Thymineless death kills cells of any type and is used in anticancer and antimicrobial treatments. We tested the idea that the more replication forks there are in the chromosome during growth, the more extensive the resulting thymineless death. We varied the number of replication forks in the Escherichia coli chromosome, as measured by the origin-to-terminus ratio, ranging it from the normal 2 to 60, and even completely eliminated replication forks in the nonreplicating chromosomes (ori/ter ratio = 1). Unexpectedly, we found that thymineless death is unaffected by the intensity of replication or by its complete absence; we also found that even nonreplicating chromosomes still disappear during thymine starvation. We conclude that thymineless death can kill E. coli independently of chromosomal replication.
KEYWORDS: chromosomal replication complexity, chromosome degradation, dnaA(Ts), replication forks, replication initiation, thymineless death
ABSTRACT
Thymineless death (TLD) is a rapid loss of viability of unclear mechanism in cultures of thyA mutants starved for thymine/thymidine (T starvation). It is accepted that T starvation repeatedly breaks replication forks, while recombinational repair restores them, but when the resulting futile breakage-repair cycle affects the small replication bubbles at oriC, the origin is degraded, killing the cell. Indeed, cells with increased chromosomal replication complexity (CRC), expressed as an elevated origin/terminus (ori/ter) ratio, die more extensively during TLD. Here we tested this logic by elevating the CRC in Escherichia coli thyA mutants before T starvation, anticipating exaggerated TLD. Unexpectedly, TLD remained unaffected by a CRC increase to either the natural limit (ori/ter ratio, ∼6) or the functional limit (ori/ter ratio, ∼16). Moreover, when we forced the CRC over the functional limit (ori/ter ratio, ∼30), TLD lessened. Thus, prior overinitiation does not sensitize cells to TLD. In contradiction with the published results, even blocking new replication initiations by the dnaA(Ts) defect at 42°C fails to prevent TLD. Using the thyA dnaA(Ts) mutant in a new T starvation protocol that excludes new initiations, we show that at 42°C, the same degree of TLD still occurs when chromosomes are demonstrably nonreplicating. Remarkably, 80% of the chromosomal DNA in these nonreplicating T-starved cells is still lost, by an unclear mechanism.
IMPORTANCE Thymineless death kills cells of any type and is used in anticancer and antimicrobial treatments. We tested the idea that the more replication forks there are in the chromosome during growth, the more extensive the resulting thymineless death. We varied the number of replication forks in the Escherichia coli chromosome, as measured by the origin-to-terminus ratio, ranging it from the normal 2 to 60, and even completely eliminated replication forks in the nonreplicating chromosomes (ori/ter ratio = 1). Unexpectedly, we found that thymineless death is unaffected by the intensity of replication or by its complete absence; we also found that even nonreplicating chromosomes still disappear during thymine starvation. We conclude that thymineless death can kill E. coli independently of chromosomal replication.
INTRODUCTION
Discovery of the medically relevant phenomenon of thymineless death (TLD) celebrates its 65th year in 2019 (1, 2), but a mechanistic understanding is still lacking (3, 4). Denying thymine or thymidine (T) to thymine auxotrophs, the condition that we collectively refer to as “T starvation,” blocks their chromosomal DNA synthesis and is still one of the most efficient ways of causing chromosome-related death in cells of diverse complexity, from microbes to humans (5). As a result, medicines that induce thymine auxotrophy in target cells are widely used to treat conditions from microbial infections to cancer (6–8), making it even more urgent to understand the nature of thymineless poisoning.
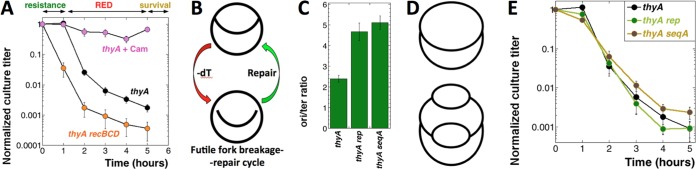
Even though TLD strikes all kinds of cells, the discovery as well as the bulk of the characterization of this complicated phenomenon was done in Escherichia coli (3, 5), in which the “classic” TLD curve shows three phases (Fig. 1A): (i) the resistance phase equivalent to up to 2 generations of normal (with thymidine [+dT]) growth; (ii) the rapid exponential death (RED) phase equivalent to ≥6 generations of +dT growth; and (iii) the survival phase (9, 10).
FIG 1.
The TLD phenomenon and its modifiers, a general explanation, and the effect of CRC doubling. (A) The three phases of TLD kinetics in the thyA mutant (at 37°C): (i) the resistance phase (no loss of CFU titer), (ii) the rapid exponential death (RED) phase (CFU titer loss of 3 orders of magnitude), and (iii) the survival phase (stabilization of the CFU titer). At time zero, the dT supplementation was removed (and chloramphenicol was added in the +Cam culture). The strains are as follows: thyA mutant, KKW58; thyA recBCD mutant, KJK63. The kinetics of the thyA recBCD double mutant, as well as the thyA mutant without dT but in the presence of chloramphenicol, are shown as examples of modified behavior during T starvation. Here and in all figures, unless indicated otherwise, all values are means of the results of 3 to 12 independent measurements ± standard error of the mean (SEM). (B) A possible scheme of the futile fork breakage-repair cycle. (C) ori/ter ratios in the rep and seqA mutants grown in the presence of dT. The strains are as follows: thyA mutant, KKW58; thyA rep mutant, KJK202; thyA seqA mutant, SRK271. (D) Schemes of the replicating chromosomes in WT cells (top) versus rep or seqA mutants (bottom). (E) TLD kinetics of the thyA rep and thyA seqA double mutants compared with the thyA single mutant. The strains are as described for panel C.
At first, TLD was explained as “unbalanced growth” due to an inability to synthesize DNA in otherwise normally growing cells (2, 6), but at least two “rebalancing” ideas to explain the resistance phase turned out to be off target (9). Instead, it was found that parallel protein synthesis inhibition by chloramphenicol or by amino acid starvation, which blocks initiation of chromosomal replication, makes E. coli cells immune to TLD (Fig. 1A) (11, 12), pointing to nascent replication bubbles as the likely main target of thymineless poisoning. More recently, rifampin was proposed to save E. coli from TLD by blocking the transcriptional activation of the replication origin (13, 14). Indeed, blocking new initiations with the dnaA(Ts) defect at nonpermissive temperatures has been reported to save affected cells (14, 15), suggesting that the newly induced forks are more susceptible to T starvation-induced damage than preexisting forks. The recently found T starvation-induced local overreplication (16) that subsequently destabilizes the chromosomal macrodomain around the replication origin (16–18) emphasized new initiations as the critical factor in TLD. Finally, a strong correlation was demonstrated between the magnitude of TLD and the replication complexity of the chromosome at the time of T removal (13), again implicating new initiations in T starvation-induced chromosomal damage.
The resulting understanding of chromosomal pathology during T starvation, although still lacking mechanistic details, frames the formation of irreparable chromosomal lesions (DNA lesions that block the chromosome cycle and cannot be repaired even when dT is restored) within the following simple logic (Fig. 1B). In T-starved cells, inhibited replication forks are unstable and disintegrate by several possible mechanisms (19, 20), yet in the presence of functional recombinational repair in thyA mutants, such disintegrated forks are rapidly reassembled (21, 22). The replication fork instability and repair during T starvation are supported by the fact that the thyA recBCD mutants, deficient in repair of disintegrated replication forks, have no resistance phase and show extreme sensitivity to T starvation (Fig. 1A) (10, 23). However, the recombinational repair proficiency of the thyA mutants does not save them during continuing T deprivation, presumably because the restored replication forks are still inhibited and will break again, going through the futile cycle of fork breakage and repair (Fig. 1B), which, eventually, leads to the formation of double-strand gaps at the smallest replication bubbles, resulting in the loss of the replication origin (16–18, 24). Importantly, although this futile fork breakage-repair cycle scheme appears to make sense, it still lacks mechanistic details linking replication fork metabolism with formation of irreparable chromosomal lesions (for example, destruction of all replication origins). To enhance experimental testing of this TLD paradigm, we sought to significantly increase replication fork numbers in the chromosome before the onset of starvation and to quantify the elicited survival and chromosomal changes.
Chromosomal replication complexity (CRC) represents the ratio of the copy number of the most replicated region in the chromosome to the copy number of the unreplicated regions (25, 26). In bacterial chromosomes, the CRC is simply expressed as the origin/terminus (ori/ter) ratio. While the CRC of eukaryotic chromosomes and in slowly growing bacterial cells rarely exceeds 2, the ori/ter ratio goes up to 8 (representing the “natural” CRC limit [25]) in rapidly growing bacteria, as well as in E. coli cells starved for thymidine (27) or for dGTP (another DNA precursor) (28). In E. coli, the CRC can be increased beyond the natural limit to an ori/ter ratio of ∼22, the “functional” CRC limit (the highest one achievable from oriC and without viability loss) (25, 29). Finally, the functional CRC limit in E. coli can be overridden by inserting an inducible plasmid replication origin into the chromosome, although the resulting ori/ter ratio increase leads to stasis and, eventually, to decreased viability, presumably due to a disoriented recombinational repair generating “pince-nez chromosomes” (25).
The logic of T starvation-induced chromosomal damage at replication forks predicts that, with more replication forks in the chromosome, TLD will be faster and more extensive (as in the thyA recBCD mutants [Fig. 1A]), due to the aggravated chromosome destruction and disorientation of recombinational repair. In this work, we brought E. coli cells to various levels of CRC and then challenged these cells with thymine starvation to test this prediction.
RESULTS
Tripling the number of replication forks does not affect TLD.
In bacteria, CRC reflects both the frequency of replication initiation and the rate of replication fork elongation. Thus, one of the simplest ways to increase CRC in E. coli is to use mutants that either cannot control overinitiation, like seqA (30), or have to overinitiate because of slow replication forks, like rep (31). When grown in rich medium, these mutants raise the CRC close to its natural limit of an ori/ter ratio of ∼8 (25), which is defined by the eclipse system that controls the interval between consecutive initiations (32, 33). In MOPS-CAA medium (morpholinepropanesulfonic acid minimal phosphate medium supplemented with 0.2% glucose and 0.2% Casamino Acids [CAA]), both seqA and rep mutants elevate the CRC from ∼2.5 to ∼5 (2-fold) (Fig. 1C), which approximately triples the number of replication forks in their chromosomes (Fig. 1D). If replication forks are indeed the points of chromosomal vulnerability during TLD, increasing their numbers should make the resulting mutants hypersensitive to thymine starvation, reducing their resistance phase and/or deepening the RED phase, similar to the effect of the recBCD inactivation (Fig. 1A). In contrast to these expectations, we detected no change in the TLD kinetics of the seqA and rep mutants (Fig. 1E). In other words, more than tripling the number of replication forks (by doubling the CRC) was without detriment to the chromosomes during T starvation, at least in these two mutants.
Effect on TLD of the 5-fold increase in the number of replication forks.
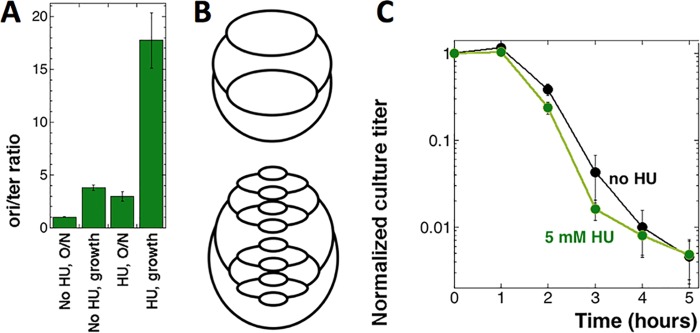
To increase the CRC beyond the natural limit, we chronically inhibited replication forks by growing wild-type (WT) cells in the presence of 5 mM hydroxyurea (HU), a specific inhibitor of the enzyme ribonucleotide reductase responsible for the biosynthesis of DNA precursors (34, 35). In the presence of 5 mM HU, E. coli cells grow slowly in rich medium and the CRC increases to its functional limit of an ori/ter ratio of ∼22, inherent to the initiation capacity of the oriC/DnaA system (25). Propagation in 5 mM HU-supplemented MOPS-CAA medium with dT (MOPS-CAA +dT medium) elevates CRC in our cells from ∼4 to ≥16 (4-fold) (Fig. 2A), increasing the number of replication forks five times (Fig. 2B). However, the TLD curve of the HU-treated strain remains essentially the same as that of the untreated control (Fig. 2C); therefore, multiple additional replication bubbles at the time of dT withdrawal again fail to exacerbate TLD.
FIG 2.
Quadrupling CRC does not change the TLD kinetics. (A) ori/ter ratios of the thyA mutant (KKW58) in either stationary or growth phase and in either the absence or the presence of 5 mM HU for at least 10 generations. The assay was done at 28°C, as HU inhibition is inefficient at 37°C. (B) Schemes of the replicating chromosomes in WT cells undergoing normal initiation without HU (top panel, CRC = 4) and after treatment with HU, which causes overinitiation to compensate for replication fork inhibition (bottom panel, CRC = 16). (C) TLD kinetics of the thyA mutant (KKW58), pregrown in either the absence or presence of 5 mM HU. Hydroxyurea was removed at time zero.
Inducible chromosomal overreplication lessens TLD.
Insertion of an inducible plasmid origin (inducible origin construct [IOC], comprising two back-to-back ColE1-type IPTG [isopropyl-β-d-thiogalactopyranoside]-driven origins) ∼5 kbp from oriC allows one to override the initiation constraints of the latter (25). Previously, conditions for maximal IOC induction proved hard to identify (suggesting that the chromosome prefers to stay within the functional CRC limit of 22), although eventually we were able to reliably bring the ori/ter ratio to ≥64 in cells grown in rich medium (25). In this work, we had to optimize the IOC induction in the MOPS-CAA medium to increase the ori/ter ratio above 16, the functional CRC limit in the MOPS-CAA medium that we achieved with HU (Fig. 2A); the induced overreplication was also expected to be modest in this semidefined medium.
The control cultures, even though deeply diluted, usually reached saturation while waiting for the induction, and saturated cultures of thyA mutants, changed into the same volume of fresh medium with dT removed (–dT medium), are immune to thymine starvation (Fig. 3A), a basic observation that, to our knowledge, has never been reported before. Therefore, we had to appropriately dilute the control thyA mutant culture at the time of T removal, so that its optical density (OD) would roughly match the OD of the induced IOC cultures.
FIG 3.
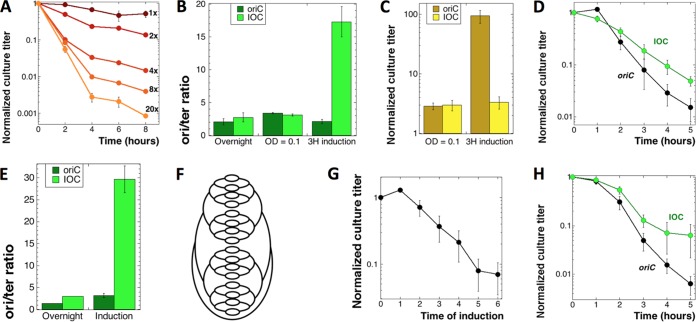
Driving CRC above the functional limit lessens TLD. Throughout this figure, “oriC” stands for our regular ΔthyA mutant (KKW58), while “IOC” stands for the ΔthyA mutant with the inducible origin construct (SRK290). (A) The extent of the dilution of stationary ΔthyA mutant cultures into fresh –dT medium determines the extent of subsequent TLD. 1× to 20×, fold dilutions into fresh medium. (B) ori/ter ratio of the oriC and IOC strains under induction condition 1. Cultures were grown overnight at 37°C in MOPS-CAA +dT medium, subcultured in the same medium, and grown to an OD600 of 0.1. At this time, IPTG was added, and induction was continued for 3 h before determination of the ori/ter ratio. (C) Culture titers upon induction under condition 1 (normalized to CFU at the time of subculture). (D) TLD kinetics of the IOC strain versus the thyA mutant under induction condition 1 (described for panel B). The cells were washed and T starved in MOPS-CAA medium without IPTG. Since during the induction the thyA culture became dense, it was diluted to an OD of 0.1 to 0.2 at the onset of starvation. (E) ori/ter ratios of the thyA and IOC strains grown under induction condition 2. Cultures of thyA and IOC strains were grown overnight at 37°C in MOPS-CAA +dT medium, subcultured in MOPS-CAA +dT medium with IPTG, and allowed to grow until the OD of IOC reached 0.1 to 0.3 and stopped increasing further. Incubation was continued for another 2 h, following which both cultures were harvested, and samples were collected to determine the ori/ter ratio. (F) The chromosomal scheme of IOC under induction condition 2 (CRC = 32). (G) Culture titers of the IOC strain upon induction under condition 2 (normalized to preinduction CFU). (H) TLD kinetics of the IOC mutant versus the thyA mutant under induction condition 2 (described for panel E). The cells were washed and starved in MOPS-CAA medium without IPTG.
Under induction condition 1, when cells were initially grown without IPTG and then induced for 3 h before the onset of starvation, we managed to increase the ori/ter ratio from 3 in the uninduced cultures to ∼18 in the induced cultures, by a factor of ∼6-fold (Fig. 3B). Under these mild induction conditions, the cultures became static, keeping their preinduction titer (Fig. 3C). We found that the TLD time course of the induced IOC thyA culture, instead of a deeper slope, developed a shallower slope (Fig. 3D).
Under induction condition 2, in which we maximized overinitiation by adding IPTG to freshly diluted cultures and shaking them for at least 6 more hours, the CRC was elevated from ∼3 in the thyA mutant cultures to ∼30 in the induced IOC cultures (Fig. 3E), increasing the number of replication forks ∼16-fold (Fig. 3F). The observed 10-fold induction was close to the maximal overinitiation factor achieved in the previous work (25). Under induction condition 2, the titer of overinitiating cultures gradually decreased (Fig. 3G), as was reported previously (25). The effect on TLD was unchanged, though; the induced overreplication again partially saved the cells from TLD (Fig. 3H). We concluded that amplifying the number of replication forks beyond the functional CRC limit not only failed to make cells more vulnerable to TLD but, in fact, made them more resistant to it.
Blocking new initiations from the origin during thymine starvation.
As explained in the introduction, the previous demonstration that TLD is blocked by preventing new initiations in the thyA dnaA(Ts) mutants by shifting them to the nonpermissive temperature upon dT removal (14, 15) dovetails with the demonstration of the spike of new replication initiations triggered by thymine starvation (16) and with the eventual origin macrodomain destruction coincident with TLD (16, 18). Our unsuccessful attempts to aggravate TLD by artificially increasing the number of replication forks in the chromosomes of prestarved cells suggested either general TLD independence of the number of replication forks or at least TLD independence of the number of preexisting forks. According to the latter idea, only the forks that are induced during T starvation make the chromosome vulnerable to TLD, the idea being supported by the initiation spike in response to T starvation, followed by the disappearance of overinitiated DNA during TLD (9, 16). The initiation spike in response to T starvation is a natural reaction of cells to inhibition of DNA synthesis during normal cell mass increase (27, 36).
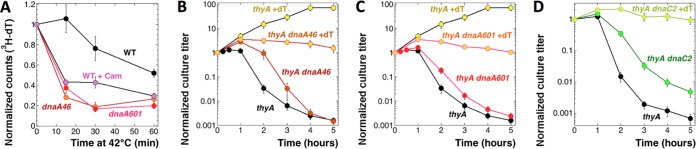
If only new initiations made the chromosome vulnerable to TLD, then blocking new initiations by shifting the dnaA(Ts) mutant to the nonpermissive temperature at the time of dT removal should prevent TLD, as has already been reported (14, 15). Our dnaA(Ts) mutants finished all DNA synthesis activities within 30 min of the shift to 42°C, as expected, paralleling the behavior of chloramphenicol-treated cells (Fig. 4A). However, we found that in our hands, both the thyA dnaA46(Ts) mutant (Fig. 4B) and the thyA dnaA601(Ts) mutant (Fig. 4C) died at 42°C in the absence of dT with kinetics only slightly slower than that of thyA DnaA+ cells and showed the same three TLD phases. Because of this surprising finding, we also tested the thyA dnaC2(Ts) mutant, which at 36°C is completely blocked for initiations, either from the origins or from forks restarted anywhere in the chromosome (37), and found a slower kinetics of TLD that still projected to the same low survival (Fig. 4D). Since some dnaA(Ts) and dnaC(Ts) mutants are known to die at nonpermissive temperatures (38, 39), we made sure that our thyA dna(Ts) mutants are stable at their nonpermissive temperatures when supplemented with thymidine (Fig. 4B to D). Also, as expected, the ori/ter ratio in the thyA dnaA601(Ts) mutant in the presence of dT at 42°C decreases from ∼2 to ∼1 within 1 h (Fig. 5A), indicating that the dnaA(Ts) defect efficiently blocks new initiations from the origin at 42°C, without interfering with the progress of existing forks. We conclude that (i) the dnaA(Ts) or dnaC(Ts) defects at the nonpermissive temperatures do not confer immunity against TLD, in contrast to previous reports (14, 15), and (ii) TLD in the dnaA(Ts) or dnaC(Ts) mutants at their nonpermissive temperatures suggests that the preexisting replication round in T-starved cells is sufficient to damage the chromosome beyond repair.
FIG 4.
Blocking replication initiations during T starvation does not save cells from TLD. (A) Kinetics of the rate of DNA synthesis at 42°C, as determined by [3H]dT incorporation. The strains are WT (AB1157), either untreated or treated with 40 µg/ml chloramphenicol (Cam), as well as dnaA46(Ts) (SRK309-1) and dnaA601(Ts) (SRK270-1) mutants (all strains are Thy+). Cultures were pregrown at 28°C to an OD of 0.4 to 0.5 before being shifted to 42°C. The variation in the initial counts between different cultures was from ∼6,000 to ∼14,000 cpm. (B) TLD kinetics of the thyA dnaA46(Ts) mutant (KJK170) versus the thyA DnaA+ parent (KKW58), both grown in the presence of thymidine at 28°C and switched to –dT medium with a simultaneous shift to 42°C at time zero. As a control for dnaA46(Ts) viability at the nonpermissive temperature, half of each culture was switched to fresh medium supplemented with thymidine (+dT) and also incubated at 42°C. (C) The same as for panel B but with the thyA dnaA601(Ts) mutant (SRK291). (D) The same as for panel B but with the thyA dnaC2(Ts) mutant (RA101) and a temperature of 36°C.
FIG 5.
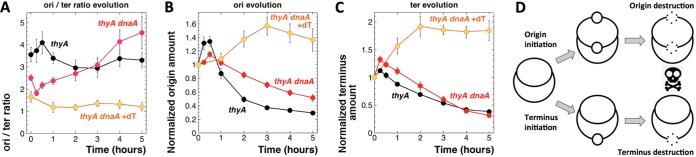
Evolution of the origin and terminus quantities in the thyA and thyA dnaA601 cultures during T starvation at 42°C. The strains are as follows: thyA mutant, KKW58; thyA dnaA601(Ts) mutant, SRK291. (A) Evolution of the ori/ter ratios in the thyA –dT, thyA dnaA601(Ts) –dT, and dnaA thyA601(Ts) +dT cultures at 42°C. In the –dT cultures, dT was removed at time zero. Experimental conditions were as described for Fig. 4B. (B and C) Evolution of the origin (B) and terminus (C) amounts in the cultures and conditions described for panel A. (D) Model of the chromosomal behavior during thymine starvation. All chromosomes initiate, but some do it in the origin, while others do it in the terminus. As a result of thymine limitation, both types of nascent bubbles become vulnerable to degradation, opening irreparable double-strand gaps in the chromosome.
T-starved cells attempt to initiate at either the origin or the terminus.
To further investigate the unexpected TLD in our dnaA(Ts) mutants at the nonpermissive temperature, we made sure that switching thyA dnaA(Ts) cultures from 28°C to 42°C upon dT removal did not relieve the initiation block (the block itself is evident from the decrease of the ori/ter ratio to 1 in the presence of dT at 42°C [Fig. 5A]). For this, we first confirmed the initiation spike triggered by T starvation in the DnaA+ thyA cells, reported before as a rapid increase in the ori/ter ratio following dT withdrawal (16). At the point of thymidine removal, the thyA mutant and the thyA dnaA mutant have similar ori/ter ratios (3.5 versus 2.5) (Fig. 5A, zero time points), yet their subsequent evolution during T starvation was clearly opposite (Fig. 5A). The general trend of the ori/ter ratio of the thyA mutant at 42°C repeated the previously observed trend at 28°C (16), by going slightly up to 4 but then coming down and stabilizing around 3. In contrast, the ori/ter ratio in the thyA dnaA mutant declined to 2 right after dT removal (Fig. 5A), apparently reflecting the initiating defect of the mutants at 42°C. Surprisingly, the ori/ter ratio in the double mutant then gradually climbed to 4.5 (Fig. 5A), suggesting either new origin initiations or significant terminus loss under the dnaA-inactive conditions. The possible replication from new initiations under the dnaA-deficient conditions might explain TLD in the thyA dnaA(Ts) mutant at 42°C.
To detect the suspected initiations directly, we monitored the evolution of the absolute origin amount in the three cultures (Fig. 5B), as described previously (16). In the presence of dT, in the thyA dnaA(Ts) mutant incubated at 42°C, the origin amount stayed the same for 1 h, reflecting no new initiations, but then slowly increased to ∼1.5-fold, likely because the accumulating cell mass increased the DnaA copy number per cell, eventually compromising the initiation defect of the mutant. Removal of thymidine in both thyA and thyA dnaA mutant strains triggered the absolute origin amount to peak modestly in 30 min and then to decline (Fig. 5B), again as was described previously at 28°C (16) and at 37°C (9), apparently reflecting the previously reported degradation of the origin macrodomain during TLD (16, 18). As anticipated, in the thyA dnaA mutant, both the original spike (∼1.2-fold) was lower than that in the thyA mutant (∼1.4-fold) and the subsequent origin disappearance was more modest (2-fold in the thyA dnaA mutant versus >3-fold in the thyA DnaA+ mutant). We conclude that the dnaA(Ts) defect at 42°C mostly inhibits, but does not completely block, new initiations and slows down the origin destruction. Surprisingly, these real differences in the origin quantities have little influence on the overall kinetics of TLD in the two strains (Fig. 4C).
Since the evolution of the absolute origin amount did not reveal the reason for the increasing ori/ter ratio in the thyA dnaA mutant, we looked at the evolution of the absolute terminus amount, expecting more modest changes, observed previously both at 28°C (16) and at 37°C (9). In the presence of dT, the absolute terminus amounts in the thyA dnaA(Ts) mutant at 42°C behaved as expected, climbing steadily and then plateauing at 2-fold the original amount (Fig. 5C), reflecting successful completion of the existing replication round with no further initiations. Unexpectedly, the evolution of the terminus amount in the absence of thymidine was almost as dramatic as the evolution of the origin amount (compare Fig. 5B and C): in both strains, the terminus amount first modestly increased but then steadily decreased to values ∼30% of the original ones.
Curiously, the relationship between the two strains was now reversed: while the thyA mutant showed a tiny terminus increase followed by a 3-fold decrease, the thyA dnaA mutant showed a higher terminus increase, followed by a 4.2-fold decrease. Perhaps the higher terminus increase in the thyA dnaA mutant was because, under T starvation conditions, more current replication forks could finish in the absence of competition from the newly initiated rounds that would happen in the thyA cells. However, real initiations at the terminus, as observed in certain mutants (40, 41), could not be excluded. Thus, the evolution of the ori/ter ratios during T starvation at 42°C was a product of the significant evolution of both the origin and the terminus under these conditions, while the opposite trends of this evolution for thyA versus thyA dnaA(Ts) strains (Fig. 5A) reflected the switch in the relative magnitudes of the two effects between the two strains.
Encouraged by these results, we formulated a new model of chromosome behavior during T starvation. In contrast to previous ideas, which placed the emphasis on the number of preexisting replication forks, the new model focused only on the initiations during T starvation. It further posited that T starvation triggers new initiations either at the origin or at the terminus, mostly at the origin in the thyA mutants while mostly at the terminus in the thyA dnaA(Ts) mutants (Fig. 5D). The nascent replication bubbles would be unstable in the absence of dT and eventually disintegrate, resulting in irreparable double-strand gaps in the chromosome, either at the origin or the terminus (whichever initiates), killing the cell. Terminus signal disappearance during TLD has been reported before (16, 24). An attractive feature of this model is that it proposed that the same event (initiation followed by disintegration and degradation) would kill independently of the chromosomal location of the new initiation. A useful aspect of the model was that it explained the evolution of the ori/ter ratio in both the thyA mutants (where ori initiates more than ter and is preferentially destroyed, driving the ori/ter ratio down) and the thyA dnaA mutants (where ter initiates more than ori and is preferentially destroyed, driving the ori/ter ratio up) (compare Fig. 5A and D).
The “awakening” T starvation protocol completely blocks DNA replication.
To test our new model, we blocked any replication activity in the chromosome at the time of dT removal by preincubating the thyA dnaA(Ts) cultures for 2 h at 42°C, still in the presence of thymidine, to run off the existing replication rounds, and then removing dT at 42°C and continuing to keep the T-starved cultures at 42°C to prevent any further initiation. The result was unexpected: while the thyA dnaA(Ts) control with dT decreased ∼6-fold in titer (similar to other experiments) and the thyA mutant without dT showed a somewhat delayed, yet typical TLD curve, the thyA dnaA(Ts) mutant without dT lost the resistance phase but then died slowly and steadily, losing 99% of its original titer by 5 h of T starvation, without signs of plateauing (Fig. 6A). Importantly, this was the first clear observation of TLD in cells that were expected to have no chromosomal DNA replication whatsoever. Unfortunately, besides generating an unexpected behavior of the thyA dnaA(Ts) mutant, this protocol was also nontrivial to perform, in that it required changing the medium in cultures while keeping them at 42°C, so we sought another way of T-starving cells with no chromosomal replication, yet normal metabolism.
FIG 6.
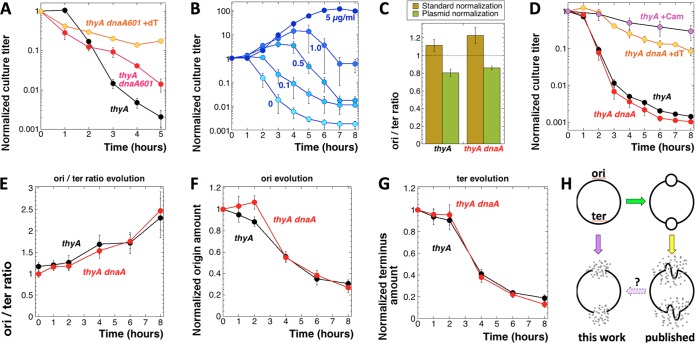
The new protocol for T starvation to ensure that dnaA(Ts) cells do not initiate or harbor preexisting forks. The strains are as follows: thyA mutant, KKW58; thyA dnaA601(Ts) mutant, SRK291. (A) TLD kinetics of the thyA dnaA601(Ts) mutant versus the thyA DnaA+ parent, both grown in the presence of thymidine at 28°C to an OD of 0.1 and shifted to 42°C for 2 h before being switched to the –dT medium and with continued incubation at 42°C. As a control for dnaA(Ts) viability at the nonpermissive temperature, half of the thyA dnaA601(Ts) culture was switched to fresh +dT medium and also incubated at 42°C. (B) The “awakening” protocol of thymine starvation: direct dilution of the overnight (15- to 18-h) culture into fresh medium supplemented with various concentrations of dT, from 0 to 5 µg/ml (the values marking the curves), at time zero and with shaking at 37°C. (C) ori/ter ratios in 48-h MOPS-CAA +dT cultures. Standard normalization, normalization to the average of 24-h stationary cultures in LB and cultures treated with chloramphenicol for 3 h; plasmid normalization, normalization to the “loading control” plasmid pSRK-OD that carries one copy each of the origin and terminus probes. The horizontal dotted line indicates the expected ori/ter ratio of 1 in a perfectly aligned chromosome. (D) TLD kinetics of the thyA dnaA(Ts) mutant versus the thyA single mutant, grown to saturation (for 48 h) in the presence of thymidine at 28°C, washed and diluted into thymineless medium prewarmed and with shaking at 42°C. As a control for dnaA(Ts) viability at the nonpermissive temperature, a part of the 48-h dnaA thyA culture was diluted into fresh +dT medium, also prewarmed and with shaking at 42°C. (E) Evolution of the ori/ter ratios in the 48-h +dT cultures of thyA and thyA dnaA(Ts) mutants at 28°C after they were diluted into the same –dT medium and with incubation at 42°C (“awakening protocol”). (F and G) Evolution of the origin (F) and terminus (G) amounts in the same cultures and conditions as described for panel E. (H) Schematic illustration of the current findings. Left column, degradation of the origin and terminus in a nonreplicating chromosome during T starvation; right column, the published findings, according to which only the chromosomal regions that initiate during T starvation are susceptible to an incomplete loss.
In an initially unrelated development, to minimize the metabolic and handling disturbances in cells growing during T starvation, we designed a new protocol, in which we pregrew thyA mutant cultures in the presence of dT for 48 h to make sure they were fully stationary, and their chromosomes were aligned, and then diluted the cultures 50-fold in fresh medium containing various concentrations of dT, from 0 to 5 µg/ml. At the time of culture dilution, the cultures could be also shifted to new conditions, for example from 28°C to 42°C. In this new protocol, the cells would grow out, as expected, in the presence of the regular amounts of dT, while showing increasingly limited outgrowth with decreasing amounts of dT, which would eventually turn into an increasingly extensive TLD (Fig. 6B). Remarkably, the TLD curve of cells awakening from the 48-h stasis in the complete absence of dT faithfully reproduced the typical TLD curve of a growing culture (with active metabolism and replication forks) changed into medium without dT (compare Fig. 6B with Fig. 1A, 2C, or 3A, 20-fold dilution). This unexpected behavior of stationary thyA cultures upon dilution into T-less medium was mentioned previously (42).
We used the “awakening” T starvation protocol to securely prevent replication initiation upon switching the cultures to a –dT medium. For this, we pregrew thyA and thyA dnaA(Ts) cultures at 28°C for 48 h into stationary phase to fully align their chromosomes and then diluted the cultures into a medium lacking dT that had been prewarmed at 42°C to initiate starvation. The ori/ter ratios of the thyA DnaA+ and thyA dnaA(Ts) cultures were around 1 after 48 h at 28°C in +dT medium (Fig. 6C), confirming chromosome alignment. After dilution into fresh +dT medium, the stationary thyA dnaA(Ts) cultures lost some titer at 42°C (Fig. 6D), similar to other 42°C incubation results in +dT medium (Fig. 6A). At the same time, neither thyA DnaA+ nor thyA dnaA(Ts) mutants showed any signs of initiation at oriC in the absence of dT at 42°C (Fig. 6F, compare time zero with 1 h), so initiation was tightly controlled. We found that in the awakening T starvation protocol, the thyA and thyA dnaA(Ts) mutant TLD curves at 42°C were essentially superimposable (Fig. 6D), confirming that new initiations from oriC were not required for T starvation to kill E. coli. At the same time, addition of chloramphenicol at the time of culture dilution without dT still prevented TLD (compare Fig. 6D and 1A), showing that the observed killing was not due to the switch to 42°C.
Destruction of the whole chromosome during T starvation at 42°C.
To measure possible initiations from either origin or terminus under the awakening protocol, we followed the evolution of origin and terminus in both thyA and thyA dnaA(Ts) mutants at 42°C, also calculating the ori/ter ratios. The ori/ter ratios in the thyA and thyA dnaA(Ts) mutants at 42°C paralleled each other, climbing to over 2.0 after 8 h of T starvation (Fig. 6E). From the results with unaligned cultures (Fig. 5), we already knew that the increase in the ori/ter ratio might reflect preferential terminus degradation rather than origin initiation. This turned out to be the case in the awakening protocol too: (i) the curves of origin evolution (Fig. 6F) or terminus evolution (Fig. 6G) at 42°C for thyA and thyA dnaA(Ts) cultures were essentially superimposable and showed no significant initiation in either strain or at either chromosomal location; (ii) both the origin and the terminus locations showed a remarkable loss over 8 h of T starvation, i.e., the origin lost ∼70% of the original amount while the terminus lost ∼80% of the original amount, explaining the gradual rise of the ori/ter ratio.
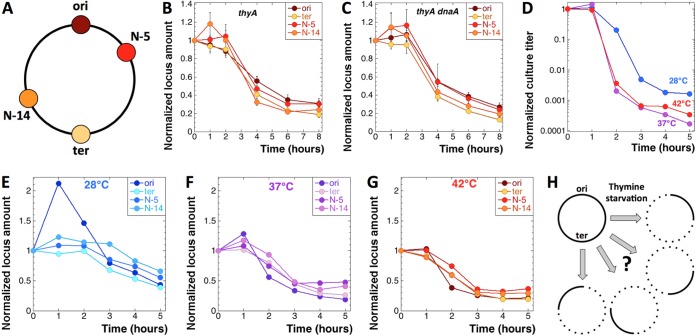
Such a strong loss at the two opposite chromosomal locations (Fig. 6H, left column) was consistent with the preferential destruction of the chromosomal domains around oriC and the terminus, or at least the two loci themselves, reported previously as occurring in T-starved cells (16, 18, 24), although the more modest scale of destruction in the previous reports never exceeded 50% of the original amount and was deemed the consequence of replication initiation (Fig. 6H, right column). To investigate whether the chromosomal destruction at 42°C was indeed specific for origin and terminus even without replication initiation, we determined the evolution at two more chromosomal locations: N-5 in the right replichore (740 kbp from oriC) and N-14 in the left replichore (1,413 kbp from oriC) (Fig. 7A) (43). When the evolution of all four chromosomal locations was compared in the awakening protocol at 42°C, either in the thyA mutant (Fig. 7B) or in the thyA dnaA(Ts) mutant (Fig. 7C), no significant differences were observed, suggesting that all chromosomal regions were lost with similar kinetics, independently of whether initiation could potentially happen at them or not. In other words, the overall chromosomal DNA amount within 8 h of T starvation at 42°C was down to approximately one-quarter to one-fifth of the original signal (Fig. 7B and C).
FIG 7.
T starvation at 42°C leads to the general destruction of even nonreplicating chromosomes. The strains are as follows: thyA mutant, KKW58; thyA dnaA601(Ts) mutant, SRK291. (A) Locations of the four chromosomal loci for which we probed: oriC, N-5, N-14, and dif (ter). (B) Evolution of the amounts of the four chromosomal loci (shown in panel A) in the thyA mutant during the T starvation awakening protocol at 42°C. (C) Same as for panel B but in the thyA dnaA(Ts) mutant. (D) TLD kinetics of the thyA mutant at 28°C, 37°C, or 42°C in the cultures used for analysis in panels E, F, and G. In contrast to all other data in the figures, the data in panels D, E, F, and G are from a single assay. (E to G) Evolution of the amounts of the four chromosomal loci, oriC, N-5, N-14, and dif (ter), in the thyA mutant during the standard T starvation protocol (growing cultures) at 28°C (E), 37°C (F), and 42°C (G). (H) General scheme of the nonreplicating chromosome changes during TLD.
Since the chromosomal destruction at 42°C turned out to be locus independent, in contrast to the previous observations at 28°C (16) and at 37°C (18), we performed the same four-point analysis of the chromosomal DNA evolution in the thyA single mutant under standard conditions (growing cultures), comparing starvation at 42°C with that at 37°C and 28°C. As expected, the time course of TLD was delayed at 28°C (Fig. 7D), while initiation at the origin was significant during the resistance phase (Fig. 7E), exactly as described previously (16). Interestingly, the behavior of the four chromosomal positions at 37°C was not much different from their behavior at 42°C (possible initiation at the origin, significant disappearance of the signal) (Fig. 7F and G). We conclude that the entire chromosome becomes vulnerable during TLD at any temperature, but at 28°C there is also significant initiation from the origin that, nevertheless, seems to have little effect on the subsequent chromosomal DNA disappearance.
DISCUSSION
Overall, our experiments failed to clearly link TLD with any number of preexisting replication bubbles or even with the nascent replication bubbles. Instead, we found two T starvation conditions [dnaA(Ts) preincubation with dT at 42°C and “awakening” at 42°C] in which cells with demonstrably no chromosomal replication suffer total TLD. Curiously, compared to T starvation at 28°C described previously (16), T starvation at 42°C according to both the classic and awakening protocols maximizes the overall loss of chromosomal DNA. Since the final survival is down to 10−3 in these cultures (Fig. 4B and C and 6D), while there is still ∼25% of the chromosomal DNA left in these cells (Fig. 7B, C, and G), we imagine that all individual chromosomes (except 1 out of 1,000) have lost most of their DNA at random, averaging 75% (Fig. 7H). The mechanism of this dramatic and apparently nonspecific chromosomal DNA loss at 42°C is currently unclear. A recent finding of reactive oxygen species accumulation in T-starved cells suggests that chromosomal DNA degradation is due to oxidation-induced double-strand breaks (44). While we are in the process of evaluating these observations and ideas, a somewhat less dramatic loss of the chromosomal DNA during TLD at 37°C (Fig. 7F) turned out to be independent of the RecBCD enzyme, the main linear DNA degradation activity in E. coli (9), making the overall scenario unlikely. Also, it remains to be tested whether this dramatic chromosomal DNA loss is the cause or the consequence of cell death during T starvation.
When just discovered, the phenomenon of thymineless death was thought to be due to an unbalanced growth, an inability to replicate DNA in otherwise growing cells (2, 6). However, a recent experimental testing of two specific rebalancing strategies as explanations for the resistance phase turned up negative (9). Interestingly, populational alignment by chloramphenicol treatment or amino acid starvation of auxotrophs promoted the conclusion that cells with no replication forks in their chromosomes develop immunity against thymineless death (11, 12). The same conclusion was derived from blocking origin initiation by inhibiting transcription (13, 14). Of course, these treatments, by blocking protein synthesis, would also “rebalance” cell growth by inhibiting it, but these experiments channeled understanding of thymineless death as the phenomenon of chromosome poisoning at inhibited replication forks (10, 11, 14, 15) (Fig. 1B).
Along the same lines, T starvation survival of the dnaA(Ts) mutants at the nonpermissive temperature was interpreted to mean preferential susceptibility of the newly initiated replication bubbles to T starvation (14, 15, 45). Indeed, cells with more replication rounds in their chromosomes at the time of T removal would experience more extensive TLD (13), again implicating replication forks as the points of T starvation-induced chromosomal damage. Moreover, since there was a spike of initiations at the origin upon thymine starvation (16), and subsequent partial destruction of exactly the same chromosomal segment centered on the original initiation spike (16, 18), thymineless chromosomal damage was thought to preferentially target nascent replication bubbles, just as we have proposed in our own intermediate model (Fig. 5D). Since it looked like the younger the replication forks, the more sensitive to T starvation they appear, the TLD paradigm was comfortably developing as “the tragedy of inhibited nascent replication bubbles.”
Here we have tested this idea, increasing or decreasing the number of replication forks in the chromosome by manipulating the chromosome replication complexity (CRC) (25) by employing four independent approaches (overinitiating mutants, HU inhibition of replication forks, inducible overinitiation, initiation-minus mutants). The conservative expectation was that cells with maximal CRC would be maximally vulnerable to TLD and the associated origin destruction. Our findings were just the opposite: one or two additional replication rounds in the chromosome did not change the kinetics of TLD (Fig. 1 and 2), while three or more additional rounds lessened TLD (Fig. 3), likely because of the preinduced replication stress.
Our attempts to block the new rounds of replication by shifting the dnaA(Ts) mutants to the nonpermissive temperature at the time of starvation were successful (Fig. 5A and B); however, blocking new initiations failed to stop TLD (Fig. 4), suggesting that a single replication round, preexisting or newly initiated, is already enough to make the chromosome fully sensitive to T starvation under these conditions. Unexpectedly, blocking any detectable initiation in strains devoid of preexisting replication forks, by preincubating the dnaA(Ts) mutant with dT at 42°C or by employing the awakening protocol, still failed to block TLD. Remarkably, when T starvation was administered at 42°C, the whole chromosome, rather than oriC or the terminus, became vulnerable and disappeared to the same extent, i.e., to approximately one-fourth of the original amount, in 8 h after T removal (Fig. 7). We conclude that (i) the chromosome is utterly destroyed when T starvation is administered at 42°C, (ii) the destruction seems to affect all chromosomal regions equally, and (iii) this chromosome destruction does not require preexisting replication forks and equally affects nonreplicating chromosomes.
The high temperature may affect the level of chromosomal destruction (Fig. 7E to G) but not the mechanism of chromosomal lesions during T starvation. In other words, if it was possible to block replication initiation at 28°C without blocking transcription or translation, we predict that T starvation would still kill cells with nonreplicating chromosomes the same as it does cells with replicating chromosomes, even though the level of chromosomal DNA degradation would be lower. Blocking RNA or protein synthesis in the T-starved cells does block TLD (reviewed in reference 5), which was assumed to mean that new initiations from the origin, which would also be blocked by inhibition of RNA or protein synthesis, were required to induce irreparable chromosomal lesions. Another observation of a similar “dual” nature is the TLD immunity of stationary cells (Fig. 3A). Our present results, i.e., that chromosomal replication is not required for TLD, strongly suggest that immunity against TLD works by blocking protein synthesis directly.
We are not the first ones who could not repeat the original observation of Bouvier and Sicard in 1975 (15) that dnaA(Ts) mutants do not undergo TLD if switched to the nonpermissive temperature upon T removal (A. Khodursky, personal communication; K. J. Kuong, unpublished data). Nakayama and colleagues also reported the same problem (23, 45), even though they were eventually able to find conditions that worked: the dnaA(Ts) defect made cells immune to TLD, if the growing cells were preincubated at 42°C for 90 min before initiation of T starvation (45). However, since they did not compensate for the continued cell growth during the chromosome alignment at 42°C by diluting their cultures upon starting T starvation, we suspect that they observed “immunity to TLD” due to culture saturation (as we now document in Fig. 3A). Essentially the same protocol in our hands, in which the cultures were diluted sufficiently, failed to block TLD and even led to the disappearance of the resistance phase (Fig. 6A). In fact, Bouvier and Sicard themselves were unable to stop thymineless death in two mutants defective in another initiation function, DnaC (15).
In conclusion, and quite shockingly to us, we found no evidence for DNA replication involvement in TLD, at least at 42°C and under the conditions used in this study. This means that T starvation has yet another target in the cell besides the chromosome, and this target is effectively protected by blocking protein synthesis. An unexpectedly high background of labeled dT incorporation in the cells with no replication forks (about 20% of the replicating cells) (Fig. 4A) suggests a replication-independent dT-utilizing process in E. coli that begs future investigation. At the same time, since T starvation kills at any temperature, while DNA loss is dramatic and indiscriminate only at 42°C, the chromosome destruction during TLD might be a secondary process unrelated to the real cell death mechanism. Future experiments will be focused on finding cellular processes other than chromosome replication affected by T starvation, how their disruption affects the chromosome, and if there is any connection between initiation of chromosomal DNA replication and TLD. Separately, the related phenomenon of dGTP starvation in E. coli (28, 46) and thymineless death in other prokaryotic or eukaryotic organisms (5) all need to be tested for their dependence on DNA replication.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains used in this study (Table 1) are all E. coli K-12 derivatives. The strains were maintained at room temperature on LB agar plates supplemented with 10 μg/ml thymidine. For T starvation, MOPS (morpholinepropanesulfonic acid) minimal phosphate medium (47) was used as described below. When required, antibiotics were added to the following final concentrations (μg/ml): ampicillin, 100; kanamycin, 50; chloramphenicol, 10 or 40; tetracycline, 10. Drug resistance-marked mutations were introduced into the strains by P1 transduction and confirmed by PCR and functional analysis (when available). We marked the dnaA601(Ts) allele of strain CM2500 by introducing a cat cassette in the intergenic region between the convergent yidX and yidA genes by the Datsenko and Wanner method (48). We confirmed the point mutations in both dnaA(Ts) alleles by sequencing. We created a plasmid containing both origin and terminus fragments (the same fragments used to generate radioactive probes for ori/ter determinations) by cloning 2.0-kb oriC- and dif-specific fragments from plasmid pEAK23-1 and pEAK17-1, respectively, into pBluescript. Briefly, pEAK23-1 was digested with EcoRI, and the 2.0-kb oriC fragment was cloned at the EcoRI site in pBluescript, creating plasmid pSRK-O. The 2.0-kb dif fragment was excised from pEAK17-1 by SpeI and XbaI and cloned in pSRK-O digested with the same enzymes. The resulting plasmid, pSRK-OD, was verified to have both oriC and dif fragments by Southern hybridization and to harbor single copies of both regions by restriction digestion.
TABLE 1.
E. coli strains and plasmids used in this study (all K-12)
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Published strains | ||
| AB1157a | Wild type for DNA metabolism | 51 |
| KKW58a | ΔthyA72 ΔdeoCABD2 | 10 |
| ER15a | ΔseqA20::kan | 52 |
| KJK63a | ΔthyA72 ΔdeoCABD2 ΔrecBCD3::kan | 10 |
| KJK202a | ΔthyA72 ΔdeoCABD2 Δrep::kan | 10 |
| L392 | MG1655 dnaC2(Ts) thr::Tn10 | 53 |
| NS373 | dnaA46(Ts) tna::Tn10 | 54 |
| SRK253a | IOC Δkup::kan | 25 |
| CM2500b | dnaA601(Ts) | 55 |
| Strains from this study | ||
| KJK170a | ΔthyA72 ΔdeoCABD2 dnaA46(Ts) tna::Tn10 | KKW58 × P1 NS373 |
| RA101a | ΔthyA72 ΔdeoCABD2 dnaC2(Ts) thr::Tn10 | KKW58 × P1 L392 |
| SRK309-1a | dnaA46(T) tna::Tn10 | AB1157 × P1 NS373 |
| SRK270 | dnaA601(Ts) yidX+ cat yidA+ | Precise insertion in CM2500 |
| SRK270-1a | dnaA601(Ts) yidX+ cat yidA+ | AB1157 × P1 SRK270 |
| SRK271a | ΔthyA72 ΔdeoCABD2 ΔseqA20::kan | KKW58 × P1 ER15 |
| SRK290a | ΔthyA72 ΔdeoCABD2 IOC Δkup::kan | KKW58 × P1 SRK253 |
| SRK291a | ΔthyA72 ΔdeoCABD2 dnaA601(Ts) yidX+ cat yidA+ | KKW58 × P1 SRK270 |
| Plasmids | ||
| pEAK17-1 | 2.0-kb ter fragment in pCR TOPO2.1 | Elena Kouzminova, this lab |
| pEAK23-1 | 2.0-kb oriC fragment in pCR TOPO2.1 | Elena Kouzminova, this lab |
| pSRK-O | 2.0-kb oriC fragment of pEAK23-1 in pBluescript | |
| pSRK-OD | 2.0-kb oriC and ter fragments in pBluescript |
Other mutations: F– λ– rac thi-1 hisG4 Δ(gpt-proA)62 argE3 thr-1 leuB6 kdgK51 rfbD1 araC14 lacY1 galK2 xylA5 mtl-1 tsx-33 supE44(glnV44) rpsL31(strR).
Other mutations: λc+ metE46 trp-3 his-4 thi-1 galK2 lacY1 or lacZ4 mtl-1 ara-9 tsx-3 ton-1 rpsL8 or rpsL9 supE44.
The standard thymine starvation assay.
T starvation assays were performed as described before (10, 16), with minor modifications. In the standard assay, strains were initially grown at appropriate temperatures (28°C or 37°C as described in the assays) in MOPS-CAA +dT medium (MOPS-minimal phosphate medium supplemented with 0.2% glucose, 0.2% Casamino Acids [CAA], 20 μg/ml thiamine, and 10 μg/ml thymidine [dT]) for 15 to 18 h. The cultures were then diluted in MOPS-CAA +dT medium to an initial OD of 0.02 to 0.03 and further shaken until they reached an OD of 0.1. At this time, the cells were pelleted by centrifugation, and cell pellets were resuspended in the same volume of sterile 1% NaCl and reprecipitated like this three times to remove traces of thymidine. The final cell pellets were suspended in the same volumes of starvation medium (MOPS-CAA-glucose-thiamine, but without dT), and cell suspensions were shaken at the temperatures described in the assays. Samples for CFU titer determination or for making agarose plugs were collected at various times of starvation, with 0 indicating the time of dT removal, up to 8 h, depending on the assay. For determination of culture viability (CFU titer), serial 10-fold dilutions of the starved culture in sterile 1% NaCl were spotted in 10-μl volumes on LB agar plus dT plates. The plates were incubated at room temperature, and colonies were counted under a stereomicroscope while still small. The CFU titers were all normalized to the titer in the same culture at time zero, the time of dT removal. The actual titers of the initial cultures varied in the range of 10 × 106 to 400 × 106 cells/ml. For making plugs, culture aliquots were centrifuged at 10,000 to 15,000 rpm, and the resulting pellets were stored at −20°C until all samples were ready. Plugs were made as described before (49).
The “awakening” thymine starvation assay.
The “awakening” thymine starvation assay method was developed to avoid manipulations with growing cultures. For this method, the strains are shaken in MOPS-CAA +dT at 28°C for 48 h. These deep stationary cultures are then harvested by centrifugation, and cell pellets are washed three times in an equal volume of 1% NaCl (as above), to remove traces of dT. After the final wash, the cells are suspended in the same volume of 1% NaCl, and this suspension is then diluted at least 20 times into the starvation medium and shaken at temperatures described in the assays. Collection and processing of aliquots for CFU determination are done exactly as described above for the standard TLD assay, except that the actual titer of the initial cultures varied much less, in the range of 20 × 106 to 80 × 106 cells/ml. For making plugs, aliquots of the starved cultures are passed through 0.2-μm nitrocellulose filters (Merck Millipore) using a vacuum manifold. Cells collected on the filters are washed once with 1% NaCl and Tris-EDTA (TE) and are retrieved by vigorous vortexing and shaking of the filters in three changes of 1 ml of TE in sterile glass tubes. The washes are combined, the cells are harvested by centrifugation, and the pellets are stored at –20°C until all samples are ready. Plugs are made as described before (49).
Inducible overinitiation and HU treatment.
Overinitiation from inducible origins was achieved by growing strains in MOPS-CAA +dT in the presence of 1 mM IPTG for various periods of time, as described for the specific assays. For HU treatment, 15- to 18-h cultures in MOPS-CAA +dT at 28°C were diluted 100-fold into MOPS-CAA +dT supplemented with 5 mM HU and shaken for another 15 h at 28°C. Such HU-pregrown cultures were further diluted 50- to 60-fold into MOPS-CAA +dT containing 5 mM HU and shaken at 28°C to an OD at 600 nm (OD600) of 0.1. At this point, aliquots were collected for making agarose plugs for ori/ter determination, or cells were harvested and T starved in the absence of HU as described above.
Quantification of chromosomal regions.
Determination of the quantity of various parts of the chromosome was done by plug-blotting as described before (25). Briefly, cell pellets collected from 0.5 to 20 ml of cultures were made into agarose plugs and lysed overnight to release cellular constituents, as described previously (49). Following the completion of lysis, the plugs were washed twice in 1 ml of TE and then shaken sequentially, for 30 min in 1 ml of 0.25% HCl, then in 0.5 M NaOH, and finally in 1 M Tris HCl (pH 8.0). The DNA from the plugs was vacuum transferred to a nylon membrane (Hybond H+; GE Biosciences) in duplicate (or quadruplicate) to create identical sets. Once the transfer was complete, the DNA was cross-linked to the membrane by UV exposure in a UVP HL-2000 Hybri-Linker. The cross-linked membrane was cut to separate the identical sets, which were then probed with origin-specific, terminus-specific, or mid-replichore-specific 32P-labeled probes, made by random-primer labeling of the corresponding 2-kbp fragments (43). The blots were exposed to phosphorimaging screens and quantified as described previously (25). The individual counts at various time points were all normalized to the counts in the same culture at time zero, the time of dT removal. The actual counts at the zero time point varied in the 15 × 103 to 300 × 103 range.
Rate of DNA replication.
The strains (all ThyA+ in this case) were grown in MOPS-CAA-glucose-thiamine (no dT) at 28°C until they reached an OD of 0.4 to 0.5. At this time, one set of samples was collected to measure the rate of DNA replication (0-h samples) before shifting the cultures to 42°C and further quantifying DNA replication at 15 min, 30 min, and 60 min. To measure the rate of DNA replication, a 200-μl aliquot of the culture was mixed with an equal volume of prewarmed-to-42°C MOPS-CAA-glucose-thiamine medium containing 1 μCi [methyl-3H]thymidine (Perkin-Elmer) and 0.4 μg of cold thymine. The reaction mixtures were incubated for 2 min at 42°C in a stationary heat block, following which the incorporation of thymidine was stopped by the addition of 5 ml of ice-cold 5% trichloroacetic acid (TCA). The TCA-precipitable material was collected on 1.6-μm glass fiber filters (G6; Fisher Scientific) using a vacuum manifold and processed as described previously (50).
ACKNOWLEDGMENTS
We are grateful to Arkady Khodursky (University of Minnesota) and to Roel Schaaper (NIEHS) for many helpful suggestions to improve the clarity of our presentation. We thank Elena Kouzminova (this laboratory) for critical reading of the manuscript.
This work was supported by grant number GM 073115 from the National Institutes of Health.
We have no conflicts of interest to declare.
REFERENCES
- 1.Barner HD, Cohen SS. 1954. The induction of thymine synthesis by T2 infection of a thymine requiring mutant of Escherichia coli. J Bacteriol 68:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen SS, Barner HD. 1954. Studies on unbalanced growth in Escherichia coli. Proc Natl Acad Sci U S A 40:885–893. doi: 10.1073/pnas.40.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khodursky A, Guzmán EC, Hanawalt PC. 2015. Thymineless death lives on: new insights into a classic phenomenon. Annu Rev Microbiol 69:247–263. doi: 10.1146/annurev-micro-092412-155749. [DOI] [PubMed] [Google Scholar]
- 4.Zaritsky A, Woldringh CL, Einav M, Alexeeva S. 2006. Use of thymine limitation and thymine starvation to study bacterial physiology and cytology. J Bacteriol 188:1667–1679. doi: 10.1128/JB.188.5.1667-1679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad SI, Kirk SH, Eisenstark A. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol 52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 6.Cohen SS. 1971. On the nature of thymineless death. Ann N Y Acad Sci 186:292–301. doi: 10.1111/j.1749-6632.1971.tb31155.x. [DOI] [PubMed] [Google Scholar]
- 7.Ladner RD. 2001. The role of dUTPase and uracil-DNA repair in cancer chemotherapy. Curr Protein Pept Sci 2:361–370. doi: 10.2174/1389203013380991. [DOI] [PubMed] [Google Scholar]
- 8.Guzzo MB, Nguyen HT, Pham TH, Wyszczelska-Rokiel M, Jakubowski H, Wolff KA, Ogwang S, Timpona JL, Gogula S, Jacobs MR, Ruetz M, Kräutler B, Jacobsen DW, Zhang GF, Nguyen L. 2016. Methylfolate trap promotes bacterial thymineless death by sulfa drugs. PLoS Pathog 12:e1005949. doi: 10.1371/journal.ppat.1005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao T, Kuzminov A. 2019. Sources of thymidine and analogs fueling futile damage-repair cycles and ss-gap accumulation during thymine starvation in Escherichia coli. DNA Repair (Amst) 75:1–17. doi: 10.1016/j.dnarep.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuong KJ, Kuzminov A. 2010. Stalled replication fork repair and misrepair during thymineless death in Escherichia coli. Genes Cells 15:619–634. doi: 10.1111/j.1365-2443.2010.01405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maaloe O, Hanawalt PC. 1961. Thymine deficiency and the normal DNA replication cycle. I. J Mol Biol 3:144–155. doi: 10.1016/S0022-2836(61)80041-7. [DOI] [PubMed] [Google Scholar]
- 12.Okagaki H, Tsubota Y, Sibatani A. 1960. Unbalanced growth and bacterial death in thymine-deficient and ultraviolet-irradiated Escherichia coli. J Bacteriol 80:762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín CM, Guzmán EC. 2011. DNA replication initiation as a key element in thymineless death. DNA Repair (Amst) 10:94–101. doi: 10.1016/j.dnarep.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Martín CM, Viguera E, Guzmán EC. 2014. Rifampicin suppresses thymineless death by blocking the transcription-dependent step of chromosome initiation. DNA Repair (Amst) 18:10–17. doi: 10.1016/j.dnarep.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Bouvier F, Sicard N. 1975. Interference of dna ts mutations of Escherichia coli with thymineless death. J Bacteriol 124:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuong KJ, Kuzminov A. 2012. Disintegration of nascent replication bubbles during thymine starvation triggers RecA- and RecBCD-dependent replication origin destruction. J Biol Chem 287:23958–23970. doi: 10.1074/jbc.M112.359687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regamey A, Harry EJ, Wake RG. 2000. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: co-ordinating chromosome replication with cell division. Mol Microbiol 38:423–434. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- 18.Sangurdekar DP, Hamann BL, Smirnov D, Srienc F, Hanawalt PC, Khodursky AB. 2010. Thymineless death is associated with loss of essential genetic information from the replication origin. Mol Microbiol 75:1455–1467. doi: 10.1111/j.1365-2958.2010.07072.x. [DOI] [PubMed] [Google Scholar]
- 19.Kuzminov A. 1995. Instability of inhibited replication forks in E. coli. Bioessays 17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 20.Michel B, Grompone G, Florès MJ, Bidnenko V. 2004. Multiple pathways process stalled replication forks. Proc Natl Acad Sci U S A 101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage l. Microbiol Mol Biol Rev 63:751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel B, Flores M-J, Viguera E, Grompone G, Seigneur M, Bidnenko V. 2001. Rescue of arrested replication forks by homologous recombination. Proc Natl Acad Sci U S A 98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama H, Nakayama K, Nakayama R, Nakayama Y. 1982. Recombination-deficient mutations and thymineless death in Escherichia coli K12: reciprocal effects of recBC and recF and indifference of recA mutations. Can J Microbiol 28:425–430. doi: 10.1139/m82-064. [DOI] [PubMed] [Google Scholar]
- 24.Fonville NC, Bates D, Hastings PJ, Hanawalt PC, Rosenberg SM. 2010. Role of RecA and the SOS response in thymineless death in Escherichia coli. PLoS Genet 6:e1000865. doi: 10.1371/journal.pgen.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan SR, Mahaseth T, Kouzminova EA, Cronan G, Kuzminov A. 2016. Static and dynamic factors limit chromosomal replication complexity in Escherichia coli, avoiding dangers of runaway overreplication. Genetics 202:945–960. doi: 10.1534/genetics.115.184697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuzminov A. 2016. Chromosomal replication complexity: a novel DNA metrics and genome instability factor. PLoS Genet 12:e1006229. doi: 10.1371/journal.pgen.1006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bird RE, Louarn J, Martuscelli J, Caro L. 1972. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol 70:549–566. doi: 10.1016/0022-2836(72)90559-1. [DOI] [PubMed] [Google Scholar]
- 28.Itsko M, Schaaper RM. 2014. dGTP starvation in Escherichia coli provides new insights into the thymineless-death phenomenon. PLoS Genet 10:e1004310. doi: 10.1371/journal.pgen.1004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charbon G, Bjørn L, Mendoza-Chamizo B, Frimodt-Møller J, Løbner-Olesen A. 2014. Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res 42:13228–13241. doi: 10.1093/nar/gku1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotman E, Khan SR, Kouzminova E, Kuzminov A. 2014. Replication fork inhibition in seqA mutants of Escherichia coli triggers replication fork breakage. Mol Microbiol 93:50–64. doi: 10.1111/mmi.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane HED, Denhardt DT. 1975. The rep mutation. IV. Slower movement of the replication forks in Escherichia coli rep strains. J Mol Biol 97:99–112. doi: 10.1016/S0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- 32.Olsson J, Dasgupta S, Berg OG, Nordström K. 2002. Eclipse period without sequestration in Escherichia coli. Mol Microbiol 44:1429–1440. doi: 10.1046/j.1365-2958.2002.02954.x. [DOI] [PubMed] [Google Scholar]
- 33.von Freiesleben U, Krekling MA, Hansen FG, Løbner-Olesen A. 2000. The eclipse period of Escherichia coli. EMBO J 19:6240–6248. doi: 10.1093/emboj/19.22.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarbro JW. 1992. Mechanism of action of hydroxyurea. Semin Oncol 19:1–10. [PubMed] [Google Scholar]
- 35.Navarra P, Preziosi P. 1999. Hydroxyurea: new insights on an old drug. Crit Rev Oncol Hematol 29:249–255. doi: 10.1016/S1040-8428(98)00032-8. [DOI] [PubMed] [Google Scholar]
- 36.Zaritsky A, Pritchard RH. 1971. Replication time of the chromosome in thymineless mutants of Escherichia coli. J Mol Biol 60:65–74. doi: 10.1016/0022-2836(71)90447-5. [DOI] [PubMed] [Google Scholar]
- 37.Withers HL, Bernander R. 1998. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J Bacteriol 180:1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirota Y, Jacob F, Ryter A, Buttin G, Nakai T. 1968. On the process of cellular division in Escherichia coli. I. Asymmetrical cell division and production of deoxyribonucleic acid-less bacteria. J Mol Biol 35:175–192. doi: 10.1016/S0022-2836(68)80046-4. [DOI] [PubMed] [Google Scholar]
- 39.Kouzminova EA, Kadyrov FF, Kuzminov A. 2017. RNase HII saves rnhA mutant Escherichia coli from R-loop-associated chromosomal fragmentation. J Mol Biol 429:2873–2894. doi: 10.1016/j.jmb.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maduike NZ, Tehranchi AK, Wang JD, Kreuzer KN. 2014. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol 91:39–56. doi: 10.1111/mmi.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimude JU, Stockum A, Midgley-Smith SL, Upton AL, Foster HA, Khan A, Saunders NJ, Retkute R, Rudolph CJ. 2015. The consequences of replicating in the wrong orientation: bacterial chromosome duplication without an active replication origin. mBio 6:e01294-15. doi: 10.1128/mBio.01294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fonville NC, Vaksman Z, Denapoli J, Hastings PJ, Rosenberg SM. 2011. Pathways of resistance to thymineless death in Escherichia coli and the function of UvrD. Genetics 189:23–36. doi: 10.1534/genetics.111.130161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouzminova EA, Kuzminov A. 2008. Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-strand breaks. Mol Microbiol 68:202–215. doi: 10.1111/j.1365-2958.2008.06149.x. [DOI] [PubMed] [Google Scholar]
- 44.Hong Y, Li L, Luan G, Drlica K, Zhao X. 2017. Contribution of reactive oxygen species to thymineless death in Escherichia coli. Nat Microbiol 2:1667–1675. doi: 10.1038/s41564-017-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayama K, Kusano K, Irino N, Nakayama H. 1994. Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulse field gel electrophoresis and evidence for involvement of homologous recombination. J Mol Biol 243:611–620. doi: 10.1016/0022-2836(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 46.Itsko M, Schaaper RM. 2016. Transcriptome analysis of Escherichia coli during dGTP starvation. J Bacteriol 198:1631–1644. doi: 10.1128/JB.00218-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bochner BR, Ames BN. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem 257:9759–9769. [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan SR, Kuzminov A. 2017. Degradation of RNA during lysis of Escherichia coli cells in agarose plugs breaks the chromosome. PLoS One 12:e0190177. doi: 10.1371/journal.pone.0190177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan SR, Kuzminov A. 2012. Replication forks stalled at ultraviolet lesions are rescued via RecA and RuvABC protein-catalyzed disintegration in Escherichia coli. J Biol Chem 287:6250–6265. doi: 10.1074/jbc.M111.322990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachmann BJ. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p 1190–1219. In Neidhardt FC. (ed), Escherichia coli and Salmonella typhimurium cellular and molecular biology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 52.Rotman E, Kuzminov A. 2007. The mutT defect does not elevate chromosomal fragmentation in Escherichia coli because of the surprisingly low levels of MutM/MutY-recognized DNA modifications. J Bacteriol 189:6976–6988. doi: 10.1128/JB.00776-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kouzminova EA, Kuzminov A. 2012. Chromosome demise in the wake of ligase-deficient replication. Mol Microbiol 84:1079–1096. doi: 10.1111/j.1365-2958.2012.08076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaus N, O'Day K, Peters W, Wright A. 1981. Isolation and characterization of amber mutations in gene dnaA of Escherichia coli K-12. J Bacteriol 145:904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen FG, Koefoed S, Atlung T. 1992. Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol Gen Genet 234:14–21. [DOI] [PubMed] [Google Scholar]