Abstract
Increasing evidences have indicated the vital roles of long noncoding RNA (lncRNA) in the atherosclerosis. However, whether lncRNA LINC00341 play pivotal roles in the vascular smooth muscle cells (VSMCs) is still unclear. This work presents the authentic functions of LINC00341 on the proliferation and migration of VSMCs and unveils the underlying mechanism. Functional experiment data demonstrated that LINC00341 expression was increased in the ox-LDL induced VSMCs with dose-dependent and time-dependent mode. Moreover, the knockdown of LINC00341 suppressed the proliferation and migration ability of VSMCs. Mechanically, we found that LINC00341 promoted the FOXO4 protein expression via sponging miR-214, which, in return, resulting in the transcription activation of LINC00341. In conclusion, the results conclude that LINC00341 promotes the proliferation and migration of VSMCs and confirm the positive feedback loop of LINC00341/miR-214/FOXO4 axis.
Keywords: Vascular smooth muscle cells, LINC00341, miR-214, FOXO4, proliferation, migration
Introduction
Atherosclerosis acts as the main causes of coronary heart disease, cerebral infarction and peripheral vascular disease [1,2]. The pathological basis of atherosclerosis is the disorder of lipid metabolic, which is characterized by the arterial lesions originated from the intima [3]. Vascular smooth muscle cells (VSMCs) function as the majority of arterial walls and its dysfunction could participate in the atherosclerosis [4,5]. The lesions of VSMCs also occur in the blood vessels of vital organs, such as heart, brain and kidney [6,7]. It could cause the ischemia and necrosis, such as myocardial infarction, cerebral infarction and renal infarction [8]. Nevertheless, the profound molecular mechanism by which VSMCs contribute to the atherosclerosis is still undetermined.
Recently, the epigenetic regulation on the human genomic expression has been adequately convinced, especially the long non-coding RNAs (lncRNAs) [9]. Human genome projects discovered that approximate 90% of the human genome is transcribed, however, there is about 2% of the transcripts encoding proteins. LncRNAs have been identified to be implicated in the cardiovascular disease, including atherosclerosis, myocardial fibrosis, lipid metabolism and vascular endothelial abnormality [10]. For example, in the vascular smooth muscle cell (VSMCs) induced by ox-LDL, UCA1 is up-regulated and antagonizes miR-26a through downregulation of its target PETN to regulate the PCNA, α-SMA and SM22-α expression [11]. LncRNA MALAT1 expression is up-regulated in the ox-LDL treated HCAECs, and MALAT1 knocking down promotes the ox-LDL-induced cytokine release and apoptosis of HCAECs via binding miR-155/SOCS1 axis [12].
In this study, we present a research about the lncRNA LINC00341 in the vascular smooth muscle cells (VSMCs) pathophysiological process in the atherosclerosis. Our data suggest that the over-expressed lncRNA LINC00341 promotes the proliferation and migration of VSMCs by sponging miR-214/FOXO4 axis. This finding reveals that lncRNA LINC00341 could act as vital regulators of post-transcriptional modification of VSMCs.
Materials and methods
VSMCs culture
All the items about the ethical approval had get approved by the Ethics Committee of Yantai Affiliated Hospital of Binzhou Medical University. For the following cellular experiments, human VSMCs were provided by ATCC Company (Rockville, MD, USA, American Type Culture Collection). The condition of VSMCs culture was set in Dulbecco’s modified Eagle’s medium (DMEM) and incubation in 5% CO2 at 37°C. Besides, more elements were supplemented, including FBS (fetal bovine serum, 10%, Grand Island, NY, USA), penicillin (1%, 100 U/ml) and streptomycin sulfate (1% 100 mg/ml). VSMCs were administrated with ox-LDL (100 mg/l) to simulate the high blood lipid environment.
Oligonucleotides transfection
The oligonucleotides small interfering RNAs (siRNAs) targeting LINC00341 and miR-214 and mimics and their control were purchased from RiboBio (Guangzhou, China). Lipofectamine 2000 (Thermo Fisher Scientific, Inc, Rockford, IL, USA) was used for transfection according to the manufacturer’s instructions. All the sequences were presented in the Table S1.
Quantitative real-time PCR (qRT-PCR)
Total RNA were extracted from VSMCs using RNAiso Plus (TaKaRa, Dalian, China). PrimeScrip RT Master Mix was performed for the reverse transcription and SuperScript First-Stand Synthesis system (Invitrogen, US) the cDNA amplification. Quantitative PCR analysis was carried out using the Hieff qPCR SYBR Green Master Mix kit (TaKaRa, Dalian, China) with ABI Prism 7300 Sequence Detection System (Applied Biosystems, Foster, Calif., USA). GAPDH was used as an endogenous control and calculated the relative expression level. All primers were listed in Table S1.
Western blot analysis
The protein was extracted from VSMCs using RIPA lysis buffer (Thermo Scientific, Rockford, IL, USA) and the purity was measured with the BCA Protein assay kit (Beyotime Institute of Biotechnology, Shanghai, China). The extraction was transferred to the PVDF membranes and blocked with the non-fat milk powder. Then, the PVDF membrane was incubated with primary antibodies (anti-FOXO4, 1:1000 dilution, Abcam) overnight at 4°C and then incubated with HRP-conjugated secondary antibodies (anti-GAPDH, 1:500 dilution) at room temperature. Blots signals were detected using an enhanced chemiluminescence (ECL) system according to the manufacturer’s instructions, and the density was determined using Image LabTM software (version 4.1; Bio-Rad, CA, USA).
CCK-8 assay
Proliferation ability analysis was performed using CCK-8 assay for the VSMCs. After the transfection, VSMCs (5×103 cells/well) were seeded in 96-well culture plates and administrated with CCK-8 agent to examine the cell viability.
Migration analysis
Migration analysis was performed using wound healing assay and transwell migration assay. For the wound healing assay, cells at 90% confluence were seeded in 6-well plates. The cell monolayer was wounded by manually scraping the cells with a 200 ul pipette tip. Migration rate was calculated with the following formula: migration distance/original distance. For the transwell experiment, Transwell chamber (8 μm, Sigma, Dallas, TX, USA) was coated with the Matrigel (50 uL). 1×104 cells was suspended in the RPMI-1640 serum-free medium (200 µl) of the upper chamber. The lower chamber was medium (600 µl) with the 10% fetal bovine serum-containing medium (Gibco; Thermo Fisher Scientific). After the incubation at 37°C, cells that migrated to the lower chamber were fixed in 4% paraformaldehyde for 30 min and stained with 1% crystal violet.
Cell cytoplasm/nucleus fraction isolation
The subcellular location analysis of VSMCs was measured using the Nuclear and Cytoplasmic Extraction Reagents.
Luciferase gene reporter assay
The 3’-UTR sequences complementary to miR-214 (wild type, wild type) of FOXO4 and LINC00341 were cloned into psi-CHECK2 vector (Promega, WI, USA). VSMCs were co-transfected with CHECK2 vector and miR-214 mimic or control using Lipofectamine 2000 (Invitrogen). For the luciferase reporter ability, it was assessed by using a Dual Luciferase Reporter Assay System (Promega) normalized to luciferase activity.
RNA immunoprecipitation (RIP)
RIP analysis was carried out using EZ-Magna RIP Kit (Millipore) according to the manufacturer’s protocol. The value was extracted and measured using qPCR.
Chromatin immunoprecipitation (ChIP)
ChIP was carried out using the EZ ChIP Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Crosslinked chromatin was sonicated into fragments and anti-FOXO4 antibody was used to precipitate DNA-protein complex. The precipitated RNA was measured using qRT-PCR normalized to IgG.
Statistical analysis
All data were indicated using SPSS 18.0 (SPSS, Chicago, IL, USA) as means ± standard error, and shown GraphPad Prism 6.0 (La Jolla, USA). The statistical approach were analyzed using independent samples t-test or chi-square (P < 0.05) for evaluating the Group difference.
Results
LncRNA LINC00341 is over-expressed in the VSMCs induced by ox-LDL and the knockdown of LINC00341 suppresses the proliferation and migration
To explore whether lncRNA LINC00341 was differently expressed in the VSMCs in the atherosclerosis environment, we administrated the gradient concentration of ox-LDL (0-150 mg/L) to VSMCs. Results showed that LINC00341 expression was increased in the administration of ox-LDL on VSMCs (Figure 1A). When VSMCs were administrated with ox-LDL (100 mg/l), LINC00341 expression was also increased with the processing time (Figure 1B). The special oligonucleotides targeting LINC00341 was transfected into VSMCs to silence the LINC00341 expression (Figure 1C). CCK-8 proliferation assay indicated that LINC00341 silencing suppressed the absorbance value in VSMCs (Figure 1D). Wound healing assay reported that LINC00341 silencing suppressed the migrated distance of VSMCs (Figure 1E). Transwell analysis illustrated that LINC00341 silencing inhibited the migration of VSMCs (Figure 1F). Overall, data concluded that lncRNA LINC00341 is over-expressed in the VSMCs induced by ox-LDL.
Figure 1.
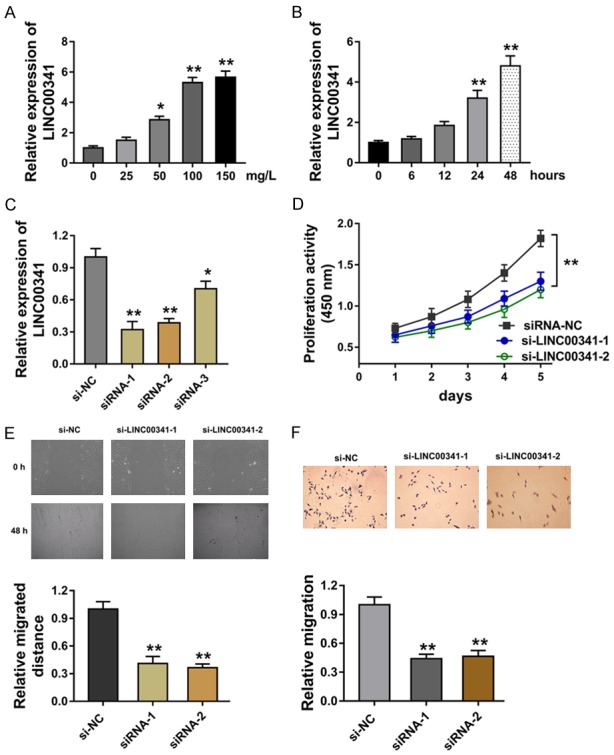
LncRNA LINC00341 is over-expressed in the VSMCs induced by ox-LDL and the knockdown of LINC00341 suppresses the proliferation and migration. A. RT-PCR measured the LINC00341 expression in the VSMCs treated with the gradient concentration of ox-LDL (0-150 mg/L). B. The LINC00341 expression in the VSMCs treated with 100 mg/L ox-LDL with the processing time. C. LINC00341 expression in the VSMCs transfected with special oligonucleotides targeting LINC00341 (si-LINC00341) or control. D. CCK-8 proliferation assay indicated the proliferation of absorbance value in VSMCs. E. Wound healing assay reported the migrated distance of VSMCs. F. Transwell analysis illustrated the migration of VSMCs. **P < 0.01 vs. si-NC control. *P < 0.05 vs. si-NC control.
miR-214 targets the lncRNA LINC00341 3’-UTR
In order to determine the in-depth mechanism that LINC00341 regulates the VSMCs biological phenotype, we performed subcellular fractionation analysis to verify the possible mechanism. Results determined that LINC00341 mainly located in the cytoplasm compared with the nucleus, indicating the significant enrichment of LINC00341 in the cytoplasm and post-transcriptional control of VSMCs (Figure 2A). Online bioinformatics tools (StarBase v2.0, RegRNA2) suggested that miR-214 could targeted the LINC00341 at the 3’-UTR (Untranslated Regions) to form the complementary pairing (Figure 2B). Luciferase gene reporter analysis and RNA immunoprecipitation (RIP) assay revealed that the LINC00341 could efficiently bind with the miR-214 by the molecular binding (Figure 2C, 2D). In the VSMCs administrated with increasing concentration of ox-LDL (0-150 mg/l), the expression of miR-214 was significantly decreased, which is negatively correlated with that of LINC00341 (Figure 2E). In the VSMCs transfected with the si-LINC00341, miR-214 expression was up-regulated (Figure 2F). Collectively, our data revealed that miR-214 targets the 3’-UTR of lncRNA LINC00341 in the VSMCs.
Figure 2.
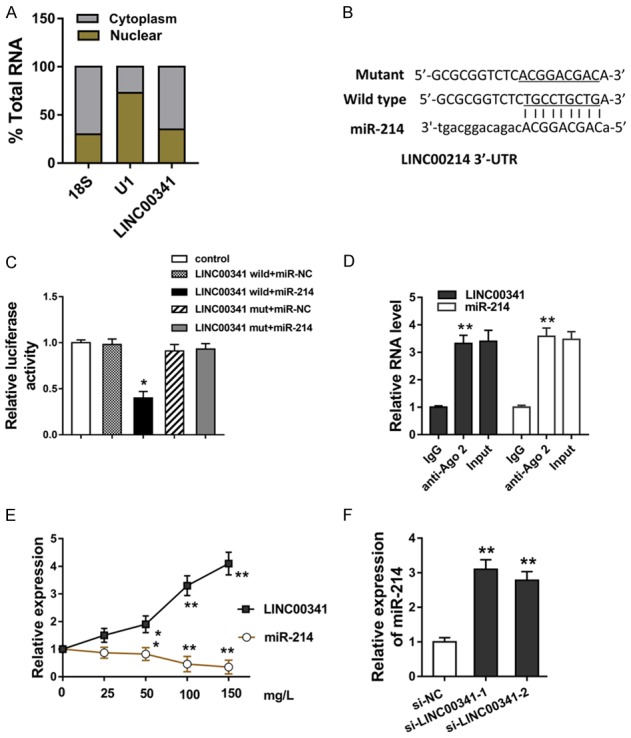
miR-214 targets the 3’-UTR of lncRNA LINC00341 in the VSMCs. A. Subcellular fractionation analysis revealed the location of LINC00341 in the cytoplasm and nucleus. B. Online bioinformatics tools (StarBase v2.0, RegRNA2) suggested the complementary pairing within LINC00341 and miR-214. C. Luciferase gene reporter analysis showed the luciferase activity within the LINC00341 (wild type or mutant) and miR-214 or control. D. RNA immunoprecipitation (RIP) assay revealed the efficient molecular binding within LINC00341 and miR-214. E. RT-PCR revealed the LINC00341 and miR-214 in the VSMCs administrated with increasing concentration of ox-LDL (0-150 mg/l). F. miR-214 expression in the VSMCs transfected with the si-LINC00341. **P < 0.01 vs. si-NC control. *P < 0.05 vs. si-NC control.
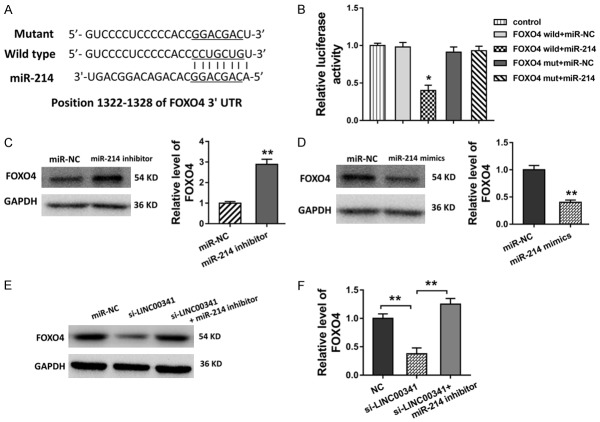
FOXO4 functions as the target of miR-214 in VSMCs
Presently, existing evidence have indicated that miRNAs could bind with their downstream protein to inhibit its expression, thereby regulating its translation expression. In silico methods revealed the potential putative binding sites within miR-214 and FOXO4 3’-UTR (Figure 3A). Luciferase gene reporter assay revealed that miR-214 binds with the FOXO4 3’-UTR with the activity, accounting for the molecular binding (Figure 3B). In the following western blot analysis performed in the VSMCs, the transfection of miR-214 inhibitor or mimics could enhance or weaken the level of FOXO4 protein (Figure 3C, 3D). Moreover, the transfection of LINC00341 siRNA decreased the FOXO4 protein level, and the transfection of miR-214 inhibitor recovered it (Figure 3E, 3F). Therefore, these results illustrated that FOXO4 functions as the target of miR-214 in VSMCs.
Figure 3.
FOXO4 functions as the target of miR-214 in VSMCs. A. The potential putative binding sites within miR-214 and FOXO4 3’-UTR predicted using in silico methods. B. Luciferase gene reporter assay revealed the molecular binding activity within miR-214 and FOXO4 3’-UTR. C, D. Western blot analysis showed the level of FOXO4 protein in the VSMCs with the transfection of miR-214 inhibitor or mimics. E, F. Western blot analysis showed the level of FOXO4 protein in the VSMCs with the transfection of LINC00341 siRNA and/or miR-214 inhibitor. **P < 0.01 vs. si-NC control.
FOXO4 targets with the promoter and accelerates the transcriptional level of LINC00341
We had confirmed the LINC00341/miR-214/FOXO4 pathway in the VSMCs, which acts as a vital regulation of ncRNA on the AS. Interestingly, we also found that FOXO4, a vital transcription factor, could bind with the promoter of LINC00341 via bioinformatics prediction tools. Firstly, the plasmid of over-expressed FOXO4 (pcDNA3.1, pcDNA-FOXO4) and corresponding control were transfected into the VSMCs to enhance its expression (Figure 4A, 4B). Subsequently, RT-PCR revealed that the transfection of plasmidfor FOXO4 (pcDNA-FOXO4) could increase the LINC00341 expression, indicating the acceleration of FOXO4 for the LINC00341 (Figure 4C). Using the online tools (JASPAR, http://jaspar.genereg.net/, RegRNA2.0, http://regrna2.mbc.nctu.edu.tw/index.html), there were three putative binding sites that FOXO4 could bind with LINC00341 (Figure 4D). Chromatin immunoprecipitation (ChIP) revealed that FOXO4 endogenous binds with the TFBS 3 binding motif of promoter region of LINC00341 (Figure 4E). Therefore, this mechanical experiments showed that FOXO4 could target with the promoter and accelerates the transcriptional level of LINC00341, forming the LINC00341-miR-214-FOXO4 positive feedback loop.
Figure 4.
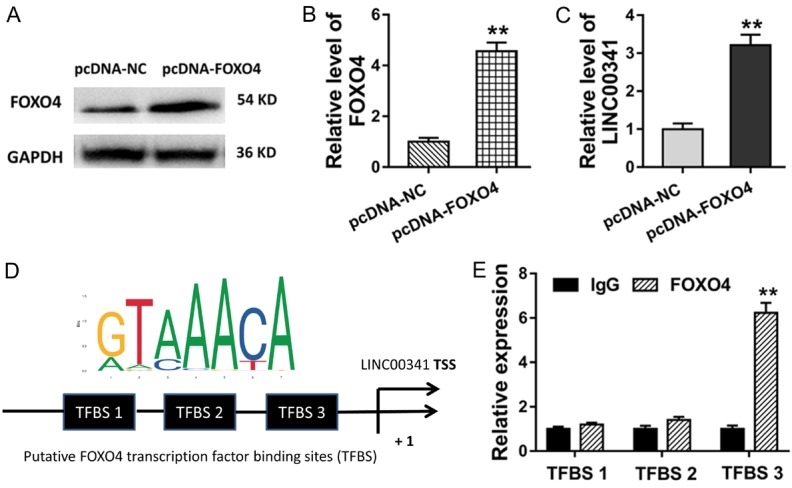
FOXO4 targets with the LINC00341 promoter and accelerates the transcriptional level. A, B. Western blot analysis showed the FOXO4 level in the VSMCs transfected plasmid of over-expressed FOXO4 (pcDNA-FOXO4) and corresponding control. C. RT-PCR revealed the LINC00341 expression in VSMCs transfected with plasmid for FOXO4 (pcDNA-FOXO4). D. The putative binding sites that FOXO4 could bind with LINC00341 predicted using the online tools (JASPAR, http://jaspar.genereg.net/, RegRNA2.0, http://regrna2.mbc.nctu.edu.tw/index.html). E. Chromatin immunoprecipitation (ChIP) revealed the binding potential for FOXO4 with the binding motif of promoter region of LINC00341. **P < 0.01 vs. si-NC control.
Discussion
In the last century, genome-wide association studies (GWAS) and human genome project has detailedly revealed the integrated gene sequence of human genome, therefore, we could deeply comprehend the profound mystery of human heredity [13,14]. However, in the post-genomic era, more and more attention have focused on the epigenetics, including DNA methylation, histone modifications, chromosomal remodeling, and noncoding RNA (ncRNA) regulation [15]. In this respect of ncRNA, long noncoding RNA (lncRNA) acts as vital regulator for the human pathogenesis and regulates the gene transcription or translation to affects its function and characteristics [16].
Long noncoding RNAs (LncRNAs) are a larger subgroup of ncRNA with more than 200 nucleotides, acting as the post-transcriptional elements [17-20]. Existing evidence have revealed that LINC00341 is associated with poor prognosis in cancers and LINC00341 inhibited cancer metastasis [21]. Besides, LINC00341 expression level is enhanced in the human umbilical vein endothelial cells and recruits EZH2 to the promoter region of the VCAM1 gene to inhibit VCAM1 expression, playing an important role in modulating endothelial function [22]. Therefore, these evidence powerfully support the assumption that LINC00341 might regulate the cellular physiological and pathological functions in the cardiovascular disease.
In this study, we found that lncRNA LINC00341 was significantly up-regulated in the VSMCs treated with the ox-LDL. Thus, LINC00341 might function as a vital regulator in the VSMCs lesion and the atherosclerosis pathogenesis. The cellular functional experiments showed that LINC00341 silencing with the oligonucleotides transfection could efficiently suppress the proliferation and migration of VSMCs. Therefore, the aberrantly expressed LINC00341 in the simulative atherosclerosis models accelerates the pathological differentiation of VSMCs.
In the pathogenesis of atherosclerosis, the proliferation and migration of VSMCs acts as the important sign phenomenon. During the atherosclerosis, VSMCs migrate from middle membrane to the inner membrane, in which lncRNAs participate in this process [23,24]. For instance, lncRNA XR007793 expression is significantly increased in the injured carotid artery of SD rats and platelet-derived growth factor-BB induced aortic smooth muscle cells. Then, the XR007793 knockdown inhibits the proliferation and migration of VSMC in vitro via directly bonding to miR-23b/FOXO4 axis [25]. Moreover, in the ox-LDL (50 mg/l) induced vascular smooth muscle cell (VSMCs), overexpressed lncRNA UCA1 antagonized miR-26a and then the downregulation of its target PETN, revealing that UCA1 acts as an endogenous sponge of miR-26a to regulate VSMCs proliferation [11].
Our study and works pay close attention to the roles of lncRNA in the VSMCs in atherosclerosis environment. We find that lncRNA LINC00341 is significantly increased in the ox-LDL induced VSMCs, furthermore, LINC00341 acts as the endogenous sponge of miR-214. It means that LINC00341 acts as the miR-214 sponge in the VSMCs. Then, FOXO4 functions as the target protein of miR-214. Interestingly, the transcription factor FOXO4 could bind with the promoter region of LINC00341 and promote its transcriptional level. Thus, this suggests the positive feedback loop of LINC00341/miR-214/FOXO4 in the VSMCs.
In conclusion, our study reveals the vital roles of LINC00341 in the proliferation and migration of VSMCs in the atherosclerosis. LINC00341 acts as the sponge of miR-214 and therefore promotes the FOXO4 expression. Interestingly, transcription factor FOXO4 activates the transcription of LINC00341 and accelerates the abundance, suggesting the positive feedback loop of LINC00341/miR-214/FOXO4 in the VSMCs.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chistiakov DA, Grechko AV, Myasoedova VA, Melnichenko AA, Orekhov AN. The role of monocytosis and neutrophilia in atherosclerosis. J Cell Mol Med. 2018;22:1366–1382. doi: 10.1111/jcmm.13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo M, Guo G, Xiao J, Sheng X, Zhang X, Tie Y, Cheng YK, Ji X. Ginsenoside Rg3 stereoisomers differentially inhibit vascular smooth muscle cell proliferation and migration in diabetic atherosclerosis. J Cell Mol Med. 2018;22:3202–3214. doi: 10.1111/jcmm.13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui Y, Li C, Zeng C, Li J, Zhu Z, Chen W, Huang A, Qi X. Tongmai Yangxin pills anti-oxidative stress alleviates cisplatin-induced cardiotoxi- city: network pharmacology analysis and experimental evidence. Biomed Pharmacother. 2018;108:1081–1089. doi: 10.1016/j.biopha.2018.09.095. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, Xu J, Guo JL, Lin CY, Luo WM, Yuan Y, Liu H, Zhang J. YAP1 up-regulation inhibits apoptosis of aortic dissection vascular smooth muscle cells. Eur Rev Med Pharmacol Sci. 2017;21:4632–4639. [PubMed] [Google Scholar]
- 5.Ren XS, Tong Y, Ling L, Chen D, Sun HJ, Zhou H, Qi XH, Chen Q, Li YH, Kang YM, Zhu GQ. NLRP3 gene deletion attenuates angiotensin II-induced phenotypic transformation of vascular smooth muscle cells and vascular remodeling. Cell Physiol Biochem. 2017;44:2269–2280. doi: 10.1159/000486061. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y, Qi X, Huang A, Li J, Hou W, Liu K. Differential and predictive value of galectin-3 and soluble suppression of tumorigenicity-2 (sST2) in heart failure with preserved ejection fraction. Med Sci Monit. 2018;24:5139–5146. doi: 10.12659/MSM.908840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Cui Y, Huang A, Li Q, Jia W, Liu K, Qi X. Additional diagnostic value of growth differentiation factor-15 (GDF-15) to N-terminal B-type natriuretic peptide (NT-proBNP) in patients with different stages of heart failure. Med Sci Monit. 2018;24:4992–4999. doi: 10.12659/MSM.910671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Nishimura Y, Kimura A, Ozawa K, Kondo T, Tanaka T, Yoshizumi M. Chemokines protect vascular smooth muscle cells from cell death induced by cyclic mechanical stretch. Sci Rep. 2017;7:16128. doi: 10.1038/s41598-017-15867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Z, Shi L. Long non-coding RNA LUCAT1 modulates methotrexate resistance in osteosarcoma via miR-200c/ABCB1 axis. Biochem Biophys Res Commun. 2018;495:947–953. doi: 10.1016/j.bbrc.2017.11.121. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, Ren K, Zhu X, Zheng Z, Yi G. Long noncoding RNAs: advances in lipid metabolism. Adv Clin Chem. 2018;87:1–36. doi: 10.1016/bs.acc.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Tian S, Yuan Y, Li Z, Gao M, Lu Y, Gao H. LncRNA UCA1 sponges miR-26a to regulate the migration and proliferation of vascular smooth muscle cells. Gene. 2018;673:159–166. doi: 10.1016/j.gene.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Sun Y, Zhong L, Xiao Z, Yang M, Chen M, Wang C, Xie X, Chen X. The suppression of ox-LDL-induced inflammatory cytokine release and apoptosis of HCAECs by long non-coding RNA-MALAT1 via regulating microRNA-155/SOCS1 pathway. Nutr Metab Cardiovasc Dis. 2018;28:1175–1187. doi: 10.1016/j.numecd.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Chen C, Gao A, Tong HH, Xie L Alzheimer’s Disease Neuroimaging Initiative. VariFunNet, an integrated multiscale modeling framework to study the effects of rare non-coding variants in genome-wide association studies: applied to Alzheimer’s disease. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2017;2017:2177–2182. doi: 10.1109/BIBM.2017.8217995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sallah N, Carstensen T, Wakeham K, Bagni R, Labo N, Pollard MO, Gurdasani D, Ekoru K, Pomilla C, Young EH, Fatumo S, Asiki G, Kamali A, Sandhu M, Kellam P, Whitby D, Barroso I, Newton R. Whole-genome association study of antibody response to Epstein-Barr virus in an African population: a pilot. Glob Health Epidemiol Genom. 2017;2:e18. doi: 10.1017/gheg.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Aragam B, Xing EP. Variable selection in heterogeneous datasets: a truncated-rank sparse linear mixed model with applications to genome-wide association studies. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2017;2017:431–438. doi: 10.1109/BIBM.2017.8217687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malhotra S, Freeberg MA, Winans SJ, Taylor J, Beemon KL. A novel long non-coding RNA in the hTERT promoter region regulates hTERT expression. Noncoding RNA. 2017;4 doi: 10.3390/ncrna4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni N, Song H, Wang X, Xu X, Jiang Y, Sun J. Up-regulation of long noncoding RNA FALEC predicts poor prognosis and promotes melanoma cell proliferation through epigenetically silencing p21. Biomed Pharmacother. 2017;96:1371–1379. doi: 10.1016/j.biopha.2017.11.060. [DOI] [PubMed] [Google Scholar]
- 18.Wei AW, Li LF. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed Pharmacother. 2017;96:953–959. doi: 10.1016/j.biopha.2017.11.145. [DOI] [PubMed] [Google Scholar]
- 19.Zhang D, Cao J, Zhong Q, Zeng L, Cai C, Lei L, Zhang W, Liu F. Long noncoding RNA PCAT-1 promotes invasion and metastasis via the miR-129-5p-HMGB1 signaling pathway in hepatocellular carcinoma. Biomed Pharmacother. 2017;95:1187–1193. doi: 10.1016/j.biopha.2017.09.045. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Zhao W, Wang Z, Xiang X, Zhang S, Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2019;23:680–688. doi: 10.1111/jcmm.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao M, Li B, Zhang S, Liu Q, Liao W, Xie W, Zhang Y. Relationship between LINC00341 expression and cancer prognosis. Oncotarget. 2017;8:15283–15293. doi: 10.18632/oncotarget.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang TS, Wang KC, Quon S, Nguyen P, Chang TY, Chen Z, Li YS, Subramaniam S, Shyy J, Chien S. LINC00341 exerts an anti-inflammatory effect on endothelial cells by repressing VCAM1. Physiol Genomics. 2017;49:339–345. doi: 10.1152/physiolgenomics.00132.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang JH, Kim EA, Park HJ, Sung EG, Song IH, Kim JY, Woo CH, Doh KO, Kim KH, Lee TJ. Methylglyoxal-induced apoptosis is dependent on the suppression of c-FLIPL expression via down-regulation of p65 in endothelial cells. J Cell Mol Med. 2017;21:2720–2731. doi: 10.1111/jcmm.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Liu S, Cao G, Sun Y, Chen W, Dong F, Xu J, Zhang C, Zhang W. Nicotine induces endothelial dysfunction and promotes atherosclerosis via GTPCH1. J Cell Mol Med. 2018;22:5406–5417. doi: 10.1111/jcmm.13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu YX, Zhang SH, Cui J, Liu FT. Long noncoding RNA XR007793 regulates proliferation and migration of vascular smooth muscle cell via suppressing miR-23b. Med Sci Monit. 2018;24:5895–5903. doi: 10.12659/MSM.908902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.