Abstract
Introduction
Vitamin D (VD) deficiency has been associated with multiple sclerosis (MS) and other autoimmune diseases (AIDs). However, the effect of the genetics of VD on the risk of MS is subject to debate. This study focuses on genes linked to the VD signaling pathway in families with MS. The evaluation of gene variants in all the members of families could contribute to an additional knowledge on the information obtained from case‐control studies that use nonrelated healthy people.
Material and Methods
We studied 94 individuals from 15 families including at least two patients with MS. We performed whole‐exome next generation sequencing on all individuals and analyzed variants of the DHCR7, CYP2R1, CYP3A4, CYP27A1, GC, CYP27B1, LRP2, CUBN, DAB2, FCGR, RXR, VDR, CYP24A1, and PDIA3 genes. We also studied PTH, FGF23, METTL1, METTL21B, and the role of the linkage disequilibrium block on the long arm of chromosome 12, through analysis of the CDK4, TSFM, AGAP2, and AVIL genes. We compared patients with MS, other AIDs and unaffected members from different family types.
Results
The study described the variants in the VD signaling pathway that appear in families with at least two patients with MS. Some infrequent variants were detected in these families, but no significant difference was observed between patients with MS and/or other AIDs and unaffected family members in the frequency of these variants. Variants previously associated with MS in the literature were not observed in these families or were distributed similarly in patients and unaffected family members.
Conclusion
The study of genes involved in the VD signaling pathway in families that include more than one patient with MS did not identify any variants that could explain the presence of the disease, suggesting that VD metabolism could probably play a role in MS more as an environmental factor rather than as a genetic factor. Our study also supports the analysis of cases and unaffected individuals within families in order to determine the influence of genetic factors.
Keywords: cubilin, CYP24A1, familial multiple sclerosis, megalin, PDIA3, VDR, vitamin D, whole‐exome sequencing
Abbreviations
- 1,25(OH)2D
1,25‐dihydroxyvitamin D
- 1,25(OH)2D3
1,25‐dihydroxyvitamin D3
- 7‐DHC
7 dehydrocholesterol
- 25(OH)D
25‐hydroxyvitamin D3
- AID
autoimmune disease
- CCDS
consensus coding sequence
- CNS
central nervous system
- DBP
vitamin D binding protein
- FGF23
phosphaturic factor fibroblast growth factor 23
- GWAS
genome‐wide association study
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MS
multiple sclerosis
- PDIA3
Protein disulfide isomerase family member 3
- PPMS
primary‐progressive multiple sclerosis
- PTH
parathyroid hormone
- RRMS
relapsing‐remitting multiple sclerosis
- RXRA
retinoid X receptor alpha
- SPMS
secondary‐progressive multiple sclerosis
- VD
vitamin D
- VDR
vitamin D receptor
- WES
whole‐exome next generation sequencing
1. INTRODUCTION
Multiple sclerosis (MS) is an inflammatory autoimmune disease of the central nervous system (CNS) that causes demyelination and axonal damage. MS etiology involves multiple factors, with environmental factors interacting with genetic predisposition. Although genome‐wide association studies (GWAS) have validated the central role of the major histocompatibility complex in the genetics of MS, susceptibility has been found to be influenced by several genes outside the short arm of chromosome 6 (Baranzini & Oksenberg, 2017; Cree, 2014; Sawcer et al., 2011). The disease is therefore considered to be polygenic. However, the variants identified through GWAS have little effect on the overall risk of MS; as a result, there is a considerable need for further research into genetic factors in MS. New techniques such as whole‐exome sequencing (WES) offer a comprehensive view of the coding region of the genome, allowing us to study complex diseases like MS with more detail.
Vitamin D (VD) is a fat‐soluble hormone which plays an essential role in calcium homeostasis and in skeletal development and maintenance. It is also thought to play a role in cancer, immune function, and autoimmune diseases (AIDs), including MS (Ascherio et al., 2014; Gianfrancesco et al., 2017; Løken‐Amsrud et al., 2012; Mowry et al., 2012; Munger & Ascherio, 2011; Munger, Levin, Hollis, Howard, & Ascherio, 2006; Salzer et al., 2012; Simpson et al., 2010; Wang, Zeng, Wang, & Guo, 2018). There are several types of VD, including ergocalciferol (D2) and cholecalciferol (D3), formed from their respective previtamins, ergosterol, and 7‐dehydrocholesterol (7‐DHC). The main natural source of VD3 in humans is production in the skin, where 7‐DHC undergoes a 2‐step reaction involving ultraviolet‐B irradiation to form pre‐VD3, followed by thermal isomerization, forming VD3. Both VD2 and VD3 can also be obtained in small amounts from a varied diet, and in larger amounts from specific foods or nutritional supplements. Dietary VD is mainly absorbed in the small intestine by means of chylomicrons, which enter the lymphatic system and drain into the superior vena cava. After entering the bloodstream, VD (obtained either through intestinal absorption or synthesis in the skin) is converted into 25‐hydroxyvitamin D (25[OH]D) in the liver, and then into 1,25‐dihydroxyvitamin D (1,25[OH]2D). Both compounds are mainly transported by vitamin D‐binding protein (DBP, encoded by the GC gene), although a small fraction circulates freely or bound to albumin. Internalization is mainly mediated by the DBP‐megalin (LRP2) complex. In some cells, however, internalization may take place through binding to other proteins or by passive diffusion of free molecules (Figure 1). It is subsequently converted to 25‐hydroxyvitamin D3 (25[OH]D3, calcifediol) by cytochrome P450 2R1 (or vitamin D 25‐hydroxylase, encoded by CYP2R1). Another cytochrome P450 enzyme, 1α‐hydroxylase (CYP27B1), converts this to the active form, 1,25‐dihydroxyvitamin D3 (1,25[OH]2D3, calcitriol). This step appears to be regulated by parathyroid hormone (PTH) and phosphaturic factor fibroblast growth factor 23 (FGF23) (Christakos, 2017). It has also been suggested that this may in turn be influenced by intronic variants located in the nearby genes METTL1 and METTL21B (methyltransferase‐like proteins). CYP24A1 may convert 25(OH)D3 into the inactive metabolite 24,25(OH)2D3, or even convert the active form of the vitamin into an inactive form, 1,24,25(OH)2D3. 1,25[OH]2D3 binds to the nuclear vitamin D receptor (VDR), interacting with retinoid X receptor alpha (RXRA) to form a heterodimer, enabling transport into the cell nucleus (Matías‐Guiu, Oreja‐Guevara, Matías‐Guiu, & Gomez‐Pinedo, 2018) (Figure 1). 1,25 [OH] 2D3 can also bind to the membrane receptor PDIA3 (ERp57 or 1,25 D3‐MARRS) that could be responsible for the actions of nongenomic mechanisms of VD.
Figure 1.
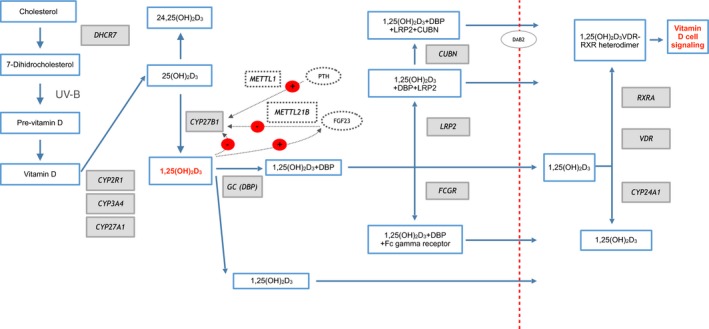
Vitamin D pathway. 7‐DHC follows a two‐stage reaction involving ultraviolet‐B irradiation and thermal isomerization. Vitamin D is converted into 25‐hydroxyvitamin D3 (25[OH]D3) by a hydroxylation mechanism mediated by vitamin D 25 hydroxylase. Subsequently, 1α‐hydroxylase, regulated by PTH and FGF23, transforms 25(OH)D3 into its active form, 1,25‐dihydroxyvitamin D3 (1,25[OH]2D3). The proteins 25(OH)D3 and 1,25 (OH)2D3 mainly circulate bound to carrier proteins (DBP), but a small fraction circulates freely or bound to albumin. The internalization mechanism is produced mainly by DBP‐megalin (LRP2). However, in some cells this process can occur by binding to other proteins and even by passive diffusion. Finally, 1,25(OH)2D3binds to the VDR nuclear receptor, interacting with the RXRA, forming a heterodimer that allows transport to the cell nucleus. Genes involved in the vitamin D pathway are shown in grey. The regulatory pathway of the gene CYP27B1 (METTL1, METTL21B, FGF23, PTH) is represented with a dotted line. Endocytosis of the complex formed by LRP2 and CUBN requires DAB2, represented in the figure within a light grey circle. The mechanism of VD binding to the PDIA3 receptor is not included in the figure
Low‐frequency genetic variants in VD signaling pathway have been linked to MS; however, their role in disease pathogenesis is controversial. We are yet to determine their influence over familial forms of MS, whether the association is due to variations in a gene, or whether the presence of variants at different points in the pathway may be associated with the disease.
The aim of the present study is to analyze the genetic variations of genes related with the VD signaling pathway in families from familial forms of MS. By studying families, genetic factors may be more easily observable and less obscured by environmental factors than in sporadic cases.
2. MATERIAL AND METHODS
2.1. Study population
We studied 15 families with at least two patients with MS according to the McDonald criteria (Polman et al., 2011), in total 94 individuals. The study protocol included a specifically designed questionnaire to gather information about patients’ personal and family histories of neurological, systemic, and autoimmune conditions. A modified version of the list of diseases created by the American Autoimmune Related Diseases Association (American Autoimmune Related Diseases Association), retrieved from http://www.aarda.org/autoimmune-information/list-of-diseases/) (February, 2016), was used to define the history of AIDs. Cases with no history of MS or other AIDs were considered unaffected members, and data was obtained through direct interrogation. We also recorded demographic and clinical variables, sex, age at onset, time since symptom onset, and clinical form (relapsing‐remitting MS [RRMS], primary‐progressive MS [PPMS], or secondary‐progressive MS [SPMS]). We gathered information on family members by interviewing them directly; families were classified according to the types of family (A or B) defined in a previous study (Pytel et al., 2017): Type A, in which all members with MS belonged to the same generation; and type B, in which MS patients were distributed between two or more generations. The software Genial Pedigree Draw (http://app.pedigreedraw.com) was used to generate pedigrees for each family included. We also analyzed the duration of daylight for the habitual place of residence for each individual studied, in order to evaluate the impact of sunlight exposure on the risk of MS; the Spanish National Geographic Institute (www.ign.es) was used as the source for this analysis. Finally, we performed WES studies of peripheral blood samples from the 94 participants.
2.2. Whole‐exome sequencing
DNA was extracted from blood samples using the MagNA Pure automated nucleic acid purification system (Roche Molecular Systems, Inc.). The Qubit™ 2.0 and NanoDrop devices (Thermo Fisher Scientific Inc.) were used to determine DNA concentration and purity. The AmpliSeq™ Exome panel (Thermo Fisher Scientific Inc.) was used for library preparation. This technique captures >97% of consensus coding sequences (>19,000 genes, >198,000 exons, >85% of alterations responsible for genetic diseases) and adjacent splicing regions (5 bp). The panel is approximately 33 Mb in size and comprises a total of 293,903 amplicons. Libraries were quantified by qPCR and subsequently prepared and enriched using the Ion Chef™ system providing a high uniformity of coverage.
Library sequencing at a mean depth of coverage of >100 reads was performed using the Ion Proton (Thermo Fisher Scientific Inc.) sequencing platform, covering >90% of amplicons with at least 20 reads. The sequences obtained were aligned against the reference genome (Genome Reference Consortium human genome 19, build 37) using the Torrent Mapping Alignment Program software. The sequences, aligned and filtered according to specific quality criteria, were analyzed with the Torrent Variant Caller tool to identify nucleotide variations with respect to the reference genome. Variant annotation was performed using the latest available version of Ion Reporter™ (Thermo Fisher Scientific Inc.). Our analysis aimed to identify SNPs and indels located in the exons and splicing junctions of the genes and causing protein‐level modifications (excluding synonymous variants), which were detected in over 40% of reads.
2.3. Variant prioritization
Observing quality controls and bioinformatic filtering, we analyzed the coding and splicing regions of genes involved in the VD signaling pathway. We evaluated the list of variants identified against database information on previously described variants (http://www.ncbi.nlm.nih.gov/SNP/, http://www.1000genomes.org, http://gnomad.broadinstitute.org, and http://evs.gs.washington.edu/EVS). Firstly, we excluded variants located in introns, intergenic regions, and untranslated regions. We also removed synonymous variants; according to a polygenic pattern, we considered variants with a minor allele frequency (MAF) below 5% (http://gnomad.broadinstitute.org/). In order to understand the possible biological functions of the variants selected, we estimated the functional effect of the genomic variations classified as pathogenic using seven prediction algorithms (SIFT, PROVEAN, PolyPhen2, Mutation Taster, Mutation Assessor, LRT, and FATHMM) included in the ALAMUT (http://www.interactive-biosoftware.com) and ANNOVAR (http://www.openbioinformatics.org/annovar/) analysis packages. Finally, we reviewed candidate genes in publications on PubMed and the Online Mendelian Inheritance in Man database.
2.4. Analysis of vitamin D signaling pathways
In order to analyze the exonic variants detected, we defined that VD signaling pathway includes the following genes: DHCR7, CYP2R1, CYP3A4, CYP27A1, GC, LRP2, CUBN, FCGR, DAB2, PTH, FGF23, METTL1, METTL21B CYP27B1, RXRA, CYP24A1, VDR and PDIA3 (Kamisli et al., 2018; Tajouri et al., 2005). Given the existence of a linkage disequilibrium (LD) block on the long arm of chromosome 12, centered on the CYP27B1, METTL1, and METTL21B genes, we also analyzed some genes in this area that may be related to autoimmunity (CDK4, TSFM, AGAP2, AVIL, CYP27B1, METTL1, and METT21B).
2.5. Sequencing data analysis
Sequencing results were evaluated using three analysis models:
Comparison of the variants detected in the overall cohort of family members, analysing the groups of individuals with MS, other AIDs, and, unaffected family members, taking into account the type of familial MS.
Prioritization of variants.
Analysis of pedigrees, assessing the role of the variants identified in each pedigree.
2.6. Statistical analysis
Descriptive analysis results are expressed as absolute frequencies and percentages (n [%]), means ± standard deviation (SD), or medians (interquartile range). The Kolmogorov‐Smirnov test was used to test quantitative data for normal distribution. The chi‐square test was used to compare independent samples with qualitative variables; the Mann‐Whitney U test was used for quantitative variables. Intergroup differences were evaluated with the Kruskal‐Wallis H test and the Dunn post hoc test. More powerful quasi‐likelihood score test (MQLS) was used, which allows testing for case‐control associations in samples with related individuals (Thornton & McPeek, 2007; Thornton, Zhang, Cai, Ober, & McPeek, 2012). Allele frequencies were tested to identify deviations from Hardy‐Weinberg equilibrium. Bonferroni method was used to correct for multiple comparisons. Statistical significance was set at p < 0.05.
3. RESULTS
3.1. Description of the families, population, and daylight exposition
We defined two types of family: type A and type B. The sample included seven type‐A families (44 individuals, 46.8%) and eight type‐B families (50; 53.2%). Fifteen patients with MS (42.8%) belonged to type‐A families and 20 (57.14%) belonged to type‐B families. Seven individuals with other AIDs (53.8%) and 22 unaffected individuals (47.8%) belonged to type‐A families and six individuals with other AIDs (46.1%) and 24 unaffected individuals (52.1%) belonged to type‐B families. Of the 94 individuals studied, 35 were diagnosed with MS, 46 had no AIDs, and 13 had other AIDs than MS (hypothyroidism in seven, hyperthyroidism in one, rheumatoid arthritis in two, type 1 diabetes mellitus in one, autoimmune uveitis in one, and systemic lupus erythematosus in one). Of the 35 patients with MS, eight had an additional AID (hypothyroidism in one, type 1 diabetes mellitus in one, ulcerative colitis in one, autoimmune uveitis in one, Guillain‐Barré syndrome in one, systemic lupus erythematosus in one, and rheumatic fever in one). Figure 2 shows the demographic and clinical characteristics of the study population, including the clinical form of MS. Regarding daylight duration (Villar‐Quiles et al., 2016), 91 individuals (96.8%) lived in Madrid from at least 2016, one (1.06%) lived in Miami (USA), one lived in Seville, and one lived in Toledo. The cumulative daylight duration for 2016 was 4,468.4 hr in Madrid, 4,447.8 hr in Miami, 4,459.8 hr in Seville, and 4,463.3 hr in Toledo. By group, 33 patients with MS lived in Madrid, one lived in Miami, and one lived in Toledo. The mean daylight duration for the MS group was 4,467.6 ± 3.5 hr. All family members with other AIDs lived in Madrid, with 4,468.4 ± 0.0 hr of daylight. Of the unaffected family members, one lived in Seville and the rest lived in Madrid, with 4,468.2 ± 1.2 hr of daylight.
Figure 2.
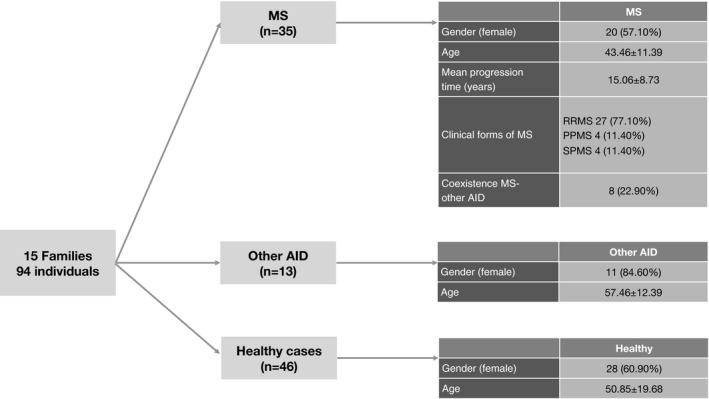
Demographic and clinical characteristics of the study population (n = 94; 15 families). RRMS, relapsing‐remitting multiple sclerosis; PPMS, primary‐progressive multiple sclerosis; SPMS, secondary‐progressive multiple sclerosis
3.2. Exonic variants of genes involved in the vitamin D signaling pathway
Analysis of the genes associated with the VD signaling pathway revealed a total of 154 different variants in the 94 individuals tested. Disregarding synonymous variants gives a final total of 77 different nonsynonymous variants (Table 1). All variants found followed the Hardy‐Weinberg equilibrium.
Table 1.
Nonsynonymous exonic variants detected in a cohort of 94 individuals belonging to 15 families including at least two patients with multiple sclerosis
| Gene | Locus | Variant (rs) | MAF | Variant effect | Reference allele | Change | MS (n = 35) | Other AID (n = 13) | Unaffected (n = 46) |
|---|---|---|---|---|---|---|---|---|---|
| CYP3A4 | chr7:99361620 | ND | NA | Missense | A | c.884T>C | 0 | 1 | 0 |
| CYP27A1 | chr2:219677022 | rs2229381 | 0.001 | Missense | C | c.524C>T | 1 | 0 | 4 |
| LRP2 | Chr2:169985338 | rs34564141 | 0.007 | Missense | C | c.13803G>A | 2 | 1 | 1 |
| LRP2 | Chr2:169997025 | rs764880181 | NA | frameshiftDeletion | TG | c.13139delC | 1 | 0 | 0 |
| LRP2 | Chr2:170003432 | rs4667591 | 0.712 | Missense | T | c.12628A>C | 35 | 12 | 45 |
| LRP2 | Chr2:170010985 | rs2075252 | 0.763 | Missense | T | c.12280A>G | 32 | 13 | 45 |
| LRP2 | Chr2:170013904 | rs79723119 | 0.008 | Missense | A | c.11996T>G | 1 | 0 | 1 |
| LRP2 | Chr2:170029657 | rs34355135 | 0.006 | Missense | C | c.11092G>A | 2 | 0 | 0 |
| LRP2 | Chr2:170038761 | rs3213760 | 0.003 | Missense | C | c.9914G>A | 1 | 1 | 0 |
| LRP2 | Chr2:170053505 | rs2228171 | 0.267 | Missense | C | c.8614G>A | 6 | 3 | 8 |
| LRP2 | Chr2:170060603 | rs17848169 | 0.029 | Missense | T | c.7894A>G | 2 | 4 | 7 |
| LRP2 | Chr2:170062977 | rs61995915 | 0.013 | Missense | T | c.7253A>G | 2 | 2 | 3 |
| LRP2 | chr2:170063250 | rs886055084 | NA | Missense | G | c.6980C>T | 0 | 0 | 2 |
| LRP2 | chr2:170070172 | rs4667596 | 0.024 | Missense | C | c.6035G>A | 1 | 0 | 1 |
| LRP2 | chr2:170097707 | rs17848149 | 0.030 | Missense | T | c.3836A>C | 1 | 0 | 2 |
| LRP2 | chr2:170113670 | rs150752263 | 0.001 | Missense | G | c.2603C>G | 2 | 0 | 1 |
| LRP2 | Chr2:170129547 | rs34291900 | 0.028 | Missense | C | c.2006G>A | 2 | 4 | 7 |
| LRP2 | chr2:170136882 | ND | NA | frameshiftDeletion | CA | c.1318delT | 1 | 0 | 0 |
| LRP2 | Chr2:170147502 | rs34693334 | 0.063 | Missense | C | c.775G>C | 2 | 0 | 4 |
| LRP2 | Chr2:170175334 | rs2229263 | 0.278 | Missense | T | c.248A>G | 19 | 7 | 19 |
| CUBN | chr10:16870912 | rs1801232 | 0.086 | Missense | G | c.10656C>A | 5 | 5 | 10 |
| CUBN | chr10:16877080 | rs7898873 | 0.023 | Missense | G | c.10295C>G | 1 | 1 | 1 |
| CUBN | chr10:16911671 | rs148491916 | <0.001 | Missense | C | c.9418G>A | 0 | 0 | 1 |
| CUBN | chr10:16918947 | ND | NA | frameshiftDeletion | AG | c.9054_9054delC | 2 | 0 | 2 |
| CUBN | chr10:16918997 | rs1801240 | 0.105 | Missense | T | c.9005A>G | 8 | 6 | 13 |
| CUBN | chr10:16919052 | rs1801239 | 0.087 | Missense | T | c.8950A>G | 7 | 5 | 12 |
| CUBN | chr10:16930419 | rs45569534 | 0.014 | Missense | C | c.8902G>C | 1 | 0 | 0 |
| CUBN | chr10:16932490 | rs1801238 | 0.028 | Missense | G | c.8635C>A | 3 | 0 | 0 |
| CUBN | chr10:16942818 | ND | NA | Missense | T | c.8216A>G | 1 | 0 | 2 |
| CUBN | chr10:16943371 | rs2796835 | 1.000 | Missense | G | c.8150C>G | 35 | 12 | 46 |
| CUBN | chr10:16948277 | rs144626884 | <0.001 | Missense | T | c.7837A>C | 1 | 0 | 2 |
| CUBN | chr10:16948390 | rs3740168 | 0.041 | Missense | G | c.7724C>G | 1 | 0 | 4 |
| CUBN | chr10:16961995 | rs2271460 | 0.016 | Missense | A | c.6788T>G | 0 | 0 | 2 |
| CUBN | chr10:16962122 | rs143291127 | <0.001 | Missense | C | c.6661G>A | 0 | 0 | 2 |
| CUBN | chr10:16967362 | ND | NA | Missense | A | c.6524T>G | 2 | 1 | 3 |
| CUBN | chr10:16967401 | rs1276712 | 0.994 | Missense | C | c.6485G>A | 35 | 13 | 46 |
| CUBN | chr10:16967586 | rs62619939 | 0.129 | Missense | C | c.6459G>C | 7 | 2 | 9 |
| CUBN | chr10:16979606 | rs2356590 | 0.063 | Missense | G | c.5911C>A | 1 | 0 | 1 |
| CUBN | chr10:16979714 | rs41289305 | 0.144 | Missense | T | c.5803A>G | 6 | 2 | 12 |
| CUBN | chr10:16982061 | rs2271462 | 0.075 | Missense | C | c.5518G>A | 1 | 0 | 1 |
| CUBN | chr10:16989271 | rs74116778 | 0.017 | Missense | C | c.5305G>A | 1 | 1 | 0 |
| CUBN | chr10:17024503 | rs1801231 | 0.767 | Missense | G | c.4675C>T | 34 | 13 | 44 |
| CUBN | chr10:17110639 | rs148869805 | 0.004 | Missense | T | c.2756A>G | 2 | 0 | 4 |
| CUBN | chr10:17113456 | rs138083522 | 0.007 | Missense | C | c.2594G>A | 1 | 1 | 1 |
| CUBN | chr10:17126383 | rs7905349 | 0.028 | Missense | G | c.2188C>T | 0 | 0 | 1 |
| CUBN | chr10:17147521 | rs1801224 | 0.613 | Missense | G | c.1165C>A | 29 | 12 | 37 |
| CUBN | chr10:17153023 | rs78201384 | 0.004 | Missense | C | c.910G>A | 2 | 1 | 2 |
| CUBN | chr10:17156151 | rs1801222 | 0.727 | Missense | A | c.758T>C | 34 | 13 | 44 |
| CUBN | chr10:17171176 | rs12259370 | 0.007 | Missense | C | c.196G>A | 1 | 1 | 0 |
| CYP24A1 | chr20:52774635 | rs6068812 | <0.001 | Missense | A | c.1226T>C | 2 | 0 | 1 |
| VDR | chr12:48272895 | rs2228570 | 0.631 | Missense | A | c.152T>C | 30 | 12 | 41 |
|
PDIA3
PDIA3 |
chr15:44055344 chr15:44038766 |
rs139812953 ND |
<0.001 NA |
Missense Missense |
A C |
c.542A>G c.29C>T |
1 0 |
1 0 |
2 2 |
| RXRA | chr9:137309155 | rs61751479 | 0.002 | Missense | G | c.762G>A | 0 | 0 | 3 |
| GC | chr4:72618296 | rs9016 | 0.998 | Missense | T | c.1391A>G | 35 | 13 | 46 |
| GC | chr4:72618311 | ND | NA | Missense | T | c.1376A>G | 1 | 0 | 0 |
| GC | chr4:72618323 | rs4588 | 0.250 | Missense | G | c.1364C>A | 22 | 9 | 25 |
| GC | chr4:72618334 | rs7041 | 0.515 | Missense | A | c.1353T>G | 31 | 10 | 41 |
| GC | chr4:72669661 | rs76781122 | 0.018 | Missense | C | c.3G>T | 2 | 0 | 4 |
| FCGR2A | chr1:161476204 | rs201218628 | NA | Missense | CA | c.187_188delCAinsTG | 8 | 3 | 12 |
| FCGR2A | chr1:161479745 | rs1801274 | 0.478 | Missense | A | c.500A>G | 32 | 10 | 40 |
| FCGR2C | chr1:161559571 | rs138747765 | 0.279 | Missense | C | c.353C>T | 12 | 6 | 17 |
| FCGR2C | chr1:161561156 | rs76016754 | 0.010 | Missense | A | c.614A>T | 2 | 0 | 0 |
| FCGR3A | chr1:161512873 | rs115866423 | 0.010 | Missense | T | c.1009A>T | 2 | 0 | 3 |
| FCGR3A | chr1:161514542 | rs396991 | 0.324 | Missense | A | c.841T>G | 22 | 10 | 26 |
| FCGR3A | chr1:161518214 | rs148181339 | NA | Missense | T | c.631A>G | 3 | 0 | 4 |
| FCGR3A | chr1:161518333 | rs10127939 | 0.054 | Missense | A | c.512T>A | 3 | 1 | 9 |
| FCGR3A | chr1:161518333 | rs10127939 | 0.054 | Missense | A | c.512T>G | 5 | 3 | 3 |
| FCGR3A | chr1:161518336 | rs77144485 | 0.091 | Missense | C | c.509G>A | 18 | 7 | 24 |
| FCGR3A | chr1:161519601 | rs770473456 | <0.001 | Missense | A | c.241T>C | 0 | 1 | 0 |
| FCGR3A | chr1:161519622 | rs773823413 | <0.001 | Missense | C | c.220G>A | 0 | 1 | 0 |
| FCGR3B | chr1:161595986 | rs200215055 | 0.001 | Missense | C | c.634G>T | 1 | 1 | 1 |
| FCGR3B | chr1:161599654 | rs5030738 | 0.040 | Missense | G | c.341C>A | 1 | 1 | 2 |
| FCGR3B | chr1:161599693 | rs448740 | 0.636 | Missense | T | c.302A>G | 34 | 11 | 40 |
| METTL21B | chr12:58174368 | rs141172155 | 0.001 | Missense | G | c.620G>A | 0 | 0 | 2 |
| FGF23 | chr12:4479549 | rs7955866 | 0.126 | Missense | G | c.716C>T | 7 | 1 | 8 |
| DAB2 | chr5:39376988 | rs3733801 | 0.133 | Missense | C | c.1901G>A | 13 | 4 | 15 |
| DAB2 | chr5:39377132 | rs700241 | 0.023 | Missense | G | c.1757C>T | 0 | 0 | 1 |
| AVIL | chr12:58204283 | rs2172521 | 1.000 | Missense | T | c.610A>G | 35 | 13 | 46 |
| AVIL | chr12:58209772 | rs753181730 | <0.001 | Missense | C | c.52G>A | 1 | 0 | 1 |
AID, autoimmune disease; ND, no data; NA, not available. Genes involved in the regulatory pathway of CYP27B1 gene have also been added at the end of this table.
3.3. Prioritization of variants
Figure 3 illustrates the filtering and prioritization process. Prioritization yielded eight exonic variants meeting the established criteria, located on the genes LRP2 (four variants), CUBN (two variants), CYP24A1 (one variant), and METTL21B (one variant).
Figure 3.
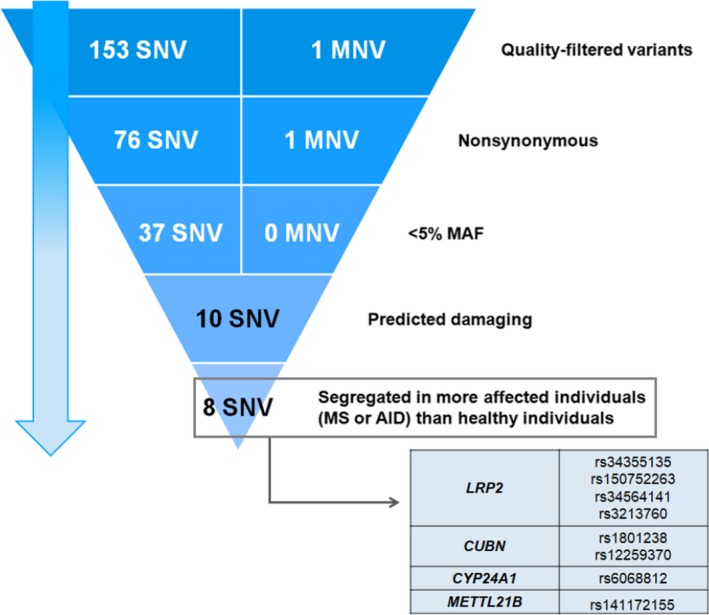
Prioritization of variants of genes involved in the vitamin D signaling pathway. SNV, single nucleotide variants; MNV, multiple nucleotide variants, repeats two or more SNV in successions
3.4. Variants affecting levels of circulating vitamin D
In the families, we detected single variants of CYP3A4, CYP27A1, CYP24A1, and five variants of the GC gene, with no significant differences between groups. We also analyzed the genes that regulate CYP27B1 expression (METTL21B, METTL1, FGF23, and PTH) (Table 1). Seven patients with MS (20%), one with an AID other than MS (7.7%), and eight unaffected individuals (17.4%) were carriers of FGF23 variant rs7955866. Regarding family type, this variant was present in six members of type‐A families (13.6%) and 10 members of type‐B families (29%). METTL21B variant rs141172155 was found in two patients with MS (5.7%) and in no other group. We observed two additional synonymous variants of the genes METTL21B (rs923829) and PTH (rs6256). Variant rs923829 was more frequent in type‐A than in type‐B families (16 [36.4%] vs. 8 [16%]). After prioritization and filtering, we detected rs6068812 variant of CYP24A1in one patient with RRMS, one with PPMS, and one unaffected individual from a type‐B family. We analyzed the variants from genes potentially involved in the 12q LD block (CDK4, TSFM, AGAP2, AVIL, CYP27B1, METTL1, and METTL21B) and compared these against variant from CYP24A1 (rs6068812). All three individuals with rs6068812 also had a variant in AVIL gene (rs2172521), which was present in all members of our cohort. Therefore, no association was observed between the 12q LD block genes variants analyzed and CYP24A1 gene variant (rs6068812). None of these variants found in the families had a significant relationship to MS or MS with AID compared to unaffected family members.
3.5. Variants affecting vitamin D internalization
We observed 18 nonsynonymous variants of LRP2, none of which was associated with MS. We also detected 29 variants of CUBN. The majority of variants observed after filtering and prioritization affected VD internalization. LRP2 variant rs34564141 was observed in two patients with RRMS associated with another AID (ulcerous colitis and Guillain‐Barré syndrome), one individual with autoimmune uveitis, and one unaffected individual from a type‐B family. LRP2 variant rs34355135 was detected in two individuals with RRMS, one of whom also had autoimmune uveitis, from a type‐B family. LRP2 variant rs3213760 was identified in one patient with RRMS and one family member with hypothyroidism from a type‐B family. LRP2 variant rs150752263 was observed in two patients with RRMS and an unaffected individual from a type‐A family. Regarding the CUBN gene, variant rs1801238 was identified in two patients with RRMS and one with PPMS (from tow type‐B families); two of these patients (siblings) had an additional AID (hypothyroidism and systemic lupus erythematosus). Variant rs12259370 was observed in one individual with RRMS and comorbid rheumatic fever and one individual with hypothyroidism from a type‐B family. We also identified a family in which the variants rs3213760 (LRP2) and rs12259370 (CUBN) were present in an individual with hypothyroidism and in no patients with MS. We found two nonsynonymous variants of DAB2 (rs3733801 and rs700241), which codes for a protein involved in VD endocytosis. No differences were detected between groups (MS, other AIDs, or unaffected individuals; type‐A or type‐B families). Variant rs3733801 was observed in 13 patients with MS (37.1%), four individuals with other AIDs (30.8%), and 15 unaffected individuals (32.6%). Regarding family type, this variant was observed in 12 members of type‐A families (27.3%) and 20 members of type‐B families (40%). Variant rs700241 was detected in one unaffected individual from a type‐B family (Table 1). None of these variants found in the families had a significant relationship to MS or MS with AID compared to unaffected family members.
3.6. VDR and RXRA variants
We observed only one variant of VDR and one variant (rs61751479) of RXRA (Table 1). The VDR variants observed were TaqI (rs731236), in 30 patients with MS (85.7%), 11 individuals with other AIDs (84.6%), and in 35 unaffected individuals (76.1%); and FokI (rs2228570) in 30 patients with MS (85.7%), 12 individuals with other AIDs (85.7%), and in 41 unaffected individuals (89.1%). Nine patients with MS (25.7%), three patients with other AIDs (23.1%), and 12 unaffected individuals (26.1%) were homozygous for the G allele of TaqI. GG homozygosity for FokI was observed in 17 patients with MS (48.6%), five patients with other AIDs (38.5%), and 15 unaffected individuals (32.6%). Analysis of the differences between family types for these variants revealed that TaqI (rs731236) was present in 43 members of type‐A families (97.7%) and 33 members of type‐B families (66.0%). No significant differences between family types were observed for FokI (rs2228570) (41 members of type‐A families [93.2%] vs. 42 members of type‐B families [84.0%]). None of these variants found in the families had to do with unaffected family members.
3.7. PDIA3 variants
Two missense variants have been detected on the gene encoding PDIA3 in our cohort. One of them, rs139812953, is only observed in a MS case, in a case with other AID and in two unaffected cases. The second one, positioned in chr15:44038766 is only present in two nonaffections.
4. DISCUSSION
Several authors have hypothesized that predisposition to MS and other AIDs may involve VD signaling pathway components in which various potentially associated variants have been described. This theory is supported by some case‐control studies which have analyzed variants at different points of the pathway; results are conflicting, however, with some studies finding no association (Agnello et al., 2017; Simon, Munger, Yang, & Ascherio, 2010). This hypothesis involves two possible underlying mechanisms: firstly, genomic variations may reduce the effectiveness of VDR function; and secondly, these variations may cause a change in gene expression in either immune or CNS cells (Lu, Taylor, & Körner, 2018). The most complex issue in the study of the VD metabolic pathway is related to the heterogeneity of the pathway depending on the cell or tissue type. Accounting for the fact that different cell types internalize VD in different ways, our WES study of the different variants in the families of patients with familial MS included variations in all subpathways of the VD signaling pathway. VDR action is also variable after translocation to the nucleus, depending on the target cell (Zella, Kim, Shevde, & Pike, 2006; Zella et al., 2010). It has been suggested that this is due to different levels of local activators and coregulators and epigenetic mechanisms (Saccone, Asani, & Bornman, 2015); this issue lies beyond the scope of the present study. The influence of specific tissue or cell type prevents us from easily drawing conclusions as to the potential role of VD based only on levels of some of its metabolites in the blood.
4.1. Genes influencing circulating vitamin D
Some variants had been described in the literature in association with MS that may act through an effect on circulating VD (Alloza et al., 2012; Australia New Zealand Multiple Sclerosis Genetics Consortium, 2009; Cortes et al., 2013; Karaky et al., 2016; Laursen et al., 2015; Manousaki et al., 2017; Orton et al., 2011; Ramagopalan et al., 2011; Ramasamy et al., 2014; Ross et al., 2014; Scazzone et al., 2018; Simon et al., 2011; Sundqvist et al., 2010; Zhuang et al., 2015). No significant intergroup differences were observed for presence of these variants in our cohort. The results obtained are shown in Table S1. We did locate the CYP24A1 variant rs6068812 in one patient with RRMS, one with PPMS, and one unaffected individual from a type‐B family; this variant has not been associated with MS in the literature. Given the very low MAF of the variant, we should consider a potential association with the disease and could be analyzed in further studies.
This stage of the VD metabolic pathway is one of those in which researchers have searched for variants influencing MS, especially in CYP27B1; it is therefore surprising that no association was found. Associations have been described between VD deficiency and numerous genetic variants (Lafi, Irshaid, El‐Khateeb, Ajlouni, & Hyassat, 2015; Li et al., 2014; Lu et al., 2012; Nissen et al., 2014; Signorello et al., 2011; Slater, Rager, Havrda, & Harralson, 2017; Wang et al., 2010; Zhang et al., 2012) (Table S1). However, these associations do not necessarily represent increased risk of MS, as no correlation has been demonstrated between the disease and plasma VD level (Ahn et al., 2010). This is probably due to the existence of confounding factors (Bu et al., 2010), including geographical (Elkum et al., 2014) or ethnic (Batai et al., 2014) variation, as well as environmental factors influencing plasma VD levels, such as sex, age, the season in which samples are taken (Engelman et al., 2013), the use of dietary vitamin D supplements, consumption of milk and cereals, obesity, and daily amount of time spent walking (Hansen et al., 2015). The biomarker used also represents a methodological bias that may affect study results: 25(OH)D and 1,25(OH)2D have been associated with different findings (Engelman et al., 2008). Regarding the regulation of CYP27B1 expression (genes METTL21B, METTL1, FGF23, and PTH), which is thought to influence VDR (Bouksila et al., 2018), METTL21B variant rs141172155 was observed only in two patients with MS from type‐B families. We did not detect any METTL1 variant associated with MS (Alcina et al., 2013). This pathway is of interest, as CYP27B1 may be influenced by processes occurring in the kidneys in MS and other AIDs (Meyer et al., 2017). GWAS data suggest that there may be a specific locus for MS in a LD block affecting 17 genes on the long arm of chromosome 12. This area as a whole may therefore be considered a risk locus. However, we did not observe related variants in our analysis. Only two exonic and nonsynonymous variants (rs2172521, rs753181730) belonging to the AVIL gene were found in the families. Neither did we observe the proposed relationship with CYP24A1, located on chromosome 20 (Gandhi et al., 2010).
4.2. Genes influencing the cellular internalization of 1,25(OH)2D
Uptake of the 25(OH)D‐DBP complex by the proximal tubules of the kidneys and by other cell types occurs by means of LRP2‐, CUBN‐, or DAB2‐mediated endocytosis; this process maintains serum VD concentrations and activates 1,25(OH)2D. Following internalization, DBP is degraded in the lysosomes, releasing 25(OH)D, which is activated by CYP27B1 to 1,25(OH)2D. Therefore, functional alterations in LRP2, CUBN, or DAB2 could lead to excessive urinary excretion of the 25(OH)D‐DBP complex and reduced activation of 1,25(OH)2D by VDR. In this way, VD internalization can be mediated by LRP2, with or without the participation of CUBN and the DAB2 adaptor protein, although vitamin D also accesses some cells freely. Zhou et al. (2017) describe an association between MS relapses and LRP2 variant rs12988804. The study was based on data from three cohorts of patients with MS, including one series of paediatric patients. Variant rs12988804 is located in the intronic region of transcript NM_004525.2, as well as in the transcript variants XM_011511183.2 and XM_011511184.2. Because intronic variants do not participate in protein synthesis, they rarely have any impact on disease; however, they are occasionally involved in pathophysiology due to their influence on splicing regions. Variant rs12988804 was not detected in our cohort. While it is a frequent variant (MAF 0.206), it is located in an intron, whereas our study focused on exonic regions. Zhou et al. signal the proximity to variant rs754235034 as the potential mechanism by which rs12988804 is associated with MS. We identified four exonic variants: rs34564141, rs34355135, and rs3213760 in type‐B families and rs150752263 in a type‐A family. All four were detected in patients with AIDs other than MS; it could therefore be the case that variants of LRP2 are associated with AIDs in general, rather than MS specifically. This is consistent with our hypothesis about the effect of family type. Similarly, we observed AID‐associated CUBN variants (rs1801238 and rs12259370) in type‐B families; one type‐B family included a patient with a non‐MS AID who displayed variants of both LRP2 (rs3213760) and CUBN (rs12259370). We also analyzed variants of DAB2, which, together with LRP2 and CUBN, is involved in vitamin D internalization. Variant rs3733801 was the most frequent, although no difference was observed in its frequency between patients with MS and the other individuals studied; it was more frequent in type‐B than in type‐A families, however. This could support the hypothesis that these families have more variants of genes related to VD internalization, which could represent a genetic predisposition to AIDs and could be analyzed in further studies.
4.3. VDR and RXRA genes variants
Vitamin D receptor variants are one of the genetic biomarkers which have aroused the most interest in evaluating the risk of AID. They have been associated with increased risk of a range of AIDs, including systemic lupus erythematosus (Carvalho et al., 2015), rheumatoid arthritis (Cavalcanti et al., 2016), autoimmune thyroid disease (Feng, Li, Chen, & Zhang, 2013), and ankylosing spondylitis (Cai et al., 2016). The most studied VDR variants are BsmI, FokI, ApaI, and TaqI, although other variants have been described (Dickinson et al., 2009). The ApaIand BsmI variants are located close to the 3′ end of the VDR gene, in the intron between exons 8 and 9, and cannot be analyzed in WES studies. The FokI (rs2228570) and TaqI (rs731236) variants, on the other hand, can be detected with this technique. The findings published in the literature are conflicting, and these variants have been related with both risk and progression of MS (Altemaimi, Alenezi, Alserri, Alroughani, & Al‐Mulla, 2015; Čierny et al., 2016; Fukazawa et al., 1999); other studies observe no association, however. Meta‐analyses have not solved this controversy, finding both negative (García Martín et al., 2013; Huang & Xie, 2012) and positive results (Tizaoui, Kaabachi, Hamzaoui, & Hamzaoui, 2015), although they do suggest that ethnicity, age, and geographic latitude may influence the associations between these variants and MS risk. We found no significant differences in rs2228570 and rs731236 frequency between patients with MS, patients with other AIDs, and unaffected family members. The only Spanish study into VDR variants in sporadic MS did find an association (García Martín et al., 2013). Variant rs731236 was significantly more frequent in members of type‐A than type‐B families, whereas rs2228570 prevalence was similar in both groups. No associations were detected for variants of the gene encoding RXRA, a transcription factor that together with VDR forms heterodimers needed for nuclear translation, similar to the findings of a recent study (Agnello et al., 2018).
4.4. PDIA3 gene variants
Protein disulfide isomerase family member 3 was recently described as a VD receptor (Doroudi, Olivares‐Navarrete, Boyan, & Schwartz, 2015). Considering that its presence in the brain is greater than VDR, it has been suggested that would have a greater role in the CNS disorders (Tohda, Urano, Umezaki, Nemere, & Kuboyama, 2012) since it is present in practically all cell types and acts in nongenomic functions of the VD. There is no previous information on the influence of variants on the PDIA3 gene in MS (Landel, Sthefan, Cui, Eyles, & Feron, 2018). We observed only two variants of PDIA3 gene in our cohort but no associations were observed in MS or AID groups. However, information on the signaling pathway of PDIA3 is scarce, so further studies will probably be needed.
4.5. Analysis of variants in families
The hypothesis of genetic regulation of vitamin D in families has been proposed in studies of twins, which observed that certain variants, particularly of CYP27B1, may influence the risk of MS (Orton et al., 2008); however, very little information is available on this subject. A CYP27A1 mutation thought to be related to MS has been described in a family including three patients with the disease (Traboulsee et al., 2017). Another published study design, using WES to study father‐mother‐child trio, includes 28 patients from eight families; however, the results are not known (García‐Rosa et al., 2017). The only published study on the subject, evaluating VDR variants in 29 patients with familial MS and comparing them to unrelated controls, found a significant difference between groups for the TaqI variant of VDR (Yucel et al., 2018). Our study is the first to analyze whole families and to compare patients to family members, thereby eliminating potential confounders. Furthermore, families were divided into the two types described above (A and B) according to the generational distribution of cases. Analysis of the variants detected in the different pedigrees demonstrates the presence in some families of variants which may appear to be linked to the disease, although this was not supported by the results of the comparative analysis of the entire cohort. Given the variants detected, we may hypothesize that type‐B families, with a probable greater genetic load and greater prevalence of other AIDs, are the best group to study in order to analyze the role of the genetic variants in each family. In a type‐B family, one variant in FCGR2C gene (rs76016754) was observed in homozygosis in an MS case, but not in other MS patients. Due to that, further cases will be necessary to clarify its significance.
4.6. Study limitations
Genetic factors have a clear influence over susceptibility to MS. Linkage analysis using single‐nucleotide variants, and especially GWAS studies, have made it possible to detect loci related to the disease. However, the alleles detected are neither necessary nor sufficient to cause MS; GWAS may also be unable to detect rare variants that may have a significant effect. While GWAS is based on the hypothesis of common variants related to the disease, and provides information on the risk associated with common genetic variability (Simón‐Sánchez & Singleton, 2008), WES is based around the idea of risk associated with rare variants, such as TYK2 variant rs55762744, which was identified with this technique (Dyment et al., 2012). Evidently, we are not restricted to the use of a single one of these approaches to research the genetic basis of MS (Jiang, Tan, Tan, & Yu, 2014); rather, they are mutually enriching.
WES has a number of disadvantages. The first of these is incomplete coverage: it does not encompass the whole genome, and yields a high number of rare variants of uncertain importance; we should also mention the ethical dilemma involved in the unexpected discovery of incidental findings of variants associated with diseases that were not the target of the study (Klein & Foroud, 2017). Secondly, identifying multiple rare variants within a single gene makes it difficult to establish a clear interpretation of the gene's role. Finally, there is a high probability of false positives due to the large quantity of sequencing data generated and the unclear likelihood of causal relationships. GWAS also has the disadvantage of detecting single‐nucleotide variants that may be associated with disease but have no known function related to the disease. This may mean that the true causal gene is a nearby, rare variant. Nevertheless, this limitation is reduced if the full genome or exome is sequenced; analysis aims to establish how the variant is related to the disease.
Familial studies search for genetic factors from different perspectives than studies of sporadic cases of MS. However, these study types cannot replace one another; rather, the data they yield are complementary. These studies offer the advantage of controls being members of the same family, preventing such confounding factors as ethnic and often geographical differences. They are also characterized by greater genetic homogeneity between individuals, and enable a better understanding of associated autoimmunity. However, they also involve certain disadvantages, such as small sample sizes (Wang et al., 2016) and reduced statistical power when analysing the cohort as a whole. Analysis of whole families, as performed in the present study rather than trios (comparing patients to first‐degree relatives), increases the power of family studies. This is of special interest in studies where factors may also have a considerable environmental influence, as is the case of vitamin D. Studying cases and controls from a single family reduces biases related to exposure to environmental factors, compared to studies with controls from the general population. However, it may not be possible to extrapolate information from familial forms of MS to patients with sporadic forms.
A limitation of our study is that the unaffected cases were classified according to the data obtained from direct interrogatory, but brain magnetic resonance was not performed. Thus, we cannot exclude the existence of subclinical MS.
As previously discussed, the great difficulty with WES is addressing and interpreting large numbers of variants. Our study was limited to exonic variants, which we can expect to be functional. Although this technique does allow the detection of intronic variants located close to exonic regions, causality is very difficult to interpret, particularly with small sample sizes. For this reason, we do not address some of the associations described in the literature.
5. CONCLUSIONS
Multiple S is considered to be a polygenic disease and it is very doubtful that the risk may depend on one or few variants. It is more likely to depend on alterations on signaling pathways genes such as the ones related to VD pathway. In order to analyze the data of the studies, it seems necessary to determine the prevalence of the variants in these pathways in the families of patients with MS and especially in the different forms of MS families. Our study aimed to know which variants appear in families of MS. Table 1 includes the 77 nonsynonymous exonic variants in genes participating in the VD pathway found in the subjects families that present more than one patient with MS and that could be related to VD functionality. The genes with the greatest presence of variants correspond to those that affect the entry mechanisms in the cells, spatially CUBN and LRP2. However, the presence of all the variants found does not differ between the cases and the not affected members in the family. When analyzing the pedigrees of each family included in the study, a relationship of these variants with MS has not been observed either.
This is the first study to address the whole family of patients with familial MS using WES and focusing their analysis into the VD metabolic pathway as a whole, analyzing the potential genetic influences over the pathway. Despite the number of studies reporting an association between genetic variants affecting the VD signaling pathway, MS and other AIDs, the information obtained is relatively limited. After comparison with unaffected family members and analyzing the pedigrees, the majority of the variants published appear not to be specific to MS. This might be due to the fact that sporadic and familial forms involve different genetic factors, or that GWAS led to false positives in comparisons between patients and controls. Our study did not reproduce the association of MS risk with a CYP27B1 variant, or with exonic variants of VDR. We did detect several variants of interest in LRP2 and CUBN, as well as CYP24A1 and METTL21B variants; these should be confirmed by further studies. We have not observed any relationship between variants in the gene that encodes the PDIA3 receptor with the studied groups.
Furthermore, we did not identify any relationship with genetic variants previously associated with MS, including in type‐B families, which we had hypothesized would display a greater influence due to genetic load; and there are two potential explanations for this. First, familial MS may not be directly influenced by the genetic alterations that affect VD signaling pathways in sporadic MS, which are detectable by GWAS. The second possible explanation is that vitamin D may be an environmental factor that influences the risk of MS but is not determined by genetic variants. This theory is supported by the variability of the cells and tissues in which the VD metabolic pathway occurs, and the fact that the pathway is influenced by local molecular and epigenetic factors, which makes it difficult for epidemiological and intervention studies to understand (uncover) VD's possible role in the disease. Although this study does not allow us to confirm either hypothesis, it indeed provides information on the variants found in the VD signaling pathway in MS families.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
ETHICS AND CONSENT
This study was approved by the Clinical Research Ethics Committee of Hospital Clínico San Carlos. All participants included in the study signing a written consent. Data were handled in observance of the Spanish legislation regarding data protection (Organic Law 15/1999 of 13 December). The project was carried out in accordance with the principles included in the Declaration of Helsinki (Recommendations Guiding Physicians in Biomedical Research Involving Human Subjects, Helsinki 1964, amended October 2013).
AUTHOR CONTRIBUTIONS
Lead researcher: JMG; study design: VP, JAMG, SA, JMG; patient assessments: JAMG, PME, JMG; family studies: VP, LTF, PME; coordination of information: VP, LTF, UGP; WES: SA, JBR, PM; database: VP, JAMG, LTF, JMG; data filtering and analysis: VP, LTF, SA, JBR, PM, JMG, JAMG; statistical analysis: VP, UGP, JMG, JAMG; analysis of results: VP, LTF, JMG, UGP, JAMG; figures and tables: VP, LTF, JMG; manuscript draft: JMG, VP, JAMG, LTF; revision of manuscript; all authors.
Supporting information
ACKNOWLEDGMENTS
The authors are grateful to our patients and their families for the information and for the DNA samples provided for the study. The authors also thank the Spanish Society of Neurology's Research Operations Office for helping in the English language revision of this paper.
Pytel V, Matías‐Guiu JA, Torre‐Fuentes L, et al. Exonic variants of genes related to the vitamin D signaling pathway in the families of familial multiple sclerosis using whole‐exome next generation sequencing. Brain Behav. 2019;9:e01272 10.1002/brb3.1272
REFERENCES
- Agnello, L. , Scazzone, C. , Lo Sasso, B. , Bellia, C. , Bivona, G. , Realmuto, S. , … Ciaccio, M. (2017). VDBP, CYP27B1, and 25‐Hydroxyvitamin D gene polymorphism analyses in a group of sicilian multiple sclerosis patients. Biochemical Genetics, 55, 183–192. 10.1007/s10528-016-9783-4 [DOI] [PubMed] [Google Scholar]
- Agnello, L. , Scazzone, C. , Lo Sasso, B. , Ragonese, P. , Milano, S. , Salemi, G. , & Ciaccio, M. (2018). CYP27A1, CYP24A1, and RXR‐αPolymorphisms, vitamin D, and multiple sclerosis: A pilot study. Journal of Molecular Neuroscience, 66, 77–84. 10.1007/s12031-018-1152-9 [DOI] [PubMed] [Google Scholar]
- Ahn, J. , Yu, K. , Stolzenberg‐Solomon, R. , Simon, K. C. , McCullough, M. L. , Gallicchio, L. , … Albanes, D. (2010). Genome‐wide association study of circulating vitamin D levels. Human Molecular Genetics, 19, 2739–2745. 10.1093/hmg/ddq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcina, A. , Fedetz, M. , Fernández, O. , Saiz, A. , Izquierdo, G. , & Lucas, M. (2013). Identification of a functional variant in the KIF5A‐CYP27B1‐METTL1‐FAM119B locus associated with multiple sclerosis. Journal of Medical Genetics, 50, 25–33. 10.1136/jmedgenet-2012-101085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloza, I. , Otaegui, D. , de Lapuente, A. L. , Antigüedad, A. , Varadé, J. , Núñez, C. , … Vandenbroeck, K. (2012). ANKRD55 and DHCR7 are novel multiple sclerosis risk loci. Genes & Immunity, 13, 253–257. 10.1038/gene.2011.81 [DOI] [PubMed] [Google Scholar]
- Altemaimi, R. A. , Alenezi, A. , Alserri, A. , Alroughani, R. , & Al‐Mulla, F. (2015). The association of vitamin D receptor polymorphisms with multiple sclerosis in a case‐control study from Kuwait. PLoS One, 10, e0144565 10.1371/journal.pone.0142265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Autoimmune Related Diseases Association (AARDA) . List of diseases ‐ AARDA Web site. Descargado de http://www.aarda.org/autoimmune-information/list-of-diseases/ Acceso en el 20 de febrero del 2016.
- Ascherio, A. , Munger, K. L. , White, R. , Köchert, K. , Simon, K. C. , Polman, C. H. , … Pohl, C. (2014). Vitamin D as an early predictor of multiple sclerosis activity and progression. JAMA Neurology, 71(3), 306–314. 10.1001/jamaneurol.2013.5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australia New Zealand Multiple Sclerosis Genetics Consortium (2009). Genome‐wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nature Genetics, 41, 824–828. 10.1038/ng.396 [DOI] [PubMed] [Google Scholar]
- Baranzini, S. E. , & Oksenberg, J. R. (2017). The genetics of multiple sclerosis: From 0 to 200 in 50 Years. Trends in Genetics, 33, 960–970. 10.1016/j.tig.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batai, K. , Murphy, A. B. , Shah, E. , Ruden, M. , Newsome, J. , Agate, S. , … Kittles, R. A. (2014). Common vitamin D pathway gene variants reveal contrasting effects on serum vitamin D levels in African Americans and European Americans. Human Genetics, 133, 1395–1405. 10.1007/s00439-014-1472-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouksila, M. , Kaabachi, W. , Mrad, M. , Smaoui, W. , El Kateb, E. C. , Zouaghi, M. K. , … Bahlous, A. (2018). FGF 23, PTH and vitamin D status in end stage renal disease patients affected by VDR Fok I and Bsm I variants. Clinical Biochemistry, 54, 42–50. 10.1016/j.clinbiochem.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Bu, F.‐X. , Armas, L. , Lappe, J. , Zhou, Y. U. , Gao, G. , Wang, H.‐W. , … Zhao, L.‐J. (2010). Comprehensive association analysis of nine candidate genes with serum 25‐hydroxy vitamin D levels among healthy Caucasian subjects. Human Genetics, 128, 549–555. 10.1007/s00439-010-0881-9 [DOI] [PubMed] [Google Scholar]
- Cai, G. , Zhang, X. , Xin, L. , Wang, L. , Wang, M. , Yang, X. , … Pan, F. (2016). Associations between vitamin D receptor gene polymorphisms and ankylosing spondylitis in Chinese Han population: A case–control study. Osteoporosis International, 27, 2327–2333. 10.1007/s00198-016-3500-3 [DOI] [PubMed] [Google Scholar]
- Carvalho, C. , Marinho, A. , Leal, B. , Bettencourt, A. , Boleixa, D. , Almeida, I. , et al. (2015). Association between vitamin D receptor (VDR) gene polymorphisms and systemic lupus erythematosus in Portuguese patients. Lupus, 4, 846–853. 10.1177/0961203314566636 [DOI] [PubMed] [Google Scholar]
- Cavalcanti, C. A. J. , de Azevêdo Silva, J. , de Barros Pita, W. , Veit, T. D. , Monticielo, O. A. , Xavier, R. M. , … Sandrin‐Garcia, P. (2016). Vitamin D receptor polymorphisms and expression profile in rheumatoid arthritis brazilian patients. Molecular Biology Reports, 43, 41–51. 10.1007/s11033-015-3937-z [DOI] [PubMed] [Google Scholar]
- Christakos, S. (2017). In search of regulatory circuits that control the biological activity of vitamin D. Journal of Biological Chemistry, 292, 17559–17560. 10.1074/jbc.H117.806901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čierny, D. , Michalik, J. , Škereňová, M. , Kantorová, E. , Sivác, S. , Javor, J. , et al. (2016). ApaI, BsmI and TaqI VDR gene polymorphisms in association with multiple sclerosis in Slovaks. Neurological Research, 38, 678–684. 10.1080/01616412.2016.1200287 [DOI] [PubMed] [Google Scholar]
- Cortes, A. , Field, J. , Glazov, E. A. , Hadler, J. , Stankovich, J. , Brown, M. A. , … Carroll, W. M. (2013). Resequencing and fine‐mapping of the chromosome 12q13‐14 locus associated with multiple sclerosis refines the number of implicated genes. Human Molecular Genetics, 22(11), 2283–2292. 10.1093/hmg/ddt062 [DOI] [PubMed] [Google Scholar]
- Cree, B. A. (2014). Multiple sclerosis genetics. Handbook of Clinical Neurology, 1(22), 193–209. 10.1016/B978-0-444-52001-200009-1 [DOI] [PubMed] [Google Scholar]
- Dickinson, J. L. , Perera, D. I. , van der Mei, A. F. , Ponsonby, A.‐L. , Polanowski, A. M. , Thomson, R. J. , … Dwyer, T. (2009). Past environmental sun exposure and risk of multiple sclerosis: A role for the Cdx‐2 Vitamin D receptor variant in this interaction. Multiple Sclerosis Journal, 15, 563–570. 10.1177/1352458509102459 [DOI] [PubMed] [Google Scholar]
- Doroudi, M. , Olivares‐Navarrete, R. , Boyan, B. D. , & Schwartz, Z. (2015). A review of 1α,25(OH)2D3 dependent Pdia3 receptor complex components in Wnt5a non‐canonical pathway signaling. The Journal of Steroid Biochemistry and Molecular Biology, 152, 84–88. 10.1016/j.jsbmb.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Dyment, D. A. , Cader, M. Z. , Chao, M. J. , Lincoln, M. R. , Morrison, K. M. , Disanto, G. , … Ramagopalan, S. V. (2012). Exome sequencing identifies a novel multiple sclerosis susceptibility variant in the TYK2 gene. Neurology, 79, 406–411. 10.1212/WNL.0b013e3182616fc4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkum, N. , Alkayal, F. , Noronha, F. , Ali, M. M. , Melhem, M. , Al‐Arouj, M. , … Abubaker, J. (2014). Vitamin D insufficiency in Arabs and South Asians positively associates with polymorphisms in GC and CYP2R1 genes. PLoS One, 9, e113102 10.1371/journal.pone.0113102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman, C. D. , Fingerlin, T. E. , Langefeld, C. D. , Hicks, P. J. , Rich, S. S. , Wagenknecht, L. E. , … Norris, J. M. (2008). Genetic and environmental determinants of 25‐hydroxyvitamin D and 1,25‐dihydroxyvitamin D levels in Hispanic and African Americans. The Journal of Clinical Endocrinology & Metabolism, 93, 3381–3388. 10.1210/jc.2007-2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman, C. D. , Meyers, K. J. , Iyengar, S. K. , Liu, Z. , Karki, C. K. , Igo, R. P. , … Millen, A. E. (2013). Vitamin D intake and season modify the effects of the GC and CYP2R1 genes on 25‐hydroxyvitamin D concentrations. The Journal of Nutrition, 143, 17–26. 10.3945/jn.112.169482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, M. , Li, H. , Chen, S.‐F. , Li, W.‐F. , & Zhang, F.‐B. (2013). Polymorphisms in the vitamin D receptor gene and risk of autoimmune thyroid diseases: A meta‐analysis. Endocrine, 43, 318–326. 10.1007/s12020-012-9812-y [DOI] [PubMed] [Google Scholar]
- Fukazawa, T. , Yabe, I. , Kikuchi, S. , Sasaki, H. , Hamada, T. , Miyasaka, K. , & Tashiro, K. (1999). Association of vitamin D receptor gene polymorphism with multiple sclerosis in Japanese. Journal of the Neurological Sciences, 166, 47–52. 10.1016/S0022-510X(99)00112-4 [DOI] [PubMed] [Google Scholar]
- Gandhi, K. S. , McKay, F. C. , Cox, M. , Riveros, C. , Armstrong, N. , Heard, R. N. , … Bahlo, M. (2010). The multiple sclerosis whole blood mRNA transcriptome and genetic associations indicate dysregulation of specific T cell pathways in pathogenesis. Human Molecular Genetics, 19, 2134–2143. 10.1093/hmg/ddq090 [DOI] [PubMed] [Google Scholar]
- García‐Martín, E. , Agúndez, J. A. G. , Martínez, C. , Benito‐León, J. , Millán‐Pascual, J. , Calleja, P. , … Jiménez‐Jiménez, F. J. (2013). Vitamin D3 receptor (VDR) gene rs2228570 (Fok1) and rs731236 (Taq1) variants are not associated with the risk for multiple sclerosis: Results of a new study and a meta‐analysis. PLoS One, 8, e65487 10.1371/journal.pone.0065487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Rosa, S. , de Amorim, M. G. , Valieris, R. , Marques, V. D. , Lorenzi, J. C. C. , Toller, V. B. , … Dias‐Neto, E. (2017). Exome sequencing of multiple‐sclerosis patients and their unaffected first‐degree relatives. BMC Research Notes, 10(1), 735 10.1186/s13104-017-3072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfrancesco, M. A. , Stridh, P. , Rhead, B. , Shao, X. , Xu, E. , Graves, J. S. , … Waubant, E. (2017). Evidence for a causal relationship between low vitamin D, high BMI, and pediatric‐onset MS. Neurology, 88, 1623–1629. 10.1212/WNL.0000000000003849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J. G. , Tang, W. , Hootman, K. C. , Brannon, P. M. , Houston, D. K. , Kritchevsky, S. B. , … Cassano, P. A. (2015). Genetic and environmental factors are associated with serum 25‐hydroxyvitamin D concentrations in older African Americans. The Journal of Nutrition, 145, 799–805. 10.3945/jn.114.202093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , & Xie, Z. F. (2012). Polymorphisms in the vitamin D receptor gene and multiple sclerosis risk: A meta‐analysis of case‐control studies. Journal of the Neurological Sciences, 313, 79–85. 10.1016/j.jns.2011.09.024 [DOI] [PubMed] [Google Scholar]
- Jiang, T. , Tan, M. S. , Tan, L. , & Yu, J. T. (2014). Application of next generation sequencing technologies in Neurology. Annals of Translational Medicine, 2, 125 10.3978/j.issn.2305-5839.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisli, O. , Acar, C. , Sozen, M. , Tecellioglu, M. , Yücel, F. E. , Vaizoglu, D. , & Özcan, C. (2018). The association between vitamin D receptor polymorphisms and multiple sclerosis in a Turkish population. Multiple Sclerosis and Related Disorders, 20, 78–81. 10.1016/j.msard.2018.01.002 [DOI] [PubMed] [Google Scholar]
- Karaky, M. , Alcina, A. , Fedetz, M. , Barrionuevo, C. , Potenciano, V. , Delgado, C. , … Matesanz, F. (2016). The multiple sclerosis‐associated regulatory variant rs 10877013 affects expression of CYP27B1 and VDR under inflammatory or vitamin D stimuli. Multiple Sclerosis Journal, 22, 999–1006. 10.1177/1352458515610208 [DOI] [PubMed] [Google Scholar]
- Klein, C. J. , & Foroud, T. M. (2017). Neurology individualized medicine: When to use next‐generation sequencing panels. Mayo Clinic Proceedings, 92, 292–305. 10.1016/j.mayocp.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Lafi, Z. M. , Irshaid, Y. M. , El‐Khateeb, M. , Ajlouni, K. M. , & Hyassat, D. (2015). Association of rs7041 and rs4588 polymorphisms of the vitamin D binding protein and the rs10741657 polymorphism of CYP2R1 with vitamin D status among Jordanian patients. Genetic Testing and Molecular Biomarkers, 19, 629–636. 10.1089/gtmb.2015.0058 [DOI] [PubMed] [Google Scholar]
- Landel, V. , Sthefan, D. , Cui, X. , Eyles, D. , & Feron, F. (2018). Differential expression of vitamin D‐associated enzymes and receptors in brain cell subtypes. The Journal of Steroid Biochemistry and Molecular Biology, 177, 129–134. 10.1016/j.jsbmb.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Laursen, J. H. , Søndergaard, H. B. , Albrechtsen, A. , Frikke‐Schmidt, R. , Koch‐Henriksen, N. , Soelberg Sørensen, P. , … Oturai, A. (2015). Genetic and environmental determinants of 25‐hydroxyvitamin D levels in multiple sclerosis. Multiple Sclerosis Journal, 21, 1414–1422. 10.1177/1352458514563590 [DOI] [PubMed] [Google Scholar]
- Li, L.‐H. , Yin, X.‐Y. , Wu, X.‐H. , Zhang, L. , Pan, S.‐Y. , Zheng, Z.‐J. , & Wang, J.‐G. (2014). Serum 25(OH)D and vitamin D status in relation to VDR, GC and CYP2R1 variants in Chinese. Endocrine Journal, 61, 133–141. 10.1507/endocrj.EJ13-0369 [DOI] [PubMed] [Google Scholar]
- Løken‐Amsrud, K. I. , Holmøy, T. , Bakke, S. J. , Beiske, A. G. , Bjerve, K. S. , Bjørnarå, B. T. , et al. (2012). Vitamin D and disease activity in multiple sclerosis before and during interferon‐βtreatment. Neurology, 79, 267–273. 10.1212/WNL.0b013e31825fdf01 [DOI] [PubMed] [Google Scholar]
- Lu, L. , Sheng, H. , Li, H. , Gan, W. , Liu, C. , Zhu, J. , … Lin, X. U. (2012). Associations between common variants in GC and DHCR44/NADSYN1 and vitamin D concentration in Chinese Hans. Human Genetics, 131, 505–512. 10.1007/s00439-011-1099-1 [DOI] [PubMed] [Google Scholar]
- Lu, M. , Taylor, B. V. , & Körner, H. (2018). Genomic effects of the vitamin D receptor: Potentially the link between vitamin D, immune cells, and multiple sclerosis. Frontiers in Immunology, 9, 477 10.3389/fimmu.2018.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manousaki, D. , Dudding, T. , Haworth, S. , Hsu, Y.‐H. , Liu, C.‐T. , Medina‐Gómez, C. , … Richards, J. B. (2017). Low‐frequency synonymous coding variation in CYP2R1 has large effects on vitamin D levels and risk of multiple sclerosis. The American Journal of Human Genetics, 101, 227–238. 10.1016/j.ajhg.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías‐Guiu, J. , Oreja‐Guevara, C. , Matias‐Guiu, J. A. , & Gomez‐Pinedo, U. (2018). Vitamin D and remyelination in multiple sclerosis. Neurologia, 33, 177–186. 10.1016/j.nrl.2016.05.001 [DOI] [PubMed] [Google Scholar]
- Meyer, M. B. , Benkusky, N. A. , Kaufmann, M. , Lee, S. M. , Onal, M. , Jones, G. , & Pike, J. W. (2017). A kidney‐specific genetic control module in mice governs endocrine regulation of the cytochrome P450 gene Cyp27b1essential for vitamin D3 activation. Journal of Biological Chemistry, 292, 17541–17558. 10.1074/jbc.M117.806901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry, E. M. , Waubant, E. , McCulloch, C. E. , Okuda, D. T. , Evangelista, A. A. , Lincoln, R. R. , … Pelletier, D. (2012). Vitamin D status predicts new brain magnetic resonance imaging activity in multiple sclerosis. Annals of Neurology, 72, 234–240. 10.1002/ana.23591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger, K. , & Ascherio, A. (2011). Prevention and treatment of MS: Studying the effects of vitamin D. Multiple Sclerosis Journal, 17, 1405–1411. 10.1177/1352458511425366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger, K. L. , Levin, L. I. , Hollis, B. W. , Howard, N. S. , & Ascherio, A. (2006). Serum 25‐Hydroxyvitamin D Levels and Risk of Multiple Sclerosis. JAMA, 296(23), 2832– 10.1001/jama.296.23.2832 [DOI] [PubMed] [Google Scholar]
- Nissen, J. , Rasmussen, L. B. , Ravn‐Haren, G. , Andersen, E. W. , Hansen, B. , Andersen, R. , … Vogel, U. (2014). Common variants in CYP2R1 and GC genes predict vitamin D concentrations in healthy Danish children and adults. PLoS One, 9, e89907 10.1371/journal.pone.0089907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton, S.‐M. , Morris, A. P. , Herrera, B. M. , Ramagopalan, S. V. , Lincoln, M. R. , Chao, M. J. , … Ebers, G. C. (2008). Evidence for genetic regulation of vitamin D status in twins with multiple sclerosis. The American Journal of Clinical Nutrition, 88, 441–447. 10.1093/ajcn/88.2.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton, S.‐M. , Ramagopalan, S. V. , Para, A. E. , Lincoln, M. R. , Handunnetthi, L. , Chao, M. J. , … Ebers, G. C. (2011). Vitamin D metabolic pathway genes and risk of multiple sclerosis in Canadians. Journal of the Neurological Sciences, 305, 116–120. 10.1016/j.jns.2011.02.032 [DOI] [PubMed] [Google Scholar]
- Polman, C. H. , Reingold, S. C. , Banwell, B. , Clanet, M. , Cohen, J. A. , Filippi, M. , … Wolinsky, J. S. (2011). Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Annals of Neurology, 69, 292–302. 10.1002/ana.22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytel, V. , Matías‐Guiu, J. A. , Torre‐Fuentes, L. , Montero, P. , Gómez‐Graña, Á. , García‐Ramos, R. , … Matías‐Guiu, J. (2017). Familial multiple sclerosis and association with other autoimmune diseases. Brain and Behavior, 8, e00899 10.1002/brb3.899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan, S. V. , Dyment, D. A. , Cader, M. Z. , Morrison, K. M. , Disanto, G. , Morahan, J. M. , … Ebers, G. C. (2011). Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Annals of Neurology, 70, 881–886. 10.1002/ana.22678 [DOI] [PubMed] [Google Scholar]
- Ramasamy, A. , Trabzuni, D. , Forabosco, P. , Smith, C. , Walker, R. , Dillman, A. , … Ryten, M. (2014). Genetic evidence for a pathogenic role for the vitamin D3 metabolizing enzyme CYP24A1 in multiple sclerosis. Multiple Sclerosis and Related Disorders, 3(2), 211–219. 10.1016/j.msard.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. P. , Bernales, C. Q. , Lee, J. D. , Sadovnick, A. D. , Traboulsee, A. L. , & Vilariño‐Güell, C. (2014). Analysis of CYP27B1 in multiple sclerosis. Journal of Neuroimmunology, 266(1‐2), 64–66. 10.1016/j.jneuroim.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone, D. , Asani, F. , & Bornman, L. (2015). Regulation of the vitamin D receptor gene by environment, genetics and epigenetics. Gene, 561, 171–180. 10.1016/j.gene.2015.02.024 [DOI] [PubMed] [Google Scholar]
- Salzer, J. , Hallmans, G. , Nyström, M. , Stenlund, H. , Wadell, G. , & Sundström, P. (2012). Vitamin D as a protective factor in multiple sclerosis. Neurology, 79, 2140–2145. 10.1212/WNL.0b013e3182752ea8 [DOI] [PubMed] [Google Scholar]
- Sawcer, S. , Hellenthal, G. , Pirinen, M. , Spencer, C. C. A. , Patsopoulos, N. A. , Moutsianas, L. , … Compston, A. (2011). Genetic risk and a primary role for cell‐mediated immune mechanisms in multiple sclerosis. Nature, 476(7359), 214–219. 10.1038/nature10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzone, C. , Agnello, L. , Ragonese, P. , Lo Sasso, B. , Bellia, C. , Bivona, G. , … Ciaccio, M. (2018). Association of CYP2R1 rs10766197 with MS risk and disease progression. Journal of Neuroscience Research, 96(2), 297–304. 10.1002/jnr.24133 [DOI] [PubMed] [Google Scholar]
- Signorello, L. B. , Shi, J. , Cai, Q. , Zheng, W. , Williams, S. M. , Long, J. , … Blot, W. J. (2011). Common variation in vitamin D pathway genes predicts circulating 25‐hydroxyvitamin D Levels among African Americans. PLoS One, 6, e28623 10.1371/journal.pone.0028623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, K. C. , Munger, K. L. , Kraft, P. , Hunter, D. J. , De Jager, P. L. , & Ascherio, A. (2011). Genetic predictors of 25‐hydroxyvitamin D levels and risk of multiple sclerosis. Journal of Neurology, 258, 1676–1682. 10.1007/s00415-011-6001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, K. C. , Munger, K. L. , Yang, X. , & Ascherio, A. (2010). Polymorphisms in vitamin D metabolism related genes and risk of multiple sclerosis. Multiple Sclerosis Journal, 16, 133–138. 10.1177/1352458509355069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón‐Sánchez, J. , & Singleton, A. (2008). Genome‐wide association studies in neurological disorders. The Lancet Neurology, 7, 1067–1072. 10.1016/S1474-4422(08)70241-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, S. Jr , Taylor, B. , Blizzard, L. , Ponsonby, A. L. , Pittas, F. , Tremlett, H. , … van der Mei, I. (2010). Higher 25‐hydroxyvitamin D is associated with lower relapse risk in multiple sclerosis. Annals of Neurology, 68, 193–203. 10.1002/ana.22043 [DOI] [PubMed] [Google Scholar]
- Slater, N. A. , Rager, M. L. , Havrda, D. E. , & Harralson, A. F. (2017). Genetic variation in CYP2R1 and GC genes associated with vitamin D deficiency status. Journal of Pharmacy Practice, 30, 31–36. 10.1177/0897190015585876 [DOI] [PubMed] [Google Scholar]
- Sundqvist, E. , Baarnhielm, M. , Alfredsson, L. , Hillert, J. , Olsson, T. , & Kockum, I. (2010). Confirmation of association between multiple sclerosis and CYP27B1. European Journal of Human Genetics, 18(12), 1349–1352. 10.1038/ejhg.2010.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajouri, L. , Ovcaric, M. , Curtain, R. , Johnson, M. P. , Griffiths, L. R. , Csurhes, P. , … Lea, R. A. (2005). Variation in the vitamin D receptor gene is associated with multiple sclerosis in an Australian population. Journal of Neurogenetics, 19, 25–38. 10.1080/01677060590949692 [DOI] [PubMed] [Google Scholar]
- Thornton, T. , & McPeek, M. S. (2007). Case–control association testing with related individuals: A more powerful quasi‐likelihood score test. The American Journal of Human Genetics, 81, 321–337. 10.1086/519497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, T. , Zhang, Q. , Cai, X. , Ober, C. , & McPeek, M. S. (2012). XM: association testing on the X‐chromosome in case‐control samples with related individuals. Genetic Epidemiology, 36(5), 438–450. 10.1002/gepi.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizaoui, K. , Kaabachi, W. , Hamzaoui, A. , & Hamzaoui, K. (2015). Association between vitamin D receptor polymorphisms and multiple sclerosis: Systematic review and meta‐analysis of case‐control studies. Cellular & Molecular Immunology, 12, 243–252. 10.1038/cmi.2014.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohda, C. , Urano, T. , Umezaki, M. , Nemere, I. , & Kuboyama, T. (2012). Diosgenin is an exogenous activator of 1, 25D3‐MARRS/Pdia3/ERp57 and improves Alzheimer's disease pathologies in 5XFAD mice. Scientific Reports, 2(1), 535 10.1038/srep00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traboulsee, A. L. , Sadovnick, A. D. , Encarnacion, M. , Bernales, C. Q. , Yee, I. M. , Criscuoli, M. G. , & Vilariño‐Güell, C. (2017). Common genetic etiology between “multiple sclerosis‐like” single‐gene disorders and familial multiple sclerosis. Human Genetics, 136, 705–714. 10.1007/s00439-017-1784-9 [DOI] [PubMed] [Google Scholar]
- Villar‐Quiles, R. N. , Matías‐Guiu, J. A. , Ortega, G. , González‐Suárez, I. , Oreja‐Guevara, C. , & Matías‐Guiu, J. (2016). Analysis of the relationship between the month of birth and risk of multiple sclerosis in a Spanish population. European Neurology, 76, 202–209. 10.1159/000449246 [DOI] [PubMed] [Google Scholar]
- Wang, C. , Zeng, Z. , Wang, B. , & Guo, S. (2018). Lower 25‐Hydroxyvitamin D is associated with higher relapse risk in patients with relapsing‐remitting multiple sclerosis. The Journal of Nutrition, Health & Aging, 22(1), 38–43. 10.1007/s12603-017-0894-3 [DOI] [PubMed] [Google Scholar]
- Wang, T. J. , Zhang, F. , Richards, J. B. , Kestenbaum, B. , van Meurs, J. B. , Berry, D. , … Spector, T. D. (2010). Common genetic determinants of vitamin D insufficiency: A genome‐wide association study. Lancet, 376, 180–188. 10.1016/S0140-6736(10)60588-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Sadovnick, A. D. , Traboulsee, A. L. , Ross, J. P. , Bernales, C. Q. , Encarnacion, M. , … Vilariño‐Güell, C. (2016). Case‐controlstudies are not familialstudies. Neuron, 92, 339–341. 10.1016/j.neuron.2016.09.053 [DOI] [PubMed] [Google Scholar]
- Yucel, F. , Kamisli, O. , Acar, C. , Sozen, M. , Tecellioğlu, M. , & Ozcan, C. (2018). Analysis of vitamin D receptor polymorphisms in patients with familial multiple sclerosis. Medical Archives, 72(1), 58–61. 10.5455/medarh.2017.72.58-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zella, L. A. , Kim, S. , Shevde, N. K. , & Pike, J. W. (2006). Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autorregulation by 1,25‐dihydroxivitamin D3. Molecular Endocrinology, 20, 1231–1247. 10.1210/me.2006-0015 [DOI] [PubMed] [Google Scholar]
- Zella, L. A. , Meyer, M. B. , Neretz, R. D. , Lee, S. M. , Markowicz, M. L. , & Pike, J. W. (2010). Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Molecular Endocrinology, 24, 128–147. 10.1210/me.2009-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, X. , Liu, Y. , Qu, H. , Qu, S. , Wang, W. , & Ren, L. (2012). The GC, CYP2R1 and DHCR84 genes are associated with vitamin D levels in northeastern Han Chinese children. Swiss Medical Weekly, 142, w13636 10.4414/smw.2012.13636 [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Graves, J. S. , Simpson, S. Jr , Charlesworth, J. C. , Mei, I. V. , Waubant, E. , … Ponsonby, A. L. (2017). Genetic variation in the gene LRP2 increases relapse risk in multiple sclerosis. Journal of Neurology, Neurosurgery, and Psychiatry, 88, 864–868. 10.1136/jnnp-2017-315971 [DOI] [PubMed] [Google Scholar]
- Zhuang, J. C. , Huang, Z. Y. , Zhao, G. X. , Yu, H. , Li, Z. X. , & Wu, Z. Y. (2015). Variants of CYP27B1 are associated with both multiple sclerosis and neuromyelitisoptica patients in Han Chinese population. Gene, 557, 236–239. 10.1016/j.gene.2014.12.045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


