Abstract
Context
Mutations in alkaline phosphatase (AlkP), liver/bone/kidney (ALPL), which encodes tissue-nonspecific isozyme AlkP, cause hypophosphatasia (HPP). HPP is suspected by a low-serum AlkP. We hypothesized that some patients with bone or dental disease have undiagnosed HPP, caused by ALPL variants.
Objective
Our objective was to discover the prevalence of these gene variants in the Vanderbilt University DNA Biobank (BioVU) and to assess phenotypic associations.
Design
We identified subjects in BioVU, a repository of DNA, that had at least one of three known, rare HPP disease-causing variants in ALPL: rs199669988, rs121918007, and/or rs121918002. To evaluate for phenotypic associations, we conducted a sequential phenome-wide association study of ALPL variants and then performed a de-identified manual record review to refine the phenotype.
Results
Out of 25,822 genotyped individuals, we identified 52 women and 53 men with HPP disease-causing variants in ALPL, 7/1000. None had a clinical diagnosis of HPP. For patients with ALPL variants, the average serum AlkP levels were in the lower range of normal or lower. Forty percent of men and 62% of women had documented bone and/or dental disease, compatible with the diagnosis of HPP. Forty percent of the female patients had ovarian pathology or other gynecological abnormalities compared with 15% seen in controls.
Conclusions
Variants in the ALPL gene cause bone and dental disease in patients with and without the standard biomarker, low plasma AlkP. ALPL gene variants are more prevalent than currently reported and underdiagnosed. Gynecologic disease appears to be associated with HPP-causing variants in ALPL.
ALPL gene variants show an association of bone, dental, and gynecologic disease in women with and without low AlkP levels are more prevalent than currently reported and are underdiagnosed.
Hypophosphatasia (HPP), first described in 1948 (1), is an inherited metabolic disorder, in which affected individuals have inadequate mineralization of bones and teeth as a result of a deficiency in the tissue-nonspecific isozyme of alkaline phosphatase (AlkP), leading to musculoskeletal and dental complications (2–8). Patients with diagnosed HPP have mutations at the gene AlkP, liver/bone/kidney (ALPL) that encodes AlkP (4, 9–11), leading to low age/sex-adjusted serum AlkP levels (6, 12–14). To date, at least 330 distinct mutations in ALPL have been linked to HPP in European, North American, Asian, and Australian individuals, some of which have a more predictable penetrance pattern than others (10). Although serum AlkP levels are not a traditional screening test, low levels in the correct clinical context often lead to a diagnosis of HPP. The true prevalence of HPP, including in undiagnosed individuals who may harbor partial, late-onset, or atypical phenotypes, is not known (7).
HPP has been classified according to age of onset of the first symptom and includes the following subgroups: perinatal lethal, infantile, juvenile, adult, and odontohypophosphatasia (4, 8, 13, 15). This delineation has been acknowledged to be arbitrary, owing to considerable overlap and variability in symptom presentation, and does not represent the phenotypic continuum or the evolution of the disease throughout the patient’s life. It has been proposed that a worse prognosis is associated with presentation at an earlier age (2, 13); however, there is increasing recognition that HPP has a wide clinical variability, and patient phenotype more likely exists within a spectrum of signs and symptoms that do not fit into a strict classification scheme (14, 15). Adults with HPP have been characterized by a wide range of orthopedic symptoms, ranging from a variety of fractures and dental symptoms to being completely asymptomatic (5, 16). Incomplete penetrance may occur, and asymptomatic individuals with a genetic mutation are considered to be carriers of the risk alleles that may manifest classically in subsequent generations (7). Patients with pediatric-onset HPP are often not diagnosed with HPP until adulthood or may be misdiagnosed with other disorders, such as osteoporosis or rheumatologic disorders, probably as a result of a general lack of knowledge of the disease among health care professionals.
AlkP is expressed in multiple tissues and is especially abundant in hepatic, skeletal, and renal tissue. Its function in bone is well described (17, 18); its effect in other tissues is less well understood. The aims of this study were to determine the frequency of certain ALPL mutations in our population; to highlight clinical characteristics, including AlkP levels, and potentially to uncover previously undocumented phenotypic associations.
Methods
The cohort studied was taken from Vanderbilt University DNA Biobank (BioVU), Vanderbilt University’s de-identified DNA biobank that pairs DNA isolated from leftover clinical blood samples with a de-identified and randomly date-shifted image of the electronic health record (EHR), as previously described (19). In 2012 to 2013, ∼40,000 subjects in BioVU were genotyped using the Infinium HumanExome BeadChip (http://genome.sph.umich.edu/wiki/Exome_Chip_Design; Illumina), which includes >240,000 mostly exonic markers, as well as single-nucleotide polymorphisms (SNPs) from the genome-wide association study catalog (20). Genotyping was performed at the Vanderbilt Technologies for Advanced Genomics Core, and genomic data were processed by the Vanderbilt Technologies for Advanced Genomics Analysis and Research Design Core. Genotyping quality was evaluated using SNP call rates and concordance rates with HapMap controls; SNPs with <99.8% call rate or <98% concordance were excluded (21).
Ethics statement
This project was determined to be nonhuman subjects research by the Vanderbilt University Medical Center Institutional Review Board (VUMC IRB #120277), as there are no personal health identifiers in the dataset. In accordance with the ethical framework previously described for BioVU studies (22), all investigators must have Institutional Review Board determination before accessing the resource and also sign a standard data-use agreement (19, 22).
Phenome-wide association study
The phenome-wide association study (PheWAS) approach has been previously described (23, 24). Conceptually, PheWAS is the obverse of the genome-wide association study; instead of searching for genetic associations with a measured phenotype, PheWAS searches for phenotypic associations with a measured genotype. PheWAS has previously been successfully applied to genomic and laboratory results with high validity to replicate known associations (19). In brief, cases are defined as hierarchical groupings of International Classification of Disease, Ninth Edition, Clinical Modification (ICD-9-CM) billing codes, termed phecodes (25). We required that a patient have two or more instances of pertinent phecodes in their longitudinal EHR to be defined as a case for a given phenotype, and we restricted our analysis to phecodes occurring in >40 individuals. The statistical test used for analysis was logistic regression, adjusted for age, sex, and race/ethnicity. Plots were created in R (https://cran.r-project.org), using the R PheWAS package (26), and the association was tested against 742 phecodes.
We conducted a PheWAS on rs121918007, thought to be the most common HPP mutation in European ancestry and also the most common SNP in our cohort, to investigate for replication of known phenotypic associations as identifying potential associations. For this analysis, we combined all European and non-European American individuals from the HumanExome BeadChip.
Manual chart review
We examined 13 ALPL SNPs passing quality control. The Illumina included only 13 variants in the ALPL gene on the HumanExome BeadChip. All of the variants were used for this study, and no variants were excluded. Three SNPs of known pathogenicity were further examined: ALPL: rs199669988 (c.529G > A), rs121918007 (c.571G > A), and rs121918002 (c.881A > C; see Table 1) (27, 28). We then performed a PheWAS of these individuals matched to a control population to identify key differences in phenotypes (23, 24, 26). Subsequently, we performed a manual record review to refine better the skeletal and nonskeletal phenotype of the variant carriers. The de-identified chart was accessed in the BioVU interface, which allows for review of the entire medical record of each patient. The search function was then used to identify key phrases within documents to flag them for further review (see Appendix for search terms). A blinded, manual abstraction of medical records was performed by two independent reviewers for the 52 female and 53 male patients with identified ALPL variants, as well as a 1:1 age-matched control group. Individuals were considered controls for a given case if they did not have any ICD-9-CM codes belonging to the specified list of disease exclusions defined for each case group.
Table 1.
ALPL Variants Identified Among Patients Included as Cases
| SNP | rsID | Molecular Consequence | Previously Reported Phenotype | Carrier Count | MAFa |
|---|---|---|---|---|---|
| exm28129 | rs199669988 | c.529G > A | Adultb | 2 | 0.01 |
| exm28131 | rs121918007 | c.571G > A | Adult, childhood, infantilec | 74 | 0.02 |
| exm28158 | rs121918002 | c.881A > C | Adult, childhood, infantilec | 29 | <0.01 |
Three known ALPL SNPs were identified in 105 patients of the 25,822 in the BioVU cohort. All variants in this study were ultimately associated with an HPP phenotype upon review of their de-identified records. All of these variants were missense mutations.
Abbreviations: MAF, minor allele frequency, the frequency of the second-most common allele in a given population to differentiate between common and rare variants; rsID, accession number assigned to refer to specific SNPs.
Highest minor allele frequency (MAF) observed in any population from 1000 genomes, Phase 3, Exome Sequencing Project, and Exome Aggregation Consortium. Genome Reference Consortium Human genome build 37.
Mornet (27).
Landrum et al. (28).
Thirteen of the 52 female patients were compound heterozygotes; the remaining 39 were heterozygotes. Records were manually abstracted to document clinical, laboratory, and radiographic findings, consistent with the known skeletal and dental phenotype of HPP, as well as additional potential gynecologic phenotypic characteristics identified by PheWAS. On average, EHR data between 2000 and 2012 were reviewed, which also include problem lists, laboratory results, imaging results, and narrative reports from the EHR.
Statistical analysis of the manual chart review of cases and controls
All statistical comparisons were made between groups matched for age and sex. Marginal comparisons between the prevalence of disease in groups with and without an ALPL mutation were made using a t test for proportions or Fisher’s exact test in cases with low number of events. As a result of the low event rates, analyses involving the adjustment for additional covariates were not used to avoid overfitting the data. P < 0.05 is significant; odds ratios (ORs) are given in cases when they were estimable. In some cases, no events were observed in the control group, making point estimates intractable.
Results
PheWAS
The ALPL SNP rs121918007 was noted in 72 patients (69% of the total SNPs under investigation), and therefore, these data were used for PheWAS. PheWAS analyzes many phenotypes compared with a single genetic variant to detect disease–gene associations using phecodes and translates the distinct ICD-9-CM codes for each individual into their respective case or control groups. The PheWAS program had a total of 6,550,738 billing codes accrued over a total of 308,706 patient-years. A total of 742 phenotypes present in at least 40 individuals were analyzed, yielding a Bonferroni correction of P = 0.05/742 = 6.7 × 10−5; the Bonferroni correction was used when conducting multiple analyses on the same dependent variable. The Bonferroni-corrected P value is calculated by dividing the original α-value (0.05) by the number of analyses on the dependent variable, producing a much stricter level of significance.
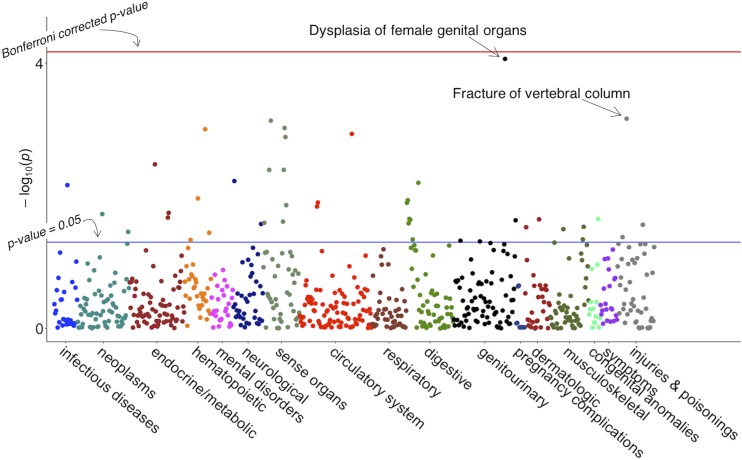
The strongest associations were dysplasia of female genital organs (uncorrected P value = 8.6 × 10−5, OR = 19.76) and fracture of the vertebral column, which is a known sequela of HPP (uncorrected P value = 6.9 × 10−4, OR = 4.54; Fig. 1.)
Figure 1.
Manhattan Plot of PheWAS for rs121918007. This figure represents 742 phenotypes tested for association with SNP rs121918007, using logistic regression, assuming an additive genetic model adjusted for age, sex, and batch. Phenotypes are grouped along the x-axis by categorization within the PheWAS code hierarchy. The upper-red line indicates Bonferroni correction P = 6.7 × 10−4 (false discovery rate = 0.1 for entire PheWAS); lower blue lines indicate P = 0.05.
Among patients with the pathogenic ALPL variants, the average serum AlkP levels were typically within the normal range, although lower compared with the cohort of matched controls (Table 2 and Fig. 2). Eighty-eight percent of subjects had AlkP levels < 50 U/L compared with 35% of controls (P < 0.001), and 55% of patients with ALPL variants had AlkP levels < 40 U/L compared with 20% of controls (P < 0.001). There was no significant difference in renal function in cases compared with controls.
Table 2.
Demographic Information
| Women With ALPL SNPs, n = 52 | Female Controls, n = 52 | P Value | Men With ALPL SNPs, n = 53 | Male Controls, n = 53 | P Value | |
|---|---|---|---|---|---|---|
| Race, % | NS | |||||
| European ancestry | 50 (96.2) | 50 (96.2) | 49 (92.5) | 49 (92.5) | ||
| African ancestry | 1 (1.9) | 1 (1.9) | 2 (3.8) | 2 (3.8) | ||
| Unknown | 1 (1.9) | 1 (1.09) | 2 (3.8) | 2 (3.8) | ||
| Ethnicity, % | NS | |||||
| Non-Hispanic | 51 (98.1) | 52 (100) | 51 (96.2) | 51 (96.2) | ||
| Hispanic | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| Unknown | 1 (1.9) | 0 (0) | 2 (3.8) | 2 (3.8) | ||
| Age, ya | 59.5 (24.1) | 59.4 (23.1) | 0.98 | 56.26 (27.2) | 54.2 (27.4) | 0.69 |
Abbreviation: NS, not significant.
Mean (standard deviation).
Figure 2.
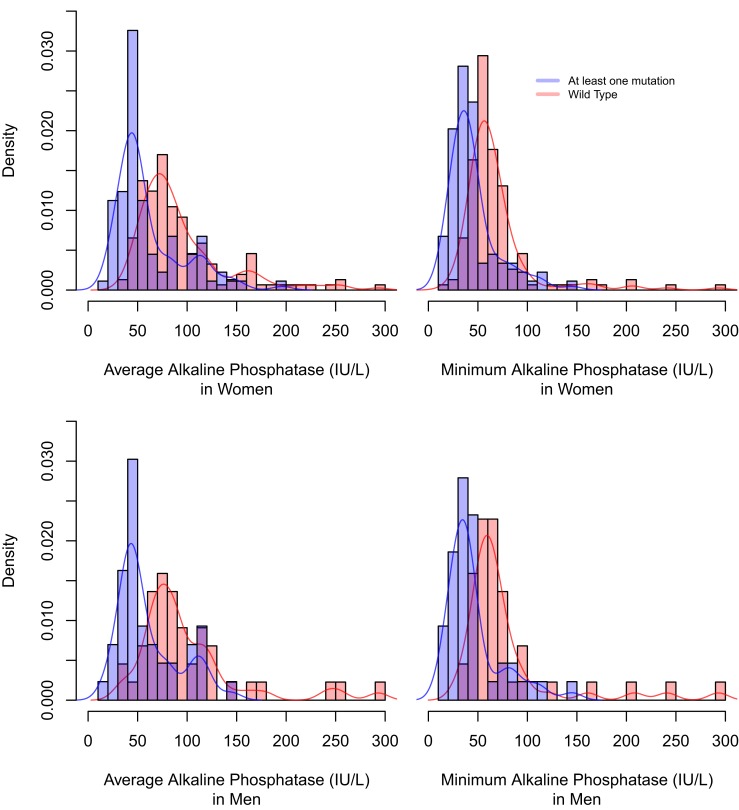
Overlapping histograms of AlkP levels in both female and male cases and controls. For female individuals, there is considerable overlap of both the average and the minimum AlkP levels, with the majority of cases and controls falling within the reference range. Although those with one of the three ALPL variants tend to have lower AlkP levels, there is substantial overlap in the bimodal distribution. For male individuals, there is also overlap between cases and controls; however, male controls tend to have higher AlkP levels, and male cases tend to have lower AlkP levels similar to women.
As depicted in the histogram in Fig. 2, there is considerable overlap of average AlkP levels with the majority of cases and controls falling within the reference range. Although cases tended to have lower AlkP levels, there is substantial overlap in the distribution with controls also having evidence of low AlkP levels. Therefore, low AlkP was not predictive of having HPP.
In the manual review, as depicted in Tables 2 and 3, patients with ALPL variants had an increased incidence of spine fractures [OR: 11.94 (2.15, 302.40); P = 0.006] and dental abnormalities, such as tooth fractures, extractions, and tooth loss (P = 0.02). We also noted an increased prevalence of presumed gynecological pathology. Women with ALPL variants were more likely to have at least one of oophorectomy, endometrial abnormality, or ovarian pathology [OR: 3.64 (1.46, 9.87); P = 0.009] and in particular, were more likely to have ovarian pathology (P = 0.005) compared with controls. Figure 3 depicts the phenotype seen in each individual SNP. Males carrying variants had an increase in scoliosis (P = 0.053).
Table 3.
Associations Based on SNP Status Compared With Controls
| Women With ALPL SNPs, n = 52 | Female Controls, n = 52 | Female rs121918007, n = 41 | Female rs121918002, n = 10 | Female rs19966998, n = 1 | Men With ALPL SNPs, n = 53 | Male Controls, n = 53 | Male rs121918007, n = 33 | Male rs121918002, n = 19 | Male rs19966998, n = 1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age, ya | 59.5 (24.1) | 59.4 (23.1) | 58.17 (24.1) | 69.1 (20.3) | 18 (NA) | 56.26 (27.2) | 54.2 (27.4) | 60.7(25.4) | 43.3 (28.6) | 46 |
| P = 0.98 | P = 0.69 | |||||||||
| Average AlkP, U/Lb | 62.9 (38.8) | 85.3 (35.4) | 61.03 (30.5) | 60.22 (57.4) | 155 (NA) | 60.4 (31.0) | 100.4 (53.8) | 63.65(32.5) | 56 (29.6) | 46 |
| P = 0.005 | P < 0.001 | |||||||||
| Minimum AlkP, U/La | 43.7 (20.3) | 58.0 (21.1) | 44.94 (19.7) | 33.89 (15.7) | 89 (NA) | 46.07 (28.37) | 77.77 (52.4) | 52.54(30.5) | 36.19 (22.9) | 36 |
| P = 0.001 | P = 0.001 | |||||||||
| AlkP ever < 50 U/L, n (%) | 36 (78.3) | 18 (34.6) | 28 (68) | 8 (80) | 0 (0) | 34 (79.0) | 9 (17.0) | 20(77) | 13 (81) | 1 (1) |
| P < 0.001 | P < 0.001 | |||||||||
| AlkP ever < 40 U/L, n (%) | 25 (54.3) | 10 (19.2) | 18 (44) | 7 (70) | 0 (0) | 24 (55.8) | 2 (3.8) | 12(46) | 11 (69) | 0 (0) |
| P = 0.001 | P < 0.001 | |||||||||
| Coexistent Bone Diagnoses | ||||||||||
| Osteoarthritis, n (%) | 18 (34.6) | 21 (42) | 11 (27) | 7 (7) | 0 (0) | 10 (18.9) | 9 (17.0) | 8 (42) | 2 (11) | 0 (0) |
| P = 0.69 | P = 1.00 | |||||||||
| Rheumatoid arthritis, n (%) | 4 (7.7) | 3 (5.8) | 3 (7) | 1 (1) | 0 (0) | 2 (3.8) | 3 (5.7) | 1 (3) | 1 (5) | 0 (0) |
| P = 1.0 | P = 1.00 | |||||||||
| Chondrocalcinosis, n (%) | 6 (11.5) | 2 (3.8) | 4 (9.8) | 2 (20.0) | 0 (0) | 2 (3.77) | 1 (1.85) | 2 (6.25) | 0 (0) | 0 (0) |
| P = 0.270 | P = 1 | |||||||||
| History of long bone fractures, n (%) | 9 (17.3) | 13 (25) | 7 (17) | 2 (2) | 0 (0) | 5 (9.4) | 11 (20.8) | 2 (6) | 3 (16) | 0 (0) |
| P = 0.47 | P = 0.128 | P = 1 | P = 0.18 | P = 0.423 | P = 0.693 | |||||
| History of spine fractures, n (%) | 11 (21.2) | 1 (1.9) | 10 (24) | 1 (1) | 0 (0) | 4 (7.5) | 2 (3.8) | 2 (6) | 1 (5) | 0 (0) |
| P = 0.006 | P = 0.025 | P = 0.67 | P = 1 | P = 1 | ||||||
| Osteoporosis, n (%) | 23 (44.2) | 14 (26.9) | 18 (44) | 5 (5) | 0 (0) | 7 (13.2) | 2 (3.8) | 3 (9) | 0 (0) | 0 (0) |
| P = 0.10 | P = 0.252 | P = 0.16 | P = 0.148 | P = 1 | ||||||
| Scoliosis, n (%) | 8 (15.4) | 5 (9.6) | 7 (17) | 1 (1) | 0 (0) | 7 (13.2) P = 0.07 | 1 (1.9) | 5 (15) P = 0.053 | 2 (11) P = 0.487 | 0 (0) |
| Coexistent Dental Diagnoses | ||||||||||
| Dental abnormalities, n (%)c | 7 (13.5) | 0 (0) | 7 (17) | 0 (0) | 0 (0) | 6 (11.3) | 2 (3.8) | 0 (0) | 2 (11) | 0 (0) |
| P = 0.02 | P = 0.012 | P = 1 | P = 0.27 | P = 1 | P = 0.487 | |||||
| Gynecological History | ||||||||||
| Total abdominal hysterectomy, n (%) | 13 (25) | 14 (26.9) | 10 (24) | 3 (3) | 0 (0) | |||||
| P = 1 | P = 1 | P = 1 | ||||||||
| Oophorectomy, n (%) | 13 (25) | 7 (13.5) | 11 (27) | 2 (2) | 0 (0) | |||||
| P = 1.21 | P = 0.16 | P = 1 | ||||||||
| Endometrial abnormalities, n (%) | 10 (19.2) | 5 (9.6) | 9 (22) | 1 (1) | 0 (0) | |||||
| P = 0.26 | P = 0.12 | P = 1 | ||||||||
| Ovarian pathology, n (%) | 9 (17.3) | 0 (0.0) | 8 (2) | 1 (1) | 0 (0) | |||||
| P = 0.005 | P = 0.005 | P = 1 | ||||||||
| Any gynecological findings, n (%)d | 21 (40.4) | 8 (15.4) | 19 (46) | 2 (2) | 0 (0) | |||||
| P = 0.009 | P < 0.001 | P = 1 | ||||||||
Long bone fractures included humeral, radial, clavicular, sternal, tibial, fibular, or femoral fractures. Spine fractures included cervical, thoracic, lumbar, or sacral fractures. Endometrial abnormalities included those with endometrial polyps, hyperplasia, hypertrophy, carcinoma, adenomyosis, or dysfunctional uterine bleeding or those that underwent endometrial ablations. Ovarian pathology included those with imaging-confirmed or self-reported ovarian cysts or cystadenomas.
Abbreviation: NA, not applicable.
Mean (standard deviation).
Reference range, 40 to 140 (U/L).
Dental abnormalities included dental fractures, extractions, or other tooth loss.
Patients with at least one of oophorectomy, endometrial abnormalities, or ovarian pathology.
Figure 3.
Venn diagram of phenotypes seen in women with pathogenic ALPL SNPs rs121918007, rs121918002, and rs19966998. This Venn diagram depicts the distribution of associations seen in women with the ALPL SNPs rs121918007, rs121918002, and rs19966998, all previously reported as pathogenic. Of these 52 total women examined, 40 had evidence of disease. Of the 13 women who were compound heterozygotes, three had gynecologic (gyn) only, two had bone only, one had dental only, and one had gyn + bone. Women with no associated HPP-related or gynecologic findings had higher AlkP.
We evaluated each of the three ALPL variants individually to determine whether there were differences in clinical presentations (Tables 2 and 3 and Fig. 3). Among the three variants evaluated, 28 of the 41 patients with rs121918007 (68%), and 8 of the 10 with rs121918002 (80%) had at least one recorded serum AlkP below 50 U/L; 18 of 41 with rs121918007 (44%) and seven of 10 (70%) had a recorded serum activity below 40 U/L. Whereas serum AlkP activity was similarly depressed in both groups with known variants, those with rs121918002 tended to have lower average (60.22 U/L vs 61.03 U/L) and lowest recorded (33.89 U/L vs 44.94 U/L) serum AlkP activities. Those with no documented features had the highest AlkP activity (Fig. 3) and may represent carriers with reduced expression of disease.
Variant rs12198007 had a statistically significant increase in dental abnormalities (P = 0.012) and history of spine fractures [OR: 6.16 (1.19, 61.96); P = 0.025] compared with controls. However, there was also an increase in ovarian cysts (P = 0.005) and a trend toward increased oophorectomy [OR: 2.61 (0.73, 10.69); P = 0.162] and endometrial abnormalities [OR: 3.51 (0.79, 21.85); P = 0.116] in variant rs12198007 compared with controls. Variant rs121918002 exhibited 10% more ovarian cysts and 20% more oophorectomy than the control group, although this was not statistically significant.
None of the individuals with these three ALPL variants had a diagnosis of HPP in the EHR, either by billing codes or by any reference in their medical history, even with evidence of low AlkP and clinically recognized skeletal and dental pathology.
Discussion
The current study demonstrates that pathogenic variants in ALPL occurred in ∼7 of 1000 unselected patients among a mostly outpatient clinical population, which is more common than previously thought (7, 8, 29). This is likely an underestimate, as the current study was based on available genotyping and not full exome sequencing of ALPL. Additionally, AlkP levels were within the reference range in some affected carriers and would escape detection by the classical biomarker, leading to a failure to diagnose patients with disease. This suggests that additional screening biomarkers and better recognition of the disease phenotype are needed for appropriate identification of this common disorder.
Given that ALPL is expressed in many tissues, in addition to skeletal tissue, it is not surprising that there are additional sequela of HPP. We used PheWAS as a tool to discover relationships between these known SNPs, identify pleiotropy, provide mechanistic insights, and foster hypothesis generation. The current analysis revealed associations between ALPL pathogenic variants and the expected diagnoses of spine fractures and periodontal disease but also identified an association with gynecologic disorders. Gynecologic and ovarian disease was associated with rs12198007 in the PheWAS and then confirmed by manual chart review across the three SNPs. Reported indications for oophorectomy were only available for six of the 13 patients and included ovarian cysts, dysfunctional uterine bleeding, endometrial cancer, ovarian tumors, torsion in an 11 year old, endometriosis, tubal pregnancy, and an ovarian cystadenoma in a 10 year old. Physiologic cysts, documented in imaging reports, were excluded. The biological significance of AlkP activity in the female reproductive tract has been described by Bentala et al. (30) and Lei et al. (31, 32) who demonstrated that AlkP expression and activity in the uterus may affect blastocyst implantation by its effects on dephosphorylation of lipopolysaccharide detoxification in the uterine epithelium. A potential mechanism of this bacterial endotoxin detoxification involves dephosphorylation of lipids by AlkPs, rendering the endotoxins nontoxic (33, 34), and explains the inherent defense barrier in the uterus during pregnancy to protect against infection by bacteria and consequent early pregnancy loss. Additionally, reproductive tract infections, including lipopolysaccharide-induced activation of the signaling pathway on stromal fibroblasts, may have a relationship with fibroid development (35, 36), and uterine fibroids are the most common benign tumor of the female reproductive tract. Effects of AlkP activity, specifically on ovarian function, have not yet been described. Given its ubiquity and high level of expression in tissues throughout the body, further study of both the molecular function and clinical impact of these ALPL variants will be necessary to define further the important skeletal and extraskeletal functions of this enzyme.
This analysis has certain limitations. The demonstrated association between genotype and phenotype cannot establish causality. The accuracy and completeness of information in the database are potential sources of unrecognized error. Data regarding subtype and severity, history of medications prescribed by non-Vanderbilt University providers, and history of fractures not documented on the medical record were not available. Although ours is a large cohort of undiagnosed subjects identified, the numbers are still relatively small, raising the possibility of underpowering for more subtle phenotypic characteristics. Finally, we only evaluated available ALPL variants present on a standard array; subsequent studies should evaluate more ALPL variants through sequencing or more comprehensive genotyping arrays.
The current study demonstrates that the presence of pathogenic ALPL variants is more common than previously estimated. None of these patients discovered via BioVU had the correct diagnosis, even with obvious skeletal and dental pathology, potentially, in part, as a result of the fact that AlkP levels were within the reference range for many patients. Therefore, a normal or low-normal AlkP level should not prohibit further assessment for HPP when suspected clinically. Additional biomarkers and better recognition of the disease phenotype are needed for appropriate identification of this disorder. Unexpectedly, we found an association of ovarian and gynecological disease with variants in the ALPL gene discovered by PheWAS and then confirmed by manual chart view. This is an association in which replication in other cohorts is necessary to confirm this finding, which might lead to a change in screening female patients known to have HPP. Given its ubiquity and high level of expression in tissues throughout the body, further study of both the molecular function and clinical impact of HPP causing ALPL variants will be necessary to define further the important skeletal, gynecologic, extraskeletal, and other functions of this enzyme. As a common, potentially debilitating, and treatable disorder of adults and children, HPP deserves increased scrutiny.
Acknowledgments
Financial Support: Support for L.B., J.L.W., and J.C.D. was provided by the National Institutes of Health Grant R01 LM010685. The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the National Institutes of Health-funded Shared Instrumentation Grant S10RR025141 and Clinical and Translational Science Awards Grants UL1TR002243, UL1TR000445, and UL1RR024975.
Disclosure Summary: K.M.D. is a clinical trial investigator for Alexion Pharmaceuticals and Mereo BioPharma Group. J.S.S. is a clinical trial investigator for Alexion Pharmaceuticals and Ultragenyx Pharmaceuticals.
Glossary
Abbreviations:
- AlkP
alkaline phosphatase
- ALPL
alkaline phosphatase, liver/bone/kidney
- BioVU
Vanderbilt University DNA Biobank
- EHR
electronic health record
- HPP
hypophosphatasia
- ICD-9-CM
International Classification of Disease, Ninth Edition, Clinical Modification
- OR
odds ratio
- PheWAS
phenome-wide association study
- SNP
single-nucleotide polymorphism
References
- 1. Rathbun JC. Hypophosphatasia; a new developmental anomaly. Am J Dis Child. 1948;75(6):822–831. [DOI] [PubMed] [Google Scholar]
- 2. Whyte MP. Hypophosphatasia - aetiology, nosology, pathogenesis, diagnosis and treatment. Nat Rev Endocrinol. 2016;12(4):233–246. [DOI] [PubMed] [Google Scholar]
- 3. Bianchi ML. Hypophosphatasia: an overview of the disease and its treatment. Osteoporos Int. 2015;26(12):2743–2757. [DOI] [PubMed] [Google Scholar]
- 4. Reibel A, Manière M-C, Clauss F, Droz D, Alembik Y, Mornet E, Bloch-Zupan A. Orodental phenotype and genotype findings in all subtypes of hypophosphatasia. Orphanet J Rare Dis. 2009;4(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braunstein NA. Multiple fractures, pain, and severe disability in a patient with adult-onset hypophosphatasia. Bone Rep. 2015;4:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taketani T, Onigata K, Kobayashi H, Mushimoto Y, Fukuda S, Yamaguchi S. Clinical and genetic aspects of hypophosphatasia in Japanese patients. Arch Dis Child. 2014;99(3):211–215. [DOI] [PubMed] [Google Scholar]
- 7. Mornet E. Hypophosphatasia [published online ahead of print September 20, 2017]. Metabolism.
- 8. Whyte MP. Hypophosphatasia: an overview for 2017. Bone. 2017;102:15–25. [DOI] [PubMed] [Google Scholar]
- 9. Mornet E. Molecular genetics of hypophosphatasia and phenotype-genotype correlations. Subcell Biochem. 2015;76:25–43. [DOI] [PubMed] [Google Scholar]
- 10. Taillandier A, Domingues C, De Cazanove C, Porquet-Bordes V, Monnot S, Kiffer-Moreira T, Rothenbuhler A, Guggenbuhl P, Cormier C, Baujat G, Debiais F, Capri Y, Cohen-Solal M, Parent P, Chiesa J, Dieux A, Petit F, Roume J, Isnard M, Cormier-Daire V, Linglart A, Millán JL, Salles JP, Muti C, Simon-Bouy B, Mornet E. Molecular diagnosis of hypophosphatasia and differential diagnosis by targeted next generation sequencing. Mol Genet Metab. 2015;116(3):215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stoll C, Fischbach M, Terzic J, Alembik Y, Vuillemin MO, Mornet E. Severe hypophosphatasia due to mutations in the tissue-nonspecific alkaline phosphatase (TNSALP) gene. Genet Couns. 2002;13(3):289–295. [PubMed] [Google Scholar]
- 12. McKiernan FE, Berg RL, Fuehrer J. Clinical and radiographic findings in adults with persistent hypophosphatasemia. J Bone Miner Res. 2014;29(7):1651–1660. [DOI] [PubMed] [Google Scholar]
- 13. Whyte MP, Zhang F, Wenkert D, McAlister WH, Mack KE, Benigno MC, Coburn SP, Wagy S, Griffin DM, Ericson KL, Mumm S. Hypophosphatasia: validation and expansion of the clinical nosology for children from 25 years experience with 173 pediatric patients. Bone. 2015;75:229–239. [DOI] [PubMed] [Google Scholar]
- 14. Caswell AM, Whyte MP, Russell RG. Hypophosphatasia and the extracellular metabolism of inorganic pyrophosphate: clinical and laboratory aspects. Crit Rev Clin Lab Sci. 1991;28(3):175–232. [DOI] [PubMed] [Google Scholar]
- 15. Berkseth KE, Tebben PJ, Drake MT, Hefferan TE, Jewison DE, Wermers RA. Clinical spectrum of hypophosphatasia diagnosed in adults. Bone. 2013;54(1):21–27. [DOI] [PubMed] [Google Scholar]
- 16. Weber TJ, Sawyer EK, Moseley S, Odrljin T, Kishnani PS. Burden of disease in adult patients with hypophosphatasia: results from two patient-reported surveys. Metabolism. 2016;65(10):1522–1530. [DOI] [PubMed] [Google Scholar]
- 17. Foster BL, Ramnitz MS, Gafni RI, Burke AB, Boyce AM, Lee JS, Wright JT, Akintoye SO, Somerman MJ, Collins MT. Rare bone diseases and their dental, oral, and craniofacial manifestations. J Dent Res. 2014;93(7Suppl):7S–19S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bloch-Zupan A. Hypophosphatasia: diagnosis and clinical signs - a dental surgeon perspective. Int J Paediatr Dent. 2016;26(6):426–438. [DOI] [PubMed] [Google Scholar]
- 19. Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(D1):D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cronin RM, Field JR, Bradford Y, Shaffer CM, Carroll RJ, Mosley JD, Bastarache L, Edwards TL, Hebbring SJ, Lin S, Hindorff LA, Crane PK, Pendergrass SA, Ritchie MD, Crawford DC, Pathak J, Bielinski SJ, Carrell DS, Crosslin DR, Ledbetter DH, Carey DJ, Tromp G, Williams MS, Larson EB, Jarvik GP, Peissig PL, Brilliant MH, McCarty CA, Chute CG, Kullo IJ, Bottinger E, Chisholm R, Smith ME, Roden DM, Denny JC. Phenome-wide association studies demonstrating pleiotropy of genetic variants within FTO with and without adjustment for body mass index. Front Genet. 2014;5:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010;3(1):42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.PheWAS Resources. Phecode map 1.2 with ICD-9 codes. Available at: https://phewascatalog.org/phecodes. Accessed 20 April 2018.
- 26. Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mornet E. The tissue nonspecific alkaline phosphatase gene mutations database. Available at: www.sesep.uvsq.fr/03_hypo_mutations.php. Accessed 15 September 2017. [DOI] [PubMed]
- 28. Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetskey M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. ClinVar: public archive of the interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mornet E, Yvard A, Taillandier A, Fauvert D, Simon-Bouy B. A molecular-based estimation of the prevalence of hypophosphatasia in the European population. Ann Hum Genet. 2011;75(3):439–445. [DOI] [PubMed] [Google Scholar]
- 30. Bentala H, Verweij WR, Huizinga-Van der Vlag A, van Loenen-Weemaes AM, Meijer DK, Poelstra K. Removal of phosphate from lipid A as a strategy to detoxify lipopolysaccharide. Shock. 2002;18(6):561–566. [DOI] [PubMed] [Google Scholar]
- 31. Lei W, Nguyen H, Brown N, Ni H, Kiffer-Moreira T, Reese J, Millán JL, Paria BC. Alkaline phosphatases contribute to uterine receptivity, implantation, decidualization, and defense against bacterial endotoxin in hamsters. Reproduction. 2013;146(5):419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lei W, Ni H, Herington J, Reese J, Paria BC. Alkaline phosphatase protects lipopolysaccharide-induced early pregnancy defects in mice. PLoS One. 2015;10(4):e0123243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rader BA. Alkaline phosphatase, an unconventional immune protein. Front Immunol. 2017;8:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pettengill M, Matute JD, Tresenriter M, Hibbert J, Burgner D, Richmond P, Luis Millán J, Ozonoff A, Strunk T, Currie A, Levy O. Human alkaline phosphatase dephosphorylates microbial products and is elevated in preterm neonates with a history of late-onset sepsis. PLoS One. 2017;12(4):e0175936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo J, Zheng L, Chen L, Luo N, Yang W, Qu X, Liu M, Cheng Z. Lipopolysaccharide activated TLR4/NF-κB signaling pathway of fibroblasts from uterine fibroids. Int J Clin Exp Pathol. 2015;8(9):10014–10025. [PMC free article] [PubMed] [Google Scholar]
- 36. Ciavattini A, Di Giuseppe J, Stortoni P, Montik N, Giannubilo SR, Litta P, Islam MS, Tranquilli AL, Reis FM, Ciarmela P. Uterine fibroids: pathogenesis and interactions with endometrium and endomyometrial junction. Obstet Gynecol Int.2013;2013:173184. [DOI] [PMC free article] [PubMed] [Google Scholar]