Abstract
Cardiovascular disease (CVD) is the number one killer in the USA, yet it is largely preventable (World Health Organization 2011). To prevent CVD, carotid intima-media thickness (CIMT) imaging, a noninvasive ultrasonography method, has proven to be clinically valuable in identifying at-risk persons before adverse events. Researchers are developing systems to automate CIMT video interpretation based on deep learning, but such efforts are impeded by the lack of large annotated CIMT video datasets. CIMT video annotation is not only tedious, laborious, and time consuming, but also demanding of costly, specialty-oriented knowledge and skills, which are not easily accessible. To dramatically reduce the cost of CIMT video annotation, this paper makes three main contributions. Our first contribution is a new concept, called Annotation Unit (AU), which simplifies the entire CIMT video annotation process down to six simple mouse clicks. Our second contribution is a new algorithm, called AFT (active fine-tuning), which naturally integrates active learning and transfer learning (fine-tuning) into a single framework. AFT starts directly with a pre-trained convolutional neural network (CNN), focuses on selecting the most informative and representative AU s from the unannotated pool for annotation, and then fine-tunes the CNN by incorporating newly annotated AU s in each iteration to enhance the CNN’s performance gradually. Our third contribution is a systematic evaluation, which shows that, in comparison with the state-of-the-art method (Tajbakhsh et al., IEEE Trans Med Imaging 35(5):1299–1312, 2016), our method can cut the annotation cost by >81% relative to their training from scratch and >50% relative to their random selection. This performance is attributed to the several advantages derived from the advanced active, continuous learning capability of our AFT method.
Keywords: Active learning, Transfer learning, Cardiovascular disease
Introduction
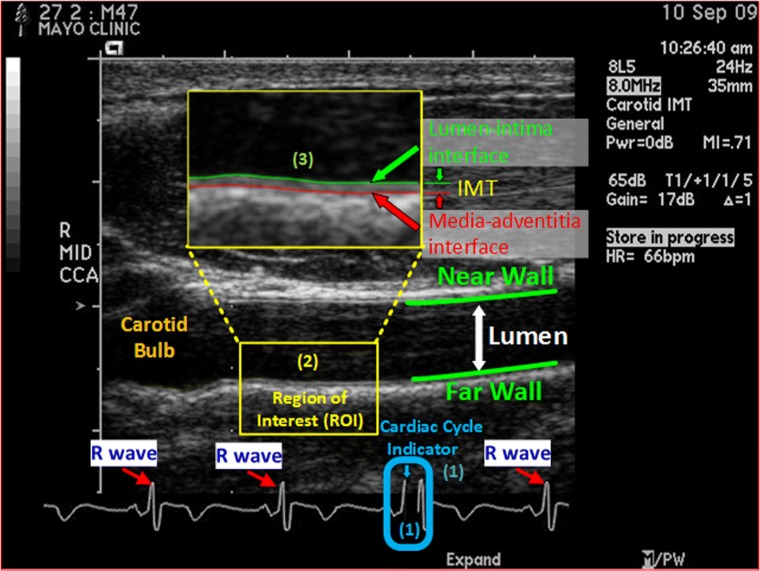
Cardiovascular disease (CVD) is the leading cause of death in the USA: every 40 s, one American dies of CVD; nearly one-half of these deaths occur suddenly and one-third of them occur in patients younger than 65 years, but CVD is preventable [1]. To prevent CVD, the key is to identify at-risk persons, so that scientifically proven and efficacious preventive care can be prescribed appropriately. Carotid intima-media thickness (CIMT) imaging, a noninvasive ultrasonography method, has proven to be clinically valuable for predicting individual CVD risk [8, 22, 31]. It quantifies subclinical atherosclerosis, adds predictive value to traditional risk factors (e.g., the Framingham Risk Score), and has several advantages over computed tomography (CT) coronary artery calcium score: safer (no radiation exposure), more sensitive in a young population, and more accessible to the primary care setting. However, the CIMT imaging protocol (see the “CIMT Imaging Protocol” section) requires to acquire four videos for each subject, and interpretation of each CIMT video involves three manual steps (illustrated Fig. 1), which are not only tedious and laborious but also subjective to large interoperator variability if guidelines are not properly followed, hindering the widespread utilization of CIMT in clinical practice. Therefore, it is highly desirable to have a system that can automate the CIMT video interpretation.
Fig. 1.
End-diastolic ultrasound frame (EUF), showing a longitudinal view of a common carotid artery in an ultrasound B-scan image. EUFs are selected based on the cardiac cycle indicator, a black line, which indicates to where in the cardiac cycle the current frame corresponds. CIMT is the distance between the lumen-intima interface (in green) and the media-adventitia interface (in red) at an EUF, and it is determined in a region of interest (ROI) approximately 1 cm distal from the carotid bulb at the EUF. In a CIMT exam, the sonographer examines the common carotid arteries on both sides of the neck from the two angles, yielding 4 CIMT ultrasound videos for each subject. Interpreting each CIMT video involves three manual steps: (1) select 3 EUFs in each video based on the cardiac cycle indicator; (2) localize an ROI in each selected EUF according to the carotid bulb; (3) trace the lumen-intima and the media-adventitia interfaces within the localized ROI and compute the minimum, maximum, and average of the distance between the traced lumen-intima and the media-adventitia interfaces. The final CIMT report of a subject is a statistical summary of all CIMT measurements on the 12 (4 × 3) EUFs from the 4 CIMT videos acquired for the subject. This figure is used with permission [28] under IEEE license number 4407260599014
The tedious and laborious manual operations also mean significant work in expert annotation when developing such systems based on machine learning. This paper is not to develop such a system but rather to present a new idea: how to minimize the cost of expert annotation for building such systems that can automate CIMT video interpretation based on deep learning. In this research, we make the following three contributions:
Our first contribution is a new concept, called Annotation Unit (AU), which naturally groups the objects to be annotated into sets, and all the objects in each set can be conveniently labeled once with as few operations as possible. This concept significantly simplifies the entire CIMT video annotation process down to six mouse clicks as detailed in the “Annotation Units” section and illustrated in Fig. 2. Our second contribution is a new algorithm, called AFT (active fine-tuning), which naturally integrates active learning and transfer learning into a single framework (see Algorithm 1) to focus on selecting the most informative and representative AU s for annotation, thereby dramatically reducing the cost of annotation in CIMT. AFT starts directly with a pre-trained CNN to seek “worthy” samples from the unannotated pool for annotation, and then fine-tunes the CNN by incorporating newly annotated samples in each iteration to enhance the CNN’s performance gradually. Compared with conventional active learning, AFT offers four advantages: (1) it starts with a completely empty labeled dataset, requiring no initial seed-labeled training samples; (2) it incrementally improves the learner through fine-tuning rather than repeatedly re-training; (3) it can automatically handle multiple classes; and (4) it is applicable to many biomedical image analysis tasks [37], including detection, classification, and segmentation. Our third contribution is a systematic evaluation of our proposed method, which demonstrates that, with AFT, the cost of annotation for CIMT can be cut by at least half in comparison with FT (fine-tuning with random selection) and by >81% relative to their training from scratch as detailed in the “Experiments” section. This result is significant for enhancing the system performance for automating CIMT video interpretation [28, 34]. Given the current performance of our system [28], it is very difficult to improve its performance by randomly annotating new CIMT videos. We must focus on annotating the most informative and representative videos; otherwise, we will have to annotate many new videos but gain very little in boosting its performance.
Fig. 2.
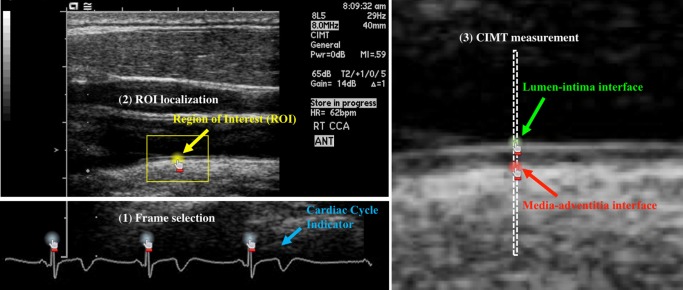
We simplify the annotation process for each CIMT video down to six mouse clicks. As illustrated, the annotation of EUF selection is made at the video level with three mouse clicks on the three R waves of an ECG signal, while for ROI localization, the annotation is made at the frame level with one mouse click at the center of the ROI. Manually tracing the lumen-intima interface or the media-adventitia interface is tedious and laborious. To reduce workload, we eliminate the tracing by two mouse clicks on the lumen-intima and media-adventitia interfaces between two vertical dashed lines, only when requested by our proposed AFT algorithm (see the “Annotation Units” section for details)
CIMT Imaging Protocol
The CIMT exams utilized in our research were performed with B-Mode ultrasound using an 8–14-MHz linear array transducer utilizing fundamental frequency only (Acuson SequoiaTM, Mountain View, CA, USA). The carotid screening protocol begins with scanning bilateral carotid arteries in a transverse manner from the proximal aspect to the proximal internal and external carotid arteries. The probe is then turned to obtain the longitudinal view of the distal common carotid artery (Fig. 1). The sonographer optimizes the 2D images of the lumen-intima and media-adventitia interfaces at the level of the common carotid artery by adjusting overall gain, time gain, compensation, and focus position. Once the parameters are optimized, the sonographer captures two CIMT videos focused on the common carotid artery from two optimal angles of incidence, and ensures that each CIMT video covers at least three cardiac cycles. The same procedure is repeated for the other side of the neck, resulting in a total of four CIMT videos for each subject.
CIMT stands for carotid intima-media thickness, but in the literature, it may refer to the imaging method, the ultrasonography examination, or the examination results. For clarify, we define some terms used in this paper. By CIMT imaging, we refer to the noninvasive ultrasonography examination procedure described above, yielding four CIMT videos for each subject. The CIMT video interpretation is a process to analyze all four CIMT videos acquired for a subject and produce a CIMT report, which includes a statistical summary of all CIMT measurements performed on the three end-diastolic ultrasound frames (EUFs) selected from each of the four CIMT videos acquired for the subject. EUFs are selected based on the cardiac cycle indicator as shown in Fig. 1, and there are 12 (= 4 × 3) EUFs for each subject. A CIMT measurement on an EUF includes the minimum, maximum, and average of the distance between the lumen-intima and the media-adventitia interfaces (see Fig. 1), thereby requiring the tracing of lumen-intima and the media-adventitia interfaces. The interpretation of each CIMT video involves three manual steps: (1) select three EUFs in each video based on the cardiac cycle indicator; (2) localize an ROI in each selected EUF according to the carotid bulb; and (3) trace the lumen-intima and the media-adventitia interfaces within the localized ROI and compute the minimum, maximum, and average of the distance between the traced lumen-intima and the media-adventitia interfaces. Certainly, we may adopt the full CIMT interpretation process to annotate CIMT videos as required by machine learning algorithms. However, to dramatically reduce the annotation efforts, we will introduce a separate CIMT video annotation process in conjunction with our proposed AFT algorithm.
Related Work
Carotid Intima-Media Thickness Video Interpretation
As discussed in the “CIMT Imaging Protocol” section, to measure CIMT, the lumen-intima and the media-adventitia interfaces must be traced first. Naturally, the earlier approaches are focused on analyzing the intensity profile and distribution, computing the gradient, or combining various edge properties through dynamic programming [19]. Recent approaches [5, 20] are mostly based on active contours (a.k.a snakes) or their variations [15]. Most recently, researchers are fucusing on developing algorithms based on machine learning for CIMT video interpretation. For example, Menchón-Lara et al. employed a committee of standard multi-layer perceptron in [24] and a single standard multi-layer perceptron with an auto-encoder in [25] for CIMT video interpretation. Shin et al. [28] presented a unified framework based on convolutional neural networks (CNNs) for automating the entire CIMT video interpretation process, and Tajbakhsh et al. [34] further demonstrated that the measurement errors are within the interobserver variation. However, none of the aforementioned publications has mentioned the cost of expert annotation in their system development. To our knowledge, we are among the first to minimize the cost of annotation by integrating active learning with the fine-tuning of CNNs for building systems that automate the CIMT video interpretation.
Transfer Learning for Medical Imaging
Gustavo et al. [3] replaced the fully connected layers of a pre-trained CNN with a new logistic layer and trained only the appended layer with the labeled data while keeping the rest of the network the same, yielding promising results for classification of unregistered multi-view mammogram. In [4], a fine-tuned pre-trained CNN was applied for localizing standard planes in ultrasound images. Gao et al. [7] fine-tuned all layers of a pre-trained CNN for automatic classification of interstitial lung diseases. In [27], Shin et al. used fine-tuned pre-trained CNNs to automatically map medical images to document-level topics, document-level sub-topics, and sentence-level topics. In [23], fine-tuned pre-trained CNNs were used to automatically retrieve missing or noisy cardiac acquisition plane information from magnetic resonance imaging and predict the five most common cardiac views. Schlegl et al. [26] explored unsupervised pre-training of CNNs to inject information from sites or image classes for which no annotations were available, and showed that such across-site pre-training improved classification accuracy compared to random initialization of the model parameters. Several researchers [9, 12, 33] have demonstrated that fine-tuning offers better performance and is more robust than training from scratch, especially in biomedical imaging tasks that labels are not easily accessible. However, none of these works involves active selection processes as our AFT method does, and they all performed one-time fine-tuning, that is, simply fine-tuned a pre-trained CNN just once with available training samples.
Integrating Active Learning with Deep Learning
Research in this area is sparse: Wang and Shang [35] may be the first to incorporate active learning with deep learning, and based their approach on stacked restricted Boltzmann machines and stacked auto-encoders. A similar idea was reported for hyperspectral image classification [17]. Stark et al. [30] applied active learning to improve the performance of CNNs for CAPTCHA recognition, while Al Rahhal et al. [2] exploited deep learning for active electrocardiogram classification. All these approaches are fundamentally different from our AFT approach in that in each iteration, they all repeatedly re-trained the learner from scratch while we fine-tune pre-trained CNNs, dramatically cutting the cost of annotation further by combining active learning with fine-tuning. Yang et al. [36] adopted active learning into fully convolutional network (FCN) [21] by extracting representative samples into training dataset but training a segmentation network requiring accurate object contour while our ROI localization is only a coarse-labeled location (a single click around the center of the ROI as shown in yellow in Fig. 2). Most recently, Zhou et al. [37] integrated active learning and deep learning based on continuous fine-tuning but their method is limited to binary classification and requires that all patches within each AU share the same label. Therefore, their method is not applicable to this CIMT application, which requires three-way classifiers.
The Proposed Method
The aim of this research is not to develop methods for automating the interpretation process, rather to investigate how to minimize the cost of expert annotation required for creating such systems that can automate CIMT video interpretation based on CNNs.
Annotation Units
We could follow the same process as illustrated in Fig. 1 [28] to create the ground truth as required to train CNNs. However, these three steps, and in particular the CIMT measurement, are not only tedious and laborious but also subjective to large inter-operator variability if guidelines are not properly followed. To accelerate the annotation process, we introduce a new concept, Annotation Unit (AU), which is defined as a set of objects that the annotator can associate with multiple labels at a time with as few operations as possible during the annotation process. The benefits of AU have two folds. First, the objects to be annotated are grouped into sets, and each set can be easily labeled with as few operations as possible. Taking CIMT measurement annotation as an example, instead of tracing the entire lumen-intima and media-adventitia interfaces within an ROI, we define an one-pixel-wide column in the ROI as an AU, so that all pixels within the column can be labeled once with two mouse clicks: one on the lumen-intima interface and one on the media-adventitia interface. Second, with the aforementioned properties, all the objects in an AU can be correctly associated with their labels once after the required operations. Using the CIMT measurement example again, after the two clicks, the first clicked pixel is associated with class 1 (lumen-intima), the second clicked pixel is with class 2 (media-adventitia), and all the rest pixels are with class 0 (background). It should be noticed that when all AU s are labeled, the interpretation quality is identical or at least similar to the standard process in [28], but our goal is to annotate as few AU s as possible during the annotation process; therefore, the annotation process may not result in a complete interpretation for a subject. In other words, the annotation process is designed for annotation (as little as possible) only, and it is not intended for clinical use, which requires a complete interpretation for each subject.
With the definition of AU, the CIMT video annotation process can be simplified down to just six mouse clicks as illustrated in Fig. 2. The annotation for EUF selection is made at the video level. With three mouse clicks on the R waves of the ECG signal, three end-diastolic ultrasound frames (EUFs) are determined and annotated as class 1, while all the rest frames are automatically labeled as class 0 (non-EUF). For ROI localization in an EUF, the annotation is made at the frame level with one mouse click on the EUF, giving the center of the ROI. Given the anatomical constraint that ROI should be approximately 1 cm distal from carotid bulb, the latter’s location can be automatically estimated. For data argumentation and classification robustness, all pixels within 15 mm from the selected center are considered as class 1 (ROI), and those within 15 mm from the estimated bulb location are as class 2, while all the rest pixels belong to class 0 automatically. For CIMT measurement, it would be too tedious and laborious for the annotator to manually trace the lumen-intima and media-adventitia interfaces. To reduce workload, two vertical dashed lines are drawn to indicate an AU (see Fig. 2) and the annotator makes two mouse clicks on the two interfaces between the two dashed lines. The top pixel and bottom pixel are regarded as the lumen-intima interface (class 1) and media-adventitia interface (class 2), respectively, while all the rest pixels between the two lines are considered as background (class 0). The optimal distance between the two dashed lines can be determined based on experiments, and we set it at one pixel (0.99 mm) currently. We summarize the objects of AU, annotation labels, and required operations per AU in each step of CIMT video annotation process in Table 1.
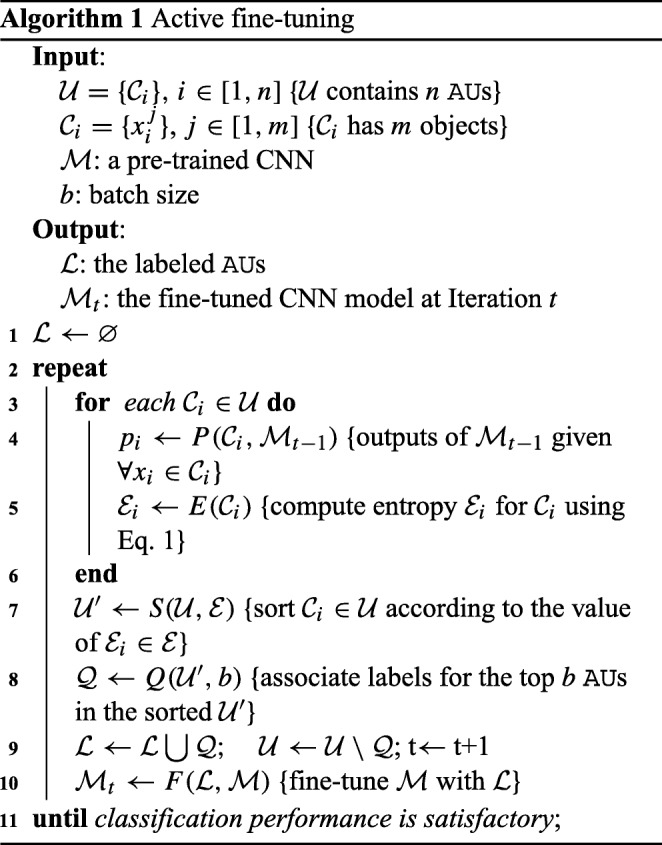
Table 1.
The AU, annotated labels, and required operations per AU in each step of CIMT video annotation process
| EUF selection | ROI localization | CIMT measurement | |
|---|---|---|---|
| AU | ECG signal | EUF frame | One-pixel-wide column in ROI |
| Labels | EUF | ROI | Lumen-intima interface |
| Non-EUF | Carotid bulb | Lumen-intima interface | |
| Background | Background | ||
| Operations | 3 clicks | 1 click | 2 clicks |
Active Fine-Tuning
Mathematically, given a set of AU s, , where n is the number of AU s, and each is associated with m objects, our AFT algorithm iteratively selects a subset of AU s for annotation as illustrated in Algorithm 1. From annotation, each object (in each selected AU) will be associated with one of Y number of possible classes. At the beginning, the labeled dataset is empty; we take a pre-trained CNN from ImageNet [6] (e.g., AlexNet) as initialization of the network and run it on to select b number of AU s for labeling. The newly labeled AU s will be incorporated into to fine-tune the CNN until the performance is satisfactory. From our experiments, we have found that continuously fine-tuning the CNN, which has been fine-tuned in the previous iteration, with enlarged datasets converges faster than repeatedly fine-tuning the original pre-trained CNN, but the latter offers better generalization. We have also found that continuously fine-tuning the CNN with only newly labeled data demands careful meta-parameter adjustments. Therefore, in this paper, our AFT fine-tunes the original pre-trained CNN with the labeled dataset enlarged with the newly labeled data in each iteration to achieve better performance. To determine the “worthiness” of an AU, we use entropy, as intuitively, entropy captures the classification certainty—higher uncertainty values denote higher degrees of information. Assuming the prediction of object in by the current CNN is , we define the entropy of as the average information furnished by all objects in from the unlabeled pool:
| 1 |
Experiments
In our experiments, we use a fully interpreted (annotated) database and simulate the active learning process (Algorithm 1) by retrieving labels for the samples selected based on selection criterion as present in Eq. 1. In this way, our approach can be validated without “physically” involving the experts in the loop.
Dataset
Due to space, we focus on the two most important tasks: ROI localization and CIMT measurement. Our AFT algorithm is implemented in Caffe [14] based on the pre-trained AlexNet [16]. In the following, we shall compare our method AFT (active fine-tuning) with the state-of-the-art method [33]: FT (fine-tuning with random selection) and LS (learning from scratch) in each task. We utilize 23 patients from UFL MCAEL CIMT research database [13]. Each patient has four videos (two on each side) [31], resulting in a total of 92 CIMT videos with 8,021 frames. Each video covers at least three cardiac cycles and thus a minimum of three EUFs. We randomly divide the CIMT videos at patient level into training, validation, and test datasets (no overlaps). The training dataset contains 44 CIMT videos of 11 patients with a total of 4,070 frames, the validation dataset contains 4 videos of 1 patient with 386 frames, and the test dataset contains 44 CIMT videos of 11 patients with 3,565 frames. From the perspective of active learning, the training dataset is the “unlabeled pool” for active selection; when an AU is selected, the label of each object will be provided. The fined-tuned CNN from each iteration is always evaluated with the test dataset, so that we can monitor the performance enhancement across AU s. Please note that we do not need many patients as we have many CIMT frames for each patient and we can generate a large number of patches for training deep models in each experiment. For example, in our ROI localization experiments, one AU practically provides 1715 labeled patches (297 as background, 709 as bulb, and 709 as ROI). Random translation and flipping data augmentation were applied when training the models.
ROI Localization
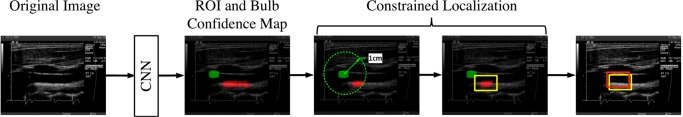
Accurate localization of the ROI is challenging because, as illustrated by Shin et al. [28] in their figure 1, no notable differences can be observed in image appearance among the ROIs on the far wall of the carotid artery. To overcome this challenge, we use the location of the carotid bulb as a contextual constraint. We choose this constraint for two reasons: (1) the carotid bulb appears as a distinct dark area in the ultrasonographic frame and thus can be uniquely identified; and (2) according to the consensus statement of the American Society of Electrocardiography for Cardiovascular Risk Assessment [31], the ROI should be placed approximately 1 cm from the carotid bulb on the far wall of the common carotid artery. The former motivates the use of the carotid bulb location as a constraint from a technical point of view, and the latter justifies this constraint from a clinical standpoint. We incorporate this constraint by simultaneously localizing both ROI and carotid bulb and then refine the estimated location of the ROI given the location of the carotid bulb. As illustrated in Fig. 3, we first determine the location of the carotid bulb as the centroid of the largest connected component within the confidence map for the carotid bulb and then localize the centroid of constrained ROI area using the following formula:
| 2 |
where M denotes the confidence map of being the ROI, C∗ is the largest connected component in M that is nearest to the carotid bulb, and I(p) is an indicator function for pixel p = [px,py] that is defined as
| 3 |
where lcb is the centroid of the carotid bulb. Basically, the indicator function excludes the pixels located farther than 1 cm from the carotid bulb location. This choice of the distance threshold is motivated by the fact that the ROI is located within 1 cm to the right of the carotid bulb.
Fig. 3.
ROI localization process (see text for details). The detected ROI, ground truth, and carotid bulb are in yellow, red, and green, respectively. The ROI is constrained by green circle with a 1-cm radius
CIMT Measurement
To automatically measure intima-media thickness, the lumen-intima and media-adventitia interfaces of the carotid artery must be detected within the ROI. Although the lumen-intima interface is relatively easy to detect, the detection of the media-adventitia interface is challenging, because of the faint image gradients around its boundary. We formulate this interface segmentation problem as a three-class classification task with the goal to classify each pixel within the ROI into thee categories: (1) a pixel on the lumen-intima interface, (2) a pixel on the media-adventitia interface, and (3) a background pixel. During testing, the trained CNN is applied to a given test ROI in a convolutional manner, generating two confidence maps with the same size as the ROI. The first confidence map shows the probability of each pixel being on the lumen-intima interface; the second confidence map shows the probability of each pixel being on the media-adventitia interface. A relatively thick high-probability band is apparent along each interface, which hinders the accurate measurement of intima-media thickness. To thin the detected interfaces, we scan the confidence map column by column, searching for the rows with the maximum response for each of the two interfaces. By doing so, we obtain a 1-pixel-thick boundary with a step-like shape around each interface. To further refine the boundaries, we use two active contour models (a.k.a., snakes) [18], one for the lumen-intima interface and one for the media-adventitia interface. The open snakes are initialized with the current step-like boundaries and then deform solely based on the probability maps generated by the CNN rather than the original image content.
Results and Discussions
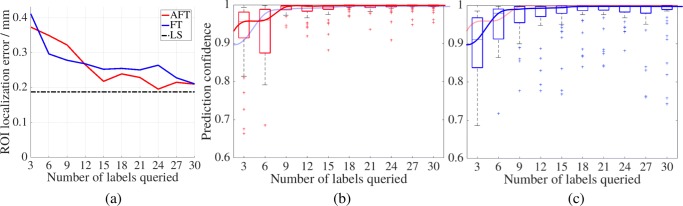
To evaluate AFT performance on ROI localization, in each iteration, we compute two criteria across all test patients: (1) the average ROI localization error (the Euclidean distance between the detected ROI and expert-annotated ROI) and (2) the predicted confidence of each expert-annotated ROI. Figure 4a shows the average ROI localization error over 30 AU s (automatically generating 51,450 labeled patches) demonstrating that our AFT dramatically reduces the labeling cost in comparison with FT and LS. Black dashed line represents the ROI localization error of LS, where the CNN was trained with the entire training dataset (132 AU s) without fine-tuning. We should note that at the earlier stage (less than 12 AU s), FT learns faster and yields better performance than AFT, a well-known phenomenon in active learning [10]. However, AFT quickly surpasses FT after a few times of fine-tuning. With only 24 AU s, AFT can nearly achieve the performance of LS with 132 AU s; with 15 AU s, AFT achieves that of FT with 30 annotations. Thereby, the cost of annotation can be cut by at least half in comparison with FT and by more than 81% in comparison with LS. To increase the robustness, the predicted confidence of each expert-annotated ROI is computed as the average of the predicted scores of all pixels within 15 pixels from the ROI center. Figure 4b, c is the box plots of the ROI confidence across all the test patients. Clearly, the more AU s used from the training dataset, the higher the ROI confidence with the test dataset. In terms of mean and standard deviation of ROI confidence, with just 9 AU s, AFT offers the same confidence as FT at 30 AU s. Moreover, ROI confidence from AFT can quickly converge to 1.0, while even using 30 AU s, FT still has many outliers.
Fig. 4.
a The average ROI localization errors of AFT, FT, and LS on the test patients over 30 AU s. The ROI confidence predicted by AFT b and FT c, respectively, on the test patients over 30 AU s. The trendlines denote active selection (in red) and random selection (in blue), and they are duplicated with different transparencies for their easy performance comparison in b and c
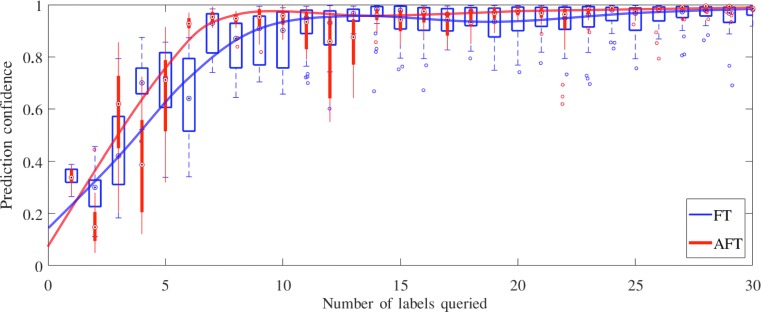
We evaluate our AFT on CIMT measurement in the same way as in ROI localization. However, due to the post-processing with snakes, AFT and FT give the similar localization errors at the lumen-intima and media-adventitia interfaces; therefore, we focus on the CIMT measurement confidence. Figure 5 is the box plot of the CIMT measurement confidence on the test dataset. AFT significantly outperforms FT, especially when a limited number of training samples are used. For example, actively selecting only 7 AU s can approximate the performance by randomly selecting 14 AU s. In addition, with only 7 AU s selected by our AFT algorithm, we can nearly achieve the accuracy offered by the entire dataset (12,144 AU s).
Fig. 5.
The CIMT measurement confidence predicted by AFT and FT, respectively, on the test patients over 30 AU s
In our experiments, we adopted the AlexNet architecture because a pre-trained AlexNet model is available in the Caffe library and its architecture strikes a nice balance in depth: it is deep enough that we can investigate the impact of AFT on the performance of pre-trained CNNs, and it is also shallow enough that we can conduct experiments quickly. Alternatively, deeper architectures, such as VGG [29], GoogleNet [32], and ResNet [11], could have been used and have shown relatively high performance for challenging computer vision tasks. However, the purpose of this work is not to achieve the highest performance for the CIMT video interpretation but to answer a critical question: How much the cost of annotation can be reduced when applying CNNs for CIMT video interpretation. For this purpose, AlexNet is a reasonable architectural choice. Nevertheless, we plan to investigate the performance of AFT on different deep architectures. Also, our algorithm aims to select the most informative and representative AU s for annotation with our proposed six-click strategy. As a result, the process will not generate full interpretations for all patients, that is, the six-click strategy is only applicable in the context of our proposed algorithm for reducing annotation efforts (as little as possible), and it is not designed and should not be used for clinical practice, where a complete interpretation is required for each patient.
Conclusions
We have developed an active fine-tuning method for CIMT video interpretation. It integrates active learning and transfer learning, offering two advantages: It starts with a completely empty labeled dataset, and incrementally improves the CNN’s performance via fine-tuning by actively selecting the most informative and representative samples. To accelerate the CIMT video annotation process, we introduced a new concept, Annotation Unit, which simplifies the CIMT video annotation process down to six mouse clicks. We have demonstrated that the cost of CIMT video annotation can be cut by at least half. This performance is attributed to the advanced active fine-tuning capability of our AFT method. In the future, we plan to explore possible algorithms in assisting sonographers to acquire high-quality CIMT videos more quickly and integrate our AFT algorithm into the process for collecting the most informative and representative CIMT videos to enhance our system performance.
Acknowledgements
This research has been supported partially by NIH under Award Number R01HL128785 and partially by ASU and Mayo Clinic through the Discovery Translation Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Contributor Information
Zongwei Zhou, Email: zongweiz@asu.edu.
Jae Shin, Email: sejong@asu.edu.
Ruibin Feng, Email: rfeng12@asu.edu.
R. Todd Hurst, Email: hurst.r@mayo.edu.
Christopher B. Kendall, Email: kendall.christopher@mayo.edu
Jianming Liang, Email: jianming.liang@asu.edu.
References
- 1.World Health Organization. Global atlas on cardiovascular disease prevention and control. Available at http://www.who.int/cardiovascular_diseases/publications (September 19, 2011)
- 2.Al Rahhal Ms, Bazi Y, AlHichri H, Alajlan N, Melgani F, Yager R. Deep learning approach for active classification of electrocardiogram signals. Inf Sci. 2016;345:340–354. doi: 10.1016/j.ins.2016.01.082. [DOI] [Google Scholar]
- 3.Carneiro G, Nascimento J, Bradley A: Unregistered multiview mammogram analysis with pre-trained deep learning models. In: (Navab N, Hornegger J, Wells WM, Frangi AF, Eds.) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Lecture Notes in Computer Science, vol. 9351, pp. 652–660. Springer International Publishing, 2015. 10.1007/978-3-319-24574-4_78
- 4.Chen H, Ni D, Qin J, Li S, Yang X, Wang T, Heng PA. Standard plane localization in fetal ultrasound via domain transferred deep neural networks. IEEE J Biomed Health Inform. 2015;19(5):1627–1636. doi: 10.1109/JBHI.2015.2425041. [DOI] [PubMed] [Google Scholar]
- 5.Delsanto S, Molinari F, Giustetto P, Liboni W, Badalamenti S, Suri JS. Characterization of a completely user-independent algorithm for carotid artery segmentation in 2-d ultrasound images. IEEE Trans Instrum Meas. 2007;56(4):1265–1274. doi: 10.1109/TIM.2007.900433. [DOI] [Google Scholar]
- 6.Deng J, Dong W, Socher R, Li LJ, Li K: Fei-fei, L.: ImageNet: A Large-Scale Hierarchical Image Database CVPR09, 2009
- 7.Gao M, Bagci U, Lu L, Wu A, Buty M, Shin HC, Roth H, Papadakis GZ, Depeursinge A, Summers RM, et al: Holistic classification of ct attenuation patterns for interstitial lung diseases via deep convolutional neural networks.. In: The 1st workshop on deep learning in medical image analysis, international conference on medical image computing and computer assisted intervention, at MICCAI-DLMIA’15, 2015
- 8.Gepner AD, Young R, Delaney JA, Tattersall MC, Blaha MJ, Post WS, Gottesman RF, Kronmal R, Budoff MJ, Burke GL, et al. A comparison of coronary artery calcium presence, carotid plaque presence, and carotid intima-media thickness for cardiovascular disease prediction in the multi-ethnic study of atherosclerosis (mesa) Circulation Cardiovascular Imaging. 2015;8(1):e002262. doi: 10.1161/CIRCIMAGING.114.002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenspan H, van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35(5):1153–1159. doi: 10.1109/TMI.2016.2553401. [DOI] [Google Scholar]
- 10.Guyon I, Cawley G, Dror G, Lemaire V, Statnikov A (2011) JMLR Workshop and conference proceedings (volume 16): Active learning challenge microtome publishing
- 11.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition.. In: The IEEE conference on computer vision and pattern recognition (CVPR), 2016
- 12.Hoo-Chang S, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst RT, Burke RF, Wissner E, Roberts A, Kendall CB, Lester SJ, Somers V, Goldman ME, Wu Q, Khandheria B. Incidence of subclinical atherosclerosis as a marker of cardiovascular risk in retired professional football players. Am J Cardiol. 2010;105(8):1107–1111. doi: 10.1016/j.amjcard.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T (2014) Caffe: Convolutional architecture for fast feature embedding. arXiv:http://arXiv.org/abs/1408.5093
- 15.Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. Int J Comput Vis. 1988;1(4):321–331. doi: 10.1007/BF00133570. [DOI] [Google Scholar]
- 16.Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks.. In: Advances in neural information processing systems, pp 1097–1105, 2012
- 17.Li J: Active learning for hyperspectral image classification with a stacked autoencoders based neural network.. In: 2016 IEEE International conference on image processing (ICIP), pp 1062–1065, 2016. 10.1109/ICIP.2016.7532520
- 18.Liang J, McInerney T, Terzopoulos D. United snakes. Med Image Anal. 2006;10(2):215–233. doi: 10.1016/j.media.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Liang Q, Wendelhag I, Wikstrand J, Gustavsson T. A multiscale dynamic programming procedure for boundary detection in ultrasonic artery images. IEEE Trans Med Imaging. 2000;19(2):127–142. doi: 10.1109/42.836372. [DOI] [PubMed] [Google Scholar]
- 20.Loizou CP, Pattichis CS, Pantziaris M, Nicolaides A. An integrated system for the segmentation of atherosclerotic carotid plaque. IEEE Trans Inf Technol Biomed. 2007;11(6):661–667. doi: 10.1109/TITB.2006.890019. [DOI] [PubMed] [Google Scholar]
- 21.Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation.. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440, 2015 [DOI] [PubMed]
- 22.Lorenz MW, Markus HS, Bots ML, Rosvall M, Sitzer M. Prediction of clinical cardiovascular events with carotid intima-media thickness. Circulation. 2007;115(4):459–467. doi: 10.1161/CIRCULATIONAHA.106.628875. [DOI] [PubMed] [Google Scholar]
- 23.Margeta J, Criminisi A, Cabrera Lozoya R, Lee DC, Ayache N. Fine-tuned convolutional neural nets for cardiac mri acquisition plane recognition. Comput Methods Biomech Biomed Eng: Imaging Visual. 2017;5(5):339–349. [Google Scholar]
- 24.Menchón-Lara RM, Bastida-Jumilla MC, González-López A, Sancho-Gómez JL: Automatic evaluation of carotid intima-media thickness in ultrasounds using machine learning.. In: Natural and artificial computation in engineering and medical applications, pp 241–249. Springer, 2013
- 25.Menchón-Lara RM, Sancho-Gómez JL. Fully automatic segmentation of ultrasound common carotid artery images based on machine learning. Neurocomputing. 2015;151:161–167. doi: 10.1016/j.neucom.2014.09.066. [DOI] [Google Scholar]
- 26.Schlegl T, Ofner J, Langs G: Unsupervised pre-training across image domains improves lung tissue classification.. In: Medical computer vision: Algorithms for big data, pp 82–93. Springer, 2014
- 27.Shin HC, Lu L, Kim L, Seff A, Yao J, Summers RM: Interleaved text/image deep mining on a very large-scale radiology database.. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 1090–1099, 2015
- 28.Shin J, Tajbakhsh N, Todd Hurst R, Kendall CB, Liang J: Automating carotid intima-media thickness video interpretation with convolutional neural networks.. In: The IEEE conference on computer vision and pattern recognition (CVPR), 2016
- 29.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition.. In: ICLR, 2015
- 30.Stark F, Hazırbas C, Triebel R., Cremers D.: Captcha recognition with active deep learning.. In: Workshop new challenges in neural computation 2015, p 94. Citeseer, 2015
- 31.Stein JH, Korcarz CE, Hurst RT, Lonn E, Kendall CB, Mohler ER, Najjar SS, Rembold CM, Post WS. Use of carotid ultrasound to identify subclinical vascular disease and evaluate cardiovascular disease risk: a consensus statement from the american society of echocardiography carotid intima-media thickness task force endorsed by the society for vascular medicine. J Am Soc Echocardiogr. 2008;21(2):93–111. doi: 10.1016/j.echo.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A: Going deeper with convolutions.. In: The IEEE conference on computer vision and pattern recognition (CVPR), 2015
- 33.Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 34.Tajbakhsh N, Shin JY, Hurst RT, Kendall CB, Liang J: Automatic interpretation of carotid intima–media thickness videos using convolutional neural networks.. In: Deep learning for medical image analysis, pp 105–131. Elsevier, 2017
- 35.Wang D, Shang Y: A new active labeling method for deep learning.. In: 2014 International joint conference on neural networks (IJCNN), pp 112–119, 2014. 10.1109/IJCNN.2014.6889457
- 36.Yang L, Zhang Y, Chen J, Zhang S, Chen DZ: Suggestive annotation: a deep active learning framework for biomedical image segmentation.. In: International conference on medical image computing and computer-assisted intervention, pp 399–407. Springer, 2017
- 37.Zhou Z, Shin J, Zhang L, Gurudu S, Gotway M, Liang J: Fine-tuning convolutional neural networks for biomedical image analysis: actively and incrementally.. In: IEEE Conference on computer vision and pattern recognition, hawaii, pp 7340–7349, 2017 [DOI] [PMC free article] [PubMed]