Abstract
Breast cancer is the second leading cause of cancer-related death among women worldwide. Emerging evidence suggests that chromobox homolog 2 (CBX2) is overexpressed in breast cancer and plays an essential role in tumor progression. However, its expression and functional roles in breast cancer development and progression require further exploration. Here, we evaluated CBX2 expression in breast cancer using mRNA expression data from the TCGA database; CBX2 expression was upregulated in breast cancer. Furthermore, upregulated CBX2 expression was significantly associated with poorer overall survival (OS) and progression-free survival (PFS) of breast cancer patients. Immunohistochemical analysis of CBX2 expression in a tissue microarray (TMA) cohort yielded concordant results. Univariate and multivariate analyses showed that elevated CBX2 expression was significantly and independently associated with poorer OS of patients in this TMA cohort. Additionally, we performed in vitro functional assays to evaluate the proliferation, migration, and invasion abilities of breast cancer cell lines wherein CBX2 was knocked down using short hairpin RNA (shRNA). CBX2 silencing inhibited cell proliferation, migration, and invasion in vitro. Furthermore, knockdown of CBX2 markedly reduced breast tumorigenesis in xenograft mouse models. Functional and pathway enrichment analyses indicated a positive correlation between high CBX2 expression and activation of the PI3K/AKT pathway, which were further confirmed by western blot and immunohistochemical analyses of mouse tumors. Our findings indicate that CBX2 is a potential prognostic biomarker and therapeutic target for breast cancer.
Keywords: CBX2, breast cancer, prognostic biomarker, PI3K/AKT pathway
Introduction
Worldwide cancer estimates indicate that about 2.1 million women were newly diagnosed with breast cancer in 2018, and that breast cancer was responsible for almost 1 in 4 cancer-related deaths among women [1]. In spite of advances in early diagnosis of the disease and adoption of improved therapeutic strategies, prognosis of breast cancer is often still poor, mainly due to aggressive biologic behavior and delayed diagnoses [2-4]. Even when diagnosed at an early stage, tumor recurrence and metastasis after surgery are persistent clinical problems in many breast cancer patients [5,6]. Thus, there is an unmet clinical need to unravel molecular mechanisms underlying breast tumorigenesis and disease progression, and to identify novel prognostic/predictive biomarkers and therapeutic targets for improving breast cancer outcomes.
Chromobox (CBX) member proteins, including CBX1-8 proteins, are essential regulators of gene expression and developmental programs [7,8]. Growing evidence suggests that aberrant expression of CBX family members plays key roles in different cancers [9]. Studies have reported that CBX6 is significantly under-expressed in glioblastoma [10]. CBX7 is overexpressed in gastric and prostate cancers [11,12], but downregulated in thyroid cancer, lung cancer, and colon cancer [13-15]. A recent study found that the expression of CBX4 was inversely correlated with the expression of Runx2. Importantly, the combination of overexpressed CBX4 and under-expressed Runx2 was strongly associated with overall survival (OS) in colorectal carcinoma [16]. Based on in silico analysis of online datasets, Liang Y. et al. reported that some CBX member proteins are overexpressed in breast cancer [17]. However, the expression and roles of CBX2 in breast cancer remain unclear.
The PI3K/AKT pathway plays a central role in regulating key biological processes in several human cancers wherein its dysregulation is implicated both in tumorigenesis and tumor progression [18-21]. A growing tide of evidence also shows that the PI3K/AKT pathway contributes to breast tumorigenesis and progression of breast cancer. Previous studies have reported that many oncogenes, such as TPX2, MEG3, and HOXA1 et al. regulate PI3K/AKT signaling pathway in breast cancer [22]. However, the molecular mechanisms through which the PI3K/AKT pathway drives breast cancer progression remain largely to be elucidated.
In our study, we evaluated the expression levels of CBX2 mRNA and protein in breast cancer. CBX2 was remarkably overexpressed in breast cancer tissues, and upregulated CBX2 expression was strongly associated with poor prognosis in breast cancer. Additionally, CBX2 silencing attenuated proliferation and invasiveness of breast cancer cells in vitro and impaired tumorigenesis in in vivo models. Moreover, results from our in silico bioinformatics analyses combined with validation experiments suggest that CBX2 plays a pivotal role in regulating the cell cycle and DNA replication, and that the PI3K/AKT signaling pathway functions downstream of CBX2 in breast cancer tumorigenesis and/or progression. Our findings uncover a crucial role for CBX2 in regulating the proliferation and invasion of breast cancer cells via the PI3K/AKT pathway and suggest that elevated CBX2 may serve as a novel prognostic biomarker and promising therapeutic target for breast cancer.
Materials and methods
Clinical samples
Formalin-fixed paraffin-embedded breast tumor resection samples (n=160) and breast non-tumor tissues (n=60) were used for the construction of tissue microarrays. Breast cancer tissues were derived from patients consecutively diagnosed with breast cancer between 2009 and 2013, for whom tissue samples and detailed clinicopathologic data were available. The clinicopathological features of the patients in our study cohort are presented in Table 1. All aspects of this study were reviewed and approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University. All patients were officially informed about the use of specimens and signed the written informed consent.
Table 1.
Correlation of clinicopathological features with CBX2 expression in breast cancer ZZU TMA cohort
| Variables | Clinicopathological features | Total (n=160) | CBX2 expression level | P-value | |
|---|---|---|---|---|---|
|
| |||||
| Low (n, %) | High (n, %) | ||||
| Age (years) | ≤50 | 95 | 44 (63.7) | 51 (56.0) | 0.324 |
| >50 | 65 | 25 (36.2) | 40 (43.9) | ||
| Her-2 status | Negative | 124 | 54 (61.1) | 70 (66.7) | 0.840 |
| Positive | 36 | 15 (38.9) | 21 (33.3) | ||
| PR status | Negative | 98 | 39 (56.5) | 59 (64.8) | 0.285 |
| Positive | 62 | 30 (43.4) | 32 (35.1) | ||
| ER status | Negative | 95 | 49 (62.0) | 46 (56.7) | 0.500 |
| Positive | 65 | 30 (37.9) | 35 (43.2) | ||
| TNBC status | TNBC | 64 | 29 (42.0) | 35 (38.4) | 0.648 |
| Non-TNBC | 96 | 40 (57.9) | 56 (61.5) | ||
| Tumor size | <20 mm | 49 | 24 (34.7) | 25 (27.4) | 0.320 |
| ≥20 mm | 111 | 45 (65.2) | 66 (72.5) | ||
| Involved lymph node | Negative | 58 | 32 (46.3) | 26 (28.5) | 0.020 |
| Positive | 102 | 37 (53.6) | 65 (71.4) | ||
| Differentiation grade | Well & Moderate | 98 | 49 (71.0) | 49 (53.8) | 0.027 |
| Poor | 62 | 20 (28.9) | 42 (46.1) | ||
| TNM stage | Stage I and II | 86 | 50 (72.4) | 36 (39.5) | 0.000 |
| Stage III and IV | 74 | 19 (27.5) | 55 (60.4) | ||
The cancer genome atlas (TCGA) dataset analysis
The mRNA expression and clinicopathologic data for a total of 1079 breast cancer tumor samples and 104 non-tumor breast tissue samples was downloaded from TCGA (http://tcga-data.nci.nih.gov/). Furthermore, we analyzed the correlation between CBX2 mRNA expression and clinical characteristics of the breast cancer patients.
Tissue microarray (TMA) construction and immunohistochemical (IHC) staining of the Zhengzhou University (ZZU) cohort
Human paraffin-embedded breast cancer TMA slides containing 160 paraffin-embedded breast cancer tissues and 60 non-tumor breast tissues were constructed using 1.5-mm diameter cores. In brief, deparaffinization, rehydration, and antigen retrieval of the tissue slides were performed according to established protocols. Subsequently, TMA slides were blocked with 5% bovine serum albumin (BSA) (Invitrogen, Carlsbad, CA, USA) for 1 hour at room temperature. Sections were incubated with antibody against CBX2 (dilution 1:100, Proteintech Group, Wuhan, China) overnight at 4°C and then probed with biotinylated goat anti-rabbit secondary antibody (AmyJet Scientific, Wuhan, China). Enzyme-labelled streptavidin was then added to the TMA slides, and the slides were incubated according to the manufacturer’s protocol. The TMA slides were then placed in the chromogenic substrate (prepared using the SignalStain® DAB Substrate Kit from CST, USA) until the desired reaction was achieved. Slides were then counterstained with hematoxylin and eosin, dehydrated, and mounted. The presence of CBX2 and its abundance (signal intensity) were evaluated semi-quantitatively as follows: scores of 1+, 2+, and 3+ represented low CBX2 expression, while scores of 4+ and 5+ represented high CBX2 expression for statistical analyses.
Cell culture
The breast cancer cell lines MDA-MB-231 and MCF-7 (Cell Bank of the Chinese Academy of Science, Shanghai, China) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Solarbio, Beijing, China) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA), at 37°C under a 5% CO2 atmosphere.
Western blotting
Protein isolation and western blotting was performed as previously described [23,24]. Briefly, equal amounts of protein lysates were resolved using 12% SDS-PAGE gels (Invitrogen) and then transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). After blocking for 1 hour, membranes were probed with specific antibodies at 4°C for 12 hours and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 2 hours at room temperature. Signal quantification and analysis were performed using an Odyssey infrared imaging system (LI-COR Biosciences). Primary antibodies used in this study were: anti-CBX2 rabbit monoclonal antibody (Proteintech Group) used at 1:2000 dilution; anti-β-actin monoclonal mouse antibody (Proteintech Group) used at a dilution of 1:5000.
Short hairpin RNA (shRNA) construction and transfection of cell lines
Stable transfectants harboring targeting CBX2 (sh-CBX2) or negative control shRNA (sh-NC) were generated in MDA-MB-231 and MCF-7 breast cancer cells via lentiviral transduction of a plasmid (synthesized by GenePharma, Shanghai, China) carrying the corresponding shRNA sequence and the gene encoding firefly luciferase. Stably transfected cell lines were selected over a period of 4 weeks using puromycin (100 μg/ml).
Cell proliferation assay
Stably transfected MDA-MB-231 and MCF-7 cells were seeded into 96-well plates in triplicates. Cell viability was determined using the Cell Counting Kit-8 (CCK-8) (Dojindo; Kumamoto, Japan) 24 hours after the cells were seeded. Absorbance was read at 450 nm using a spectrophotometer (Molecular Devices, USA).
To evaluate the effects of CBX2 knockdown on DNA replication in proliferating cells, stably transfected MDA-MB-231 and MCF-7 breast cancer cells harboring CBX2 knockdown (sh-CBX2) or negative control shRNA (sh-NC) were incubated in DMEM containing 50 μM 5-ethynyl-2’-deoxyuridine (EdU; RiboBio, Guangzhou, China) in triplicate. DNA was stained using Hoechst 33342 stain and cells were visualized using an inverted fluorescence microscope (Olympus, Tokyo, Japan). For each EdU experiment, five random fields were imaged at 100× magnification. Images were processed and analyzed using the ImageJ software. The percentage of EdU-positive cells among the total number of cells in each field was determined.
Transwell cell migration and invasion assay
For the transwell cell migration and invasion assays, 1×104 indicated cells were resuspended in serum-free medium and then seeded into the upper chamber of a transwell system (Costar, Corning, NY, USA). Medium containing 10% FBS was placed into the bottom chamber and served as the chemo-attractant. After incubation for 24 hours, the transwell insert was removed, and the side of the insert facing the upper compartment was carefully cleansed with a cotton swab to remove culture medium and cells that had not migrated through the insert. Cells that had migrated through the filter pores to the underside of the insert were fixed, stained with 0.1% crystal violet, and counted using a microscope (Olympus).
Tumor xenograft experiments
BALB/c nude mice (4 weeks old, male) were obtained from Vital River Laboratory Technology (Beijing, China). MCF-7 cells (1×107 cells) stably transfected with sh-CBX2 or sh-NC were inoculated subcutaneously into the lower flank of the mice. Tumor growth was measured every week using non-invasive bioluminescent imaging. After 5 weeks, mice were sacrificed. Tumors were weighed, and tissue specimens were harvested for IHC staining.
Statistical analysis
All statistical analyses were performed by using SPSS 23.0 and GraphPad Prism 7 software. χ2 tests or Fisher’s exact tests were used to analyze the relationship between CBX2 expression and outcomes of breast cancer patients. Kaplan-Meier survival analyses and log-rank tests were conducted for overall survival (OS) and progression-free survival (PFS) analyses. Student’s t-tests were used for comparisons between two groups. At least three independent experiments were repeated. Data are presented as means ± SD and P<0.05 was considered to be statistically significant.
Results
High expression of CBX2 is associated with poor prognosis of breast cancer patients in the TCGA dataset
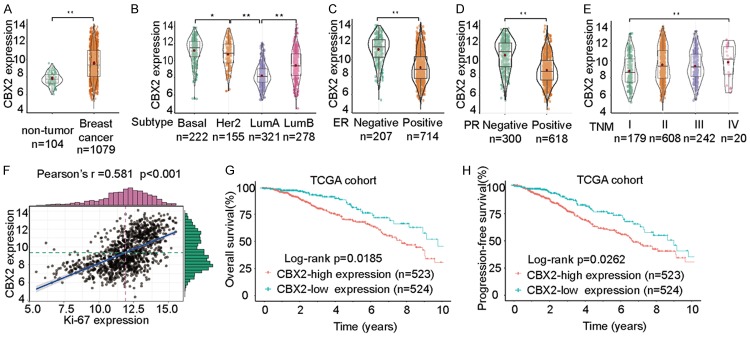
To evaluate how CBX2 expression in breast cancer compares to its expression level in breast non-tumor tissues, we analyzed microarray data from the TCGA dataset. Our results showed that CBX2 mRNA expression was higher in breast cancer tissues compared to normal tissues (Figure 1A). CBX2 was particularly upregulated in the Basal-like and Her2-enriched breast cancer subtypes (Figure 1B). We then analyzed CBX2 expression based on the ER/PR status of breast tumors in the TCGA dataset and found that CBX2 mRNA expression was elevated in ER- and PR-negative breast cancer tissues (Figure 1C and 1D). The expression of CBX2 was much higher in late TNM stages (III-IV stages) than in TNM stage I-II samples (Figure 1E). Additionally, we uncovered a significant positive correlation between the expression of CBX2 and Ki-67 (Figure 1F). The OS and PFS of breast cancer patients with high CBX2 expression were markedly worse than those of patients with low CBX2 expression (Figure 1G and 1H). Taken together, these findings suggest that CBX2 mRNA is significantly upregulated in breast cancer tissues compared to non-tumor tissues, and CBX2 overexpression is associated with unfavorable prognosis of breast cancer patients.
Figure 1.
CBX2 is upregulated in breast tumors and CBX2 expression is associated with prognosis of breast cancer patients in TCGA dataset. A. Comparison of CBX2 mRNA expression between breast cancers and normal tissues in the TCGA dataset. B. Comparison of CBX2 mRNA expression in breast cancers of different molecular subtypes. CBX2 was upregulated in Basal-like and Her2-enriched subtypes compared to LumA and LumB subtypes. C, D. Elevated CBX2 mRNA expression was observed in ER- and PR-negative breast cancer tissues. E. CBX2 mRNA expression in breast tumor tissues according to TNM stage. F. Analysis of the correlation between CBX2 expression and Ki-67 expression. G, H. Kaplan-Meier survival curves showing stratification of breast cancer patients in TCGA dataset based on CBX2 expression (red: high CBX2 expression; green: low CBX2 expression). Breast cancer patients with high CBX2 expression had markedly shorter OS and PFS time than those with low CBX2 expression. *P<0.05, **P<0.01.
CBX2 is overexpressed in breast cancer tissues and is associated with poorer prognosis of patients in the ZZU cohort
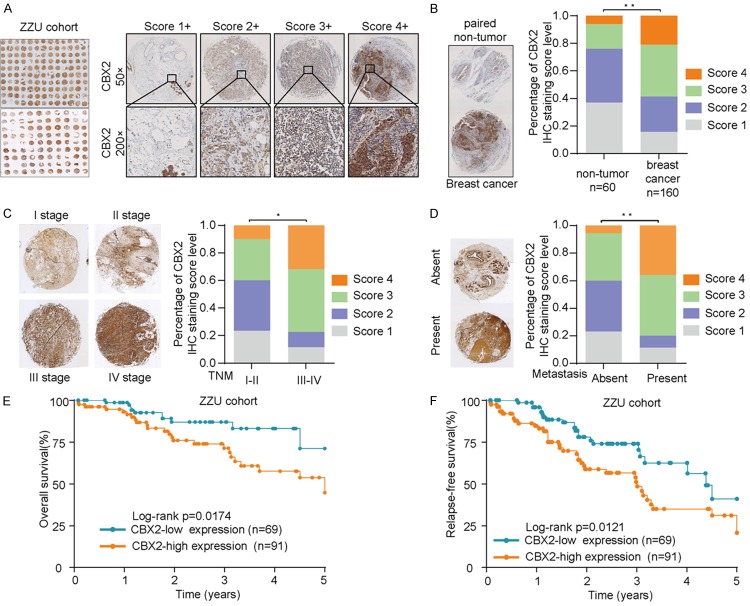
We next analyzed CBX2 protein expression in breast cancer and tumor-adjacent normal tissues in our ZZU cohort TMA via IHC staining/scoring (Figure 2A). CBX2 expression was markedly elevated in breast cancer tissues in comparison with adjacent normal tissues (Figure 2B). Additionally, our results indicated that CBX2 expression was significantly increased in breast cancer patients with advanced TNM stages (III-IV stages) (Figure 2C) and in those who presented with distant metastasis (Figure 2D). Furthermore, Kaplan-Meier analysis revealed that breast cancer patients whose tumors showed low expression of CBX2 had a notably longer OS and relapse-free survival (RFS) time compared to patients whose tumors showed an above-threshold expression of CBX2 (Figure 2E and 2F). Details pertaining to relationships between CBX2 expression and clinicopathological characteristics of breast cancer patients in the ZZU TMA are shown in Table 1. Upregulated CBX2 expression was significantly correlated with breast cancer TNM stage, lymph node involvement, and differentiation grade (Table 1). Furthermore, univariate and multivariate analyses showed that advanced TNM stages, lymph node involvement, and high CBX2 expression were independent prognostic factors for OS and RFS (Table 2). Collectively, our data suggest that CBX2 expression is significantly upregulated in breast cancer and upregulated CBX2 expression is associated with poor clinical outcomes in breast cancer.
Figure 2.
CBX2 is overexpressed in breast cancer tissues and high CBX2 expression is associated with poorer overall survival. A. Representative micrographs showing CBX2 immunohistochemical staining patterns with different staining scores in breast cancer tissues in the ZZU TMA. B. Representative images showing CBX2 staining in breast cancer tissues and paired non-tumor samples in the ZZU TMA (left) and comparison of score distributions between breast cancer tissues and paired non-tumor samples in the ZZU TMA (right). CBX2 was overexpressed in breast cancer tissues compared with non-tumor breast tissues. C. Representative images showing CBX2 staining in tumor samples from patients with different TNM classifications (left) and comparison of score distributions based on TNM stage (right). CBX2 expression was notably high in breast cancer patients with advanced TNM stage. D. Representative images showing CBX2 staining in samples from patients with or without distant metastasis at presentation (left) and comparison of score distributions based on presence/absence of metastases (right). E, F. Kaplan-Meier survival curves showing stratification of patients in the ZZU TMA cohort based on CBX2 expression (red: high CBX2 expression; green: low CBX2 expression). Breast cancer patients with high CBX2 expression had markedly shorter OS and RFS time than those with low CBX2 expression. *P<0.05, **P<0.01.
Table 2.
Analysis of associations between clinicopathological features and clinical outcomes of patients in the breast cancer TMA cohort
| Clinicopathological features | Univariate analysis | Multivariate analysis | ||||
|
| ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
|
| ||||||
| Univariate and multivariate analyses of overall survival in breast cancer patients (n=160) | ||||||
|
| ||||||
| Age (>50 vs <50) | 1.120 | 0.719-1.439 | 0.692 | |||
| Her-2 status (Negative vs positive) | 0.953 | 0.498-1.042 | 0.549 | |||
| PR status (Negative vs positive) | 1.503 | 0.621-2.482 | 0.732 | |||
| ER status (Negative vs positive) | 1.388 | 0.939-2.181 | 0.529 | |||
| TNBC status (Negative vs positive) | 1.063 | 0.729-1.875 | 0.495 | |||
| Tumor size (>20 mm vs <20 mm) | 1.249 | 1.029-2.054 | 0.260 | |||
| Involved lymph node (Negative vs positive) | 2.160 | 1.584-4.827 | 0.028 | 2.307 | 1.784-2.981 | 0.024 |
| Differentiation grade (Well & Moderate vs Poor) | 2.279 | 1.651-3.742 | 0.014 | 1.625 | 1.244-2.129 | 0.079 |
| TNM stage (I & II vs III & IV) | 2.679 | 1.951-4.841 | 0.000 | 2.532 | 1.986-3.821 | 0.002 |
| CBX2 expression level | 2.104 | 1.751-3.926 | 0.003 | 1.909 | 1.604-3.921 | 0.021 |
|
| ||||||
| Univariate and multivariate analyses of relapse-free survival in breast cancer patients (n=160) | ||||||
|
| ||||||
| Age (>50 vs <50) | 1.232 | 0.778-2.212 | 0.761 | |||
| Her-2 status (Negative vs positive) | 1.146 | 0.942-1.567 | 0.659 | |||
| PR status (Negative vs positive) | 1.728 | 0.821-3.011 | 0.842 | |||
| ER status (Negative vs positive) | 1.607 | 0.939-2.905 | 0.651 | |||
| TNBC status (Negative vs positive) | 1.163 | 0.910-1.754 | 0.545 | |||
| Tumor size (>20 mm vs <20 mm) | 1.327 | 0.729-2.154 | 0.286 | |||
| Involved lymph node (Negative vs positive) | 2.592 | 1.781-4.211 | 0.034 | 2.730 | 2.112-3.908 | 0.025 |
| Differentiation grade (Well & Moderate vs Poor) | 2.734 | 1.957-4.609 | 0.017 | 1.472 | 1.104-2.894 | 0.129 |
| TNM stage (I & II vs III & IV) | 3.214 | 2.011-4.942 | 0.000 | 3.021 | 2.809-4.391 | 0.006 |
| CBX2 expression level | 2.524 | 1.902-3.947 | 0.004 | 2.919 | 2.201-3.783 | 0.021 |
CBX2 silencing suppresses breast cancer cell proliferation and invasion in vitro
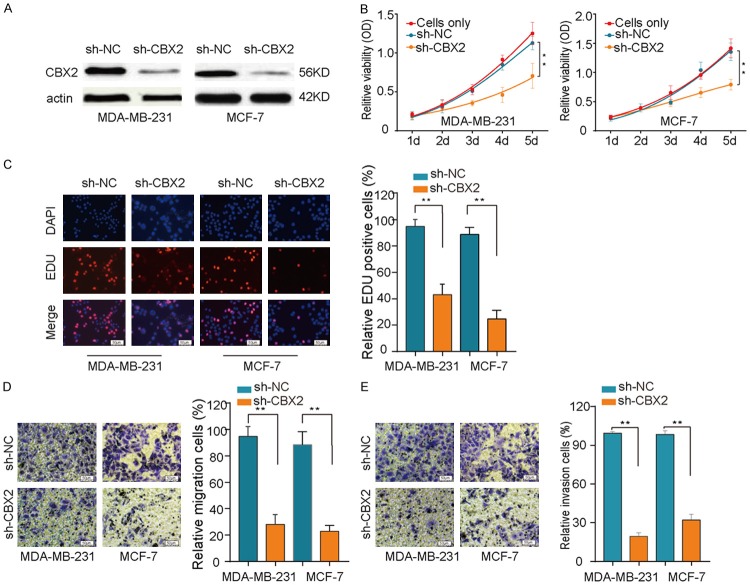
To better understand how upregulated expression of CBX2 impacts the proliferation and invasion of breast cancer cells, we examined the effects of lentivirus-mediated CBX2 knockdown in breast cancer cell lines. The efficiency of CBX2 knockdown in breast cancer cells was confirmed by western blot analysis (Figure 3A). To assess the role of CBX2 in the proliferation of breast cancer cells, we performed the CCK-8 assay. Our results showed that CBX2 silencing significantly reduced proliferation of MDA-MB-231 and MCF-7 cells (Figure 3B). Consistent with a reduction in proliferation, our EDU assay suggested that CBX2 knockdown decreased the growth of breast cancer cells (Figure 3C). Our transwell migration and invasion assays indicated that the migration and invasion abilities of breast cancer cells were remarkably impaired by CBX2 knockdown (Figure 3D and 3E). These results provided evidence that CBX2 likely functions as an oncogene by promoting proliferation and invasion in breast cancer cells.
Figure 3.
CBX2 knockdown inhibits proliferation and invasion of breast cancer cells in vitro. A. Western blots showing efficiency of shRNA-mediated CBX2 knockdown in MDA-MB-231 and MCF-7 cells. B. CCK8 assay showing that CBX2 silencing attenuated proliferation of MDA-MB-231 and MCF-7 breast cancer cells. C. Representative fluorescence micrographs (left) and bar graphical quantitation of percentage of EDU-positive cells (right) from the EDU assay demonstrating that CBX2 knockdown suppresses proliferation of MDA-MB-231 and MCF-7 cells. D, E. Representative micrographs (left) and bar graphical quantitation of migrated/invaded cells (right) from the transwell assay showing that CBX2 silencing caused a remarkable suppression of cell migration and invasion in MDA-MB-231 and MCF-7 cells. **P<0.01.
Downregulation of CBX2 inhibits breast tumorigenesis in vivo
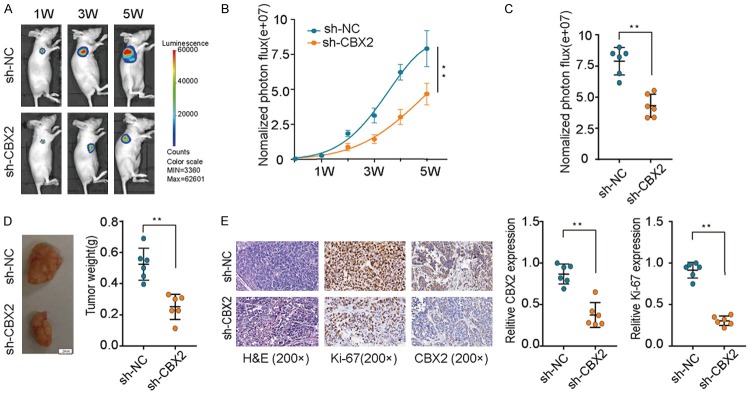
To gain insights into the impact of CBX2 silencing on breast tumorigenesis in vivo, sh-NC and sh-CBX2 cells were injected into the flanks of nude mice and tumor growth was monitored over time. Tumor growth in the sh-CBX2 group was dramatically reduced compared to that in the sh-NC group (Figure 4A-C). Tumor weight was also significantly decreased in the sh-CBX2 group (Figure 4D) compared to the sh-NC group. Moreover, IHC analysis of the expression of CBX2 and Ki-67 in mouse tumor tissues revealed that the expression of both CBX2 and Ki-67 proteins was markedly suppressed in the sh-CBX2 group (Figure 4E). Collectively, our findings demonstrated that CBX2 silencing inhibits breast tumorigenesis; thus, CBX2 can serve as a potential target for breast cancer therapy.
Figure 4.
CBX2 knockdown suppresses breast cancer tumorigenesis in vivo. A, B. Breast cancer cells stably transfected with sh-CBX2 or sh-NC were injected subcutaneously into nude mice (n=12). Images of tumors that formed in the nude mice were acquired using a live imaging system to detect the luciferase signal. C. The luciferase activity of the sh-CBX2 tumors was lower than that of the sh-NC group. D. Tumor weights in sh-CBX2 group were markedly lower than those of sh-NC group. E. Representative images of mouse (xenograft) tumor sections stained with hematoxylin and eosin and immunohistochemically stained for Ki-67 and CBX2. **P<0.01.
CBX2 may play essential roles in regulating cell cycle and DNA replication, and the PI3K/AKT pathway functions downstream of CBX2
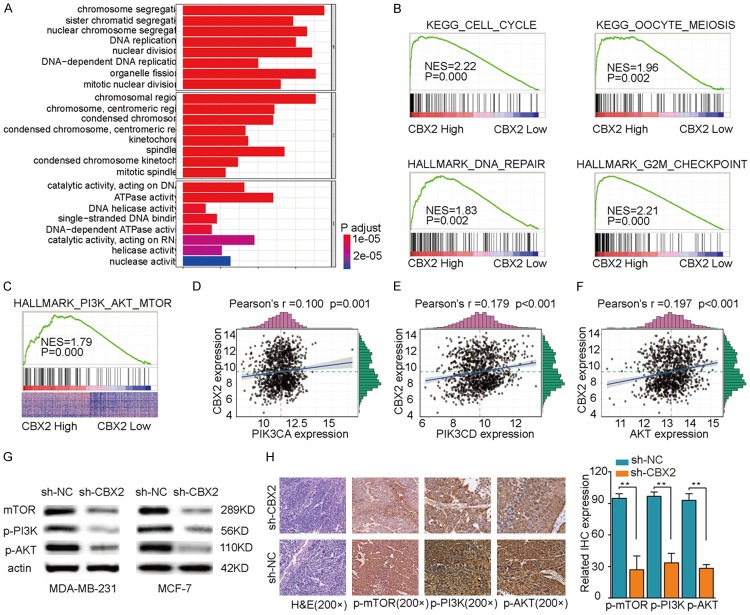
To further our understanding of the downstream molecular mechanisms by which CBX2 drives breast tumorigenesis and/or progression, we performed clusterProfile GO and KEGG analyses using gene expression data from the TCGA dataset. These bioinformatics analyses showed that cell cycle and DNA replication pathways were markedly enriched in CBX2-high expression samples (Figure 5A and 5B), suggesting that CBX2 may contribute to breast cancer development/progression by regulating cell cycle and DNA replication. Moreover, we found a striking positive correlation between the expression of CBX2 and the activation of the PI3K/AKT signaling pathway based on Gene Set Enrichment Analysis (GSEA) of the TCGA data (Figure 5C). There was a highly significant positive correlation between the expression of CBX2 and the expressions of PIK3CA, PIK3CD, and AKT (Figure 5D-F). To corroborate the findings from our in silico analyses, we examined the expressions of p-mTOR, p-PI3K, and p-AKT in tumor tissues derived from our xenograft models by western blotting and IHC. As shown in Figure 5G and 5H, the expressions of p-mTOR, p-PI3K, and p-AKT were significantly decreased in the sh-CBX2 group compared to the levels observed in the sh-NC group. Taken together, these findings indicated that (a) CBX2 potentially drives breast tumorigenesis and/or progression by regulating the cell cycle and DNA replication in breast cancer cells, and (b) the PI3K/AKT signaling pathway functions downstream of CBX2.
Figure 5.
Functional and pathway enrichment analysis. A. Analysis of correlation between expressions of CBX2 and cell cycle-related genes. B. KEGG and GO enrichment analysis of correlation between differentially expressed CBX2 genes and cell cycle, as well as DNA replication pathway-related genes. C. GO enrichment analysis of correlation between differentially expressed CBX2 genes and PI3K/AKT pathway related genes. D-F. Analyses of correlations between expression of CBX2 and expressions of PIK3CA, PIK3CD, and AKT. G. Western blot analysis showing that the expressions of p-mTOR, p-PI3K, and p-AKT were significantly decreased in the sh-CBX2 group compared to the sh-NC group. H. Representative images of immunohistochemically stained mouse tumor sections (left) and bar graphical quantitation of IHC expression scores (right) showing that the expressions of p-mTOR, p-PI3K, and p-AKT were significantly decreased in the sh-CBX2 group compared to the sh-NC group. **P<0.01.
Discussion
Breast cancer continues to be one of the most commonly diagnosed malignancies among women worldwide [25]. Advanced breast cancer with metastasis remains the most common cause of breast cancer-associated death [26]. Therefore, there is an urgent need to better understand the molecular underpinnings of breast tumorigenesis and progression, to identify new biomarkers to enable the personalization of breast cancer management, and to find novel and effective therapeutic targets for breast cancer treatment.
A growing body of evidence indicates that aberrant expression of CBX family members contributes to the progression of various types of cancer. For instance, Zeng J. et al. reported that CBX4 protein expression was upregulated in breast cancer and high expression of CBX4 was associated with increased metastasis, advanced TNM stage, and poor OS of breast cancer patients [27]. Zheng H. et al. reported that CBX6 protein expression was frequently elevated in hepatocellular carcinoma (HCC). In addition, elevated CBX6 expression was strongly associated with larger tumor sizes and poor OS of patients with HCC [28]. Concordant with these results, we found that CBX2 was significantly upregulated in breast cancer tissues both at the mRNA and protein levels. Additionally, upregulated CBX2 expression was associated with ER-negative and PR-negative status of breast tumors and with late TNM stages and metastasis. Moreover, the OS and PFS of patients with low expression of CBX2 were significantly better than those of patients with high expression of CBX2. The associations between CBX2 expression and clinical outcomes were upheld in both univariate and multivariate analyses, thereby indicating that CBX2 expression can serve as an independent prognostic factor. These results suggest that CBX2 has a critical role in breast cancer development and/or progression, and CBX2 might serve as a potential novel prognostic biomarker and potential therapeutic target for breast cancer.
Previous studies have identified CBX2 as a positive regulator of cell growth and metastasis in several cancer types [29-31]. However, the functional role of CBX2 in driving breast cancer progression remained undefined. We investigated the biological function of CBX2 in breast cancer and showed that knockdown of CBX2 inhibits proliferation of breast cancer cells in vitro. The migration and invasion abilities of breast cancer cells were also markedly impaired in CBX2 knockdown breast cancer cells in vitro. Moreover, we observed that tumorigenesis was suppressed in xenografts of breast cancer cells wherein CBX2 was silenced, indicating that CBX2 may function as an essential regulator of breast tumorigenesis and/or tumor growth. Consistent with our findings, one study showed that CBX4 silencing suppressed proliferation, colony formation, and migration abilities of colorectal cancer cells in vitro, and tumorigenicity in vivo [32]. Collectively, these finding suggest that CBX2 may play an oncogenic role in breast cancer, and silencing of CBX2 may be a potential therapeutic strategy for breast cancer.
Next, we explored the molecular mechanisms underlying CBX2’s role in driving breast cancer development and/or progression. Through bioinformatics analysis based on the TCGA dataset, we found that CBX2 was involved in regulating cell cycle progression and DNA replication. Our GSEA results supported a model wherein CBX2 promoted signaling via the PI3K/AKT pathway. Results from our in vitro and in vivo experiments showed significant downregulation of PIK3CA, PIK3CD, and p-mTOR upon CBX2 silencing; these findings suggest that CBX2 directly regulates tumor growth via the PI3K/AKT pathway. In support of our results, deregulation of the PI3K/AKT pathway has been previously associated with cancer progression in multiple cancer types, including gallbladder cancer, colorectal cancer, endometrial carcinoma, and breast cancer [33-37]. Zhu M. et al. have reported that overexpression of the long non-coding RNA (lncRNA) MEG3 suppresses breast tumorigenesis via the PI3K/AKT pathway [38]. Altogether, these data powerfully indicate that CBX2 participates in breast cancer cell proliferation, migration, and invasion via regulation of the PI3K/AKT pathway.
Conclusions
Our findings provide significant insights into the progression of breast cancer. Our study has demonstrated that high expression of CBX2 is associated with breast cancer development and poor prognosis, and that CBX2 regulates the PI3K/AKT pathway. Thus, CBX2 overexpression in breast cancer potentially drives breast cancer growth and invasion by enhancing signaling via the PI3K/AKT pathway. CBX2 merits further study as a potential therapeutic target for advanced breast cancer.
Acknowledgements
This study was sponsored by grants from the Henan Provincial Department of Science and Technology research project (182102310362) and the Key Scientific Research Project of Colleges and Universities in Henan Province (19B320042). The funding sources had no role in the design of this study, nor any role during its execution, analyses, data interpretation, or decision to submit results. The study was approved by the Human Ethics Committee of the First Affiliated Hospital of Zhengzhou University. All patients provided written informed consent and the project was in accordance with the Helsinki Declaration of 1975. Patient clinical information is kept in the databases of the First Affiliated Hospital of Zhengzhou University and utilized for research. All the in vivo experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the First Affiliated Hospital of Zhengzhou University.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sprague BL, Gangnon RE, Hampton JM, Egan KM, Titus LJ, Kerlikowske K, Remington PL, Newcomb PA, Trentham-Dietz A. Variation in breast cancer-risk factor associations by method of detection: results from a series of case-control studies. Am J Epidemiol. 2015;181:956–69. doi: 10.1093/aje/kwu474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lázár G, Bursics A, Farsang Z, Harsányi L, Kósa C, Maráz R, Mátrai Z, Paszt A, Pavlovics G, Tamás R. Modern surgical treatment of breast cancer. 3rd breast cancer consensus conference. Magy Seb. 2016;69:117–32. doi: 10.1556/1046.69.2016.3.5. [DOI] [PubMed] [Google Scholar]
- 4.Ren K, Yin Y, He F, Shao Y, Wang S. Prognostic role of derived neutrophil-to-lymphocyte ratio in surgical triple-negative breast cancer. Cancer Manag Res. 2018;10:4891–4898. doi: 10.2147/CMAR.S180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagini S. Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med Chem. 2017;17:152–163. doi: 10.2174/1871520616666160502122724. [DOI] [PubMed] [Google Scholar]
- 6.Ganz PA, Goodwin PJ. Goodwin, breast cancer survivorship: where are we today? Adv Exp Med Biol. 2015;862:1–8. doi: 10.1007/978-3-319-16366-6_1. [DOI] [PubMed] [Google Scholar]
- 7.Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci U S A. 2008;105:16572–7. doi: 10.1073/pnas.0805317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the polycomb group cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Klauke K, Radulović V, Broekhuis M, Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin K, Bystrykh L, de Haan G. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–62. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 10.Connelly KE, Martin EC, Dykhuizen EC. CBX chromodomain inhibition enhances chemotherapy response in glioblastoma multiforme. Yale J Biol Med. 2016;89:431–440. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XW, Zhang L, Qin W, Yao XH, Zheng LZ, Liu X, Li J, Guo WJ. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J Exp Clin Cancer Res. 2010;29:114. doi: 10.1186/1756-9966-29-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickl JM, Tichy D, Kuryshev VY, Tolstov Y, Falkenstein M, Schüler J, Reidenbach D, Hotz-Wagenblatt A, Kristiansen G, Roth W, Hadaschik B, Hohenfellner M, Duensing S, Heckmann D, Sültmann H. Ago-RIP-Seq identifies polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget. 2016;7:59589–59603. doi: 10.18632/oncotarget.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallante P, Federico A, Berlingieri MT, Bianco M, Ferraro A, Forzati F, Iaccarino A, Russo M, Pierantoni GM, Leone V, Sacchetti S, Troncone G, Santoro M, Fusco A. Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Res. 2008;68:6770–8. doi: 10.1158/0008-5472.CAN-08-0695. [DOI] [PubMed] [Google Scholar]
- 14.Cacciola NA, Sepe R, Forzati F, Federico A, Pellecchia S, Malapelle U, De Stefano A, Rocco D, Fusco A, Pallante P. Restoration of CBX7 expression increases the susceptibility of human lung carcinoma cells to irinotecan treatment. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:1179–86. doi: 10.1007/s00210-015-1153-y. [DOI] [PubMed] [Google Scholar]
- 15.Pallante P, Terracciano L, Carafa V, Schneider S, Zlobec I, Lugli A, Bianco M, Ferraro A, Sacchetti S, Troncone G, Fusco A, Tornillo L. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer. 2010;46:2304–13. doi: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D, Wang G, Qin G, Xu RH, Kang T. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76:7277–7289. doi: 10.1158/0008-5472.CAN-16-2100. [DOI] [PubMed] [Google Scholar]
- 17.Liang YK, Lin HY, Chen CF, Zeng Prognostic values of distinct CBX family members in breast cancer. Oncotarget. 2017;8:92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabuchi S, Kuroda H, Takahashi R, Sasano T. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol Oncol. 2015;137:173–9. doi: 10.1016/j.ygyno.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Guo H, German P, Bai S, Barnes S, Guo W, Qi X, Lou H, Liang J, Jonasch E, Mills GB, Ding Z. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. 2015;42:343–53. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. 2015;1855:104–21. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Xue C, He Y, Hu Q, Yu Y, Chen X, Chen J, Ren F, Li J, Ren Z, Cui G, Sun R. Downregulation of PIM1 regulates glycolysis and suppresses tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10:5101–5112. doi: 10.2147/CMAR.S184381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Ciruelos Gil EM. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat Rev. 2014;40:862–71. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 23.He Y, Chen X, Yu Y, Li J, Hu Q, Xue C, Chen J, Shen S, Luo Y, Ren F, Li C, Bao J, Yan J, Qian G, Ren Z, Sun R, Cui G. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Mol Carcinog. 2018;57:772–783. doi: 10.1002/mc.22799. [DOI] [PubMed] [Google Scholar]
- 24.He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G, Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10:3857–3865. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Xu L, Liu Y, Fu S, Tu J, Hu Y, Xiong Q. LncRNA MALAT1 promotes relapse of breast cancer patients with postoperative fever. Am J Transl Res. 2018;10:3186–3197. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Rao X, Yao W, Zou X. Downregulation of MiR-196b-5p impedes cell proliferation and metastasis in breast cancer through regulating COL1A1. Am J Transl Res. 2018;10:3122–3132. [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng JS, Zhang ZD, Pei L, Bai ZZ, Yang Y, Yang H, Tian QH. CBX4 exhibits oncogenic activities in breast cancer via notch1 signaling. Int J Biochem Cell Biol. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Zheng H, Jiang WH, Tian T, Tan HS, Chen Y, Qiao GL, Han J, Huang SY, Yang Y, Li S, Wang ZG, Gao R, Ren H, Xing H, Ni JS, Wang LH, Ma LJ, Zhou WP. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:18872–18884. doi: 10.18632/oncotarget.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clermont PL, Sun L, Crea F, Thu KL, Zhang A, Parolia A, Lam WL, Helgason CD. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. Br J Cancer. 2014;111:1663–72. doi: 10.1038/bjc.2014.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clermont PL, Crea F, Chiang YT, Lin D, Zhang A, Wang JZ, Parolia A, Wu R, Xue H, Wang Y, Ding J, Thu KL, Lam WL, Shah SP, Collins CC, Wang Y, Helgason CD. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. doi: 10.1186/s13148-016-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuang W, Deng Q, Deng C, Li W, Shu S, Zhou M. Hepatocyte growth factor induces breast cancer cell invasion via the PI3K/Akt and p38 MAPK signaling pathways to up-regulate the expression of COX2. Am J Transl Res. 2017;9:3816–3826. [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Kang T. Role of CBX4 in the colorectal carcinoma metastasis-response. Cancer Res. 2017;77:2550–2551. doi: 10.1158/0008-5472.CAN-17-0594. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Teng S, Zhang Y, Zhang W, Zhang X, Xu K, Yao H, Yao J, Wang H, Liang X, Hu Z. TROP2 promotes proliferation, migration and metastasis of gallbladder cancer cells by regulating PI3K/AKT pathway and inducing EMT. Oncotarget. 2017;8:47052–47063. doi: 10.18632/oncotarget.16789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang DW, Lee BH, Suh YA, Choi YS, Jang SJ, Kim YM, Choi KY, Min DS. Phospholipase D1 inhibition linked to upregulation of ICAT blocks colorectal cancer growth hyperactivated by wnt/beta-catenin and PI3K/Akt signaling. Clin Cancer Res. 2017;23:7340–7350. doi: 10.1158/1078-0432.CCR-17-0749. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Wang Y, Wang S, Liu Y, Zhang J, Xu Y, Zhang Z, Bao W, Wu S. LSD1 sustains estrogen-driven endometrial carcinoma cell proliferation through the PI3K/AKT pathway via di-demethylating H3K9 of cyclin D1. Int J Oncol. 2017;50:942–952. doi: 10.3892/ijo.2017.3849. [DOI] [PubMed] [Google Scholar]
- 36.Guerrero-Zotano A, Mayer IA, Arteaga CL. PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016;35:515–524. doi: 10.1007/s10555-016-9637-x. [DOI] [PubMed] [Google Scholar]
- 37.Bao J, Yu Y, Chen J, He Y, Chen X, Ren Z, Xue C, Liu L, Hu Q, Li J, Cui G, Sun R. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9:1045. doi: 10.1038/s41419-018-1020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem Biophys. 2019;661:22–30. doi: 10.1016/j.abb.2018.10.021. [DOI] [PubMed] [Google Scholar]