Abstract
The biosynthetic potential of soil-dwelling actinomycetes to produce diverse bioactive molecules that are useful as drug seeds has been achieved in the laboratory by modifying culture conditions. Availability of a small molecule that can induce secondary metabolism in these microbes can greatly facilitate the exploration of bioactive natural products. In this manuscript, through the screening of natural products and chemical modification, we demonstrated that the presence of the β-carboline compound, BR-1, enhanced reveromycin A production in Streptomyces sp. SN-593. BR-1 induced reveromycins production at the wide range of concentrations without affecting cell growth. Our study indicates that BR-1 might serve as an alternative to activate specialized metabolite biosynthesis without genetic engineering.
Introduction
Soil dwelling actinomycetes are characterized by their ability to produce diverse specialized metabolites (SMs), that account for majority of drugs presently in clinical use1. These bacteria utilize autoregulators such as A-factor2, 2-alkyl-4-hydroxymethylfuran-3-carboxylic acids3, PI factor4, avenolide5, L-factor6, VB-A7, IM-28, SCB19, and SRB110 to induce morphogenesis and regulate SM production. As opposed to autoregulators, chemical signals derived from extra-species/environmental stimuli such as hormaomycin11, goadsporin12, promomycin13, antibiotic-remodelling compounds (ARCs)14, and rare earth elements such as scandium15 can also induce morphogenesis and SM production in Streptomyces species. Activation of SM gene clusters by small molecule elicitors has also been observed for Gram-negative proteobacteria Burkholderia thailandensis16.
Classically, induction of SM production in Streptomyces has been achieved by modifying culture medium and recently ribosomal engineering17,18, expression of key regulatory genes19,20, and heterologous expression21 have also been used for the same. By using the classical approach of culture medium modification, we found that reveromycin (RM) production by Streptomyces sp. SN-593 was enhanced by adding tomato juice to the culture medium22. This observation led us to speculate that naturally existing extracellular chemicals can activate secondary metabolism. Such chemicals can up-regulate SM biosynthesis and facilitate the isolation of novel natural products without genetic engineering. We first tried to purify the active principle from tomato juice. However, all attempts failed regardless of extended efforts. A major obstacle impeding the identification of natural chemical signals is their low-level presence in natural environments. Therefore, screening a natural product library is an alternative approach for finding core chemical structure that enhances SM production. Given that RM-A has antifungal activities against pathogenic plant fungi23 and unique biological activity inducing the morphological reversion of srcts-NRK cells from spherical transformed cells to flat normal cells24, we used both assay systems to screen for compounds that enhance the production of RMs. By the screening of small molecules from the RIKEN Natural Products Depository (NPDepo)25, we identified a β-carboline lead compound that enhanced RM production. Based on structure activity relationship study, we succeeded to create novel chemical signals that activate RM production at sub μM concentration.
Results and Discussion
Screening for biomediators
We defined extracellular chemical signals as biomediators to distinguish them from autoregulators. After the failed attempt of purification from tomato juice, we utilized the antifungal activity of RM-A to screen overproduced RMs after the treatment of NPDepo compounds. The activity of putative hits was further analysed by the srcts-NRK cells assay followed by liquid chromatography-mass spectrometry (LC-MS) analysis to quantify the production of RM-A (1) and its derivative RM-B (2). RIKEN NPDepo harbours approximately 26,000 compounds. Of this, 3,155 compounds were screened using an antifungal assay with Pyricularia oryzae, the hits (136 compounds) were further examined in the srcts-NRK cell morphological assay. We also performed liquid chromatography-mass spectrometry (LC-MS) analysis to quantify the production of RM-A (1) and its derivative RM-B (2). Finally, we succeeded in identifying a β-carboline compound NPD2639 (3) as a lead compound (Fig. 1).
Figure 1.
LC-MS analysis of RMs after treatment with NPD2639. Streptomyces sp. SN-593 was analysed after treatment with 5% tomato juice (i), 1 µg ml−1 NPD2639 (ii), or 0.1% DMSO (iii).
Structural insights into the biomediator activity of β-carbolines
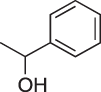
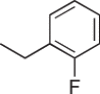
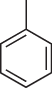
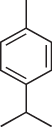
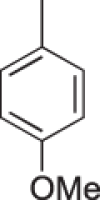
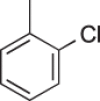
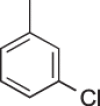
To elucidate the structure activity relationship of the biomediator, we first investigated the biomediator activity of the β-carboline core structure using natural products such as harman, harmol, and harmine. These compounds did not induce the production of RMs (Table 1). This result suggested a functional role for the 3-chlorophenyl group attached to 1 position and the amide group attached to the 3 position of the β-carboline core, in terms of biomediator activity. To investigate the structure–activity relationship, we selected 150 β-carboline compounds from the RIKEN NPDepo library25 with regard to the substituents at the 1-phenyl group (R1) and the 3-amide moiety (R2) (Fig. 2). The biomediator activity was evaluated in the srcts-NRK cells assay followed by LC-MS analysis of RM production (Table 2). The hydroxyethyl derivative (4) and 3-bromophenyl derivatives (8s and 8t) also retained biomediator activity. However, the 2 or 4-substituted phenyl derivative at R1 and compounds with substituents other than hydroxyalkylcarboxamide at R2 did not have strong biomediator activity. Moreover, the hydroxyethyl carboxamide derivatives at R2 (4 and 8t) had better biomediator activity compared to their hydroxypropyl carboxamides (3 and 8s) (Table 2). Based on these results, we speculated that the chain length of the hydroxyalkyl group attached to the carboxamide moiety was key for biomediator activity.
Table 1.
Natural β-carboline compounds and their biomediator activity. RMs produced after the treatment of 1 μg ml−1 compounds was quantified by LC analysis. The data shown are expressed as the mean ± SD from 3 experiments.

| |
|---|---|
| R | RMs mg l−1 |
| H (harman) | 12 ± 7 |
| OH (harmol) | 19 ± 5 |
| OMe (harmine) | 14 ± 3 |
Figure 2.

Substructure search for similar compounds in an NPDepo chemical library.
Table 2.
Structure–activity relationships of β-carboline compounds.

|
R2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

|
|

|

|

|

|

|

|

|
|

|
|
||
| R1 |
|
(+) | (+) | (−) | (+) | ||||||||
|
(−) | (+) | |||||||||||
|
(−) | ||||||||||||

|
(−) | (−) | (+) | (−) | (−) | (−) | (−) | (−) | |||||
|
(−) | (+) | (+) | (+) | (+) | ||||||||

|
(+) | (−) | |||||||||||
|
(+++) NPD2683 (4) | (++) NPD2639 (3) | (−) | (−) | (++) | (−) | (−) | ||||||

|
(+++) (8t) | (++) (8s) | (+) | (+) | (+) | (+) | (+) | (−) | |||||

|
(−) | (+) | (+) | (+) | |||||||||
A srcts-NRK cell assay and HPLC analysis were performed to evaluate the RM production after the treatment with 1 μg ml−1 compounds from substructure search. The biomediator activity of 41 out of 150 β-carbolines is summarized, enabling a focus on the key structures involved. (−), no activity in srcts-NRK cell assays, and RM production was not measured by HPLC; (+), positive in srcts-NRK cell assays, and RM production was less than 40 mg l−1; (++), positive in srcts-NRK cell assays, and RM production was 40–50 mg l−1; (+++), positive in srcts-NRK cell assays, and RM production exceeded 50 mg l−1.
β-carboline structure optimization
To optimize the structure of seed compound 3 for inducing RM production, we synthesized derivatives with regard to the amide group and the aromatic substituents (Table 3–5, Fig. S1). First, while retaining the 3-chlorophenyl group at R1, we modified the R2 group to afford 8a–8d. The acetate (8c) and methyl ether (8d) groups did not exhibit biomediator activity. Whereas the N-hydroxycarboxamide (8b) retained biomediator activity, the maximum yield of RMs (72 ± 12 mg/l) was obtained with the simple carboxamide (8a).
Table 3.
Structure–activity relationships of β-carboline compounds that enhanced the production of RMs: Modifications of the substituents attached to the phenyl group or pyridine ring. Streptomyces sp. SN-593 was cultured separately in the presence of 1 µg ml−1 of each compound. RM production was measured by LC analysis. The data shown are expressed as the mean ± SD from 3 experiments.

| |||
|---|---|---|---|
| Compound | R1 | R2 | RMs mg l−1 |
| 3 (NPD2639) | 3-Cl | CONH(CH2)3OH | 50 ± 6 |
| 4 (NPD2683) | 3-Cl | CONH(CH2)2OH | 61 ± 10 |
| 8a | 3-Cl | CONH2 | 72 ± 12 |
| 8b | 3-Cl | CONHOH | 57 ± 7 |
| 8c | 3-Cl | CONH(CH2)3OAc | 16 ± 6 |
| 8d | 3-Cl | CONH(CH2)3OMe | 2 ± 2 |
| 8e (BR-1) | H | CONH2 | 84 ± 5 |
| 8f | 3-OMe | CONH2 | 57 ± 4 |
| 8g | 3-F | CONH2 | 68 ± 1 |
| 8h | 3-Br | CONH2 | 63 ± 4 |
| 8i | 3-NO2 | CONH2 | 22 ± 3 |
| 8j | 4-Cl | CONH2 | 25 ± 5 |
| 8k | 4-Cl | CONHOH | 18 ± 9 |
| 8l | 3,5-diF | CONH2 | 37 ± 4 |
| 8m | 3,5-diCl | CONH2 | 22 ± 6 |
| 8n | 3,5-diBr | CONH2 | 18 ± 6 |
| 8o | H | H | 2 ± 1 |
| 8p | H | CONH(CH2)3OH | 39 ± 15 |
| 8q | H | CONH(CH2)2OH | 22 ± 6 |
| 8r | H | COOH | 0.85 ± 0.23 |
| 7a | H | COOMe | 0.35 ± 0 |
| 8s | 3-Br | CONH(CH2)3OH | 48 ± 26 |
| 8t | 3-Br | CONH(CH2)2OH | 60 ± 23 |
Table 5.
Structure–activity relationships of β-carboline compounds that enhanced the production of RMs: Modification of tetrahydro β-carboline compounds. Streptomyces sp. SN-593 was cultured separately in the presence of 1 µg ml−1 of each compound. RM production was measured by LC analysis. The data shown are expressed as the mean ± SD from 3 experiments.

| ||||
|---|---|---|---|---|
| Compound | R1 | R2 | R3 | RMs mg l−1 |
| 12a | H | COOH | H | 0.69 ± 0.59 |
| 12b | H | CONH2 | H | 2.16 ± 1.24 |
| 12c | H | COOH | CH2OH | 4.54 ± 1.82 |
| 12d | H | CONH2 | CH2OH | 4.16 ± 0.25 |
| 12e |
|
CONH2 | H | 4.52 ± 1.27 |
| 12f |
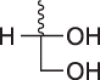
|
COOH | H | 2.41 ± 0.18 |
To examine the activity of substituents in the phenyl ring, we next modified the 3-chlorophenyl group of 8a with various aromatic rings, including 3-H (8e), 3-OMe (8f), 3-F (8g), 3-Br (8h), and 3-NO2 (8i). Except for 8i, the other compounds (8e–h) retained biomediator activity, thereby highlighting the importance of substituents at position 3. In agreement, when we replaced the 3-chlorophenyl group of 8a and 8b with the 4-chlorophenyl group present in 8j and 8k, the biomediator activity decreased. Because 3-substituted phenyl derivatives retained activity, we expected activity with the 3,5-diflurophenyl (8l), 3,5-dichlorophenyl (8m), and 3,5-dibromophenyl (8n) derivatives. However, we found that all these derivatives had reduced biomediator activity. To examine the role of the carboxamide moiety of 8e, we replaced it with hydrogen (8o), hydroxypropyl carboxamide (8p), hydroxyethyl carboxamide (8q), carboxylic acid (8r), or carboxylate (7a) moieties. The biomediator activities of 8p and 8q were partially retained, but 8o, 8r, and 7a showed abolished activity, revealing the importance of the carboxamide moiety for the activity (Table 3). In addition, we replaced the phenyl group with cyclohexane (11a), furan (11b), or pyridine (11c) and found that these groups abolished the biomediator activity (Table 4). Then, to evaluate the effect of the β-carboline core, we also synthesized tetrahydro β-carboline compounds (12a–f). Interestingly, they had abolished biomediator activity (Table 5). Based on this structure–activity-relationship study, 8e, named BR-1 (Biomediator that induce Reveromycin), was the most potent biomediator.
Table 4.
Structure–activity relationships of β-carboline compounds that enhanced the production of RMs: Replacement of the phenyl group with other substituents. Streptomyces sp. SN-593 was cultured separately in the presence of 1 µg ml−1 of each compound. RM production was measured by LC analysis. The data shown are expressed as the mean ± SD from 3 experiments.

| ||
|---|---|---|
| Compound | R3 | RMs mg l−1 |
| 11a |
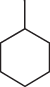
|
32 ± 12 |
| 11b |
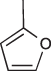
|
9 ± 5 |
| 11c |

|
18 ± 11 |
Biological activity of BR-1
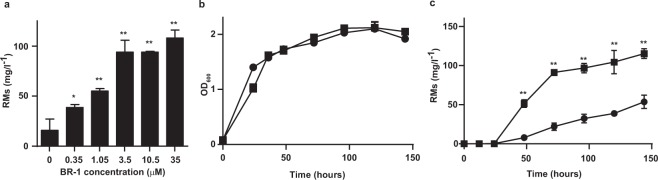
We examined the dose dependent and time dependent biomediator activity of BR-1. The lowest concentration of BR-1 that induced RM production was 0.1 µg/ml (0.35 μM), and it produced RMs over a wide range of concentrations (0.35 μM–35 μM) (Fig. 3a). There is a general notion that a reduction in growth rate, if not growth cessation, is an important signal for triggering secondary metabolism26. We found that cell growth of Streptomyces sp. SN-593 was not affected by treatment with BR-1, suggesting that the induction of RM production was neither due to an increased cell density nor due to a reduced growth rate (Fig. 3b). Moreover, BR-1 induced RM production from day 2 and continued through day 6, compared to non-treated samples (Fig. 3c).
Figure 3.
Effect of BR-1 on RM production and cell growth in Streptomyces sp. SN-593. (a) Dose-dependent RM production by BR-1. (b) Cell growth in SY medium (•: DMSO, ▪: 3.5 µM BR-1). (c) Time-dependent RM production after BR-1 treatment (•: DMSO, ▪: 3.5 µM BR-1). RM production was quantified by HPLC (data expressed as the mean ± SD from 3 experiments). *p < 0.05; **p < 0.0001 by 2-way analysis of variance.
The RM biosynthetic gene cluster in Streptomyces sp. SN-593 harbors 21 genes, including the genes involved in biosynthesis of polyketide core structure, post-PKS modification, and transcriptional regulators. Among the regulators, RevQ and RevU belong to the Streptomyces antibiotic regulatory protein and LuxR family regulators, respectively. These two families of regulators are well known to positively control secondary metabolites biosynthesis26. Previously, we have shown that the constitutive expression of revQ gene by aphII promoter resulted in about 5-fold increase in the amount of RMs produced27. While the Streptomyces sp. SN-593 produces RMs at basal level in SY-B medium (Figs 1, 3a,c), the presence of 3.5 µM BR-1 enhanced RMs production about 6-fold at day 3. The fold increment of RMs production after BR-1 treatment was almost similar to that of transformed cells which constitutively expressed transcriptional regulators27. Based on the similar observation, we speculated that BR-1 response goes through the pathway specific regulator associated with the RM biosynthetic gene cluster.
In summary, we identified β-carboline compounds as the biomediators of RMs production. Considering that β-carboline alkaloids are widely distributed in the environment28–30, our study highlights the possible presence of biomediator-microbe communication in nature and these chemicals may trigger a variety of responses, including the production of bioactive molecules in microbial communities. Furthermore, our study indicates that these signals might serve as an excellent approach to activate SM biosynthesis without genetic engineering. In future, SM production by biomediator and its application to approaches such as single cell multiplexed activity metabolomics might link with the discovery of effector molecules to human cells31.
Materials and Methods
Chemical library
A chemical library from the RIKEN Natural Products Depository (NPDepo) (http://www.npd.riken.jp) was used.
Culture medium and biomediator treatment
Streptomyces sp. SN-593 was cultured using several media, including synthetic medium21, SK222, MS22, RM-PM22, SY22, and SY-B medium (1% soluble starch and 0.1% yeast extract). Pyricularia oryzae Kita1 was cultured in oatmeal agar (7.25% oatmeal agar), YG medium (2% glucose and 0.5% yeast extract), and PD medium (2.4% potato dextrose broth and 0.2% agar).
Wild-type Streptomyces sp. SN-59323 was used to screen for biomediators. Spores were prepared on MS plates. A loopful of Streptomyces sp. SN-593 spores was grown in 70 ml SK2 medium in a 500-ml cylindrical flask at 28 °C at 150 rpm to an OD600 of 6–8, 1 ml of which was diluted in 100 ml SY-B medium. A 1-ml aliquot of this mixture was prepared on a 2.2-ml well of a 96-well plate (4titude, reorder # 4ti-0130). One microliter of each NPDepo compound, dissolved in dimethyl sulphoxide (DMSO) at 1 mg ml−1, was added per well. Cells were cultured at 28 °C at 1000 rpm (TAITEC, BioShaker M-BR-024). After 3 days, 0.5 ml of acetone was added to each SY-B culture and centrifuged at 5,000 × g for 10 min (Allegra® X-15R, Beckman Coulter). The supernatant (200 µl, acetone fraction) was dried and dissolved in water (20 µl) to prepare biomediator-treated broth (BTB). Enhanced RM production was evaluated with P. oryzae and srcts-NRK cells assay. To quantify RM production, the acetone fraction was analysed by LC-MS. Because of non-enzymatic RM-A conversion (1) into RM-B (2), which is a 5,6-spiroacetal derivative of 122, both 1 and 2 were quantified by LC-MS analysis.
P. oryzae screening system
Two agar blocks (~1 mm2) of P. oryzae Kita1 grown on oatmeal agar plates were mixed with 10 ml YG medium in a 50-ml Falcon tube, vortexed 2 min, and incubated at 27 °C at 150 rpm. After 3 days, the culture was vortexed for 2 min, diluted 50-fold in PD medium, and 200-µl aliquots were added in each well of a 96-well plate. Because RM-A inhibited P. oryzae growth at 5 µg ml−1, the optimum amount of BTB was added to each well, and the cells were cultured at 28 °C for 2 days to study growth inhibition.
srcts-NRK cells assay system
RMs induce the morphological reversion of srcts-NRK cells from spherical transformed cells to flat normal cells when cultured at 32 °C in Eagle’s minimal essential medium supplemented with 10% calf serum. RM-A, RM-C, and RM-D exhibited EC5O values of ~1.58 µg ml−1 for the reversal32. Cell aliquots (1.6 × 104/200 µl) were seeded in separate wells of a 96-well plate and incubated at 32 °C for 5 h. Five microliters of BTB was added to each well, and the cells were incubated for 2 days. Then, phenotypic changes of cells were assessed microscopically.
LC-MS analysis
Analysis of metabolites was performed by ESI-MS analysis using a Waters Alliance high-performance liquid chromatography (HPLC) system equipped with a mass spectrometer (Q-Trap; Applied Biosystems)22. The HPLC system consisted of an XTerra®MSC18 (5-μm, 2.1 mm internal diameter × 150 mm length) column maintained at 0.2 ml min−1. Solvent A was 0.05% aqueous formic acid and solvent B was acetonitrile. The sample was injected into the column after pre-equilibration with 30% solvent B; the column was developed with a linear gradient from 30% to 100% solvent B over 20 min and maintained in 100% solvent B for 20 min. Mass spectra were collected in ESI-negative mode.
Chemical synthesis of β-carboline derivatives
First, β-carboline derivatives 8a–d related to the 3–substituent were designed and synthesized33 from L-tryptophan and 3-chlorobenzaldehyde. Synthetic route (Fig. S1) involved the key step of the Pictet–Spengler reaction followed by oxidation with trichloroisocyanuric acid to give methyl β-carboline 3-carboxylate 7d. The 3-carboxylate 7d was then treated with various amines to give the desired β-carboline 3-carboxamides (3, 4, and 8a, b). The hydroxypropyl derivative of 3 was converted to acetate 8c and methyl ether 8d. Because carboxamide 8a derived from ammonium hydroxide was the most active compound among 3, 4, and 8a–d, a series of methyl β-carboline 3-carboxylates (7a–j) were synthesized and converted to the corresponding carboxamides 8e–j and 8l–n, having an amide group derived from ammonium hydroxide. The chemical structures of all β-carboline derivatives were confirmed by 1H NMR and high-resolution-MS data (Supporting Text).
Supplementary information
Acknowledgements
We thank H. Takahashi and M. Ueki for testing the culture conditions of the producer strain; Y. Futamura for help in the srcts-NRK cells assay; T. Nogawa and M. Uramoto for metabolite analysis; and E. Oowada, H. Takagi, and A. Okano for technical assistance. This work was supported by the JSPS KAKENHI (Grant 24658088, 17H06412, 18H03945), JSBBA Innovative Research Program Award, and in part by the NARO Bio-oriented Technology Research Advancement Institution (Research program on development of innovative technology, 28011A).
Author Contributions
S.P., S.T., and H.O. designed the experiments. S.P. performed biomediator screening. T.S. and T.H. performed chemical synthesis and derivatization of β-carboline compounds. S.T., S.P., and T.S. wrote the manuscript. S.T. and H.O. integrated the overall research project. All authors discussed the results and commented on the manuscript.
Data Availability
The data that support the finding of this study are available from the corresponding authors upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shunji Takahashi, Email: shunjitaka@riken.jp.
Hiroyuki Osada, Email: hisyo@riken.jp.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42268-w.
References
- 1.Baltz RH. Antimicrobials from actinomycetes: Back to the future. Microbe. 2007;2:125–131. [Google Scholar]
- 2.Hara O, Beppu T. Mutants blocked in streptomycin production in Streptomyces griseus: The role of a-factor. J. Antibiot. 1982;35:349–358. doi: 10.7164/antibiotics.35.349. [DOI] [PubMed] [Google Scholar]
- 3.Corre C, Song L, O’Rourke S, Chater KF, Challis GL. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. USA. 2008;105:17510–17515. doi: 10.1073/pnas.0805530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Recio E, Colinas Á, Rumbero Á, Aparicio JF, Martin JF. PI factor, a novel type quorum-sensing Inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 2004;279:41586–41593. doi: 10.1074/jbc.M402340200. [DOI] [PubMed] [Google Scholar]
- 5.Kitani S, et al. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA. 2011;108:16410–16415. doi: 10.1073/pnas.1113908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grafe U, et al. Isolation and structure of novel autoregulators from Streptomyces griseus. J. Antibiot. 1982;35:609–614. doi: 10.7164/antibiotics.35.609. [DOI] [PubMed] [Google Scholar]
- 7.Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J. Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 8.Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J. Ferment. Bioeng. 1989;68:170–173. doi: 10.1016/0922-338X(89)90131-1. [DOI] [Google Scholar]
- 9.Takano E, et al. Purification and structural determination of SCB1, a γ-Butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2) J. Biol. Chem. 2000;275:11010–11016. doi: 10.1074/jbc.275.15.11010. [DOI] [PubMed] [Google Scholar]
- 10.Arakawa K, Tsuda N, Taniguchi A, Kinashi H. The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. Chembiochem. 2012;13:1447–1457. doi: 10.1002/cbic.201200149. [DOI] [PubMed] [Google Scholar]
- 11.Andres N, Wolf H, Zahner H. Hormaomycin, a new peptide lactone antibiotic effective in inducing cytodifferentiation and antibiotic biosynthesis in some Streptomyces species. Z. Naturforsch. C. 1990;45:850–855. doi: 10.1515/znc-1990-7-817. [DOI] [Google Scholar]
- 12.Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in Streptomycetes I. purification and characterization. J. Antibiot. 2001;54:1036–1044. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- 13.Amano S, et al. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot. 2010;63:486–491. doi: 10.1038/ja.2010.68. [DOI] [PubMed] [Google Scholar]
- 14.Craney A, Ozimok C, Pimentel-Elardo S, Capretta A, Nodwell J. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012;19:1020–1027. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Kawai K, Wang G, Okamoto S, Ochi K. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 2007;274:311–315. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 16.Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proceedings of the National Academy of Sciences. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2) J. Bacteriol. 1996;178:7276–7284. doi: 10.1128/jb.178.24.7276-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosaka T, et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotech. 2009;27:462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 19.Laureti L, et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panthee S, et al. Furaquinocins I and J: novel polyketide isoprenoid hybrid compounds from Streptomyces reveromyceticus SN-593. J. Antibiot. 2011;64:509–513. doi: 10.1038/ja.2011.41. [DOI] [PubMed] [Google Scholar]
- 21.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi S, et al. Reveromycin A biosynthesis uses RevG and RevJ for stereospecific spiroacetal formation. Nat. Chem. Biol. 2011;7:461–468. doi: 10.1038/nchembio.583. [DOI] [PubMed] [Google Scholar]
- 23.Osada H, Koshino H, Isono K, Takahashi H, Kawanishi G. Reveromycin A, a new antibiotic which inhibits the mitogenic activity of epidermal growth factor. J. Antibiot. 1991;44:259–261. doi: 10.7164/antibiotics.44.259. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi H, et al. Reveromycins, new inhibitors of eukaryotic cell growth. I. Producing organism, fermentation, isolation and physico-chemical properties. J. Antibiot. 1992;45:1409–1413. doi: 10.7164/antibiotics.45.1409. [DOI] [PubMed] [Google Scholar]
- 25.Kato N, Takahashi S, Nogawa T, Saito T, Osada H. Construction of a microbial natural product library for chemical biology studies. Curr. Opin. Chem. Biol. 2012;16:101–108. doi: 10.1016/j.cbpa.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Bibb MJ. Regulation of secondary metabolism in. Streptomycetes. Curr. Opin. Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Osada, H., Takahashi, S., Kawatani, M., Sakaki, Y. & Toyoda, A. Process for producing reveromycin A or a synthetic intermediate thereof, process for producing compounds containing a spiroketal ring and novel antineoplastics, fungicides and therapeutic agents for bone disorders. (US Pat. 8980587 B2).
- 28.Huang HB, et al. Antimalarial beta-carboline and indolactam alkaloids from marinactinospora thermotolerans, a deep sea isolate. J. Nat. Prod. 2011;74:2122–2127. doi: 10.1021/np200399t. [DOI] [PubMed] [Google Scholar]
- 29.Aroonsri A, Kitani S, Ikeda H, Nihira T. Kitasetaline, a novel β-carboline alkaloid from Kitasatospora setae NBRC 14216T. J. Biosci. Bioeng. 2012;114:56–58. doi: 10.1016/j.jbiosc.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, et al. Discovery of McbB, an enzyme catalyzing the β-carboline skeleton construction in the marinacarboline biosynthetic pathway. Angew. Chem. Int. Ed. Engl. 2013;52:9980–9984. doi: 10.1002/anie.201303449. [DOI] [PubMed] [Google Scholar]
- 31.Earl DC, et al. Discovery of human cell selective effector molecules using single cell multiplexed activity metabolomics. Nature Communications. 2018;9:39. doi: 10.1038/s41467-017-02470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi H, et al. Reveromycins, new inhibitors of eukaryotic cell growth. II. Biological activities. J. Antibiot. 1992;45:1414–1419. doi: 10.7164/antibiotics.45.1414. [DOI] [PubMed] [Google Scholar]
- 33.Tang J-G, et al. Synthesis of analogues of flazin, in particular, flazinamide, as promising anti-HIV agents. Chem. Biodivers. 2008;5:447–460. doi: 10.1002/cbdv.200890044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the finding of this study are available from the corresponding authors upon request.