Abstract
Circular RNA hsa_circ_0002024 has been reported to be underexpressed in bladder cancer (BC). However, the biological role of hsa_circ_0002024 in BC and its underlying molecular mechanisms remain unclear. In this study, the expression levels of hsa_circ_0002024 and miR-197-3p were examined by quantitative real-time polymerase chain reaction. The proliferation abilities of EJ and T24 cells were assessed by Cell Counting Kit-8 and 5-ethynyl-20-deoxyuridine assays. Cell migration and invasion were evaluated by transwell migration and invasion assays. Luciferase reporter assay and rescue experiments were conducted to elucidate the underlying mechanism of hsa_circ_0002024. We found that the expression of hsa_circ_0002024 was downregulated, but that of miR-197-3p was upregulated in BC tissues and cell lines. Upregulation of hsa_circ_0002024 suppressed the proliferation, migration, and invasion of EJ and T24 cells. Hsa_circ_0002024 was confirmed as a direct target of miR-197-3p. In addition, we found that restoration of miR-197-3p expression could abolish hsa_circ_0002024-mediated inhibition of BC cell proliferation, migration, and invasion. In conclusion, our data demonstrated that hsa_circ_0002024 suppresses cell proliferation, migration, and invasion in BC by sponging miR-197-3p.
Keywords: Bladder cancer, hsa_circ_0002024, miR-197-3p, metastasis
Introduction
Bladder cancer (BC) is the ninth most common malignancy worldwide, with a high prevalence [1]. BC is one of the leading causes of cancer-related deaths among men and woman, resulting in huge economic losses [2,3]. On the basis of clinical pathology, BC can be divided into 3 types: transitional cell carcinoma, squamous cell carcinoma, and adenocarcinoma. Usually, BC is clinically characterized by blood in the urine, pain with urination, and lower back pain. Typically, it develops in individuals with a family history of BC, smoking, obesity, exposure to certain chemicals, and frequent bladder infections, which are thought to be the major causes of BC [4-7]. Clinically, patients with BC are treated with surgery, radiation therapy, immunotherapy, chemotherapy, or combined therapy [8]. Metastasis is the main reason of recurrence and death in BC. It may be curable before the tumor spreads to another part of the body. However, patients with metastatic BC have a poor 5-year survival rate. Despite extensive study, the pathogenesis of BC remains unclear.
It is well accepted that more than 90% of the human genome is made up of non-coding RNA (ncRNAs) [9]. In general, ncRNAs are classified into long ncRNAs (lncRNAs; ≥ 200 nucleotides) and small ncRNAs (< 200 nucleotides) according to their size (7). microRNAs (miRNAs) have emerged as a class of small ncRNAs containing about 22 nucleotides, which negatively regulate target gene expression by binding to the complementary sequences of target genes [10]. miRNAs have been shown to be involved in many cellular processes, such as cell proliferation, differentiation, and apoptosis [11]. A number of studies have demonstrated that miRNAs are aberrantly expressed in cancers and play a crucial role in promoting or repressing carcinogenesis [12]. miR-197-3p belongs to the miR-197 family and has been reported to be upregulated in BC and implicated in LINC00641-mediated cell growth inhibition in T24 and J82 cells [13]. Nevertheless, only a limited number of studies have focused on the regulatory mechanism of miR-197-3p in BC.
Circular RNAs (circRNAs) are a novel kind of ncRNA whose 3’ heads and 5’ tails bind together to form a covalently closed continuous loop [14]. CircRNAs are particularly abundant in eukaryotic cells [15]. They are the major products of “back-splicing” reaction, in which the donor splice site and acceptor splice site are joined together [16]. Owing to their unique closed loop structure, circRNAs are resistant to exonuclease-mediated degradation, indicating their ideal role as a reliable and stable disease biomarker. CircRNAs have been shown to serve as key regulators of gene expression. Due to many miRNA binding sites, circRNAs function as an “miRNA sponge” to suppress the expression and function of miRNAs [17]. A circRNA sponge for miR-7 has been reported to inhibit the activity of miR-7, thereby upregulating the expression of miR-7 target genes [18]. In addition, circRNAs are capable of cis-regulating the expression of their parent genes, binding to proteins, and encoding proteins [19,20]. Emerging evidence suggests that aberrant expression of circRNAs is closely linked to the pathogenesis of various malignancies [21]. The expression of hsa_circ_0002024, a novel circRNA, has been documented to be downregulated in bladder cancer [22]. However, the functional role of hsa_circ_0002024 in BC and its underlying mechanism have not yet been studied.
In this study, hsa_circ_0002024 was underexpressed, but miR-197-3p was overexpressed in BC tissues and cell lines. Upregulation of hsa_circ_0002024 expression suppressed the proliferation, migration, and invasion of EJ and T24 cells via sponging miR-197-3p. Our findings reveal that hsa_circ_0002024-miR-197-3p interaction is responsible for the progression of BC, thereby highlighting potential therapeutic strategies for the treatment of BC.
Materials and methods
Patient samples
Clinical frozen tumor and normal samples of 20 patients with pathologically confirmed BC were obtained from the Department of Urology, The Third People’s Hospital of Linyi. This study was reviewed and approved by the Ethics Committee of The Third People’s Hospital of Linyi.
Cell culture
BC cell lines (EJ, 5637, T24, and UMUC-2) and normal human urothelial cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were grown in Roswell Park Memorial Institute 1640 (RPMI-1640) medium (Solarbio, Beijing, China) supplemented with 10% fetal bovine serum (FBS; Solarbio) and maintained at 37°C in a 5% CO2 atmosphere.
Transient transfection
miR-197-3p, hsa_circ_0002024 overexpression vector (over-circ), empty vector (vector), small interfering RNA targeting hsa_circ_0002024 (si-circ), and si-negative control (si-NC) were designed and synthesized by GenePharma (Shanghai, China). EJ and T24 cells were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from BC tissues and cells was purified using TRIzol reagent (Invitrogen). cDNA synthesis was carried out using SuperScript First-Stand Synthesis System (Invitrogen). qRT-PCR assay was performed in triplicate with the MyIQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). TaqMan miRNA-assay (Applied Biosystems, Foster City, CA, USA) was utilized to evaluate the expression of miR-197-3p. The relative expression levels of hsa_circ_0002024 and miR-197-3p were determined using 2-ΔΔCt method with β-actin as the loading control for hsa_circ_0002024 and U6 for miR-197-3p.
Detection of cell proliferation capacity
For Cell Counting Kit-8 (CCK-8) assay, EJ and T24 cells were seeded in 96-well plates and transfected with vector, over-circ, or miR-197-3p. After transfection, cells were cultured with 10 μl CCK-8 reagent (Solarbio) for 2 h and the absorbance was measured using a microplate reader (Benchmark; Bio-Rad).
For 5-ethynyl-20-deoxyuridine (EdU) assay, EJ and T24 cells were seeded in 96-well plates after transfection, followed by incubation with EdU (50 μM; Beyotime, Shanghai, China) for 2 h. Cells were then fixed in 4% paraformaldehyde, followed by staining with 1× Apollo reaction cocktail and Hoechst 33342. Images were captured with a microscope (Olympus BX51 TRF; Tokyo, Japan) and analyzed by ImageJ software (NIH Image, Bethesda, MD, USA).
Transwell migration and invasion assay
After transfection, EJ and T24 cells were starved for 24 h. Cells were harvested, dissociated, and resuspended in serum-free medium. Transwell chambers (Corning, Steuben County, NY, USA) were pre-coated with fibronectin for migration assay or with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) for invasion assay. EJ and T24 cells were seeded in the upper chambers and RPMI-1640 medium containing 10% FBS was added into the lower chamber. After 24 h of incubation at 37°C, cells migrating or invading through the membrane were stained with 0.1% crystal violet (Solarbio) for 20 min. The number of migrating or invading cells was counted visually in 5 randomly selected fields using a microscope.
Luciferase reporter assay
Wild-type hsa_circ_0002024 (WT-circ) and mutant-type hsa_circ_0002024 (Mut-circ) were synthesized and subcloned into the pmirGLO vector (Promega, Madison, WI, USA). EJ and T24 cells were co-transfected with pmirGLO-WT-circ or pmirGLO-Mut-circ and miR-197-3p or miR-NC using Lipofectamine 2000. After 48 h of transfection, cells were harvested and the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega), following the manufacturer’s specifications.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 software (IBM, NY, USA). Results are expressed as the mean ± standard error of the mean (SEM). Student’s t-test (two groups) and one-way analysis of variance (multiple groups) were applied to determine differences among groups. Results were considered statistically significant when the P-value was 0.05 or less.
Results
Expression of hsa_circ_0002024 in BC tissues and cell lines
Expression levels of hsa_circ_0002024 were markedly lower in BC tissues compared with those in adjacent normal tissues, as shown by qRT-PCR (Figure 1A). Correspondingly, qRT-PCR results showed that hsa_circ_0002024 expression was obviously decreased in BC cell lines (EJ, 5637, T24, and UMUC-2) compared with that in normal human urothelial cells (Figure 1B).
Figure 1.
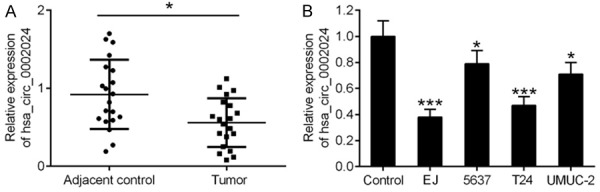
Expression of hsa_circ_0002024 in BC tissues and cell lines. A. qRT-PCR analysis of hsa_circ_0002024 expression in tissues samples. B. qRT-PCR analysis of hsa_circ_0002024 expression in BC cell lines (EJ, 5637, T24, and UMUC-2) and normal human urothelial cells (control). *P < 0.05 and ***P < 0.001.
Expression of miR-197-3p in BC tissues and cell lines
Given the putative miRNA regulatory function of circRNAs, the CircInteractome database (https://circinteractome.nia.nih.gov/) was employed to predict the interaction between validated circRNAs and potential miRNA targets. We found that miR-595, miR-197-3p, miR-646, miR-1261, miR-224, miR-361-3p, and miR-576-3p were the potential miRNA targets of hsa_circ_0002024. We determined the expression levels of these miRNAs in BC tissues and their adjacent normal tissues by qRT-PCR. The greatest difference was observed in the expression of miR-197-3p (Figure 2A). Similarly, higher levels of miR-197-3p expression were observed in BC cell lines (EJ, 5637, T24, and UMUC-2) compared to that in normal human urothelial cells (Figure 2B).
Figure 2.
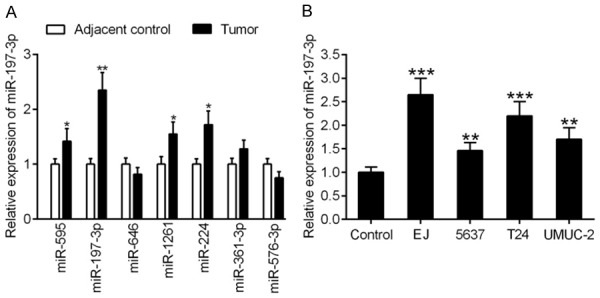
Expression of miR-197-3p in BC tissues and cell lines. A. Validation of differential expression levels of miR-595, miR-197-3p, miR-646, miR-1261, miR-224, miR-361-3p, and miR-576-3p using qRT-PCR analysis in BC tissues and their adjacent controls. B. qRT-PCR analysis of miR-197-3p expression in BC cell lines (EJ, 5637, T24, and UMUC-2) and normal human urothelial cells (control). **P < 0.01 and ***P < 0.001.
Upregulation of hsa_circ_0002024 suppresses the proliferation of EJ and T24 cells
To investigate the functional significance of hsa_circ_0002024 in BC, we transfected EJ and T24 cells with vector and over-circ for 48 h, which resulted in upregulation of hsa_circ_0002024 (Figure 3A). Upregulation of hsa_circ_0002024 expression in EJ and T24 cells led to a marked decrease in cell viability, as demonstrated by CCK-8 assay (Figure 3B and 3C). In line with this, a reduction in the number of EdU-positive cells was observed in EJ and T24 cells transfected with over-circ (Figure 3D).
Figure 3.
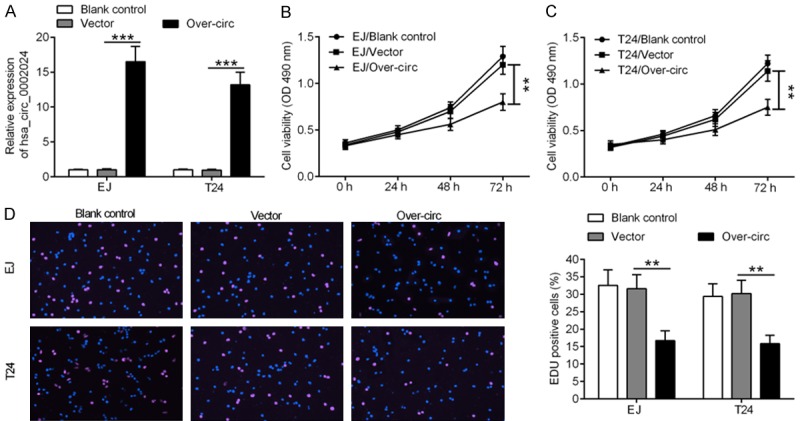
Upregulation of hsa_circ_0002024 suppresses the proliferation of EJ and T24 cells. A. Upregulation of hsa_circ_0002024 expression in EJ and T24 cells after transfection with vector or over-circ. B and C. Both EJ and T24 cells showed marked reduction of cell viability after transfection with over-circ. D. EdU assay showed a reduced number of EdU-positive cells in EJ and T24 cells transfected with over-circ. **P < 0.01 and ***P < 0.001.
Upregulation of hsa_circ_0002024 suppresses the migration and invasion of EJ and T24 cells
To determine the impact of hsa_circ_0002024 on the migration and invasion of EJ and T24 cells, transwell migration and invasion assays were performed in EJ and T24 cells after transfection. Transfection with over-circ inhibited the migration of EJ and T24 cells as compared with the vector group (Figure 4A). In parallel, a significant reduction in cell invasion was also observed in EJ and T24 cells transfected with over-circ (Figure 4B).
Figure 4.
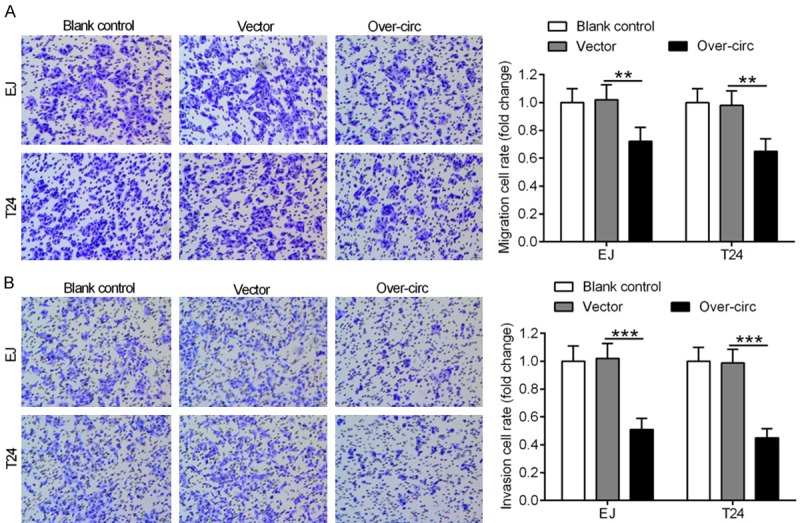
Upregulation of hsa_circ_0002024 suppresses the migration and invasion of EJ and T24 cells. A. Graphical representation and quantification of transwell migration assay showed that upregulation of hsa_circ_0002024 expression strikingly inhibited the migration of EJ and T24 cells. B. Graphical representation and quantification of transwell invasion assay showed that upregulation of hsa_circ_0002024 expression strikingly inhibited the invasion of EJ and T24 cells. **P < 0.01 and ***P < 0.001.
Hsa_circ_0002024 reduces the expression of miR-197-3p
Correlation analysis indicated a notable negative correlation between hsa_circ_0002024 and miR-197-3p expression in BC tissues (Figure 5A). Of note, hsa_circ_0002024 harbored a single predicted binding site for miR-197-3p, as shown by bioinformatics analysis (Figure 5B). Knockdown of hsa_circ_0002024 resulted in significant elevation of miR-19-3p expression in EJ and T24 cells, while overexpression of hsa_circ_0002024 led to a marked reduction of miR-197-3p expression in EJ and T24 cells (Figure 5C and 5D). To validate the interaction between hsa_circ_0002024 and miR-197-3p, luciferase reporter assay was performed in EJ and T24 cells. Luciferase reporter assay in miR-197-3p-overexpressing EJ and T24 cells showed that transfection with miR-197-3p suppressed the luciferase activity of WT-circ. However, transfection with miR-NC had no effect. Meanwhile, the luciferase activity of Mut-circ was unaltered in miR-197-3p-overexpressing or miR-NC-transfected EJ and T24 cells (Figure 5E and 5F).
Figure 5.
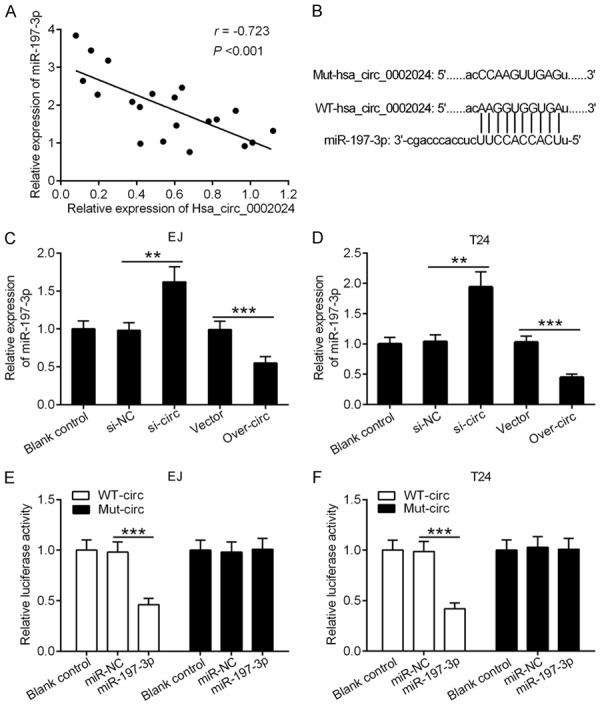
Hsa_circ_0002024 reduces the expression of miR-197-3p. A. qRT-PCR analysis of hsa_circ_0002024 and miR-197-3p expression in BC tissues. B. The complimentary binding site for miR-197-3p in hsa_circ_0002024 sequence is shown. C and D. qRT-PCR analysis of miR-197-3p expression after transfecting EJ and T24 cells with si-NC, si-circ, vector, or over-circ. E and F. Luciferase reporter assay showed the luciferase activity of EJ and T24 cells after co-transfection with WT-circ or Mut-circ and miR-197-3p and miR-NC. **P < 0.01 and ***P < 0.001.
Restoration of miR-197-3p expression reverses hsa_circ_0002024-mediated inhibition of BC cell proliferation, migration, and invasion
As miR-197-3p directly binds to hsa_circ_0002024, and hsa_circ_0002024 negatively regulates the expression of miR-197-3p, we next investigated whether miR-197-3p participates in hsa_circ_0002024-mediated inhibition of BC cell proliferation, migration, and invasion. Co-transfection with over-circ with miR-197-3p obviously increased the viability of EJ and T24 cells as compared to transfection with over-circ alone (Figure 6A and 6B). Likewise, EJ and T24 cells co-transfected with over-circ with miR-197-3p showed increased number of EdU-positive cells relative to EJ and T24 cells transfected with over-circ alone (Figure 6C). Moreover, a significant reduction in cell migration and invasion was also observed in EJ and T24 cells co-transfected with over-circ and miR-197-3p as compared to cells transfected with over-circ alone (Figure 6D and 6E).
Figure 6.
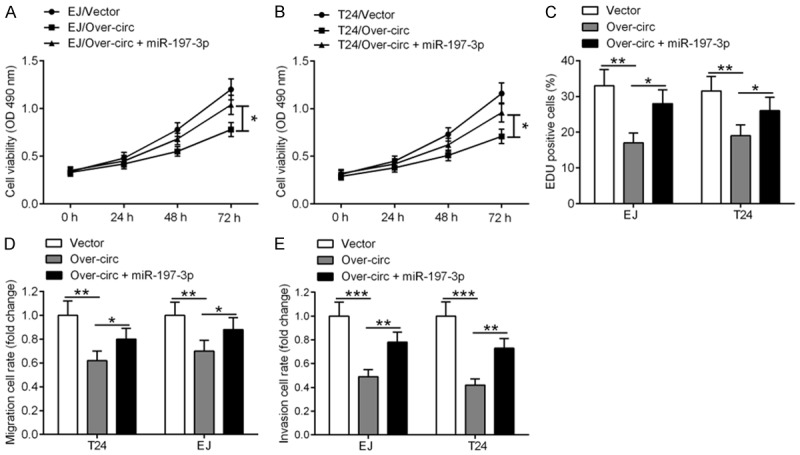
Restoration of miR-197-3p expression reverses hsa_circ_0002024-mediated inhibition of BC cell proliferation, migration, and invasion. EJ and T24 cells were transfected with over-circ alone or together with miR-197-3p. A-C. CCK-8 and EdU assays showed that restoration of miR-197-3p expression reversed hsa_circ_0002024-mediated inhibition of BC cell proliferation. D and E. Transwell migration and invasion assays revealed that overexpression of miR-197-3p blocked the inhibitory effects of hsa_circ_0002024 on the migration and invasion of EJ and T24 cells. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
For more than 30 years, circRNAs have been acknowledged as products of aberrant splicing, gene rearrangement, or non-linear reverse splicing [23]. Along with the development of high-throughput sequencing technology, circRNAs have been implicated in disease development, physiological processes, and substance metabolism via modulating gene expression [14]. In recent years, circRNAs have become a hot topic in cancer research. Compared with miRNAs and lncRNAs, circRNAs are considered to be promising therapeutic targets for cancer treatment in terms of their sequence conservation, biological stability, and tissue specificity. In our study, we identified that hsa_circ_0002024 expression was downregulated in BC tissues and cell lines, which suggests that hsa_circ_0002024 may play a potential role in the pathological mechanism of BC. Further, we reported that hsa_circ_0002024 functions as a miRNA sponge in two BC cell lines.
Several lines of evidence have indicated that circRNAs serve a crucial role in the occurrence and development of human diseases. A few earlier studies reported that circRNAs act as oncogenes or tumor suppressors in the progression of human cancers. For example, circCCDC66 was shown to be overexpressed in polyps and colon cancer, and functioned as an oncogene by promoting cell proliferation, migration, and invasion in colon cancer [24]. In triple-negative breast cancer, circEPSTI1 acted as a competing endogenous RNA to protect BCL11A mRNA from miR-4753 attack, thereby promoting triple-negative breast cancer cell proliferation and inhibiting cell apoptosis [25]. Additionally, a recent study discovered that circNT5E inhibits the proliferation, migration, and invasion of U87 and U251 cells by sponging miR-422a [26]. Of note, a previous pioneer study showed that hsa_cir_000204 was differently expressed in BC tissues; however, the pathological role of hsa_cir_000204 in BC remains unclear. Our current study reported that upregulation of hsa_cir_000204 inhibited the proliferation of EJ and T24 cells, as evidenced by the reduced number of EdU-positive cells. In parallel, overexpression of hsa_cir_000204 obviously repressed the migration and invasion of EJ and T24 cells. These findings indicate that hsa_cir_000204 acts as an oncogenic circRNA in BC.
It is well known that miRNAs play an important role in carcinogenesis [12]. miR-197, a member of the miR-197 family, is known to play a vital role in the progression of human cancers through abnormal expression. In non-small cell lung cancer, the expression of miR-197 has been positively correlated with tumor size and non-small cell lung cancer histotype, which are independent indicators of poor prognosis [27]. miR-197 depletion exerts an oncosuppressive effect on NIH-H460 and A549 cells [28]. In colorectal cancer, the expression level of miR-197 is decreased and knockdown of miR-197 promotes the proliferation, migration, and invasion of HCT116 cells [29]. Moreover, miR-197-3p has been shown to be overexpressed in basal cell carcinoma [30]. In thyroid cancer, miR-197-3p is highly expressed and suppression of miR-197-3p expression was shown to inhibit the proliferation, migration, and invasion of K1, SW579, and 8505C cells [31]. Hence, these studies demonstrated that miR-197-3p is involved in the progression of multiple cancers. However, little is known about the function of miR-197-3p and its regulatory mechanism in BC. In our study, we identified that miR-197-3p expression was upregulated in BC tissues and cell lines. With luciferase reporter assay, hsa_circ_0002024 was confirmed as a direct target of miR-197-3p. In addition, we found that restoration of miR-197-3p expression could abolish hsa_circ_0002024-mediated inhibition of BC cell proliferation, migration, and invasion, suggesting that hsa_circ_0002024 may act as a miR-197-3p sponge to repress BC cell proliferation, migration, and invasion. Our results provide novel insights into the potential therapeutic targets of BC. In our future study, we aim to further validate these in vitro findings in an in vivo model of BC.
In conclusion, we found that hsa_circ_0002024 suppressed cell proliferation, migration, and invasion in BC by sponging miR-197-3p. Our study identified a new circRNA-miRNA regulatory network in BC, thereby providing a better understanding of BC pathogenesis, and suggesting that hsa_circ_0002024 may serve as a therapeutic target for the treatment of BC.
Disclosure of conflict of interest
None.
References
- 1.Rudman SM, Crawley D. Epidemiology of bladder cancer. Urologic Clinics of North America. 2017;3:511–522. [Google Scholar]
- 2.Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahab RA, Essa HH, Eltaher A, Aboziada M, Shehata S. Concurrent chemoradiotherapy with paclitaxel and cisplatin in muscle-invasive bladder cancer. Ann Oncol. 2016:27. [Google Scholar]
- 4.Masaoka H, Matsuo K, Ito H, Wakai K, Nagata C, Nakayama T, Sadakane A, Tanaka K, Tamakoshi A, Sugawara Y, Mizoue T, Sawada N, Inoue M, Tsugane S, Sasazuki S. Cigarette smoking and bladder cancer risk: an evaluation based on a systematic review of epidemiologic evidence in the Japanese population. Jpn J Clin Oncol. 2016;46:273–283. doi: 10.1093/jjco/hyv188. [DOI] [PubMed] [Google Scholar]
- 5.Turati F, Bosetti C, Polesel J, Serraino D, Montella M, Libra M, Facchini G, Ferraroni M, Tavani A, La Vecchia C, Negri E. Family history of cancer and the risk of bladder cancer: a case-control study from Italy. Cancer Epidemiol. 2017;48:29–35. doi: 10.1016/j.canep.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Sun JW, Zhao LG, Yang Y, Ma X, Wang YY, Xiang YB. Obesity and risk of bladder cancer: a dose-response meta-analysis of 15 cohort studies. PLoS One. 2015;10:e0119313. doi: 10.1371/journal.pone.0119313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volanis D, Kadiyska T, Galanis A, Delakas D, Logotheti S, Zoumpourlis V. Environmental factors and genetic susceptibility promote urinary bladder cancer. Toxicol Lett. 2010;193:131–137. doi: 10.1016/j.toxlet.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 8.DeGeorge KC, Holt HR, Hodges SC. Bladder cancer: diagnosis and treatment. Am Fam Physician. 2017;96:507–514. [PubMed] [Google Scholar]
- 9.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng. 2010;12:1–27. doi: 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- 12.Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10:396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Hong S, Liu Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun. 2018;503:1825–1829. doi: 10.1016/j.bbrc.2018.07.120. [DOI] [PubMed] [Google Scholar]
- 14.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dragomir M, Calin GA. Circular RNAs in cancer-lessons learned from microRNAs. Front Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Yuan W, Tao J, Li P, Yang C, Deng X, Zhang X, Tang J, Han J, Wang J, Li P, Lu Q, Gu M. Identification of circular RNA signature in bladder cancer. J Cancer. 2017;8:3456–3463. doi: 10.7150/jca.19345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Non-coding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, Zhang S, Chen X, Li N, Li J, Jia R, Pan Y, Liang H. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78:4812–4825. doi: 10.1158/0008-5472.CAN-18-0532. [DOI] [PubMed] [Google Scholar]
- 27.Mavridis K, Gueugnon F, Petit-Courty A, Courty Y, Barascu A, Guyetant S, Scorilas A. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer. 2015;112:1527–35. doi: 10.1038/bjc.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 2014;21:774–82. doi: 10.1038/cdd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Liu Z, Ning X, Huang L, Jiang B. The long noncoding RNA HOTAIR promotes colorectal cancer progression by sponging miR-197. Oncol Res. 2017;26:473–481. doi: 10.3727/096504017X15105708598531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sand M, Bechara FG, Gambichler T, Sand D, Friedlander MR, Bromba M, Schnabel R, Hessam S. Next-generation sequencing of the basal cell carcinoma miRNome and a description of novel microRNA candidates under neoadjuvant vismodegib therapy: an integrative molecular and surgical case study. Ann Oncol. 2016;27:332–338. doi: 10.1093/annonc/mdv551. [DOI] [PubMed] [Google Scholar]
- 31.Liu K, Huang W, Yan DQ, Luo Q, Min X. Overexpression of long intergenic noncoding RNA LINC00312 inhibits the invasion and migration of thyroid cancer cells by down-regulating microRNA-197-3p. Biosci Rep. 2017;37 doi: 10.1042/BSR20170109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


