Abstract
Phospholipase C (PLC)-mediated hydrolysis of the limited pool of plasma membrane (PM) phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] requires replenishment from a larger pool of phosphatidylinositol (PtdIns) via sequential phosphorylation by PI 4-kinases and PIP 5-kinases. Since PtdIns is synthesized in the ER and PtdIns(4,5)P2 is generated in the PM, it has been postulated that PtdIns transfer proteins (PITPs) provide the means for this lipid transfer function. Recent studies identified the large PITP protein, Nir2 as important for PtdIns transfer from the ER to the PM. It was also found that, Nir2 was also required for the transfer of phosphatidic acid (PtdOH) from the PM to the ER. In Nir2-depleted cells, activation of PLC leads to PtdOH accumulation in the PM and PtdIns synthesis becomes severely impaired. In quiescent cells Nir2 is localized to the ER via interaction of its FFAT domain with ER-bound VAP-A and –B proteins. After PLC activation, Nir2 also binds to the PM via interaction of its C-terminal domains with diacylglycerol and PtdOH. Through these interactions, Nir2 functions in ER-PM contact zones. Mutations in VAP-B that have been identified in familial forms of amyotrophic lateral sclerosis (ALS or Lou-Gehrig’s disease) cause aggregation of the VAP-B protein, which then impairs its binding to several proteins, including Nir2. These findings have shed new lights on the importance of non-vesicular lipid transfer of PtdIns and PtdOH in ER-PM contact zones with a possible link to a devastating human disease.
Keywords: Phosphatidylinositol, lipid transfer, endoplasmic reticulum, amyotrophic lateral sclerosis, Phospholipase C
Lipid transport between various membrane compartments is essential for the maintenance of the unique lipid composition of biological membranes. Lipids can rapidly diffuse from one membrane to another during membrane fusion, but there are other mechanisms that support non-vesicular transfer of lipids between membranes [1]. It is increasingly recognized that such transfer processes take place in dynamic contact sites where two membranes belonging to different organelles are found in close proximity [2–5]. Since most lipids are synthesized in the ER, these transport processes are very important to distribute the lipids to other membranes where their functions are critical. In fact, it has been shown for several lipid classes that synthesis and transport are tightly coupled at membrane contact sites. [2].
A special case of example is the transport of phosphatidylinositol (PtdIns) between various membranes. It has been described long time ago that a protein, called Sec14 in yeast is essential for secretory transport out of the Golgi [6] and that Sec14 is a phosphatidylcholine (PtdCho)-PtdIns transfer protein [7]. In mammalian systems proteins capable of binding PtdIns and transfer the lipid between artificial membranes have been found [8, 9] and it was shown that these proteins are required for sustained PLC activity [10] and for priming of exocytic vesicles [11]. Although these features of the so- called classical (or Class I) PITPs (see Fig. 1A) made them prime candidates for functioning as proteins supplying the plasma membrane (PM) with PtdIns, this has never been formally proven in intact cell systems. In fact, it has been proposed that Sec14 and Class I PITPs are not simple lipid transfer proteins but their functions are dedicated to specific signaling processes [12], and that they can present lipids to PI kinases [13, 14].
Figure 1.
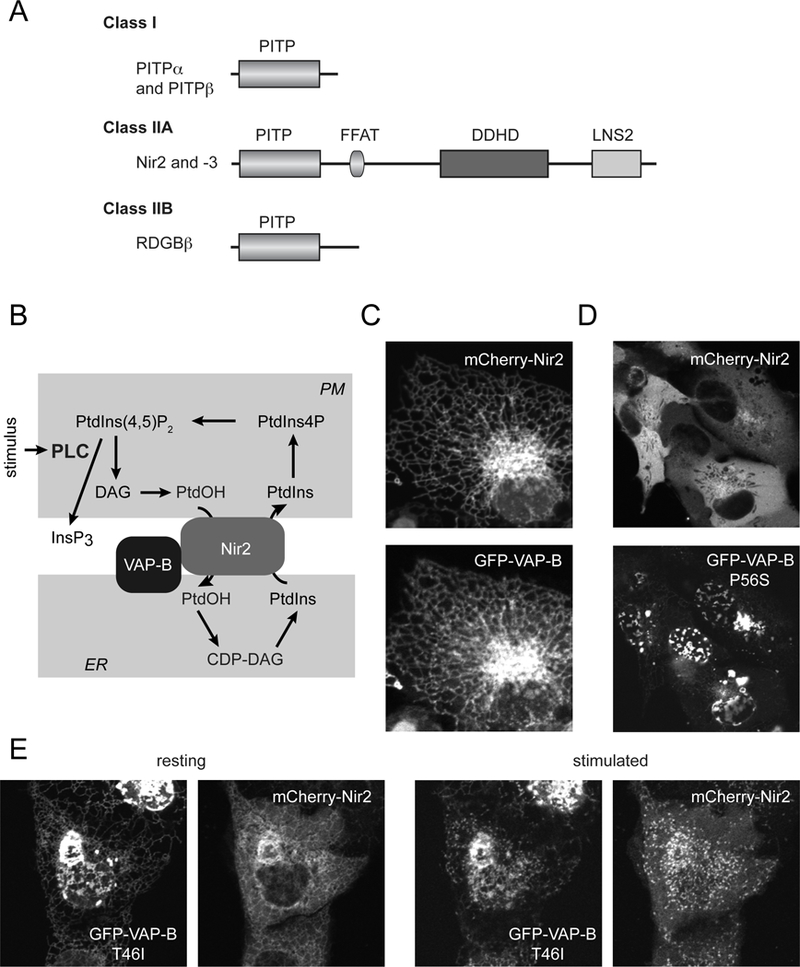
(A) Classification of PITPs. Class I PITPs comprise of PITPα and PITPβ, the latter having two splice variants. The Class IIA PITPs, Nir2 and Nir3 are homologues of the Drosophila RDGB protein (also called PITPnm1 and −2). These are larger proteins having additional domains, such as the FFAT motif (double phenylalanine in an acidic stretch), the DDHD domain and the LNS2 domain, the latter also present in lipins and binds PtdOH. Class IIB PITP is more homologous to the PITP domain of Nir2/3 than to the Class I group. (B) The proposed model of Nir2 action during PLC activation. Nir2 is recruited to the PM via its LNS2 domain (with the help of a short sequence preceding the LNS2 domain reminiscent of DAG binding sequences) and anchored to the ER via interaction via its FFAT domain with VAP proteins. Nir2 then transfers PtdIns from the ER to the PM and PtdOH in the other direction. (C) Cellular localization of Nir2 and VAP-B in resting HEK293 cells. Note the co-localization of the two proteins in the tubular ER. (D) Localization of the mutant VAP-B-P56S and Nir2 in quiescent HEK293 cells. Note the mutant VAP-B aggregates at the nuclear envelope and the lack of recruitment of Nir2 to the mutant VAP-B protein. (E) Localization of the mutant VAP-B-T46I and Nir2 in quiescent (left) or Angiotensin II-stimulated (right) HEK293 cells. Note that this mutant still attracts Nir2 to the ER even though it also shows a tendency to aggregate. After stimulation, the two proteins shows small clusters that correspond to ER-PM contact sites. The T46I mutation of the VAP-B is less disruptive than the P56S.
Another protein possessing a PITP domain was discovered in Drosophila when the rdgB mutation was mapped to a gene encoding a larger protein possessing an N-terminal PITP domain [15, 16]. RdgB mutant flies develop light-induced retinal degeneration and given the importance of PLC activation in invertebrate phototransduction, RDGB was assumed to be important for the transfer of PtdIns to the photoreceptor membrane to maintain PtdIns(4,5)P2 levels [17]. Mammalian homologues of RDGB, named PITPnm have been cloned [18] and they were also found as interacting partners of the Pyk2 tyrosine kinase and named Nir1, −2 and −3 [19]. Nir2 and Nir3 but not Nir1 contains a PITP domain and now they are called Class II PITPs (Fig. 1A). Subsequent studies found Nir2 critical for Golgi function by maintaining DAG levels [20] and PtdIns4P synthesis [21], but it was also found to support cytokinesis [22]. It is not clear at present how these functions of Nir2 relate to the role of the same protein at the ER- PM contact sites (see below). Nir3 knockout mice show no obvious phenotype but Nir2 knockout mice die at early embryonic age [23].
Several recent studies have shown that Nir2 assumes an important function as a PtdIns transfer protein in mammalian cells. First, it was shown that Nir2-depletion negatively impacts PM PtdIns(4,5)P2 levels and PtdIns(3,4,5)P3-dependent signaling from growth factor receptors [24]. Nir2 was also shown to be recruited to ER-PM contact sites after stimulation and was necessary for the maintenance of PtdIns(4,5)P2 levels during signaling from Gq-coupled receptors [25, 26]}. These results were consistent with a role of Nir2 as a PITP functioning between the ER and the PM. However, there were signs already that Nir2 might do more than plainly deliver PtdIns to the PM. First, rdgB mutant flies can be rescued by the PITP domain of RDGB but not by PITPα or by a chimera in which PITPα was used in place of the RDGB PITP domain [27]. More recently, the small RDGBβ protein, which is a simple PITP domain more homologous to the PITP domain of Class II than to the Class I PITPs (Fig. 1A), was shown to bind and transfer PtdOH in addition to PtdIns [28]. Similar PtdOH binding properties for the PITP domains of fly RDGB and human Nir2 were presented in the same study [28]. Nir2 was also shown to bind PtdOH and be recruited to the PM via its C-terminal LNS2 domain [29]. Finally, our recent studies using intact cells have demonstrated that Nir2 functions as a PtdOH transfer protein that transfers PtdOH from the PM during PLC activation. We showed that PtdIns synthesis is greatly impaired in Nir2 depleted cells when stimulated by Gq-coupled receptors because of defective recycling of PtdOH from the PM [26]. Our studies also found that it is the PITP domain that is required for both PtdIns and PtdOH transfer while the C-terminal part anchors the protein to the PM when DAG and PtdOH are formed [26]. These studies established Nir2 as an important component of the phosphoinositide cycle that delivers PtdIns to the PM and at the same time it transports PtdOH in the other direction. This exchange occurs in ER-PM contact sites and used primarily when PLC activity is consuming PtdIns(4,5)P2 at high rate producing PtdOH that has to be recycled to support this lipid cycle (Fig. 1B). This situation of high volume lipid cycling occurs in the fly eye during phototransduction and that is why RDGB is so important in this location. To underline this point, a most recent study has demonstrated that RDGB is not a simple PtdIns transfer protein but it also transfers PtdOH from the photoreceptor membranes to the subrhabdomeric cisternae. This latter study also found that the PtdIns and PtdOH transfer function both reside within the PITP domain [30]. This study on RDGB together with our findings on Nir2 demonstrates that this lipid exchange function is conserved during evolution.
Most likely, Nir2 is not the only PtdOH transfer protein in a cell. Its sister protein, Nir3 has been shown to work similarly to Nir2 in the context of PtdIns transfer (although its PtdOH transfer function has not yet been investigated) with a different threshold. Nir3 is already found in ER-PM junctions at lower level of PLC activation [31]. A PtdOH transfer protein has also been identified within the mitochondria, which transfers PtdOH between the outer and inner mitochondrial membrane [32–35]. It is conceivable that PtdOH transfer proteins are dedicated to specific metabolic pathways in a way it has been suggested for PITP proteins [36].
It is important to emphasize that Nir2 works in contact sites between the ER and the PM. As pointed out above, such contacts are also important for the transfer of other lipids between the ER and the PM, the latest example being PS [37, 38]. For Nir2, as for several other lipid transfer proteins, the ER anchors are the VAP-A and VAP-B proteins [39]. Several lipid transfer proteins can bind to VAPs via their short conserved sequences, called FFAT (double phenylalanine in an acidic stretch) motifs [40]. Mutations in the VAP-B protein have been identified in amyotrophic lateral sclerosis (ALS or Lou Gehrig Disease) patients [41–43]. Two of these mutations (T46I, P56S) affect the MSP domain that binds FFAT motifs. Although VAP-B interacts with a number of proteins through its various domains [44] it was of interest to us how the patient mutations affect its interaction with the Nir2 protein. We generated both the T46I and P56S mutant forms of VAP-B fused to green fluorescent protein (GFP) and co-expressed them with mCherry-Nir2. We found that the P56S mutation caused a major problem with the distribution of the protein: the normal ER tubular distribution was replaced with patchy distribution mostly located on the nuclear envelope (Fig. 1D). Importantly, while the wild-type VAP-B has attracted the mCherry-Nir2 causing the two proteins to co-localize in the tubular ER (Fig. 1C), the P56S mutant VAP-B failed to recruit the Nir2, which was found in the cytosol (Fig. 1D). In contrast, the T46I mutation had a much milder effect: this mutant already showed the patchy nuclear envelope enrichment, but it was still localized to the tubular ER and had Nir2 associated with it. Moreover, PLC activation still caused the clustering of both proteins into puncta formed in ER-PM contacts (Fig. 2E). We also found that immunoprecipitates of VAP-B P56S did not contain mCherry Nir2 whereas wild-type VAP-B showed clear association (not shown). This was in good agreement with the study that described the same difference in the cellular distribution of the two mutant VAP-Bs shown here in Fig. 2D, E [43] and a similar failure of the P56S mutant to attract an expressed OSBP protein [44]. The conclusion of the latter study was that proteins that use FFAT domains to interact with VAP-B would not bind to the aggregation-prone P56S mutant, while other interacting proteins may be co-aggregated with the mutant VAP-B protein. Which of the multiple defects are most important for the development of the ALS pathology remains to be seen.
In summary, recent developments in the field of non-vesicular lipid transfer highlighted the importance of these processes in establishing and maintaining the unique lipid composition of membranes. Identification of the Nir2 (and possibly the Nir3) protein as PtdIns and PtdOH transfer devices in ER-PM contact zones solved a long-standing puzzle of how various steps in the “PI cycle” are organized between the ER and the PM. More research is under way to find the link between these processes and various neurodegenerative diseases.
ACKNOWLEDGEMENT
Confocal imaging was performed at the Microscopy & Imaging Core of the National Institute of Child Health and Human Development, NIH with the kind assistance of Drs. Vincent Schram and James T. Russell. This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health. EW was supported by a scholarship from the Hungarian American Enterprise Scholarship Fund (HAESF). NE was sponsored by the NIH Summer Intern Program.
REFRENCES
- 1.Prinz WA (2010) Lipid trafficking sans vesicles: where, why, how? Cell. 143, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahiri S, Toulmay A and Prinz WA (2015) Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol. 33, 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lev S (2010) Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 11, 739–750 [DOI] [PubMed] [Google Scholar]
- 4.Stefan CJ, Manford AG and Emr SD (2013) ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol. 25, 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henne WM, Liou J and Emr SD (2015) Molecular mechanisms of inter-organelle ER-PM contact sites. Curr Opin Cell Biol. 35, 123–130 [DOI] [PubMed] [Google Scholar]
- 6.Bankaitis VA, Malehorn DE, Emr SD and Greene R (1989) The Saccharomyces cerevisiae SEC 14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J.Cell Biol. 108, 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bankaitis VA, Mousley CJ and Schaaf G (2010) The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci. 35, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmkamp GM Jr., Harvey MS, Wirtz KW and Van Deenen LL (1974) Phospholipid exchange between membranes. Purification of bovine brain proteins that preferentially catalyze the transfer of phosphatidylinositol. J Biol Chem. 249, 6382–6389 [PubMed] [Google Scholar]
- 9.Dickeson SK, Lim CN, Schuyler GT, Dalton TP, Helmkamp GM Jr. and Yarbrough LR (1989) Isolation and sequence of cDNA clones encoding rat phosphatidylinositol transfer protein. J Biol Chem. 264, 16557–16564 [PubMed] [Google Scholar]
- 10.Kauffmann-Zeh A, Thomas GM, Ball A, Prosser S, Cunningham E, Cockcroft S and Hsuan JJ (1995) Requirement for phosphatidylinositol transfer protein in epidermal growth factor signaling. Science. 268, 1188–1190 [DOI] [PubMed] [Google Scholar]
- 11.Hay JC and Martin TFJ (1993) Phosphatidylinositol transfer protein required for ATP- dependent priming of Ca2+-activated secretion. Nature. 366, 572–575 [DOI] [PubMed] [Google Scholar]
- 12.Mousley CJ, Davison JM and Bankaitis VA (2012) Sec14 Like PITPs Couple Lipid Metabolism with Phosphoinositide Synthesis to Regulate Golgi Functionality. Subcell Biochem. 59, 271–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR and Bankaitis VA (2008) Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 29, 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panaretou C, Domin J, Cockcroft S and Waterfield MD (1997) Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150/PtdIns 3-kinase complex. J.Biol.Chem. 272, 2477–2485 [DOI] [PubMed] [Google Scholar]
- 15.Vihtelic TS, Hyde DR and O’Tousa JE (1991) Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics. 127, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vihtelic TS, Goeb M, Milligan S, O’Tousa JE and Hyde DR (1993) Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J.Cell Biol. 122, 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trivedi D and Padinjat R (2007) RdgB proteins: functions in lipid homeostasis and signal transduction. Biochim Biophys Acta. 1771, 692–699 [DOI] [PubMed] [Google Scholar]
- 18.Aikawa Y, Hara H and Watanabe T (1997) Molecular cloning and characterization of mammalian homologues of the Drosophila retinal degeneration B gene. Biochem Biophys Res Commun. 236, 559–564 [DOI] [PubMed] [Google Scholar]
- 19.Lev S, Hernandez J, Martinez R, Chen A, Plowman G and Schlessinger J (1999) Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol Cell Biol. 19, 2278–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litvak V, Dahan N, Ramachandran S, Sabanay H and Lev S (2005) Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat.Cell Biol. 7, 225–234 [DOI] [PubMed] [Google Scholar]
- 21.Aikawa Y, Kuraoka A, Kondo H, Kawabuchi M and Watanabe T (1999) Involvement of PITPnm, a mammalian homologue of Drosophila rdgB, in phosphoinositide synthesis on Golgi membranes. J Biol Chem. 274, 20569–20577 [DOI] [PubMed] [Google Scholar]
- 22.Litvak V, Tian D, Carmon S and Lev S (2002) Nir2, a human homolog of Drosophila melanogaster retinal degeneration B protein, is essential for cytokinesis. Mol Cell Biol. 22, 5064–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Peng YW, Shang J, Pawlyk BS, Yu F and Li T (2001) The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 107, 35–41 [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M, Laufman O and Lev S (2013) The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 14, 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC and Liou J (2013) Feedback regulation of receptor-induced ca(2+) signaling mediated by e-syt1 and nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5, 813–825 [DOI] [PubMed] [Google Scholar]
- 26.Kim YJ, Guzman-Hernandez ML, Wisniewski E and Balla T (2015) Phosphatidylinositol- Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev Cell. 33, 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milligan SC, Alb JG Jr., Elagina RB, Bankaitis VA and Hyde DR (1997) The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol. 139, 351–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garner K, Hunt AN, Koster G, Somerharju P, Groves E, Li M, Raghu P, Holic R and Cockcroft S (2012) Phosphatidylinositol Transfer Protein, Cytoplasmic 1 (PITPNC1) Binds and Transfers Phosphatidic Acid. J Biol Chem. 287, 32263–32276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M, Laufman O and Lev S (2013) The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yadav S, Garner K, Georgiev P, Li M, Gomez-Espinosa E, Panda A, Mathre S, Okkenhaug H, Cockcroft S and Raghu P (2015) RDGBalpha, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J Cell Sci. 128, 3330–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang CL and Liou J (2015) Phosphatidylinositol 4,5-Bisphosphate Homeostasis Regulated by Nir2 and Nir3 Proteins at Endoplasmic Reticulum-Plasma Membrane Junctions. J Biol Chem. 290, 14289–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B and Langer T (2012) Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338, 815–818 [DOI] [PubMed] [Google Scholar]
- 33.Miliara X, Garnett JA, Tatsuta T, Abid Ali F., Baldie H, Perez-Dorado I, Simpson P, Yague E, Langer T and Matthews S (2015) Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes. EMBO Rep. 16, 824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe Y, Tamura Y, Kawano S and Endo T (2015) Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria. Nat Commun. 6, 7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu F, He F, Yao H, Wang C, Wang J, Li J, Qi X, Xue H, Ding J and Zhang P (2015) Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex. EMBO Rep. 16, 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ile KE, Schaaf G and Bankaitis VA (2006) Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol. 2, 576–583 [DOI] [PubMed] [Google Scholar]
- 37.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F and De Camilli P (2015) INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 349, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser von Filseck J, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W and Drin G (2015) INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 349, 432–436 [DOI] [PubMed] [Google Scholar]
- 39.Peretti D, Dahan N, Shimoni E, Hirschberg K and Lev S (2008) Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 19, 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loewen CJ and Levine TP (2005) A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem. 280, 14097–14104 [DOI] [PubMed] [Google Scholar]
- 41.Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P and Zatz M (2004) A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 75, 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitne-Neto M, Ramos CR, Pimenta DC, Luz JS, Nishimura AL, Gonzales FA, Oliveira CC and Zatz M (2007) A mutation in human VAP-B--MSP domain, present in ALS patients, affects the interaction with other cellular proteins. Protein expression and purification. 55, 139–146 [DOI] [PubMed] [Google Scholar]
- 43.Chen HJ, Anagnostou G, Chai A, Withers J, Morris A, Adhikaree J, Pennetta G and de Belleroche JS (2010) Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J Biol Chem. 285, 40266–40281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW and Gygi SP (2015) The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]


