Abstract
This study aimed to investigate the effect of non-small cell lung cancer (NSCLC) cell-derived exosome on cell proliferation and apoptosis in normal lung fibroblast cells and NSCLC cells, and whether it regulates cell functions through delivering alpha-smooth muscle actin (ASMA). NSCLC exosomes were extracted from A549 cells, then cocultured with normal lung fibroblasts (HLF1 cells) and NSCLC cells (A549 cells). Blank ShRNA and ASMA ShRNA plasmids were transferred into HLF1 cells/A549 cells with or without NSCLC exosomes, which were divided into 4 groups accordingly: Negative control (NC) group, SH group, Exosome group and Exosome+SH group. Western blot, immunofluorescence, qPCR, CCK-8 and AV/PI were used to detect protein level, gene expression, cell proliferation and cell apoptosis, respectively. In HLF1 cells, cell proliferation was enhanced while cell apoptosis rate was inhibited in Exosome group compared with NC group; and cell proliferation was attenuated while cell apoptosis rate was raised in Exosome+SH group than Exosome group in rescue experiment; the expressions of apoptotic markers C-caspase3 and Bcl-2 also revealed the same trends. Additionally, in A549 cells, cell proliferation was also increased while cell apoptosis was inhibited in Exosome group compared with NC group; and cell proliferation was reduced while cell apoptosis rate was elevated in Exosome+SH group than Exosome group in rescue experiment. In conclusion, NSCLC derived exosomes promote cell proliferation and inhibit cell apoptosis in both normal lung fibroblasts and NSCLC cells by delivering ASMA.
Keywords: Non-small cell lung cancer (NSCLC), exosome, proliferation, apoptosis, ASMA
Introduction
Lung cancer is the most frequent and lethal cancer worldwide and is also first among all cancers in China regarding its incidence and mortality with an estimated 7.33 million new lung cancer cases and 6.10 million lung cancer-related deaths occurring in China in 2015 [1,2]. Non-small cell lung cancer (NSCLC), accounting for roughly 85% of all lung cancer cases, can be treated by surgical resection, radiation therapy, chemotherapy, molecular targeted therapy and immunotherapy [3]. Through accumulating efforts to explore the pathology and genetics of NSCLC, the treatment landscape of this most frequent tumor has largely improved because precision medicine has been greatly progressed accordingly [3-5]. Nonetheless, the prognosis of NSCLC patients is still not satisfactory. Approximately 65% of NSCLC cases are advanced cases at diagnosis; however the proportion of advanced patients responding to platinum-based chemotherapy, which is the mainstay of treatment for patients who cannot undergo surgical resections, is reported to merely range from 25% to 30%, with 1-year survival rate being only 30% to 40% [6,7]. Thus, deeper investigation into the pathogenesis and the exploration of new treatment targets of NSCLC are extremely necessary.
Exosomes are a category of extracellular vesicles (EVs) with two layers of lipid membrane that are secreted by most types of cells via exocytosis, measuring 30-150 nm and containing/transferring various biomolecules which include DNA, RNA, proteins and lipids that are developed from its excreting cells [8-14]. Recent studies have revealed quite promising functions of exosomes in tumorigenesis, which consist of activating angiogenic responses, promoting evasion of the host immune system and increasing the permeability of vasculature to metastatic cancer cells [15-17]. There are also studies illustrating the effects of cancer cells derived exosomes on the activities of normal cells, such as T cells and alveolar progenitor type II cells, and cancer cells, for example, the myeloma cells and breast cancer cells [18,19]. However, the regulatory role of cancer cell-secreted exosomes in the development and progression of NSCLC is still unknown. Thus, we aimed to investigate the effect of NSCLC cells derived exosomes on cell proliferation and apoptosis in normal lung fibroblast cells and NSCLC cells, and whether it regulates cell functions through delivering alpha-smooth muscle actin (ASMA).
Materials and methods
Cells culture
The normal lung fibroblast cell line HLF1 and NSCLC cell line A549 were purchased from Cell Resource Center of Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China). HLF1 cells were cultured in 90% F12K medium (SIGMA, USA) with 10% Exosome-depleted FBS Media Supplement (SBI, USA), and A549 cells were cultured in 90% F12K medium (SIGMA, USA) with 10% Exosome-depleted FBS Media Supplement (SBI, USA). In addition, the A549 cells were cultured for 72 h, and 106 A549 cells were used for exosomes extraction.
A549 exosome extraction and validation
Exosomes were extracted from A549 cells medium using Total Exosome Isolation Reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The supernatant was discarded after the ultracentrifugation, and the remaining fluid was used for A549 exosomes extraction. Subsequently, 10 μL of A549 exosomes was diluted by polybutylene succinate (PBS) solution, and then the diluted A549 exosomes solution was added to the copper wire mesh, which was then put in room temperature for 1 min. Afterward, 3% phosphate tungsten was added for negative staining under room temperature for 5 min, which was then washed by the distilled water for once, and then the transmission electron microscopy (Hitachi, Japan) was used to determine the morphology of exosomes. And a BCA Protein Assay Kit (Sangon Biotech, China) was used for the quantification of the exosomes. In order to validate the successful isolation of exosomes, the total protein was extracted from exosome using Exosome Immunoprecipitation Reagent (Protein A) (Invitrogen, USA), and exosome markers CD9, CD63, CD81, TGF-β1, Flotillin-1 and erythroblast enhancing factor 2 (EEF2) expressions were determined by Western blot, and sample from the fetal bovine serum (FBS) free F12K medium was negative control NC (NC) group. Additionally, the exosomes are defined as nanosized particles measuring 30-50 nm made up of two layers of lipid membrane secreted by cells via exocytotic process, and the protein markers for exosomes include CD9, CD63, CD86, TGF-β, Flotillin-1 and EEF2.
Coculture of A549 exosome with HLF1 cells and subsequent assays
First, 200 μg A549 exosome was added in the HLF1 cells as follows: 200 ug/ml A549 exosomes were added in 90% F12K medium (SIGMA, USA) with 10% Exosome-depleted FBS Media Supplement (SBI, USA) to culture HLF1 cells as Exosome group, while 90% F12K medium (SIGMA, USA) with 10% Exosome-depleted FBS Media Supplement (SBI, USA) without exosome addition was used to culture HLF1 cells as NC group. Second, after culture, cell proliferation was detected by CCK8 at 0 h, 24 h, 48 h and 72 h, cell apoptosis rate was detected by AV/PI at 72 h, cells pro-apoptotic marker (C-Caspase3) and anti-apoptotic marker (Bcl-2) expressions were detected by Western blot at 72 h, and ASMA expression was detected by quantitative polymerase chain reaction (qPCR) and immunofluorescence at 72 h. In addition, in the above assays, each group was assessed in triplicate.
Transfection and subsequent assays in HLF1 cells
To explore whether A549 exosomes regulated HLF1 cell functions through modulating ASMA expression, ShRNA plasmids pGPU6 (NTCC, CHINA) was used for transfection. Blank ShRNA plasmids, ASMA ShRNA plasmids were transferred into HLF1 cells with or without subsequently adding A549 exosomes and were accordingly divided into 4 groups: NC group (blank ShRNA), SH group (ASMA ShRNA), Exosome group (A549 exosome and blank ShRNA) and Exosome+SH (A549 exosomes and ASMA ShRNA) group. After culturing, ASMA expression was detected by quantitative polymerase chain reaction (qPCR) and immunofluorescence at 72 h, cell proliferation was detected by CCK8 at 0 h, 24 h, 48 h and 72 h, and cell apoptosis rate was detected by AV/PI at 72 h.
Effect of A549 exosome and ASMA on regulating A549 cell functions
To further explore the effect of A549 exosomes and ASMA on regulating NSCLC cells, we repeated experiments (similar to those performed with HLF1 cells) in A549 cells. First, 200 ug/ml A549 exosomes were added to A549 cells as Exosome group, and A549 cells without exosome addition served as NC group, and cell proliferation, cell apoptosis rate, apoptotic markers and expression of ASMA were subsequently detected. Second, blank ShRNA plasmids, ASMA ShRNA plasmids were also transferred into A549 cells with or without subsequently adding A549 exosome and were accordingly divided into 4 groups: NC group, SH group, Exosome group and Exosome+SH group, and ASMA expression, cell proliferation and cell apoptosis rate were subsequently detected.
Western blot analysis
After the extraction of total protein, the concentration of total protein was evaluated by a BCA kit (Pierce Biotechnology, USA), and the standard curve was made for the calculation of total protein concentration. Subsequently, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis was performed to separate the total protein, which was then transformed to the polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, USA) with the same size as the SDS-PAGE gel. After blocking the PVDF membrane with 5% skim milk powder Tris-Buffered Saline Tween (TBST) for 2 h, the primary antibodies were added and incubated in 37°C for 1-3 h or under 4°C overnight, and then the secondary antibody was added, which was incubated under 37°C for 1 h. Then the bands were exposed to X-ray and visualized using an enhanced chemiluminescence (ECL) kit (Millipore, Bedford, USA). The antibodies used in Western blot were listed in Table 1.
Table 1.
Antibodies using in Western blot assay
| Antibody | Company (Country) | Dilution |
|---|---|---|
| Primary Antibody | ||
| CD9 Antibody | Affinity (China) | 1:1000 |
| CD63 Antibody | Affinity (China) | 1:1000 |
| CD81 Antibody | Affinity (China) | 1:1000 |
| TGF beta1 Antibody | Affinity (China) | 1:1000 |
| Cleaved Caspse-3 antibody | CST (USA) | 1:1000 |
| Caspase-3 Rabbit mAb | CST (USA) | 1:1000 |
| Bcl-2 Rabbit mAb | CST (USA) | 1:1000 |
| GAPDH Rabbit mAb | CST (USA) | 1:1000 |
| Secondary Antibody | ||
| Anti-rabbit IgG, HRP-linked Antibody | CST (USA) | 1:2000 |
qPCR
Cells were digested using 0.25% Trypsin (Gibco, USA) and collected, and total RNA was extracted by TRIzol Reagent (Invitrogen, USA). Then RNA was reversely transcribed into cDNA using a ReverTra Ace® qPCR RT Master Mix (Toyobo, Japan), and qPCR was carried out using a QuantiNova SYBR Green PCR Kit (Qiagen, German). The primers for qPCR were listed in Table 2, and the result was calculated using 2-ΔΔCt with GAPDH used as internal reference.
Table 2.
Primers using in qPCR
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| ASMA | 5’ TGTGCGACGAAGACGAGAC 3’ | 5’ GTAGAAGGTGTGGTGCCAGAT 3’ |
| GAPDH | 5’ GAGTCCACTGGCGTCTTCAC 3’ | 5’ ATCTTGAGGCTGTTGTCATACTTCT 3’ |
CCK-8 assay
Ten microliters of CCK-8 reagent and 90 μL RPMI 1640 medium were added to the plates, which were then incubated with 5% CO2 at 37°C for 2 h. After the incubation, the optical density (OD) value was assessed by a microplate reader (BioTek, USA) for the evaluation of cell proliferation ability.
AV/PI staining
At room temperature, the cells were digested with 0.25% trypsin (Gibco, USA) and washed twice with 1 mL precooled PBS and were then centrifuged at 1000 rpm for 5 mins. Afterward, the cells were resuspended in 100 μL binding buffer, and 5 μL AV (Invitrogen, USA) was added, and the mixture was subsequently added in the flow cytometry tube with 385 μL binding buffer. Finally, 10 μL PI was added and incubated for 15 min. Then, the flow cytometry assay was performed.
Immunofluorescence
After being washed with PBS, the cell slides were fixed with 4% formaldehyde (Invitrogen, USA), and then permeabilized with 0.2% Triton X-100 (Invitrogen, USA) and blocked with 5% BSA (Sangon, China). The cells slides were then incubated with Mouse Anti-Alpha Skeletal Muscle Actin antibody (Abcam, USA) at a dilution of 1:100 at 4°C overnight, and subsequently being washed with PBS and incubated with Goat Anti-Mouse IgG H&L antibody (Alexa Fluor® 647) (CST, USA) at a dilution of 1:800 at 37°C for 30 mins. Then, cells slides were incubated with DAPI (Sangon, China) for 5 mins, washed with PBS, and blocked with 95% glycerol (Sangon, China). The immunofluorescence images were obtained using a laser scanning focusing microscope (Nikon, Japan).
Nanoparticle tracking analysis (NTA)
A549 exosome was dissolved by the PBS solution, and then the Malvern particle size analyzer (Malvern, the UN) was used for the detection of the particle sizes and their distribution of A549 exosomes.
Statistics
Statistical analyses were performed using SPSS 21.0 software (IBM, USA) and graphs were drawn using GraphPad Prism 5.01 software (GraphPad Software, USA). Data were presented as mean ± standard error (SEM), comparison was determined by t test. P<0.05 was considered as significant.
Results
Isolation and validation of A549 exosomes
As presented in Figure 1, electron microscopy showed the morphology of exosomes extracted from A549 cells (Figure 1A), and a subsequent Western blot assay revealed that the protein levels of exosome markers CD9, CD63, CD81, TGF-β1, Flotillin-1 and EEF2 were elevated in Exosome group compared with NC group (sample from serum-free medium) (Figure 1B). In addition, we evaluated the ASMA expression in A549 exosomes and found that ASMA protein expression was upregulated in A549 exosomes compared with that in NC group (Figure 1C). And the particle size distribution of the A549 exosomes assessed by NTA was presented in Figure S1.
Figure 1.
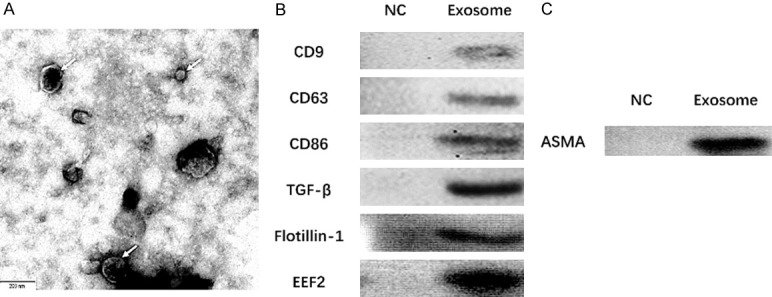
Exosomes extracted from A549 cells, and the ASMA protein expression in A549 exosomes. Morphology of exosomes extracted from A549 cells was seen under an electron microscope (A), and western blot assay disclosed that the expressions of exosome markers CD9, CD63, CD81, TGF-β1 Flotillin-1 and EEF2 were elevated in Exosome group than those in NC group (sample from serum-free medium) (B). ASMA protein expression was upregulated in A549 exosomes (C). ASMA, alpha-smooth muscle actin; NC, negative control.
Effect of A549 exosomes on HLF1 cell proliferation and apoptosis
The HLF1 cell proliferation was enhanced at 48 h (P<0.05) and 72 h (P<0.01) in Exosome group than those in NC group (Figure 2A). As for cell apoptosis, the cell apoptosis rate was decreased at 72 h in Exosome group compared with NC group (P<0.01) (Figure 2B, 2D), and pro-apoptotic protein C-Caspase 3 expression at 72 h was decreased (P<0.05) while anti-apoptotic protein Bcl-2 expression at 72 h was elevated (P<0.05) in Exosome group than those in NC group (Figure 2C).
Figure 2.
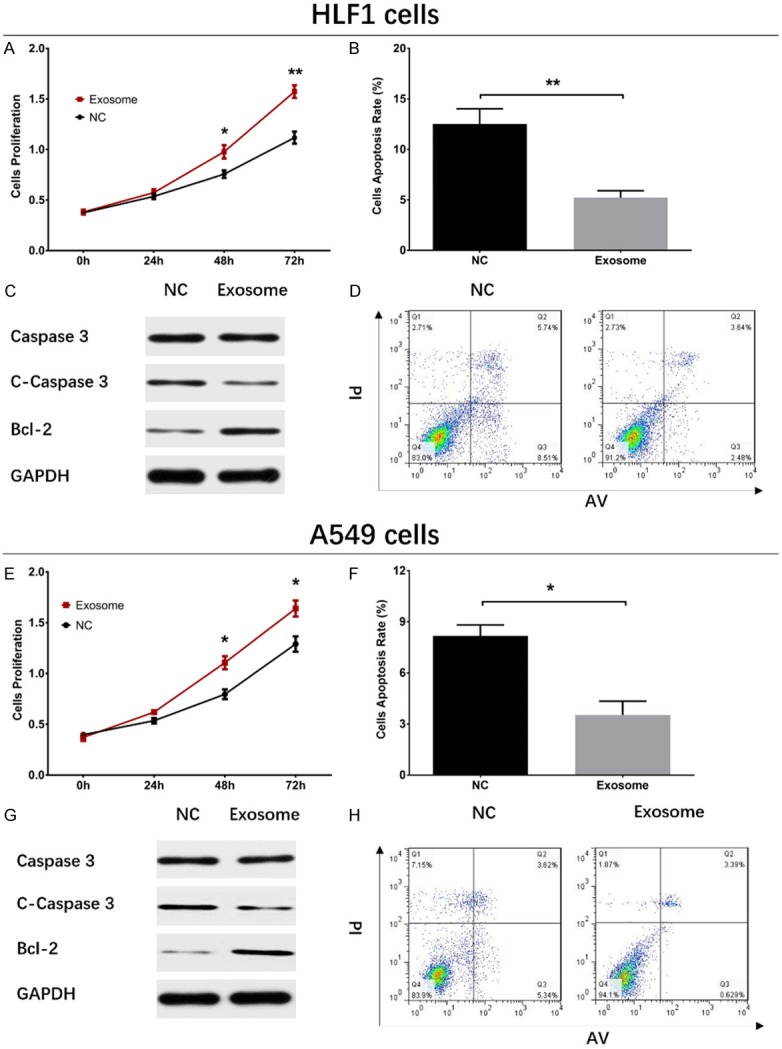
Impacts of A549 exosomes on cell proliferation and apoptosis in HLF1 cells and A549 cells. In HLF1 cells, cell proliferation was promoted (A), cell apoptosis rate declined (B, D), Caspase 3 level did not vary (C), C-Caspase 3 expression was decreased, and Bcl-2 expression was increased in Exosome group compared with NC group. In A549 cells, cell proliferation increased in Exosome group compared with NC group (E), while cell apoptosis rate was decreased (F, H), C-Caspase 3 level was declined (G) and Bcl-2 level was increased in Exosome group than that in NC group. Comparison between two groups was determined by t test. P<0.05 was considered significant. *P<0.05, **P<0.01. NC, negative control.
ASMA expression in HLF1 cells after coculture with A549 exosomes
As shown in Figure 3, the ASMA mRNA expression after a 72 h coculture in Exosome group was elevated compared with that in NC group (P<0.001) (Figure 3A), and immunofluorescence assay also revealed that the ASMA protein expression (red fluorescence) was increased in Exosome group compared with NC group (Figure 3B).
Figure 3.

ASMA expression in HLF1 cells and A549 cells. ASMA mRNA expression increased in Exosome group than that in NC group (A), and immunofluorescence assay disclosed increased ASMA protein expression (red fluorescence) in Exosome group compared with NC group as well (B). The ASMA mRNA expression was increased in Exosome group compared with NC group (C), and the protein expression of ASMA (red fluorescence) in Exosome group was also elevated than that in NC group (D). Comparison between two groups was determined by t test. P<0.05 was considered significant. *P<0.05, ***P<0.001. ASMA, alpha-smooth muscle actin; NC, negative control.
Effect of A549 exosomes and ASMA on HLF1 cell proliferation and apoptosis
The ASMA mRNA expression at 72 h after transfection was decreased in SH group compared with NC group (P<0.01), and it was also reduced in Exosome+SH group compared with that in Exosome group (P<0.01) (Figure 4A). And the ASMA protein expression (red fluorescence) was down-regulated in SH group than that in NC group and was also decreased in Exosome+SH group compared with Exosome group (Figure 4B). As shown in Figure 5A, the cell proliferation was reduced in SH group compared with NC group at 48 h (P<0.05) and 72 h (P<0.01) and was also lower in Exosome+SH group compared with that in Exosome group at 48 h (P<0.05) and 72 h (P<0.01). In addition, cell apoptosis rate at 72 h was increased in SH group compared with NC group (P<0.01), and it was elevated in Exosome+SH group compared with Exosome group (P<0.01) (Figure 5B, 5C). These results indicated that A549 exosomes promoted cell proliferation and repressed cell apoptosis of normal lung fibroblasts through delivering ASMA.
Figure 4.
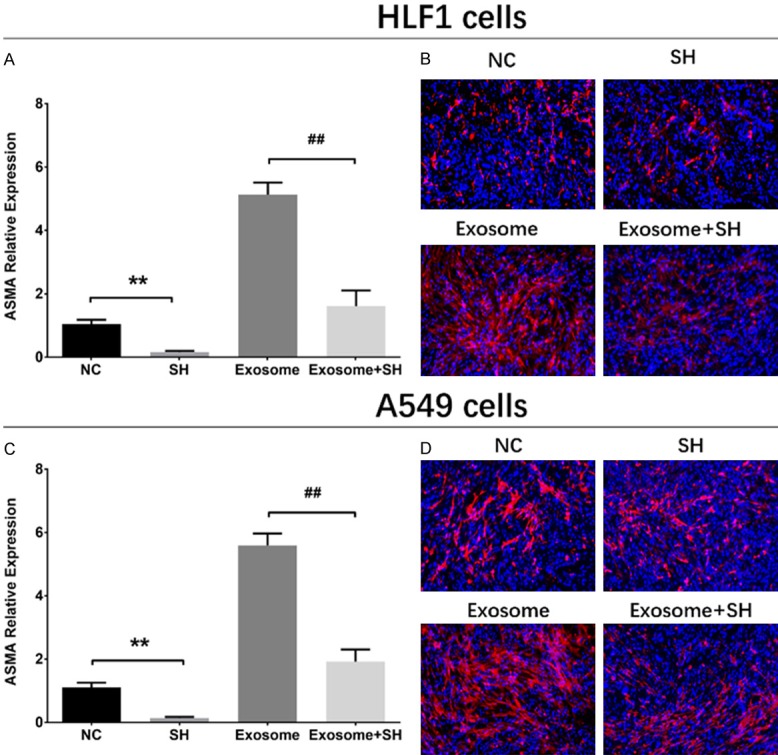
ASMA expressions in rescue experiments. In HLF1 cells, ASMA mRNA expression (A) and protein level (red fluorescence) (B) were decreased in SH group compared with NC group, and were also reduced in Exosome+SH group than that in Exosome group (A, B). In A549 cells, ASMA mRNA (C) and protein (D) expressions were inhibited in SH group compared with NC group, and were also decreased in Exosome+SH group than that in Exosome group. Comparison between two groups was determined by t test. P<0.05 was considered significant. Comparison between NC group and SH group: **P<0.01. Comparison between Exosome group and Exosome+SH group: ##P<0.01. ASMA, alpha-smooth muscle actin; NC, negative control.
Figure 5.
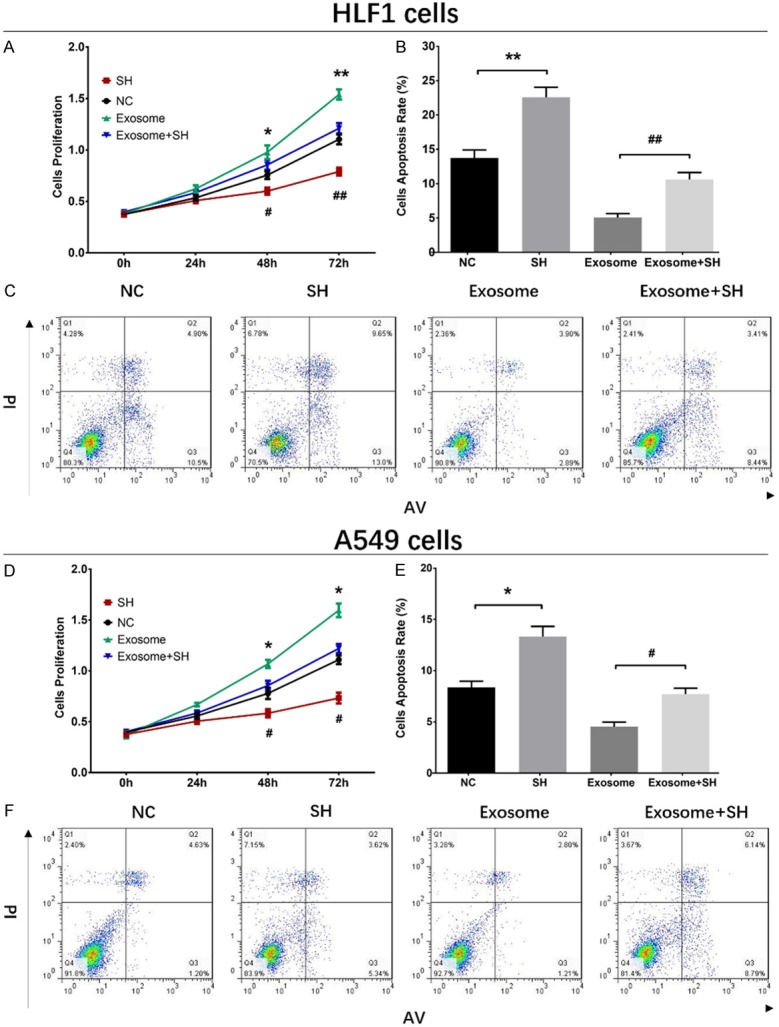
Impacts of A549 exosomes and ASMA on cell proliferation and apoptosis in HLF1 cells and A549 cells. Cell proliferation was reduced in SH group than that in NC group and was also decreased in Exosome+SH group compared with Exosome group (A). And cell apoptosis was increased in SH group compared with NC group, which was also elevated in Exosome+SH group compared with Exosome group (B, C). Cell proliferation was reduced in SH group compared with NC group, but was increased in Exosome group compared with Exosome+SH group (D). Cell apoptosis was elevated in SH group compared with NC group, while was decreased in Exosome group than that in Exosome+SH group (E, F). Comparison between two groups was determined by t test. P<0.05 was considered significant. Comparison between NC group and SH group: *P<0.05; **P<0.01. Comparison between Exosome group and Exosome+SH group: #P<0.05; ##P<0.01. NC, negative control.
Effects of A549 exosomes on A549 cell proliferation and apoptosis
To further explore the effect of A549 exosomes on NSCLC cell functions, A549 exosomes were added in A549 cells and cell proliferation and apoptosis were detected, which disclosed that the cell proliferation at 48 h (P<0.05) and 72 h (P<0.05) was increased in Exosome group compared with NC group (Figure 2E). Additionally, the cell apoptosis rate at 72 h was decreased in Exosome group compared with NC group (P<0.05) (Figure 2F, 2H), and the expression of pro-apoptotic protein C-Caspase 3 was reduced while the level of anti-apoptotic protein Bcl-2 was increased in Exosome group compared with those in NC group (Figure 2G).
ASMA expression in A549 cells after coculture with A549 exosomes
At 72 h of coculture of A549 cells and A549 exosomes, the ASMA mRNA expression was enhanced in Exosome group compared with that in NC group (P<0.05) (Figure 3C), and ASMA protein expression (red fluorescence) was also upregulated in Exosome group compared with NC group (Figure 3D).
Effects of A549 exosomes and ASMA on A549 cell proliferation and apoptosis
The ASMA mRNA expression at 72 h after transfections was reduced in SH group compared with NC group (P<0.01) and was also reduced in Exosome+SH group compared with that in Exosome group (P<0.01) (Figure 4C). Moreover, the ASMA protein level (red fluorescence) was decreased in SH group compared with that in NC group, and it was downregulated in Exosome+SH group compared with Exosome group (Figure 4D). These data indicated that the transfections were successful. The proliferation of A549 cells was suppressed at 48 h (P<0.05) and 72 h (P<0.05) in SH group compared with NC group, and it was also inhibited in Exosome+SH group at 48 h (P<0.05) and 72 h (P<0.05) compared with that in Exosome group (Figure 5D). In addition, the cell apoptosis rate of A549 cells at 72 h was increased in SH group compared with that in NC group (P<0.05), and it was also upregulated in Exosome+SH group compared with Exosome group (P<0.05) (Figure 5E, 5F). These results suggested that NSCLC cell-derived exosomes promoted cell proliferation and inhibited cell apoptosis of NSCLC cells via transferring ASMA as well.
Discussion
The results of our experiments revealed the following: (1) NSCLC derived exosomes presented with the potential to promote normal lung fibroblasts to cancer-like cells by enhancing cell proliferation and repressing cell apoptosis via delivering ASMA; (2) NSCLC derived exosomes also promoted NSCLC cell proliferation and inhibited apoptosis by transferring ASMA.
As an emerging research focus, exosome has been found to participate in various pathological processes, which include infection, autoimmune disorders, neurodegenerative diseases, pregnancy complications, obesity and various cancers [20-26]. And the development of the exosomes extraction techniques has also contributed to the investigation of the functions of exosomes in cancers. The engagement of cancer cells derived exosomes in the progression of cancers includes delivering oncogenic matters and regulating gene expression of the receiver cells to promote metastasis, vascularization and drug-resistance and so on [27-30]. In regard to oncogenesis, exosomes derived from cancer cells are initially found to have the ability to modulate the cell functions of normal human cells. For example, in HCC, the eukaryotic translation initiation factor 3 subunit C (EIF3C) induced exosomes secretion by HCC cells subsequently have an oncogenic effect through augmenting tumor angiogenesis via enhancing tube formation of human umbilical vein endothelial cells (HUVECs) and vessel growth [31]. Another fundamental research illustrates that the Rab27a-dependent secreted exosomes, along with cytokines and metalloproteinases, elicit an aggregation of neutrophil immune cells, contributing to the establishment of metastatic carcinoma (4T1) in breast cancer [32]. Mesenchymal stem cells (MSC) absorb tumor cell-secreted exosomes that interact with surface receptors on MSCs and subsequently generate factors that promote tumor growth and change the functions of nontumor cells in several cancers [33]. These findings indicate that cancer cell-derived exosomes present with oncogenic effect in normal cells by promoting the tube formation, vessel growth, immune cells aggregation via transferring or interacting with oncogenic factors in various cancers [23-25]. In our study, we used the exosome isolation reagent for exosomes extraction, the convenience and efficiency of which have been demonstrated in previous studies [34,35]. Then the CCK-8 and AV/PI experiments results disclosed that A549 exosomes promoted cell proliferation while inhibited cell apoptosis of normal lung fibroblasts, which could be explained by the fact that cancer cell-derived exosomes are found to play an oncogenic role in normal human cells through transferring various proteins and genes, or interacting with multiple factors, such as EIF3C and Rab27a etc [31-33].
Regarding the detailed mechanism by which cancer cell-derived exosomes on regulating normal cell functions, emerging studies have revealed that they mediate normal cell functions by delivering different oncogenetic factors in various cancers [36]. For instance, prostate cancer cell-secreted exosomes promote the expressions of RANKL and metalloproteinases in cancer-associated fibroblasts (CAFs) by transferring miR-100, miR-21 and miR-139, which subsequently contributes to prostate cancer progression [37]. Moreover, the fibroblasts internalized breast cancer cells, colorectal cancer cells and leukemia cells secreted exosomes promote the formation of a pre-metastatic environment by enhancing cell proliferation of fibroblast and lifespan [38]. Based on these data, we hypothesized that NSCLC-derived exosomes likewise promote normal lung fibroblasts viability through delivering oncogene factors. Thus, we performed rescue experiments focusing on ASMA, and observed that NSCLC-derived exosomes enhanced cell proliferation while repressed cell apoptosis of normal lung fibroblast functions by transferring ASMA, this result provided a very novel insight into the mechanism by which NSCLC cells derived exosomes in modulating normal lung fibroblast functions in NSCLC. In addition, in our rescue experiments, we transferred the ASMA ShRNA before the A549 exosomes were added, which is because that if the ASMA ShRNA was transferred after the A549 exosomes, there might be interferences of our results.
For the purpose of further exploring whether NSCLC cell-derived exosomes could advocate the progression of NSCLC by regulating NSCLC cell functions, we repeated the experiments in NSCLC cells and found that NSCLC-derived exosomes also promoted cell proliferation and inhibited cell apoptosis of NSCLC cells through delivering ASMA. Similarly, cancer cell-derived exosomes are reported to play a critical role in tumorigenesis by regulating the cancer cell functions. A previous study illuminates that cancer cell-derived exosomes regulate metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) transfer to advocate cell proliferation in breast cancer cells [39]. Another study shows that exosomes secreted by gastric cancer cells with high CD97 expression enhance cell proliferation and invasion in gastric cancer cells through the MAPK signaling pathway [40]. In oral squamous cell carcinoma, the cancer-associated fibroblasts derived exosome promotes cancer cell proliferation and metastasis by delivering miR-34a-5p [41]. Additionally, a recent study reports that the treatment of murine melanoma cells (B16Bl6 cells) with the exosomes produced by B16BL6 cells promotes cell proliferation and represses cell apoptosis, which is in line with our results [42]. Those studies provide some possible explanations of NSCLC cell-derived exosomes in regulating cancer cell functions. ASMA has been found to play a role in regulating cancer cell functions as well. A study elucidates that ASMA is a target of docetaxel-conjugate nanoparticles that inhibit breast cancer metastasis [43]. The study conducted by Fujita H et al. evaluates the impacts of pancreatic cancer cells (PSCs) activated by ASMA on the malignant behaviors of PSCs, and find that PSCs cell proliferation, invasion and colony formation are enhanced by PSCs activated by ASMA [44]. In this study, we repeated the experiments that were done in normal lung fibroblasts in the NSCLC cells, and found that NSCLC cell-derived exosomes promoted cell proliferation and inhibited cell apoptosis of NSCLC cells through transferring ASMA, which might be resulted from that: (1) similar to the other cancer cell-derived exosomes, the NSCLC cell-derived exosomes can promote cell proliferation while repress cell apoptosis of NSCLC cells by transferring oncogenic factors, such as transferring the miR-34a-5p [39,40,42,45]; (2) it is also possible that NSCLC cell-derived exosomes mediated NSCLC cell functions through multiple pathways, for instance, the MAPK pathway [30]; (3) ASMA could promote the cancerous acts of cancer cells, for example, ASMA activates cancer cells and subsequently enhances the cancer cell proliferation and invasion [43,44].
In conclusion, NSCLC derived exosomes promote cell proliferation and inhibit cell apoptosis in both normal lung fibroblasts and NSCLC cells by delivering ASMA.
Acknowledgements
This work was supported by New Frontier Technology Joint Research Project of Shanghai Shen Kang Hospital Development Center (SHDC12016113).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res. 2015;21:2227–2235. doi: 10.1158/1078-0432.CCR-14-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao AS, Scagliotti GV, Bunn PA Jr, Carbone DP, Warren GW, Bai C, de Koning HJ, Yousaf-Khan AU, McWilliams A, Tsao MS, Adusumilli PS, Rami-Porta R, Asamura H, Van Schil PE, Darling GE, Ramalingam SS, Gomez DR, Rosenzweig KE, Zimmermann S, Peters S, Ignatius Ou SH, Reungwetwattana T, Janne PA, Mok TS, Wakelee HA, Pirker R, Mazieres J, Brahmer JR, Zhou Y, Herbst RS, Papadimitrakopoulou VA, Redman MW, Wynes MW, Gandara DR, Kelly RJ, Hirsch FR, Pass HI. Scientific advances in lung cancer 2015. J Thorac Oncol. 2016;11:613–638. doi: 10.1016/j.jtho.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Novello S, Barlesi F, Califano R, Cufer T, Ekman S, Levra MG, Kerr K, Popat S, Reck M, Senan S, Simo GV, Vansteenkiste J, Peters S ESMO Guidelines Committee. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27:v1–v27. doi: 10.1093/annonc/mdw326. [DOI] [PubMed] [Google Scholar]
- 8.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 9.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 10.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Kramer-Albers EM, Lim SK, Llorente A, Lotvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-’t Hoen EN, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sanchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BW, Vazquez J, Vidal M, Wauben MH, Yanez-Mo M, Zoeller M, Mathivanan S. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DK, Kang B, Kim OY, Choi DS, Lee J, Kim SR, Go G, Yoon YJ, Kim JH, Jang SC, Park KS, Choi EJ, Kim KP, Desiderio DM, Kim YK, Lotvall J, Hwang D, Gho YS. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013:2. doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Yeo RWY, Lai RC, Sim EWK, Chin KC, Lim SK. Mesenchymal stromal cell exosome-enhanced regulatory T-cell production through an antigen-presenting cell-mediated pathway. Cytotherapy. 2018;20:687–696. doi: 10.1016/j.jcyt.2018.02.372. [DOI] [PubMed] [Google Scholar]
- 13.Bagheri HS, Mousavi M, Rezabakhsh A, Rezaie J, Rasta SH, Nourazarian A, Avci CB, Tajalli H, Talebi M, Oryan A, Khaksar M, Kazemi M, Nassiri SM, Ghaderi S, Bagca BG, Rahbarghazi R, Sokullu E. Low-level laser irradiation at a high power intensity increased human endothelial cell exosome secretion via Wnt signaling. Lasers Med Sci. 2018;33:1131–1145. doi: 10.1007/s10103-018-2495-8. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z, Wang L, Dong L, Wang X. Emerging role of exosome signalling in maintaining cancer stem cell dynamic equilibrium. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13676. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, Morgelin M, Bourseau-Guilmain E, Bengzon J, Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124:3748–3757. doi: 10.1182/blood-2014-05-576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, Tetta C, Bussolati B, Camussi G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 18.Quan Y, Wang Z, Gong L, Peng X, Richard MA, Zhang J, Fornage M, Alcorn JL, Wang D. Exosome miR-371b-5p promotes proliferation of lung alveolar progenitor type II cells by using PTEN to orchestrate the PI3K/Akt signaling. Stem Cell Res Ther. 2017;8:138. doi: 10.1186/s13287-017-0586-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philley JV, Kannan A, Griffith DE, Devine MS, Benwill JL, Wallace RJ Jr, Brown-Elliott BA, Thakkar F, Taskar V, Fox JG, Alqaid A, Bains H, Gupta S, Dasgupta S. Exosome secretome and mediated signaling in breast cancer patients with nontuberculous mycobacterial disease. Oncotarget. 2017;8:18070–18081. doi: 10.18632/oncotarget.14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobb RJ, Lima LG, Moller A. Exosomes: key mediators of metastasis and pre-metastatic niche formation. Semin Cell Dev Biol. 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salomon C, Guanzon D, Scholz-Romero K, Longo S, Correa P, Illanes SE, Rice GE. Placental exosomes as early biomarker of preeclampsia: potential role of exosomal MicroRNAs across gestation. J Clin Endocrinol Metab. 2017;102:3182–3194. doi: 10.1210/jc.2017-00672. [DOI] [PubMed] [Google Scholar]
- 24.Elfeky O, Longo S, Lai A, Rice GE, Salomon C. Influence of maternal BMI on the exosomal profile during gestation and their role on maternal systemic inflammation. Placenta. 2017;50:60–69. doi: 10.1016/j.placenta.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 25.Salomon C, Scholz-Romero K, Sarker S, Sweeney E, Kobayashi M, Correa P, Longo S, Duncombe G, Mitchell MD, Rice GE, Illanes SE. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes. 2016;65:598–609. doi: 10.2337/db15-0966. [DOI] [PubMed] [Google Scholar]
- 26.Buzas EI, Gyorgy B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10:356–364. doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 27.Lobb RJ, Hastie ML, Norris EL, van Amerongen R, Gorman JJ, Moller A. Oncogenic transformation of lung cells results in distinct exosome protein profile similar to the cell of origin. Proteomics. 2017;17 doi: 10.1002/pmic.201600432. [DOI] [PubMed] [Google Scholar]
- 28.Lobb RJ, van Amerongen R, Wiegmans A, Ham S, Larsen JE, Moller A. Exosomes derived from mesenchymal non-small cell lung cancer cells promote chemoresistance. Int J Cancer. 2017;141:614–620. doi: 10.1002/ijc.30752. [DOI] [PubMed] [Google Scholar]
- 29.Yousafzai NA, Wang H, Wang Z, Zhu Y, Zhu L, Jin H, Wang X. Exosome mediated multidrug resistance in cancer. Am J Cancer Res. 2018;8:2210–2226. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2018 doi: 10.1038/s41388-018-0619-z. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Lee HY, Chen CK, Ho CM, Lee SS, Chang CY, Chen KJ, Jou YS. EIF3C-enhanced exosome secretion promotes angiogenesis and tumorigenesis of human hepatocellular carcinoma. Oncotarget. 2018;9:13193–13205. doi: 10.18632/oncotarget.24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Thery C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 33.Whiteside TL. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol. 2018;35:69–79. doi: 10.1016/j.smim.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Pol E, Boing AN, Gool EL, Nieuwland R. Recent developments in the nomenclature, presence, isolation, detection and clinical impact of extracellular vesicles. J Thromb Haemost. 2016;14:48–56. doi: 10.1111/jth.13190. [DOI] [PubMed] [Google Scholar]
- 35.Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 36.Mao J, Liang Z, Zhang B, Yang H, Li X, Fu H, Zhang X, Yan Y, Xu W, Qian H. UBR2 enriched in p53 deficient mouse bone marrow mesenchymal stem cell-exosome promoted gastric cancer progression via Wnt/beta-catenin pathway. Stem Cells. 2017;35:2267–2279. doi: 10.1002/stem.2702. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez CA, Andahur EI, Valenzuela R, Castellon EA, Fulla JA, Ramos CG, Trivino JC. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget. 2016;7:3993–4008. doi: 10.18632/oncotarget.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutkin A, Uziel O, Beery E, Nordenberg J, Pinchasi M, Goldvaser H, Henick S, Goldberg M, Lahav M. Tumor cells derived exosomes contain hTERT mRNA and transform nonmalignant fibroblasts into telomerase positive cells. Oncotarget. 2016;7:59173–59188. doi: 10.18632/oncotarget.10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291–299. doi: 10.2147/OTT.S155134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang W, Wu YL, Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol. 2015;21:6215–6228. doi: 10.3748/wjg.v21.i20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li YY, Tao YW, Gao S, Li P, Zheng JM, Zhang SE, Liang J, Zhang Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. 2018;36:209–220. doi: 10.1016/j.ebiom.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumoto A, Takahashi Y, Nishikawa M, Sano K, Morishita M, Charoenviriyakul C, Saji H, Takakura Y. Accelerated growth of B16BL6 tumor in mice through efficient uptake of their own exosomes by B16BL6 cells. Cancer Sci. 2017;108:1803–1810. doi: 10.1111/cas.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami M, Ernsting MJ, Undzys E, Holwell N, Foltz WD, Li SD. Docetaxel conjugate nanoparticles that target alpha-smooth muscle actin-expressing stromal cells suppress breast cancer metastasis. Cancer Res. 2013;73:4862–4871. doi: 10.1158/0008-5472.CAN-13-0062. [DOI] [PubMed] [Google Scholar]
- 44.Fujita H, Ohuchida K, Mizumoto K, Nakata K, Yu J, Kayashima T, Cui L, Manabe T, Ohtsuka T, Tanaka M. alpha-smooth muscle actin expressing stroma promotes an aggressive tumor biology in pancreatic ductal adenocarcinoma. Pancreas. 2010;39:1254–1262. doi: 10.1097/MPA.0b013e3181dbf647. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Wu XH, Wang D, Luo CL, Chen LX. Bladder cancer cell-derived exosomes inhibit tumor cell apoptosis and induce cell proliferation in vitro. Mol Med Rep. 2013;8:1272–1278. doi: 10.3892/mmr.2013.1634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


