target gene promoter-reporter assay. The cyclin D1 promoter is the very first to be identified as a target of transcriptional activation by KLF8 [9]. The cyclin D1 promoter luciferase report was co-transfected with wild-type or mutant KLF8 in the presence or absence of NEDD4 (Figure 5).
Figure 5.
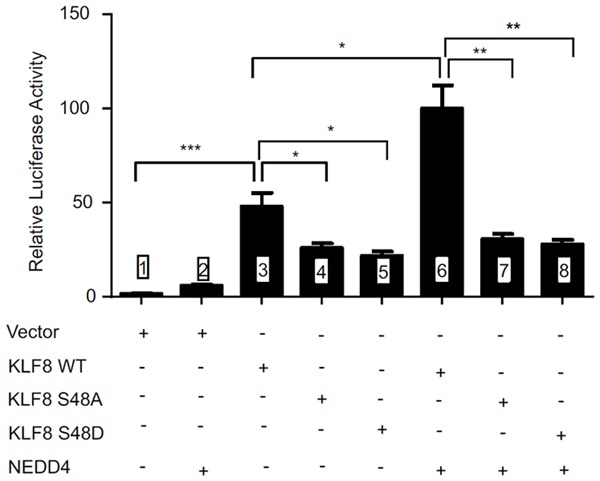
NEDD4 co-expression enhances the transcriptional activity of KLF8 in the pS48 dependent manner. luciferase activity in NIH3T3 cells co-transfected with the plasmid vectors encoding wild type (WT) KLF8, its mutants (S48A or S48D) or nothing (vector), alone or together with NEDD4 expressing vector, along with the cyclin D1 promoter luciferase reporter plasmid [9,44]. The reporter activity was performed as described in the Materials and Methods section [44]. *P<0.05, **P<0.01, ***P<0.005.
As expected, the promoter activity was strongly activated by wild-type KLF8 alone, and was further enhanced in the presence of NEDD4 (compare lanes 1, 3 & 6). By contrast, the promoter activity dropped by almost 50% when the S48 is mutated, whether S48A or S48D (compare lanes 3, 4 & 5), and overexpression of NEDD4 did not change the promoter activity associated with the KLF8 mutants (compare lanes 4 & 5 with 7 & 8). These results suggest that the ubiquitylation of KLF8 by NEDD4 promotes the transcriptional activity of KLF8 and this effect depends upon the S48.
Ubiquitylation of KLF8 by NEDD4 occurs likely in the nucleus and does not affect the nuclear localization of KLF8
It is well known that ERK phosphorylates its substrate proteins in the nucleus [49] and NEDD4 is localized both outside and inside the nucleus [50,51]. Our previous studies suggest that KLF8 may be ubiquitylated both in the cytoplasm [12] and the nucleus [22]. To determine the subcellular location where NEDD4 ubiquitylates KLF8, we performed immunofluorescent staining of KLF8 or its mutants, transfected alone or together with NEDD4 for their co-localization in the HEK293 cell (Figure 6). In the absence of NEDD4 co-expression, all three forms of KLF8 protein are exclusively localized in the nucleus (Figure 6A). In the presence of NEDD4 expression albeit being predominantly cytoplasmic, all KLF8 forms remain localized in the nucleus (Figure 6B).
Figure 6.
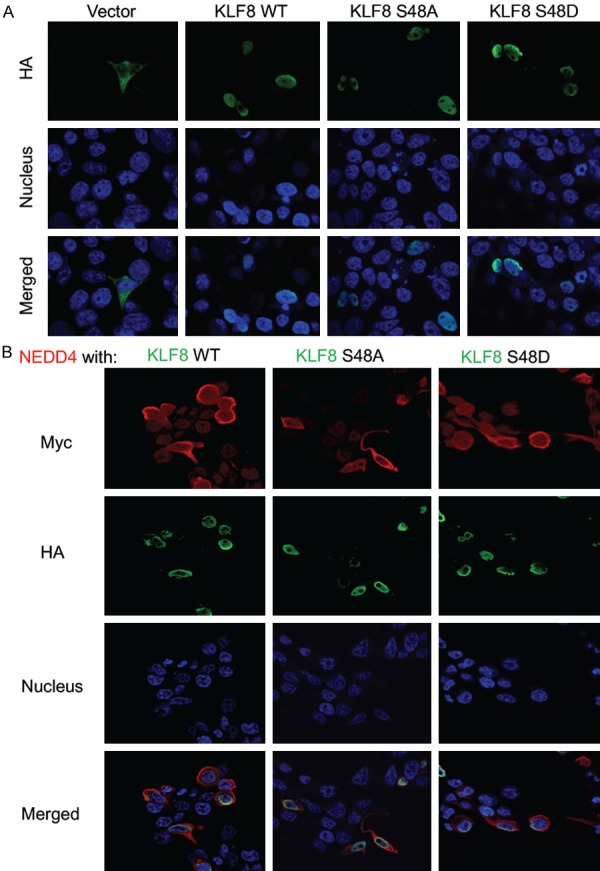
NEDD4 regulates KLF8 likely in the nucleus. HEK293 cells were transfected with the plasmids encoding HA-KLF8, wild type (WT) or mutants (S48A or S48D), alone (A) or together with the plasmid encoding Myc-NEDD4 (B). After 24 hours, the cells were fixed, permeabilized, and stained with an antibody for HA or Myc and the Hoechst dye for the nucleus followed by fluorescein-conjugated secondary antibody, and photographed by fluorescent microscopy.
These results indicate that the ubiquitylation of KLF8 takes place in the nucleus by a small fraction of NEDD4 and this modification does not affect the nuclear presence of KLF8.
Discussion
Our work identified NEDD4 as a novel E3 ubiquitin ligase for KLF8 or KLF8 as a novel substrate of NEDD4-mediated ubiquitylation in the nucleus (Figure 7).
Figure 7.
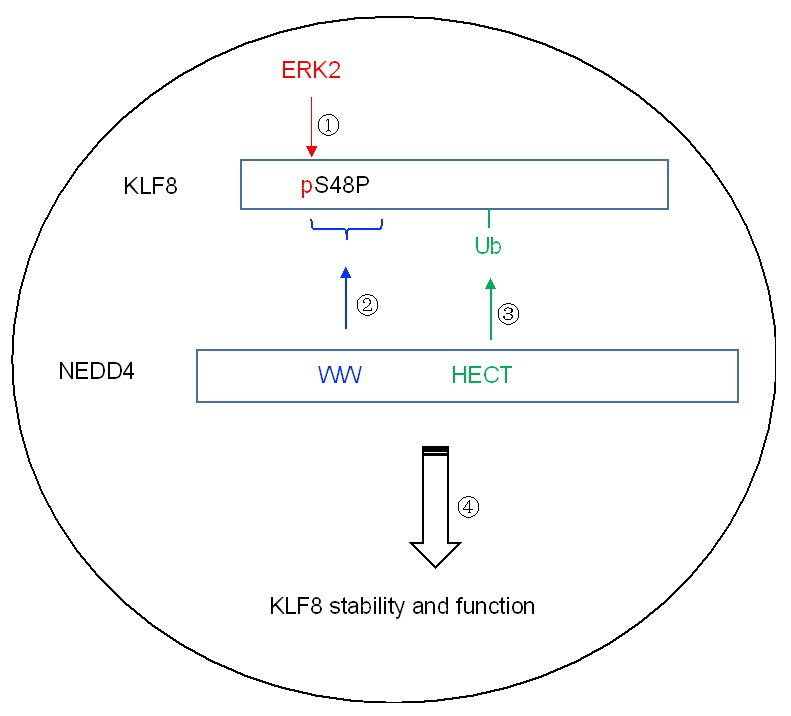
Model of mechanisms of regulation of KLF8 via phosphorylation and ubiquitylation. In the nucleus, ERK2 phosphorylates KLF8 which creates the pS48P motif. This motif recruits NEDD4 by binding to the WW domain which enables NEDD4 to add a ubiquitin to KLF8. This modification of the phosphorylated form of KLF8 plays a role in retaining the overall stability and function of KLF8 protein in the nucleus.
Once phosphorylated by ERK2, the phosphorylated form of KLF8 is able to recruit NEDD4 so that NEDD4 is able to ubiquitylate it. The overall outcome of this modification is enhanced stability and transcriptional activity of KLF8.
To ubiquitylate a substrate protein, NEDD4 needs to briefly interact with the substrate molecule. This interaction is mostly mediated with its WW domain that finds its interaction motif, such as a proline-rich motif and a phospho-serine (or phospho-threonine) followed by a proline residual in the substrate molecule [47,48]. The phosphorylation of KLF8 by ERK2 takes place at the serine 48 site [22] with a proline residual to the right (Figure 7). The S48 phosphorylation would create a perfect binding motif (pS48P) for the WW domains of NEDD4. Given that both the S48 residual and ERK activity are required for the interaction between KLF8 and NEDD4 - as well as the ubiquitylation of KLF8 by NEDD4 (Figures 2 and 3) - it is very likely that the interaction takes place via direct binding between the pS48P site in KLF8 and the WW domains in NEDD4, and that the phosphorylation at the S48 site by ERK is the priming event for the interaction and subsequent ubiquitylation of KLF8. Importantly, both the S48P site in KLF8 and the WW domains in NEDD4 are well conserved across the species [22,24], suggesting an evolutionally broad significance of the ubiquitylation of KLF8 by NEDD4.
Our results showed that NEDD4 does not bind, ubiquitylate or activate KLF8 if the S48P site remains in an unphosphorylated state (Figures 2, 3 and 5). Interestingly, the ubiquitylation by NEDD4 does not reduce the lifespan of KLF8 protein, but rather prolongs it (Figure 4). These seemingly paradoxical results again support our previous finding that the phosphorylated form of KLF8 plays a unique protective role for retaining its unphosphorylated form to the functional level in the nucleus [22]. In other words, NEDD4 targets the phosphorylated form of KLF8 for ubiquitylation and degradation in order to protect the unphosphorylated, functional form of KLF8 in the nucleus.
Typically, ubiquitylated protein is subject to degradation in the proteasome, and phosphorylation is frequently a priming event for ubiquitylation. Indeed, our recent work has demonstrated that the phosphorylated form of KLF8 protein suffers from rapid degradation whereas the co-present. unphosphorylated form of KLF8 does not [22]. However, in the absence of the phosphorylated form, the unphosphorylated KLF8 is degraded rapidly [22]. This suggests that the phosphorylated KLF8 is ubiquitylated and degraded to protect the unphosphorylated form of KLF8 from degradation. We do not know if the unphosphorylated form of KLF8 can be ubiquitylated by the same ubiquitin E3 ligase NEDD4. The answer is probably not as the S48A KLF8 mutant showed little or no ubiquitylation when co-expressed with NEDD4 (Figure 3). The degradation of the unphosphorylated KLF8 could even be caused by a mechanism not related to ubiquitylation. It will be interesting to find out what protein is responsible for degrading the unphosphorylated KLF8 and how this process is interfered with when phosphorylated KLF8 is modified by NEDD4.
Protein ubiquitylation can take place anywhere in the cell. Our previous studies indicated that KLF8 can be ubiquitylated both in the cytoplasm [12,45] and the nucleus [22]. Our data showed that NEDD4 is broadly distributed in the cell (Figure 6B), whereas the KLF8 protein remains in the nucleus under the experimental conditions whether it is the wild-type or the S48 phosphorylation defective or mimicking mutant (Figure 6). These results indicate that a small fraction of NEDD4 molecules in the nucleus are responsible for the ubiquitylation of KLF8. This makes sense considering that the nucleus is also the functioning subcellular compartment for ERKs. We do not rule out the possibility that KLF8 protein transported into the cytoplasm in the PARP-1 defective cell [45] may also be modified by the same NEDD4 E3 ubiquitin ligase, which will be interesting to test in the future.
Previously, we discovered a switch between acetylation and sumoylation of KLF8 critical for its transcriptional activity [8,21]. In this current work, we uncovered the coordination between a kinase/phosphorylation and an E3 ubiquitin ligase/ubiquitylation - a novel mechanism that regulates KLF8 expression at the posttranslational level - and for the first time tied KLF8 signaling and NEDD4 signaling together. Interestingly, both pathways contribute much to the progression of various cancer types. An interesting example is that both KLF8 and NEDD4 work to maintain an aberrantly high level of EGFR in multiple cancer types albeit by distinct mechanisms [2,38].
Conclusions
In summary, this work identified KLF8 as a novel ubiquitylation substrate for the E3 ubiquitin ligase NEDD4. It also revealed a novel posttranslational cross-talk between phosphorylation and ubiquitylation critical for the stability and transcriptional activity of KLF8 in the nucleus. These findings provide further insight into the molecular mechanism underlying the aberrant overexpression of KLF8 and NEDD4 in cancer and supports the future development of NEDD4-KLF8 axis targeted anticancer therapies.
Acknowledgements
This work was supported partially by a grant from National Institute of Health (R01 CA123977) and institutional funds to JZ and grants from National Natural Science Foundation of China (NSFC) WY (No. 81472558). AS was a joint Ph.D. student co-mentored by JZ and QL and received a scholarship award for study overseas from the China Scholarship Council, the Ministry of Education of the P.R. China (No. 201608320269).
Disclosure of conflict of interest
None.
References
- 1.Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–7193. doi: 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- 2.Li T, Lu H, Mukherjee D, Lahiri SK, Shen C, Yu L, Zhao J. Identification of epidermal growth factor receptor and its inhibitory microRNA141 as novel targets of Kruppel-like factor 8 in breast cancer. Oncotarget. 2015;6:21428–21442. doi: 10.18632/oncotarget.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, Lu H, Shen C, Lahiri SK, Wason MS, Mukherjee D, Yu L, Zhao J. Identification of epithelial stromal interaction 1 as a novel effector downstream of Kruppel-like factor 8 in breast cancer invasion and metastasis. Oncogene. 2014;33:4746–4755. doi: 10.1038/onc.2013.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu H, Hu L, Yu L, Wang X, Urvalek AM, Li T, Shen C, Mukherjee D, Lahiri SK, Wason MS, Zhao J. KLF8 and FAK cooperatively enrich the active MMP14 on the cell surface required for the metastatic progression of breast cancer. Oncogene. 2014;33:2909–2917. doi: 10.1038/onc.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee D, Lu H, Yu L, He C, Lahiri SK, Li T, Zhao J. Kruppel-like factor 8 activates the transcription of C-X-C cytokine receptor type 4 to promote breast cancer cell invasion, transendothelial migration and metastasis. Oncotarget. 2016;7:23552–23568. doi: 10.18632/oncotarget.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Lu H, Urvalek AM, Li T, Yu L, Lamar J, DiPersio CM, Feustel PJ, Zhao J. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011;30:1901–1911. doi: 10.1038/onc.2010.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Oncogene. 2007;26:456–461. doi: 10.1038/sj.onc.1209796. [DOI] [PubMed] [Google Scholar]
- 8.Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem. 2006;281:16664–16671. doi: 10.1074/jbc.M513135200. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Bian ZC, Yee K, Chen BP, Chien S, Guan JL. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol Cell. 2003;11:1503–1515. doi: 10.1016/s1097-2765(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 10.Bian T, Jiang D, Liu J, Yuan X, Feng J, Li Q, Zhang Q, Li X, Liu Y, Zhang J. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem Biophys Res Commun. 2017;492:461–467. doi: 10.1016/j.bbrc.2017.08.074. [DOI] [PubMed] [Google Scholar]
- 11.Lahiri SK, Zhao J. Kruppel-like factor 8 emerges as an important regulator of cancer. Am J Transl Res. 2012;4:357–363. [PMC free article] [PubMed] [Google Scholar]
- 12.Lu H, Hu L, Li T, Lahiri S, Shen C, Wason MS, Mukherjee D, Xie H, Yu L, Zhao J. A novel role of Kruppel-like factor 8 in DNA repair in breast cancer cells. J Biol Chem. 2012;287:43720–43729. doi: 10.1074/jbc.M112.418053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, Luo X, Yan Q, Zhang W, Wu Y, Zhang M, Zhao J, Peng Y, Chen Y, Zhang Y, Chen C, Cheng T, Chen C, Liu S, Bai Y, Wang J. Suppression of KLF8 induces cell differentiation and sensitizes colorectal cancer to 5-fluorouracil. Oncol Rep. 2015;34:1221–1230. doi: 10.3892/or.2015.4094. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Urvalek AM, Liu J, Zhao J. Activation of KLF8 transcription by focal adhesion kinase in human ovarian epithelial and cancer cells. J Biol Chem. 2008;283:13934–13942. doi: 10.1074/jbc.M709300200. [DOI] [PubMed] [Google Scholar]
- 15.Shi X, Luo X, Yan Q, Zhang W, Wu Y, Zhang M, Zhao J, Peng Y, Chen Y, Zhang Y, Chen C, Cheng T, Liu S, Bai Y, Wang J. Suppression of KLF8 induces cell differentiation and sensitizes colorectal cancer to 5-fluorouracil. Oncol Rep. 2015;34:1221–1230. doi: 10.3892/or.2015.4094. [DOI] [PubMed] [Google Scholar]
- 16.Yan Q, Zhang W, Wu Y, Wu M, Zhang M, Shi X, Zhao J, Nan Q, Chen Y, Wang L, Cheng T, Li J, Bai Y, Liu S, Wang J. KLF8 promotes tumorigenesis, invasion and metastasis of colorectal cancer cells by transcriptional activation of FHL2. Oncotarget. 2015;6:25402–25417. doi: 10.18632/oncotarget.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Wang X, Urvalek AM, Li T, Xie H, Yu L, Zhao J. Transformation of human ovarian surface epithelial cells by Kruppel-like factor 8. Oncogene. 2014;33:10–18. doi: 10.1038/onc.2012.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang T, Cai SY, Zhang J, Lu JH, Lin C, Zhai J, Wu MC, Shen F. Kruppel-like factor 8 is a new Wnt/beta-catenin signaling target gene and regulator in hepatocellular carcinoma. PLoS One. 2012;7:e39668. doi: 10.1371/journal.pone.0039668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi R, Chen B, Zhao J, Zhan X, Zhang L, Liu X, Dong Q. Kruppel-like factor 8 ameliorates Alzheimer’s disease by activating beta-catenin. J Mol Neurosci. 2014;52:231–241. doi: 10.1007/s12031-013-0131-4. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Kim HJ, Lee YJ, Lee MY, Choi H, Kim JW. Kruppel-like factor KLF8 plays a critical role in adipocyte differentiation. PLoS One. 2012;7:e52474. doi: 10.1371/journal.pone.0052474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urvalek AM, Lu H, Wang X, Li T, Yu L, Zhu J, Lin Q, Zhao J. Regulation of the oncoprotein KLF8 by a switch between acetylation and sumoylation. Am J Transl Res. 2011;3:121–132. [PMC free article] [PubMed] [Google Scholar]
- 22.Lahiri SK, Lu H, Mukherjee D, Yu L, Zhao J. ERK2 phosphorylates Kruppel-like factor 8 protein at serine 48 to maintain its stability. Am J Cancer Res. 2016;6:910–923. [PMC free article] [PubMed] [Google Scholar]
- 23.Boase NA, Kumar S. NEDD4: the founding member of a family of ubiquitin-protein ligases. Gene. 2015;557:113–122. doi: 10.1016/j.gene.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 25.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol Biol Cell. 2009;20:1816–1832. doi: 10.1091/mbc.E08-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blot V, Perugi F, Gay B, Prevost MC, Briant L, Tangy F, Abriel H, Staub O, Dokhelar MC, Pique C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 28.Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, Machluf Y, Asao H, Jovin T, Sugamura K, Yarden Y. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 2002;3:740–751. doi: 10.1034/j.1600-0854.2002.31006.x. [DOI] [PubMed] [Google Scholar]
- 29.Lin Q, Wang J, Childress C, Sudol M, Carey DJ, Yang W. HECT E3 ubiquitin ligase Nedd4-1 ubiquitinates ACK and regulates epidermal growth factor (EGF)-induced degradation of EGF receptor and ACK. Mol Cell Biol. 2010;30:1541–1554. doi: 10.1128/MCB.00013-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S, Weissman AM. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- 31.Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem. 2005;280:27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 32.Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8:1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 33.Lin Q, Dai Q, Meng H, Sun A, Wei J, Peng K, Childress C, Chen M, Shao G, Yang W. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J Cell Sci. 2017;130:3839–3850. doi: 10.1242/jcs.207068. [DOI] [PubMed] [Google Scholar]
- 34.Sun A, Wei J, Childress C, Shaw JH 4th, Peng K, Shao G, Yang W, Lin Q. The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy. Autophagy. 2017;13:522–537. doi: 10.1080/15548627.2016.1268301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abriel H, Kamynina E, Horisberger JD, Staub O. Regulation of the cardiac voltage-gated Na+ channel (H1) by the ubiquitin-protein ligase Nedd4. FEBS Lett. 2000;466:377–380. doi: 10.1016/s0014-5793(00)01098-x. [DOI] [PubMed] [Google Scholar]
- 36.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, Jiang X. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao G, Wang R, Sun A, Wei J, Peng K, Dai Q, Yang W, Lin Q. The E3 ubiquitin ligase NEDD4 mediates cell migration signaling of EGFR in lung cancer cells. Mol Cancer. 2018;17:24. doi: 10.1186/s12943-018-0784-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persaud A, Alberts P, Amsen EM, Xiong X, Wasmuth J, Saadon Z, Fladd C, Parkinson J, Rotin D. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol Syst Biol. 2009;5:333. doi: 10.1038/msb.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leon S, Erpapazoglou Z, Haguenauer-Tsapis R. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol Biol Cell. 2008;19:2379–2388. doi: 10.1091/mbc.E08-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q, Zhang S, Chen G, Zhou H. E3 ubiquitin ligase Nedd4 inhibits AP-1 activity and TNF-alpha production through targeting p38alpha for polyubiquitination and subsequent degradation. Sci Rep. 2017;7:4521. doi: 10.1038/s41598-017-04072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun A, Yu G, Dou X, Yan X, Yang W, Lin Q. Nedd4-1 is an exceptional prognostic biomarker for gastric cardia adenocarcinoma and functionally associated with metastasis. Mol Cancer. 2014;13:248. doi: 10.1186/1476-4598-13-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urvalek AM, Wang X, Lu H, Zhao J. KLF8 recruits the p300 and PCAF co-activators to its amino terminal activation domain to activate transcription. Cell Cycle. 2010;9:601–611. doi: 10.4161/cc.9.3.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Pestell R, Guan JL. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Mol Biol Cell. 2001;12:4066–4077. doi: 10.1091/mbc.12.12.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu H, Wang X, Li T, Urvalek AM, Yu L, Li J, Zhu J, Lin Q, Peng X, Zhao J. Identification of poly (ADP-ribose) polymerase-1 (PARP-1) as a novel Kruppel-like factor 8-interacting and -regulating protein. J Biol Chem. 2011;286:20335–20344. doi: 10.1074/jbc.M110.215632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol. 1998;143:1997–2008. doi: 10.1083/jcb.143.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 48.Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- 49.Zehorai E, Yao Z, Plotnikov A, Seger R. The subcellular localization of MEK and ERK--a novel nuclear translocation signal (NTS) paves a way to the nucleus. Mol Cell Endocrinol. 2010;314:213–220. doi: 10.1016/j.mce.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Staub O, Abriel H, Plant P, Ishikawa T, Kanelis V, Saleki R, Horisberger JD, Schild L, Rotin D. Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination. Kidney Int. 2000;57:809–815. doi: 10.1046/j.1523-1755.2000.00919.x. [DOI] [PubMed] [Google Scholar]
- 51.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]


