Abstract
To develop a convolutional neural network (CNN) algorithm that can predict the molecular subtype of a breast cancer based on MRI features. An IRB-approved study was performed in 216 patients with available pre-treatment MRIs and immunohistochemical staining pathology data. First post-contrast MRI images were used for 3D segmentation using 3D slicer. A CNN architecture was designed with 14 layers. Residual connections were used in the earlier layers to allow stabilization of gradients during backpropagation. Inception style layers were utilized deeper in the network to allow learned segregation of more complex feature mappings. Extensive regularization was utilized including dropout, L2, feature map dropout, and transition layers. The class imbalance was addressed by doubling the input of underrepresented classes and utilizing a class sensitive cost function. Parameters were tuned based on a 20% validation group. A class balanced holdout set of 40 patients was utilized as the testing set. Software code was written in Python using the TensorFlow module on a Linux workstation with one NVidia Titan X GPU. Seventy-four luminal A, 106 luminal B, 13 HER2+, and 23 basal breast tumors were evaluated. Testing set accuracy was measured at 70%. The class normalized macro area under receiver operating curve (ROC) was measured at 0.853. Non-normalized micro-aggregated AUC was measured at 0.871, representing improved discriminatory power for the highly represented Luminal A and Luminal B subtypes. Aggregate sensitivity and specificity was measured at 0.603 and 0.958. MRI analysis of breast cancers utilizing a novel CNN can predict the molecular subtype of breast cancers. Larger data sets will likely improve our model.
Keywords: Breast MRI, Molecular subtype, CNN
Introduction
Breast cancer is the most common cancer afflicting women worldwide and the second most common cause of cancer deaths among women in the USA [1]. These tumors are highly heterogeneous with multiple identified subtypes demonstrating a wide spectrum of intrinsic biologic diversity with associated biologic response, prognosis, and therefore variable treatment regimens [2].
Mammography is the gold standard for breast cancer screening as it is the most cost-effective imaging modality; however, magnetic resonance imaging (MRI) has gained popularity in recent years as the most sensitive imaging technique, excelling in diagnosis, preoperative planning, and prognostication of breast cancers [3–5].
Given disease heterogeneity, tissue sampling is the gold standard with immunohistochemistry (IHC) used as surrogate genetic testing to determine breast cancer subtype. Based on gene expression, cancer cells express various receptors, such as estrogen receptor (ER), progesterone receptor (PR), and the human epidermal growth factor receptor (HER2) [2]. Mainly, four intrinsic breast cancer subtypes have been described: luminal A (hormone receptor positive, HER2 negative), luminal B (hormone receptor positive, HER2 positive), HER2-enriched type (hormone receptor negative, HER2 positive), and triple-negative subtype (hormone receptor negative, HER2 negative) [2, 6, 7].
Despite advantages with the use of IHC surrogates, the range of agreement between its use in predicting breast cancer subtype and explicit genetic testing is between 41 and 100% [8]. Given the wide spectrum of prognosis and indicated treatment strategies based on tumor subtype, a need exists for more accurate diagnosis to aid in an individualized treatment plan [9–11].
Due to rapid advancements in quantitative radiology methods (i.e., radiomics), tumor biology and genetics can be evaluated in a more precise, predictive, and cost-effective way. Quantitative radiomics extracts data from routine medical imaging and analyzes high fidelity complex imaging features, unperceivable to the human eye [12]. Radiogenomics is the process of linking the radiomics to the hidden genotypic configuration of a tumor or tissue [13]. In simple terms, radiogenomics of breast cancer using MRI works on the principle of analyzing various intrinsic features including dynamic contrast enhancement (DCE) kinetics, which often define tumor heterogeneity, to predict molecular subtype [14–17].
Advances in computer technology have brought machine learning to the forefront of undertaking complex clinical challenges. Application of initial machine learning techniques to evaluate radiogenomics of breast MRI has shown promising results [18–26]. However, the currently available published literature is based on data using human extracted imaging phenotypic features to guide machine learning and subsequently predicting molecular subtypes using semi-automated methods.
Recently, a subset of machine learning named Convolutional Neural Networks (CNN) has made great strides in medical imaging analysis. As oppose to traditional machine learning, which primarily relies on human extracted feature analysis, neural networks depend on the input of raw data and allow the computer to automatically construct predictive statistical models through increasingly complex layers and self-optimization [27].
The purpose of this study is to develop a CNN algorithm to predict various molecular subtypes of breast cancer using a breast MRI tumor dataset.
Materials and Methods
An Institutional Review Board approved retrospective study was performed. Two hundred sixteen patients with known breast cancer diagnosis who underwent preoperative MRI prior to any treatment and who had available IHC staining pathology data were identified for this study. Subtypes were classified by IHC staining surrogates as luminal A (ER and/or PR+, HER2-), luminal B (ER and/or PR+, HER2+), HER2 (ER and PR-, HER2+), or basal (ER-, PR-, HER2-) (30–32). Tumors were considered HER-2 positive only if scored 3+ by IHC or if HER-2 amplification yielded a ratio ≥ 2.0 on the basis of fluorescence in situ hybridization (FISH). [6, 28–30].
MRI Acquisition
MRI was performed on a 1.5-T (121 cases, 56%) or 3.0-T (95 cases, 44%) commercially available system (Signa Excite, GE Healthcare) using an eight-channel breast array coil. The imaging sequence included a triplane localizing sequence followed by a sagittal fat-suppressed T2-weighted sequence (TR/TE, 4000–7000/85; section thickness, 3 mm; matrix, 256 × 192; FOV, 18–22 cm; no gap). A bilateral sagittal T1-weighted fat-suppressed fast spoiled gradient-echo sequence (17/2.4; flip angle, 35°; bandwidth, 31–25 Hz) was then performed post rapid bolus injection (gadobenate dimeglumine/Multihance; Bracco Imaging; 0.1 mmol/kg) delivered through an IV catheter. Image acquisition started after contrast material injection and was obtained consecutively with each acquisition time of 120 s. Section thickness was 2–3 mm using a matrix of 256 × 192 and a field of view of 18–22 cm. Frequency was in the anteroposterior direction. After the examination, post-processing was performed including subtraction of the unenhanced images from the first contrast-enhanced images on a pixel-by-pixel basis and reformation of sagittal images to axial images.
Data Annotation
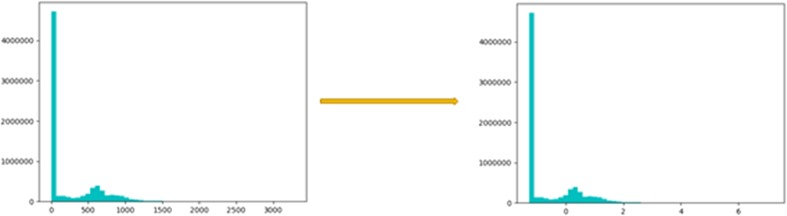
After the breast MRIs were obtained, a fellowship trained breast imaging radiologist subsequently reviewed the MRI images. Anonymized first post-contrast MRI DICOM images were downloaded to a password protected external hard-drive and loaded into 3D Slicer 4.0 (www.slicer.org), an open-source software platform for medical image informatics and analysis. Binary three-dimensional segmentations were applied to the input images. The molecular subtype of each patient’s breast cancer as obtained from the EMR was recorded as the ground truth class label. Histogram normalization of the MRI data was performed to center the non-air pixels around 0 with unit standard deviation (Fig. 1).
Fig. 1.
Results of MRI normalization: Histogram normalization of the MRI data was performed to center the non-air pixels around 0 with unit standard deviation
Image Processing
Data augmentation employed by this study involved several real-time modifications to the source images at the time of training. These modifications included random affine transformation of the original image, which alters each mass slightly utilizing a rigid transformation effectively making the same mass appear as a unique input to the network. Given a three-dimensional affine matrix, random affine warping was performed by utilizing random rotation by ± 30°, 90°, and 90° across the Z, Y, and X axes respectively. Additionally, a random shear value of 0.1 was applied to each axis. These parameters were confirmed on visual inspection as applying enough of a warp to simulate a different lesion without making the lesion appear unrealistic (Fig. 2). The choice to apply data augmentation to 50% of the example images was made to prevent inducing bias of the network towards recognition of augmented data over real data. Additional augmentation included addition of a random Gaussian noise matrix, random contrast jittering, and random brightness. The network was thus allowed to learn to marginalize random noise introduced by minor warps in the input volume as well as slight differences in acquisition parameters. Network inputs consisted of 32 × 32 pixel bounding boxes containing the size normalized lesions.
Fig. 2.
Results of data augmentation: Images of a single input example nodule with multiple random affine warps applied for data augmentation. This alters each mass slightly utilizing a rigid transformation effectively making the same mass appear as a unique input to the network. Affine parameters were confirmed on visual inspection as applying enough of a warp to simulate a different lesion without making the lesion appear unrealistic
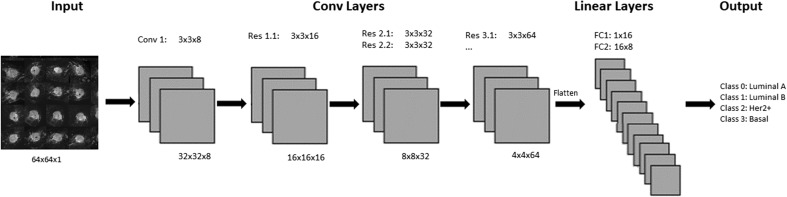
CNN Architecture
The CNN was implemented by a series of 3 × 3 convolutional kernels to maximize computational efficiency while preserving nonlinearity [31] (Fig. 3). After an initial standard convolutional layer, a series of residual layers are utilized in the network. Originally described by He et al. [32], residual neural networks can stabilize gradients during back propagation, leading to improved optimization and facilitating greater network depth. Downsampling of feature map size was implemented by means of a concatenated average and max pooling operation to decrease size by 75%. All nonlinear functions utilize the rectified linear unit (ReLU) which allows training of deep neural networks by stabilizing gradients on backpropagation [33]. Additionally, batch normalization was used between the convolutional and ReLU layers to enhance network training by stabilizing the loss landscape [34]. Upon downsampling, the number of feature channels is doubled, preventing a representation bottleneck. Dropout with a keep probability of 50% was applied to the first fully connected layer to limit overfitting and add stochasticity to the training process [35].
Fig. 3.
Network architecture: Dimensions of all of the intermediate layers of the convolutional neural network. The top row of numbers contains filter types and sizes utilized. The bottom row displays the size of the input feature maps. Each successive decrease in feature map size is accomplished by applying a concatenated average and max pooling operation as the first layer in each residual operation. “Conv” filter types correspond to a standard convolutional layer followed by batch normalization and ReLu nonlinearity. “Res” filter types correspond to residual layers with two embedded convolutional operations followed by batch normalization and ReLu nonlinearity
In addition to the customized network described above (Table 1), several additional network architectures were tested. This includes the following: (1) a ResNet 52 network architecture, initialized both randomly and with pre-trained weights from Imagenet; (2) custom built networks, initialized from random weights, and with varying numbers of convolutional layers based on the Inception v4 architecture; (3) 100 layer network based on a randomly initialized Dense Net architecture. Performance for the networks was best when initializing weights randomly across the board. Additionally, three-dimensional networks were tested by alternating between using inception style layers, residual style layers, and hybrid wide residual layers. We found for this project than when using greater than 14 hidden layers in two-dimensional residual networks, or greater than 8 3D inception style layers, overfitting occurred. Additionally, three-dimensional networks universally suffered from overfitting even with as low as 4 hidden layers, likely due to the small training set size.
Table 1.
The Network architecture: Dimensions of all of the intermediate layers of the convolutional neural network. The first column contains the input layer names. The second column displays the size of the input feature map. The middle column describes the type of filter applied followed by a column describing the filter size if applicable. The final column displays the name of the output layer, which serves as the input for the next layer. Residual layers contain two feature maps per layer
| Input layer | Input layer dimensions | Filter type | Filter size | Output layer |
|---|---|---|---|---|
| Input | 64 × 64 × 3 | Convolutional | 3 × 3 × 8 | Hidden layer 1 |
| Hidden layer 1 | 32 × 32 × 8 | Residual | 3 × 3 × 16 | Hidden layer 2/3 |
| Hidden layer 2/3 | 16 × 16 × 16 | Residual | 3 × 3 × 32 | Hidden layer 4/5 |
| Hidden layer 4/5 | 8 × 8 × 32 | Residual | 3 × 3 × 32 | Hidden layer 5/6 |
| Hidden layer 5/6 | 8 × 8 × 32 | Residual | 3 × 3 × 64 | Hidden layer 6/7 |
| Hidden layer 6/7 | 4 × 4 × 64 | Residual | 3 × 3 × 64 | Hidden layer 8/9 |
| Hidden layer 8/9 | 4 × 4 × 64 | Residual | 3 × 3 × 64 | Hidden layer 10/11 |
| Hidden layer 10/11 | 4 × 4 × 64 | Linear | × 16 | Hidden layer 12 |
| Hidden layer 12 | 1 × 16 | Linear | 16 × 8 | Hidden layer 13 |
| Hidden layer 13 | 1 × 8 | Softmax | 8 × 4 | Classification |
Training was implemented using the parameterized Adam optimizer, combined with the Nesterov accelerated gradient described by Dozat [36–38]. Parameters were initialized to equalize input and output variance utilizing the heuristic described by Glorot et al. [39]. L2 regularization was implemented to prevent overfitting of data by limiting the squared magnitude of the kernel weights. Final hyperparameter settings included a learning rate set to 1e-3, keep probability for dropout of 50%, moving average weight decay of 0.999, and L2 regularization weighting of 1e-4. The class imbalance was addressed by doubling the input of underrepresented classes and utilizing a class sensitive cost function. Parameters were tuned based on a 20% validation group. A class balanced holdout set of 40 patients was utilized as the testing set. Software code for this study was written in Python using the TensorFlow module (1.4). Experiments and CNN training were performed on a Linux workstation with NVIDIA Titan X Pascal GPU with 12 GB on chip memory, i7 CPU and 32 GB RAM.
Results
Subtypes were classified by IHC staining surrogates as luminal A (ER and/or PR+, HER2-), luminal B (ER and/or PR+, HER2+), HER2 (ER and PR-, HER2+), or basal (ER -, PR -, HER2-) (30–32). Using this classification, 74 Luminal A, 106 Luminal B, 13 HER2+, and 23 basal breast tumors were evaluated.
Testing set accuracy was measured at 70%. The class normalized macro area under receiver operating curve (ROC) was measured at 0.853. Non-normalized micro-aggregated AUC was measured at 0.871, representing improved discriminatory power for the highly represented Luminal A and Luminal B subtypes. Aggregate sensitivity and specificity was measured at 0.603 and 0.958.
Discussion
The CNN algorithm used in our study achieved an overall accuracy of 70% in predicting breast cancer subtype. This feasibility study applied radiogenomics using a novel CNN algorithm to predict tumor subtype based on a breast MRI data set. Future advances with a larger data set will likely improve the predictive strength of our model, with the potential to rival accuracy of standard genetic testing in determining breast cancer subtype.
Preoperative breast MRI has become increasingly prevalent as its role in diagnosis and treatment planning of breast cancer expands [3]. With the increasing ubiquity of breast MRI, several prior studies have applied radiogenomics to predict subtype using semi-automated techniques. Agner et al. demarcated triple-negative cancers from other molecular subtypes on DCE-MRI using a computer-aided diagnosis (CAD) system [20]. They retrospectively studied 76 breast lesions using quantitative feature extraction with a feed forward feature selection and linear discriminate analysis. Triple-negative tumors were found to be more heterogeneous in both texture and enhancement, with a higher degree of tumor tissue compactness. The area under the receiver operating characteristic (ROC) curve for subtype determination was 0.73 (95% CI 0.59, 0.87) in triple-negative versus non-triple-negative subtype, 0.74 (95% CI 0.60, 0.88) in triple-negative versus ER- and HER2-positive subtype, 0.77 (95 CI 0.63, 0.91) in triple-negative versus ER-positive subtype, and 0.74 (95% CI 0.58, 0.89) for triple-negative versus HER2-positive subtype. While this model improves on discriminating aggressive triple-negative breast cancer subtype, it relies on human MRI feature extraction.
Tumor enhancement dynamics association with luminal type B molecular subtype was explored by Mazurowski et al. [21]. Forty-eight patients with breast cancer were collected from the Cancer Genome Atlas and Cancer Imaging Archive. Twenty-three imaging features were extracted using computer vision algorithms after initial lesion delineation by a trained breast radiologist. Luminal B subtypes were found to have higher lesion enhancement to background parenchymal enhancement ratio (P = 0.0015).
Similarly, computational MR imaging features of luminal type A and B molecular subtypes were retrospectively evaluated using semi-automatically extracted imaging features by Grimm et al. [22]. Initial annotations were made by fellowship trained breast imagers and subsequently 56 features were extracted using computer vision algorithms. Tumor to fibroglandular tissue and the peak enhancement were shown to be main factors in discriminating subtype, with multivariate models able to predict luminal A (P = 0.0007) and luminal B (P = 0.0063), but not HER2 positive (P = 0.2465) or basal subtype (P = 0.1014).
In a retrospective review of 60 breast cancers, Fan et al. studied 90 features derived from DCE-MRI and selected an optimal set of 24 features for subtype classification using trained multi-class logistic regression classifier computer software [17]. The prediction model demonstrated high accuracy in overall classification (AUC = 0.869) and in discriminating between luminal A, luminal B, HER2, and triple-negative subtypes (AUC = 0.867, 0.786, 0.888, and 0.923 respectively). Although significant strides have been made in the improvement of breast cancer subtype prediction based on MRI features, these semi-automated techniques rely on specific human extracted feature analysis.
MRI contrast enhancement patterns have shown promise in determining breast cancer subtype. A retrospective study including 186 patients evaluated the distribution pattern of kinetic parameters of DCE-MRI across breast cancer molecular subtypes. No significant difference in delayed phase kinetics was identified. However, a significantly decreased percentage of washout pattern was observed in ER/PR positive/HER2 negative and triple-negative cancers [25]. Blaschke et al. retrospectively reviewed 112 patients with newly diagnosed invasive ductal carcinoma who underwent DCE-MRI [26]. Kinetic analyses showed significantly increased contrast uptake in HER2-positive molecular subtype, with > 100% uptake at early phase in HER2-positive versus luminal A/B (93.8 ± 0.92 vs. 77.3 ± 7.2; P < 0.01) and HER2-positive versus triple-negative (93.8 ± 0.92 vs. 81.3 ± 8.2; P < 0.05). While significant patterns in kinetic analyses of breast tumors have demonstrated predictive strength, these methods continue to rely on limited human extracted MRI features.
Although the previously mentioned studies have shown promising results in breast MRI data analysis, these methods are dependent on feature engineering using semi-automated feature extraction. Feature engineering works by implementing domain knowledge to build feature extractors, which simplify the complex data and create more comprehensible patterns to be applied to algorithms. These methods are limited in their function, as they are dependent on accurate human extraction of crucial features.
In contrast, CNN algorithms are trained in a manner that allows automatic extraction of features from the input feed that are crucial to the defined problem domain. This process improves its ability to study the input features in an end-to-end manner, using complex, stacked layers to predict a desired output. Therefore, CNN feature extraction is not a variable with each new MRI and thus results are consistent.
To date, only one other study utilized CNN to classify molecular subtypes using an MRI data set [40]. Study by Zhu et al. used VGGNet pre-trained on ImageNet. They used the feature maps of several convolution layers and fully connected layers, by training supervised machine learning algorithm on those feature maps. Their evaluation was limited to classifying one molecular subtype (Luminal A) versus the rest (grouped into one) yielding AUC of 0.64. Our study evaluated four molecular subtype classification yielding higher AUC of 0.85.
There are a few limitations of our study. This is a feasibility study from a single institution, with a relatively small sample size. It has been shown that the performance of a CNN increases logarithmically with increasing data sets [41]. In addition, patients in this study underwent MRI imaging at different magnetic field strengths (1.5 or 3.0 T), potentially affecting the image quality. However, selection bias is likely negligible given that the choice of patients undergoing MRI on a 1.5 or 3.0 T magnet was randomly determined purely based on availability of the scanner. Lack of external validation poses another limitation, which we plan to overcome in our future projects using external validation datasets as well as prospective validation studies. Lastly, defining various molecular subtypes using genetic analysis is an economically and technically challenging process. Alternative means using IHC as a surrogate is widely used including in our study. However, the range of agreement between predicting these subtypes using IHC and standard genetic testing can be variable and ranges between 41 and 100% [42]. Thus, this is a potential limitation given that our study used IHC surrogate to define molecular subtype instead of genetic analysis.
In conclusion, MRI analysis of breast cancers utilizing a novel CNN can predict the molecular subtype of breast cancers. Larger data sets will likely improve our model. Personalized breast cancer risk stratification may be possible with a CNN plus MRI data instead of gene expression profiling.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that there is no conflict of interest.
Footnotes
Work originated from Columbia University Medical Center.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Richard Ha, Phone: 212-305-1948, Email: rh2616@columbia.edu.
Peter Chang, Phone: 714.509.2523, Email: changp6@uci.edu.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Morris EA. Diagnostic breast MR imaging: current status and future directions. Magn Reson Imaging Clin N Am. 2010;18:57–74. doi: 10.1016/j.mric.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Liberman L, Morris EA, Dershaw DD, et al. MR imaging of the ipsilateral breast in women with percutaneously proven breast cancer. AJR Am J Roentgenol. 2003;180(4):901–910. doi: 10.2214/ajr.180.4.1800901. [DOI] [PubMed] [Google Scholar]
- 5.Schelfout K, Van Goethem M, Kersschot E, et al. Contrast-enhanced MR imaging of breast lesions and effect on treatment. Eur J Surg Oncol. 2004;30(5):501–507. doi: 10.1016/j.ejso.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiechmann L, Sampson M, Stempel M, et al. Presenting features of breast cancer differ by molecular subtype. Ann Surg Oncol. 2009;16(10):2705–2710. doi: 10.1245/s10434-009-0606-2. [DOI] [PubMed] [Google Scholar]
- 8.Morrow M, Waters J, Morris E. MRI for breast cancer screening, diagnosis, and treatment. Lancet. 2011;378:1804–1811. doi: 10.1016/S0140-6736(11)61350-0. [DOI] [PubMed] [Google Scholar]
- 9.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22(8):1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzger-Filho O, Sun Z, Viale G, et al. Patterns of recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international Breast Cancer Study Group Trials VIII and IX. J Clin Oncol. 2013;31(25):3083–3090. doi: 10.1200/JCO.2012.46.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 12.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo MD, Jamshidi N. Behind the numbers: Decoding molecular phenotypes with radiogenomics—guiding principles and technical considerations. Radiology. 2014;270(2):320–325. doi: 10.1148/radiol.13132195. [DOI] [PubMed] [Google Scholar]
- 14.Holli-Helenius K, Salminen A, Rinta-Kiikka I, et al. MRI texture analysis in differentiating luminal A and luminal B breast cancer molecular subtypes - a feasibility study. BMC Med Imaging. 2017;17(1):69. doi: 10.1186/s12880-017-0239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, Giger ML, Lan L, Bick U. Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Med Phys. 2004;31:1076–1108. doi: 10.1118/1.1695652. [DOI] [PubMed] [Google Scholar]
- 16.Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging (Bellingham) 2015;2:041007. doi: 10.1117/1.JMI.2.4.041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan M, Li H, Wang S, et al. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS One. 2017;12(2):e0171683. doi: 10.1371/journal.pone.0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhooshan N, Giger ML, Jansen SA, et al. Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology. 2010;254(3):680–690. doi: 10.1148/radiol.09090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhooshan N, Giger M, Edwards D, et al. Computerized three-class classification of MRI-based prognostic markers for breast cancer. Phys Med Biol. 2011;56(18):5995–6008. doi: 10.1088/0031-9155/56/18/014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agner SC, Rosen MA, Englander S, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology. 2014;272:91–99. doi: 10.1148/radiol.14121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazurowski MA, Zhang J, Grimm LJ, et al. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- 22.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging. 2015;42(4):902–907. doi: 10.1002/jmri.24879. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto S, Han W, Kim Y, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275(2):384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 24.Ashraf AB, Daye D, Gavenonis S, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology. 2014;272(2):374–384. doi: 10.1148/radiol.14131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi K, Abe H, Newstread G, et al. Intratumoral heterogeneity of the distribution of kinetic parameters in breast cancer: comparison based on the molecular subtypes of invasive breast cancer. Breast Cancer. 2015;22(5):496–502. doi: 10.1007/s12282-013-0512-0. [DOI] [PubMed] [Google Scholar]
- 26.Blaschke E, Abe H. MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J Magn Reson Imaging. 2015;42(4):920–924. doi: 10.1002/jmri.24884. [DOI] [PubMed] [Google Scholar]
- 27.LeChun Y, Bengio T, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 28.Ha R, Jin B, Mango V, et al. Breast cancer molecular subtype as a predictor of the utility of preoperative MRI. AJR Am J Roentgenol. 2015;204(6):1354–1360. doi: 10.2214/AJR.14.13666. [DOI] [PubMed] [Google Scholar]
- 29.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen PL, Taghian AG, Katz MS, et al. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol. 2008;26(14):2373–2828. doi: 10.1200/JCO.2007.14.4287. [DOI] [PubMed] [Google Scholar]
- 31.LeCun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proceed IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 32.He K, Zhang X, Ren S, et al: Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp 770–778
- 33.Nair, V, Hinton GE: Rectified linear units improve restricted boltzmann machines. In: Proceedings of the 27th international conference on machine learning (ICML-10), 2010, pp 807–814
- 34.Ioffe S, Szegedy C: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning, 2015
- 35.Srivastava N, Hinton G, Krizhevsky A, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(1):1929–1958. [Google Scholar]
- 36.Kingma DP, Jimmy BA: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014
- 37.Nesterov Y: Gradient methods for minimizing composite objective function. 2007
- 38.Dozat T: Incorporating nesterov momentum into adam. 2016
- 39.Glorot X, Bengio Y: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, 2010, pp 249–256
- 40.Zhu Z, Albadawy E, Saha A, et al: Breast cancer molecular subtype classification using deep features: preliminary results. In: Proceedings Volume 10575, Medical imaging 2018: computer-aided diagnosis; 105752X. 2018
- 41.Sun C, Shrivastaval A, Singh S, et al: Revisiting unreasonable effectiveness of data in deep learning Era. arXiv preprint arXIV:1707.02968. 2017
- 42.Guiu S, Michiels S, André F, et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol. 2012;23(12):2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]