Abstract
While nearly comprehensive proteome coverage can be achieved from bulk tissue or cultured cells, the data usually lacks spatial resolution. As a result, tissue based proteomics averages protein abundance across multiple cell types and/or localizations. With proteomics platforms lacking sensitivity and throughput to undertake deep single-cell proteome studies in order to resolve spatial or cell type dependent protein expression gradients within tissue, proteome analysis has been combined with sorting techniques to enrich for certain cell populations. However, the spatial resolution and context is lost after cell sorting. Here, we report an optimized method for the proteomic analysis of neurons isolated from post-mortem human brain by laser capture microdissection (LCM). We tested combinations of sample collection methods, lysis buffers and digestion methods to maximize the number of identifications and quantitative performance, identifying 1500 proteins from 60 000 μm2 of 10 μm thick cerebellar molecular layer with excellent reproducibility. To demonstrate the ability of our workflow to resolve cell type specific proteomes within human brain tissue, we isolated sets of individual Betz and Purkinje cells. Both neuronal cell types are involved in motor coordination and were found to express highly specific proteomes to a depth of 2800 to 3600 proteins.
Keywords: LC−MS/MS, laser capture microdissection, tissue proteomics, spatially resolved proteomics, brain
Introduction
Mass spectrometry-based proteomics can now generate data of a similar depth to that of RNA-seq.1,2 However, most approaches require relatively large amounts of starting material in the range of 10s to 100s of micrograms, therefore requiring the use of bulk tissue when investigating ex vivo samples. The bulk analysis of tissue consisting of multiple cellular phenotypes will result in the analyte profiles generated being an average of those phenotypes, likely concealing important features of cellular subpopulations present within the bulk sample.3 Analyses on, or approaching, the single cell level allow for the detection of these changes either through the analysis of individual cells or through analysis of cells displaying distinct phenotypes (“Pheno-proteome”). Single cell proteomics of muscle fibers4 and oocytes5 have been described previously; the study of these large or multinucleated cells is aided by the large amounts of protein present in these cells.
Advances have been made in all steps of the proteomic workflow to facilitate the analysis of trace sample amounts. For example, Hughes et al. developed a workflow for the processing of trace samples in a single reaction vessel, which utilizes protein/peptide binding to paramagnetic beads to remove detergent, digest protein, and cleanup peptides.6 Recently, Zhu et al. described a semiautomatic nanodroplet-based platform capable of performing protein digestion in <200 nL of volume (nanoPOTS).7 In combination with identification transfer from a larger sample using MaxQuant’s “match between runs” (MBR) feature, ∼2700 proteins could be quantified for samples containing 10–14 cells. The same group also demonstrated the nanoPOTS platform’s ability to analyze single cells collected by fluorescence-activated cell sorting (FACS), and they were able to detect ∼670 proteins from a single HeLa cell.8 Both studies used a custom platform with a narrower C18 column and lower flow-rate than is typically used in proteomics experiments (30 μm i.d. and 60 nL/min respectively), resulting in gains in sensitivity.9
Laser-capture microdissection (LCM) is used as a complementary method to FACS for specific cell-type enrichment from tissue sections.10 It has the advantage that it can be used to isolate specific healthy or diseased cytoarchitectonically defined features of heterogeneous tissues, including cell types for which no unique FACS-compatible cell markers are available or which are too large or complex for flow-sorting, such as large neurons of the human brain. LCM enrichment has therefore the potential to generate cell type-specific data for unbiased “bottom-up” proteomics of distinct cellular phenotypes and heterogeneous regions of tissue.11−15 The method employs a laser that is fired along the optical path of a conventional light microscope to ablate a predefined track through the tissue section, isolating the area of interest from the surrounding tissue. The system then fires a defocused pulse of the laser at the dissected area, catapulting it into a collection tube in a contact-free manner.
Highly resolved spatial proteomics has the potential to define the complex cellular and molecular architecture of the human brain in health and disease.16,17 Most of our approximately 20 000 genes are expressed in the brain in a temporospatial manner, and many demonstrate alternative splicing in different brain regions, hinting at the existence of a very complex, highly compartmentalized brain proteome. However, how RNA expression maps onto protein signaling networks in the human brain remains largely unknown. Remarkably, we do not even know the proteomic signature of some of the most distinct neuronal classes in the human brain, such as von Economo neurons in the anterior cingulate gyrus, Betz cells in the primary motor cortex or Purkinje cells of the cerebellum, and how their proteomes differ from each other and other neurons in the same anatomical region.
Identifying the defining proteomic signature of these (and other) cells is relevant not only for basic neuroscience research (for example, through the unbiased identification of cell-type specific protein markers for generation of antibody-based investigative tools), but also for a better understanding of neurological disease. For example, classic neurodegenerative disorders such as Parkinson’s and Alzheimer’s, or Amyotrophic Lateral Sclerosis (ALS) and spinocerebellar ataxia (SCA) are characterized by a phenomenon called “selective vulnerability”.18−21 It refers to the observation that degeneration in a specific disease is cell type and neuronal network specific, not diffuse.22 What drives this selective vulnerability remains unknown; however, it is clear that dysfunction of protein homeostasis, leading to protein aggregation in specific cell types, is a defining feature of most of these disorders.23 It is therefore reasonable to speculate that a better understanding of the specific proteome of certain neurons, its compartmentalization, and pattern of post-translational modification may provide crucial insights into the phenomenon of selective vulnerability.
The purpose of the current work was to develop a workflow for cell type specific microproteomics of the human brain using LCM of two easily identifiable classes of human neurons as a paradigm: the neocortical Betz cells and the cerebellar Purkinje cells. Both cell types define distinct cytoarchitectonic regions of the human brain, namely, the primary motor cortex (Betz cells) and cerebellar cortex (Purkinje cells). Both are among the largest cells of the human central nervous system and originate from distinct germinal zones of the developing brain. They are the main efferent cells of their respective cortical layer yet differ in their primary neurotransmitter use: glutamate in Betz cells and GABA in Purkinje cells. Our choice was further informed by the observation that a few Purkinje cell specific protein targets are well-known and therefore could function as internal controls that we would expect to find in the Purkinje cell proteome. In other words, we hypothesized that Betz and Purkinje cells are morphologically, topographically, and functionally so distinct that they would serve as an ideal testbed for the demonstration of proof-of-principle for our LCM-based microproteomics method development.
Here, we present the successful development of a workflow that allows us to perform robust proteomic analyses from as few as 100 pooled LCM-captured cell sections, allowing us to use simple principle component analysis of the LCM-enriched Betz and Purkinje cell proteomes to clearly distinguish these cell types and identify known Purkinje cell markers only in the Purkinje cell data set. We conclude that our approach is likely to be highly informative for the identification of factors contributing to selective vulnerability in human neurodegenerative diseases associated with protein dyshomeostasis, and has the potential to contribute to the development of a “multiomics” atlas of the human brain, such as the Human Cell Atlas project.
Methods
Tissue Retrieval and Processing
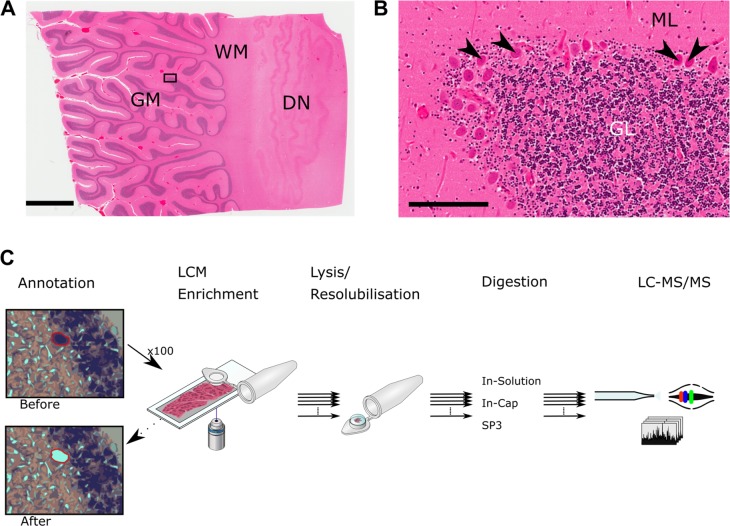
Brain tissue was retrieved 3 days post-mortem by the Oxford Brain Bank; a research ethics committee (REC) approved, HTA regulated research tissue bank (REC reference 15/SC/0639). After retrieval, the brain was divided into the two hemispheres. One was fixed in 10% Neutral Buffered Formaldehyde, the other hemisphere was sliced in the coronal plane and individual slices of motor cortex or cerebellum were immediately placed into liquid nitrogen vapor and stored at −80 °C. This method substantially reduces freezing artifacts resulting in frozen tissue sections with excellent cytoarchitecture (Figure 1), which is critical for LCM.
Figure 1.
Cerebellar cortex and overview of LCM-proteomics workflow. (A) Image of a cerebellar cortex tissue slice stained with H&E. Rectangle indicates area shown in (B). GM: gray matter. WM: white matter. DN: dentate nucleus. Scale bar represents 4 mm. (B) Higher magnification of area indicated in (A). Arrowheads indicate Purkinje cells. ML: molecular layer. GL: granular layer. Scale bar represents 200 μm. (C) Schematic overview of the LCM workflow. Different collection methods, lysis buffers, and digestion protocols were tested.
Relevant tissue blocks of frozen cerebellum and motor cortex were acclimatized to −20 °C and mounted onto a cryostat block using OCT Compound (Cell Path, ARG1180), careful consideration was taken to ensure cut sections were not contaminated with OCT. Sections were cut at 10 μm and mounted onto UV irradiated (254 nm, 30 min) 1.0 PEN membrane slides (Zeiss) at −18 °C. Sections were then air-dried for several minutes and placed onto a Shandon Linistain for automated H&E staining. Briefly, sections were fixed in 70% denatured alcohol, hydrated, stained with Harris’ hematoxylin, incubated in 0.4% acid alcohol, placed in Scot’s tap water and stained with eosin containing 0.25% acetic acid with regular washing steps in between. Stained sections were then dehydrated in increasing concentrations of denatured alcohol and air-dried without coverslips and stored at −80 °C until processing by laser-capture microdissection.
Laser-Capture Microdissection
Areas of tissue and multiple, individual sections of Purkinje and Betz cells were isolated from the prepared cerebellum and motor cortex slides using a laser-capture microscope equipped with laser pressure catapulting (PALM Microbeam, Zeiss). All annotations of areas/cells for capture were performed manually on the microscope. Purkinje and Betz cells were annotated based on histological landmarks and according to their size, location and morphology. Cutting and catapulting of areas/cells of interest were performed along annotations automatically using the 40× objective lens and the following settings in the control software for cutting: Energy 43, Focus 55; and catapulting Energy 20, Focus −15. After isolation, samples were immediately placed in dry ice and later stored at −80 °C.
Proteomic Sample Processing
Tissue Lysis
Protein was extracted from tissue collected in the LCM cap by the addition of 10 μL of lysis buffer followed by both brief centrifugation and incubation at room temperature for 30 min, or by omitting the centrifugation step and incubating in an inverted position for 30 min at room temperature.
In-Cap Digestion
Protein extracts in the LCM cap were diluted to either 1 M Urea or 10% TFE with 50 mM ammonium bicarbonate, 25 ng of trypsin was added, and samples incubated at 37 °C overnight—all steps taking place in the LCM cap. Peptides were acidified to 1% formic acid (v/v) and purified using C18 solid-phase extraction cartridges (SOLA HRP, Thermo Scientific). Peptides were dried to completeness under vacuum, resuspended in 6 μL 2% acetonitrile, 0.1% trifluoroacetic acid and stored at −20 °C until analysis.
In-Solution Digestion
Protein lysates contained within the LCM cap (either after collecting dry or directly into buffer) were reduced and alkylated in-cap with 5 mM DTT and 20 mM iodoacetamide (final concentrations, see Table 1). Lysates were then transferred to 200 μL PCR tubes. Proteins were cleaned-up prior to digestion by precipitation with methanol-chloroform.24 The protein pellet was resuspended in 6 M urea 0.1 M tris pH 7.8, this was then diluted to 1 M urea with ultrapure water, 25 ng trypsin added and incubated at 37 °C overnight. Peptides were acidified to 1% formic acid (v/v) and purified using C18 solid-phase extraction cartridges (SOLA HRP, Thermo Scientific). Peptides were dried to completeness under vacuum, resuspended in 6 μL 2% acetonitrile, 0.1% trifluoroacetic acid, and stored at −20 °C until analysis.
Table 1. Protein Preparation Steps for Array of Lysis Buffers Used in SP3 and In-Solution Digestion.
| protein solubilization system | collection method | collection buffer | protein recovery and reduction | protein alkylation |
|---|---|---|---|---|
| 6 M urea | Buffer | 20 μL 6 M urea, 0.1 M tris | Add DTT to 5 mM; Incubate 30 min, RT | Add IAA to 20 mM; 30 min, RT |
| Cap | – | Add 10 μL 6 M urea, 0.1 M tris, 5 mM DTT; Incubate 30 min, RT | ||
| 6 M urea, 30% acetonitrile | Buffer | 20 μL 6 M urea, 100 mM Tris, 30% acetonitrile (v/v) | Add DTT to 5 mM; Incubate 30 min, RT | |
| Cap | – | Add 10 μL 6 M urea, 0.1 M tris, 5 mM DTT, 30% acetonitrile (v/v); Incubate 30 min, RT | ||
| 50% Trifluoroethanol | Buffer | 20 μL Ultrapure water | Add 20 μL trifluoroethanol. Add DTT to 5 mM; Incubate 60 min, 60 °C | |
| Cap | – | Add 10 μL 50% trifluoroethanol (v/v), 0.1 M tris, 5 mM DTT; Incubate 60 min, 60 °C | ||
| RIPA | Buffer | 20 μL RIPA | Add DTT to 5 mM; Incubate 30 min, RT | |
| Cap | – | Add 10 μL RIPA (Pierce #89900), 5 mM DTT; Incubate 30 min, RT |
Single-Pot Solid-Phase-Enhanced Sample Preparation (SP3)
Paramagnetic beads were prepared as described.6 Protein lysates contained within the LCM cap (either after collecting dry or directly into buffer) were reduced and alkylated in-cap with 5 mM DTT and 20 mM iodoacetamide (final concentrations, see Table 1). Lysates were then transferred to 200 μL PCR tubes. Two μL of beads were mixed with the samples. Acetonitrile was added to the samples to a final concentration of 70% (v/v) and briefly vortexed. This mixture was incubated for 18 min with orbital shaking at 1000 rpm. Beads were then immobilized on a magnet for 2 min and the supernatant transferred to fresh tubes containing 2 μL of fresh beads and the incubation step repeated to capture any initially unbound protein. Beads were washed twice with 70% (v/v) ethanol in water and once with 100% acetonitrile, all on the magnet. Beads were resuspended in 50 mM ammonium bicarbonate containing 25 ng trypsin and digested overnight at 37 °C. After digestion, the beads were resuspended by bath sonication. Acetonitrile was added to 95% (v/v) and beads incubated at 1000 rpm for 18 min to bind peptide, then beads were immobilized on the magnet for 2 min and the supernatant discarded. Peptides in the initially unbound tubes were eluted from the beads with 6 μL 2% DMSO and combined with their other corresponding tube. Combined beads were pipet mixed and then immobilized on the magnet for 5 min, and peptides transferred to LC–MS vials containing 600 nL 1% trifluoroacetic acid; vials were stored at −20 °C until analysis.
LC–MS/MS
Peptides were analyzed by nano-UPLC–MS/MS using a Dionex Ultimate 3000 coupled online to an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). For all samples, a 75 μm × 500 mm C18 EASY-Spray column (Thermo Scientific) with 2 μm particles was used at a flow rate of 250 nL/min. Peptides were separated using a 60 min linear gradient from 2% buffer B to 35% buffer B (A: 5% DMSO, 0.1% formic acid in water; B: 5% DMSO, 0.1% formic acid in acetonitrile). For the initial Purkinje cell experiments (Figure 2), the mass spectrometer was operated with the following settings (“Universal method”25): MS scans were acquired in the Orbitrap between 400 and 1500 m/z at a resolution of 120 000 and an AGC target of 4 × 105. Selected precursors between charge states 2+ and 7+ and above the intensity threshold of 5 × 103 were selected for collision-induced dissociation (CID) fragmentation, with a collision energy of 35%, an AGC target of 4 × 103, a maximum injection time of 300 ms, a dynamic exclusion window of 60 s and a maximum duty cycle of 3 s. MS/MS spectra were acquired in the ion trap using the rapid scan mode. The option to “use all available parallelizable time” was enabled.
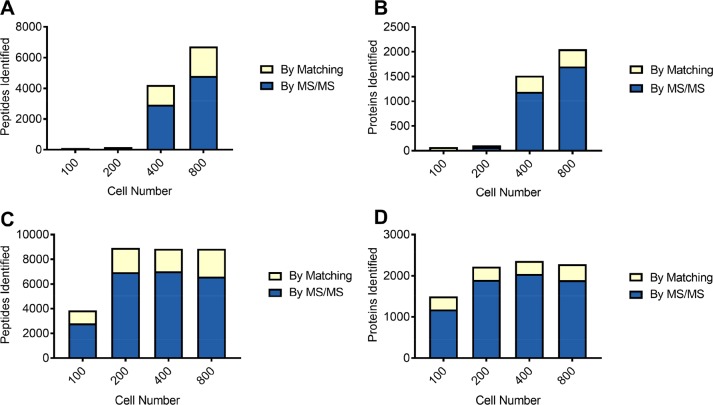
Figure 2.
Comparison between retrieving cells from the LCM cap and digesting in situ in the LCM cap. As a pilot experiment, 100, 200, 400, and 800 Purkinje cells were collected using LCM and processed after centrifuging to collect the cells from the cap prior to digestion (A,B) or digested directly in the LCM cap (C,D). “By matching” refers to identifications matched from a sample of cerebellar cortex using MaxQuant’s “match between runs”. Digesting directly in the LCM cap results in an increase in peptide and protein identifications, especially at low cell numbers. (A) Number of peptides identified from Purkinje cells recovered from the LCM cap. (B) Number of proteins identified from Purkinje cells recovered from the LCM cap. (C) Number of peptides identified from Purkinje cells digested in the LCM cap. (D) Number of proteins identified from Purkinje cells digested in the LCM cap.
For the molecular layer optimization experiments and Betz/Purkinje comparison, the following settings were used: MS1 scans were acquired as above. Selected precursors between charge states 2+ and 7+ and above the intensity threshold of 5 × 103 were selected for higher energy collisional dissociation (HCD) fragmentation, with a normalized collision energy of 28%, an AGC target of 4 × 103, a maximum injection time of 80 ms, a dynamic exclusion window of 30 s and a maximum duty cycle of 1 s. MS/MS spectra were acquired in the ion trap using the rapid scan mode.
Data Analysis
The raw data files were searched against the Uniprot human database (Retrieved 17/01/2018, 929527 sequences) using MaxQuant26,27 version 1.6.2.10 and its built-in contaminant database using tryptic specificity and allowing 2 missed cleavages. Peptide and protein identifications are reported using the protein parsimony principles as described by Nesvizhskii and Aebersold28 and results were filtered to a 1% false discovery rate at the PSM and Protein level. At least 1 unique peptide was required to identify a protein group. Cysteine carbamidomethylation was set as a fixed modification as appropriate, methionine oxidation and protein N-terminal acetylation were set as variable modifications and the “match between runs” option was used, all other settings were left as default. Protein quantification was performed with the MaxLFQ algorithm within MaxQuant.29 Downstream analysis was performed in Perseus30 and using R.31
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE32 partner repository with the data set identifier PXD012101.
Immunohistochemistry
Sections from FFPE blocks were cut at 6 μm and mounted onto superfrost microscope slides. Once air-dried and baked at 56 °C for 20 min, sections were deparaffinized and rehydrated with incubation in xylene, 100%, 90%, 70% denatured alcohol and tap water. Rehydrated sections were incubated for 30 min in 10% hydrogen peroxide to remove endogenous peroxidases. Antigen retrieval was performed by autoclave heating (121 °C for 10 min) in Sodium Citrate Buffer (pH 7.6). Mouse anti-calbindin-D28k (Clone: CB300) was diluted in TBS/T (1:1000) and incubated overnight at 4 °C. Staining visualization was performed using the Aligent’s Dako Envision Kit (Aligent, Cat. number k5007), briefly, a cocktail of mouse/rabbit secondary antibodies conjugated to HRP were incubated for 1 h at room temperature, a 2% DAB working solution was prepared with a ×50 concentrated 3,3′-diaminobenzidine stock and diluted with Dako’s substrate buffer, sections were incubated for 5 min and counterstained with Harris’ hematoxylin for 1 min.
Results and Discussion
Tissue heterogeneity is a generally overlooked factor in clinical proteomics studies, as large amounts of input material are often needed. Here, we set out to develop a sensitive and quantitative protocol capable of generating proteomic data at a spatial resolution. We chose to perform this work in the human cerebellum due to the presence of Purkinje cells, which are easily identifiable after H&E staining due to their defined location within a monolayer and large size relative to other neurons. In some cases, tissue was collected from the molecular layer of the cerebellum, which is adjacent to the Purkinje cell layer. This was done after our initial Purkinje cell titration curves were established in order to not waste valuable cell-type specific material during further optimization steps.
We set out to define an approximate minimum area that could be isolated and produce enough identifications to be of use. To do this, we first isolated the cell sections of 100, 200, 400, and 800 Purkinje cell bodies from 10 μm thick slices of H&E stained human cerebellum using a laser-capture microscope, 100 sections of Purkinje cell bodies cover approximately 60 000 μm2 resulting in an approximate tissue volume of 600 pL. The samples were recovered from the cap by centrifugation in 6 M urea and digested after dilution to 1 M urea. From these numbers of cell sections, we identified (union of identifications by MS/MS and match between runs) 108, 181, 4221, and 6728 peptides (Figure 2A) from 73, 107, 1517, and 2051 proteins respectively (Figure 2B). In the 400 and 800 cell section samples, protein identified by matching to the library sample accounted for 326 and 348 proteins. Following the recovery of protein from the LCM caps by centrifugation, some material was still visible on the LCM cap. Unsure of whether this material contained unrecovered protein, we repeated the titration and omitted the centrifugation steps, simply digesting proteins in-place without recovery from the cap, which we refer to descriptively as the “In-Cap” method in order to distinguish these samples from a more conventional in-solution digestion method below. We opted to omit reduction and alkylation in this case in order to minimize sample volume and handling steps. In this experiment, we identified 3852, 8921, 8855, and 8858 peptides (Figure 2C) from 1497, 2224, 2360, and 2282 (Figure 2D) proteins, respectively. Within the proteins identified using this method, we detected 15 proteins annotated as involved in Purkinje cell development or function, along with almost 590 proteins involved in general neurological cell function. The digest efficiency was good, 75% of peptides had no missed cleavages, 22% had one missed cleavage, and 3% had two missed cleavages. The lack of reduction and alkylation is noticeable in the data, as less than 1% of peptides detected contained a cysteine residue, as compared to 8% in the entire data collected for this work. There was little to no increase in peptide and protein identifications when collecting 400 and 800 cell sections when compared to collecting only 200 cell sections. As the protein and peptide identifications obtained from 100 cell sections were lower than this plateau, we decided to use this amount of tissue for further optimization steps. These results showed that coupling laser-capture microdissection with a simple digestion method is capable of high sensitivity.
After demonstrating that a useful number of proteins can be identified from as little as 100 Purkinje cell sections with the simple In-Cap digestion method, we performed a series of optimization experiments where we investigated the effects of the following parameters: (i) collection method, (ii) lysis buffer used, and (iii) digestion protocol. Given the multiple combinations of factors, we decided to perform these experiments on an area of the cerebellum molecular layer that is approximately equivalent to the areas of 100 sections of Purkinje cells (60 000 μm2). This removed the need for manual annotation of thousands of individual cells; dramatically reducing the amount of time needed to perform the laser-capture microdissection on unfixed tissue and allowed for the collection of all samples from the same tissue slice. We tested three methods of protein digestion: (i) a scaled-down version of our standard in-solution digestion protocol, (ii) a modified version of SP3,33 and (iii) the “in-cap” method used to generate the Purkinje cell data. In combination with these digestion methods, we tested the use of four lysis buffers: (i) 6 M Urea, (ii) 6 M Urea in 30% acetonitrile,34 (iii) 50% trifluoroethanol (TFE),35 and (iv) RIPA buffer. Finally, we also compared collecting the material into a polymer-filled LCM cap or directly into the buffers listed above.
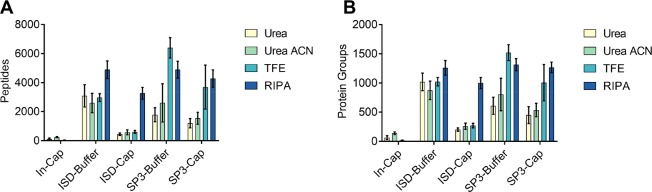
For this tissue type and amount, the various sample preparation approaches performed quite differently, we detected between 38–6399 peptides corresponding to 17–1521 proteins on average (Figure 3). In the context of protein and peptide identifications, the combination of sample collection into buffer, protein extraction with 50% TFE and processing with SP3 generating the most peptide and protein identifications. The mean (n = 3) number of identifications for this method were 6399 peptides and 1521 proteins. The methods generating the next highest numbers of identifications were Buffer-RIPA-SP3 (1316 proteins), Cap-RIPA-SP3 (1268 proteins) and Buffer-RIPA-In-Solution (1260 proteins). The overlap in protein identifications between the two methods with the most identifications (Buffer-TFE-SP3 and Buffer-RIPA-SP3) was 74% (Figure S1) when considering proteins identified in at least two replicates per condition.
Figure 3.
Comparison of protein identifications of methods used for protein retrieval and digestion. To determine an optimal method of material collection, lysis and digestion, 60 000 μm3 of the molecular layer (the area of approximately 100 Purkinje cells) was collected to test different combinations of collection methods, lysis buffers and digestion methods. (A) Mean number of peptides identified in each combination. (B) Mean number of proteins identified in each combination. All bars are mean values (n = 3) and error bars represent standard deviation. In-Cap refers to the in situ digestion method. Cap and Buffer refer to collecting material onto both a dry LCM cap and recovering in the indicated lysis buffer, or collecting directly into the indicated lysis buffer. ISD: in-solution digestion.
To further investigate the larger proportion (and number) of unique identifications in the TFE sample, the properties of these proteins were investigated. Figure S2 shows that protein recovery with RIPA was slightly biased toward recovering proteins of a lower molecular weight, whereas the distribution of proteins identified in TFE samples more closely resembles the distribution of proteins in the reviewed SwissProt database (20 336 entries). Next, the isoelectric points of the uniquely identified proteins were compared to the SwissProt database (Figure S3). Proteins uniquely identified in TFE samples tended to be more acidic than those identified by RIPA alone, and RIPA uniquely identified more proteins with a basic isoelectric point. Grand average of hydropathy36 (GRAVY) indices are a measure of the hydropathy of an amino acid sequence, where values that are more negative indicate more hydrophilic sequences. These indices were calculated for each of the proteins uniquely identified in either the TFE or RIPA samples and their distributions compared to the distribution of all proteins in the Swiss-Prot database (Figure S4). TFE shows a slight bias toward hydrophilic proteins (median GRAVY value = −0.41), and RIPA shows a slight bias toward more hydrophobic proteins (median GRAVY value = −0.32).
In general, collecting samples directly into a liquid performed better than collecting directly onto the LCM polymer cap and recovering protein afterward. When 50% TFE was used to recover proteins from the LCM cap, the number of identifications decreased and became more variable in comparison to collecting into buffer, likely still due to incomplete recovery of protein from the cap as indicated by a reduction in the summed intensity of identified peptides (Figure S5). The SP3 method outperformed the In-Cap method in all cases, whereas the In-Solution method only outperforms the In-Cap method when sample is collected into liquid. The In-Cap method could not be performed with RIPA buffer due to this method not containing a detergent removal step.
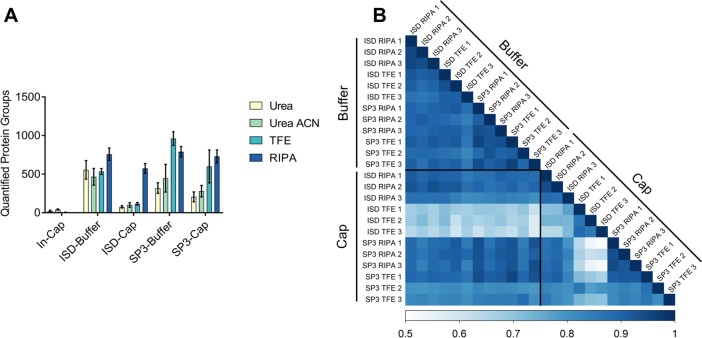
As well as identification level performance, we also investigated the quantitative performance of the methods tested. As would be expected, the number of proteins quantifiable using MaxQuant’s MaxLFQ algorithm follows the same pattern as the number of protein identifications across each method combination. The highest number of quantified proteins was 958 (median, n = 3) in the Buffer-TFE-SP3 samples (Figure 4A). Samples processed with either the in-solution digestion protocol or SP3 in combination with TFE or RIPA buffer all had high Pearson correlation coefficients within replicates (n = 3) (Figure 4B). Samples processed with RIPA buffer had the lowest median % CVs and the method with the lowest median % CV overall was Cap-RIPA-SP3 (19.0%).
Figure 4.
Quantitative performance of the tested methods. (A) The number of protein groups quantified using MaxQuant’s MaxLFQ algorithm (default settings). (B) Pearson correlation matrix of samples where Trifluoroethanol (TFE) or RIPA buffer, scale is truncated for clarity. Cap and Buffer refer to collecting material onto both a dry LCM cap and recovering in the indicated lysis buffer, or collecting directly into the indicated lysis buffer. ISD: in-solution digestion.
While using TFE as a protein extraction buffer resulted in the highest numbers of protein and peptide identifications, it is comparatively more difficult to work with than RIPA buffer. This is due to TFE’s high vapor pressure causing it to evaporate readily during sample collection and protein extraction, where previously published methods recommend heating at 60 °C for 60 min. Additionally, it does pose a higher safety risk in comparison to the other extraction buffers used here. Considering both these points, we believe the best method to use is to collect samples directly into RIPA buffer and then process them using the SP3 method. This method is among the highest performing when considering identification and quantification quality and alleviates the difficulties of working with the trifluoroethanol, which should translate well to high throughput automated processing of samples in future studies. For a proof-of-principle experiment, we tested the hypothesis that microproteomics from as few as 150 pooled Purkinje and Betz neurons can distinguish these cell types with simple statistical analysis, and that known Purkinje cell specific proteins would only be recovered from the Purkinje cell fraction.
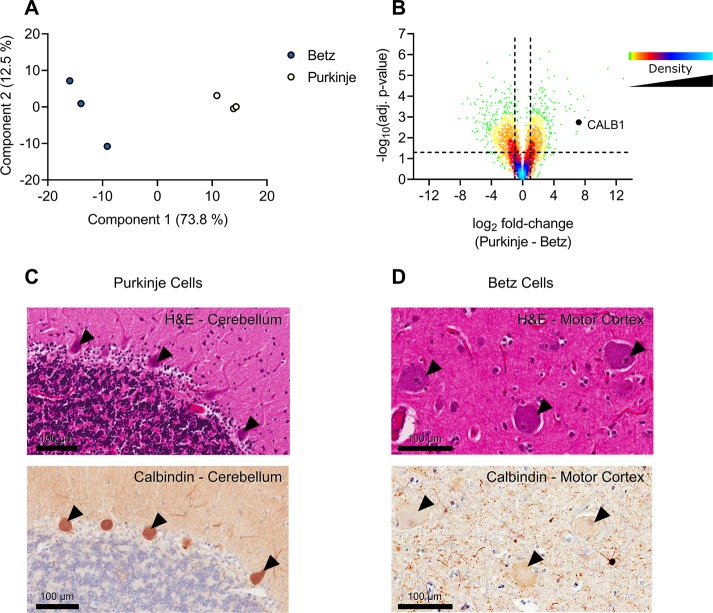
With our microproteomic approach we identified on average 3551 and 2874 proteins (2718 and 2025 with at least two peptides) from the Betz cell and Purkinje cell fractions, respectfully (n = 3). We found 526 proteins that were present only in all Betz cell replicates and 126 proteins that were found only in all the Purkinje cell replicates. By comparing the average LFQ intensity of the proteins within these cells using a two-sample t test we generated a volcano plot that clearly delineated proteins that are differentially expressed in each cell type. The substantial differences in protein expression in Betz and Purkinje cells are both expected and remarkable. 36% of all proteins detected between the two cell types are regulated more than 2-fold and with a p-value of <0.05. These differences are highly relevant as they can be linked directly to the physiological differences between Betz and Purkinje cells. The approach here performs extremely well in comparison to the existing literature, particularly when considering a metric of proteins per unit of tissue volume isolated and accounting for the LC–MS/MS acquisition time used (Table S1).
Encouragingly, some of the highest fold change proteins identified are known Purkinje cell markers, i.e., CALB1, ITPR1, PRKCG, and ALDOC. To validate these findings and to ensure our proteomic data had successfully identified proteins which were differentially expressed across the two cell types we back mapped the identified proteins onto the FFPE (formalin-fixed paraffin embedded) region counterparts using immunohistochemistry (Figure 5). As there are currently no known specific markers for the Betz cell, we aim to do further experiments and explore our data to gain further insights into its proteome.
Figure 5.
Analysis of Betz and Purkinje cells. Analysis of Betz and Purkinje cell proteomes. (A) Principal component analysis of Betz and Purkinje cells based on MaxQuant LFQ values. (B) Volcano plot of the Purkinje cell proteome against the Betz cell proteome. Positive log2 fold-changes indicate the mean LFQ value is higher in Purkinje cells. Y-axis values are permutation-corrected FDRs. Horizontal dashed line represents 5% FDR threshold. Vertical dashed lines represent ±2-fold-change. Color gradient represents density of data points. (C) Cerebellum stained with H&E (top) or for calbindin 1 (bottom); arrowheads indicate examples of positively stained Purkinje cells. Scale bar represents 100 μm. (D) Motor cortex stained with H&E (top) or for calbindin 1 (bottom); arrowheads indicate negatively stained Betz cells. Scale bar represents 100 μm.
Conclusions
In this work, we present, to our knowledge for the first time, a sample preparation regime that allows the combination of laser capture microdissection with high-end proteomics workflows in order to detect protein expression differences with cell type resolution in the cellular and spatial context of human brain tissue. When applied to cell sections of individual neuronal cells, this optimized procedure facilitates the detection of protein expression profiles that can distinguish different neuron types from within the same brain tissue. Cell type resolved proteomics now allows adding the proteome dimension to the phenotype of a specific cell type, defining the “Pheno-Proteome”. The here described methodology lays the groundwork to draw conclusions about cell physiology and function from deep proteomic data in the physiological and spatial context of complex primary tissues such as the human brain.
Acknowledgments
We would like to thank Benedikt Kessler and members of the Kessler lab for useful discussions and suggestions. S.D. is supported by a Nuffield Department of Medicine Prize DPhil studentship. R.F. is supported by the Kennedy Trust Fund. We acknowledge the Oxford Brain Bank, supported by the Medical Research Council (MRC, MR/L022656/1) and Brains for Dementia Research (BDR) (Alzheimer Society and Alzheimer Research UK). This research project was funded by the NIHR Oxford Biomedical Research Centre (to O.A., BRC-1215-20008). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health. This work uses data provided by patients and collected by the NHS as part of their care and support and would not have been possible without access to this data. The NIHR recognizes and values the role of patient data, securely accessed and stored, both in underpinning and leading to improvements in research and care.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00981.
Author Contributions
§ S.D. and C.S. contributed equally.
Author Contributions
∥ O.A. and R.F. contributed equally.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE32 partner repository with the data set identifier PXD012101.
Supplementary Material
References
- Davis S.; et al. Expanding Proteome Coverage with CHarge Ordered Parallel Ion aNalysis (CHOPIN) Combined with Broad Specificity Proteolysis. J. Proteome Res. 2017, 16, 1288–1299. 10.1021/acs.jproteome.6b00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen D. B.; et al. An Optimized Shotgun Strategy for the Rapid Generation of Comprehensive Human Proteomes. Cell Syst. 2017, 11, 214–221. 10.1016/j.cels.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht H.; Slavov N. Transformative Opportunities for Single-Cell Proteomics. J. Proteome Res. 2018, 17, 2565–2571. 10.1021/acs.jproteome.8b00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M.; et al. Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO Rep. 2015, 16, 387–95. 10.15252/embr.201439757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant-Klun I.; Leicht S.; Hughes C.; Krijgsveld J. Identification of Maturation-Specific Proteins by Single-Cell Proteomics of Human Oocytes. Mol. Cell. Proteomics 2016, 15, 2616–27. 10.1074/mcp.M115.056887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C. S.; et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 2018, 9, 882. 10.1038/s41467-018-03367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; et al. Proteomic Analysis of Single Mammalian Cells Enabled by Microfluidic Nanodroplet Sample Preparation and Ultrasensitive NanoLC–MS. Angew. Chem., Int. Ed. 2018, 57, 12370–12374. 10.1002/anie.201802843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; et al. Subnanogram proteomics: Impact of LC column selection, MS instrumentation and data analysis strategy on proteome coverage for trace samples. Int. J. Mass Spectrom. 2018, 427, 4–10. 10.1016/j.ijms.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina V.; et al. Laser-capture microdissection. Nat. Protoc. 2006, 1, 586–603. 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- Waanders L. F.; et al. Quantitative proteomic analysis of single pancreatic islets. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 18902–7. 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur D.; et al. Microproteomic analysis of 10,000 laser captured microdissected breast tumor cells using short-range sodium dodecyl sulfate-polyacrylamide gel electrophoresis and porous layer open tubular liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2011, 1218, 8168–8174. 10.1016/j.chroma.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman R. B. H.; et al. Optimized nLC–MS workflow for laser capture microdissected breast cancer tissue. J. Proteomics 2012, 75, 2844–2854. 10.1016/j.jprot.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Clair G.; et al. Spatially-Resolved Proteomics: Rapid Quantitative Analysis of Laser Capture Microdissected Alveolar Tissue Samples. Sci. Rep. 2016, 6, 39223. 10.1038/srep39223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczak K.; et al. Spatial tissue proteomics quantifies inter- and intra-tumor heterogeneity in hepatocellular carcinoma. Mol. Cell. Proteomics 2018, 17, 810. 10.1074/mcp.RA117.000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen R. R.; Rozowsky J. S.; Gerstein M. B.; Nairn A. C. Decoding neuroproteomics: integrating the genome, translatome and functional anatomy. Nat. Neurosci. 2014, 17, 1491–9. 10.1038/nn.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosp F.; et al. Spatiotemporal Proteomic Profiling of Huntington’s Disease Inclusions Reveals Widespread Loss of Protein Function. Cell Rep. 2017, 21, 2291–2303. 10.1016/j.celrep.2017.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden G. A.; La Spada A. R. Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration. Cerebellum 2008, 7, 138–149. 10.1007/s12311-008-0027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington A.; et al. Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013, 125, 95–109. 10.1007/s00401-012-1058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier D. J.; Obeso J. A.; Halliday G. M. Selective neuronal vulnerability in Parkinson disease. Nat. Rev. Neurosci. 2017, 18, 101–113. 10.1038/nrn.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akila Parvathy Dharshini S.; Taguchi Y.; Michael Gromiha M. Exploring the selective vulnerability in Alzheimer disease using tissue specific variant analysis. Genomics 2018, 10.1016/j.ygeno.2018.05.024. [DOI] [PubMed] [Google Scholar]
- Fu H.; Hardy J.; Duff K. E. Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1350–1358. 10.1038/s41593-018-0221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W. S. Selective vulnerability to neurodegenerative disease: the curious case of Prion Protein. Dis. Models & Mech. 2014, 7, 21–29. 10.1242/dmm.012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D.; Flügge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–3. 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Eliuk S.et al. A ‘Universal’ Data-Dependent Mass Spectrometry Method That Eliminates Time-Consuming Method Optimization for Achieving Maximal Identifications from Each Sample; Thermo Fisher Scientific, 2016; https://www.thermofisher.com/content/dam/tfs/ATG/CMD/cmd-documents/sci-res/posters/ms/events/ismc2014/PN-49014-Universal-MS-Method-PN49014-EN.pdf.
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–72. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J.; et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Aebersold R. Interpretation of Shotgun Proteomic Data. Mol. Cell. Proteomics 2005, 4, 1419–1440. 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- Cox J.; et al. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S.; et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018.
- Vizcaíno J. A.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielaff M.; et al. Evaluation of FASP, SP3 and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. acs.jproteome.7b00433 2017, 16, 4060. 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- Dapic I.; Uwugiaren N.; Jansen P. J.; Corthals G. L. Fast and Simple Protocols for Mass Spectrometry-Based Proteomics of Small Fresh Frozen Uterine Tissue Sections. Anal. Chem. 2017, 89, 10769–10775. 10.1021/acs.analchem.7b01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J. Proteome Res. 2005, 4, 2397–2403. 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J.; Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE32 partner repository with the data set identifier PXD012101.