Abstract
3βHSD1 enzymatic activity is essential for synthesis of potent androgens from adrenal precursor steroids in prostate cancer. A germline variant in HSD3B1, the gene that encodes 3βHSD1, encodes for a stable enzyme, regulates adrenal androgen dependence, and is a predictive biomarker of poor clinical outcomes after gonadal testosterone deprivation therapy. However, little is known about HSD3B1 transcriptional regulation. Generally, it is thought that intratumoral androgen synthesis is upregulated after gonadal testosterone deprivation, enabling development of castration-resistant prostate cancer. Given its critical role in extragonadal androgen synthesis, we sought to directly interrogate the transcriptional regulation of HSD3B1 in multiple metastatic prostate cancer cell models. Surprisingly, we found that VCaP, CWR22Rv1, LNCaP, and LAPC4 models demonstrate induction of HSD3B1 upon androgen stimulation for approximately 72 hours, followed by attenuation around 120 hours. 3βHSD1 protein levels mirrored transcriptional changes in models harboring variant (LNCaP) and wild-type (LAPC4) HSD3B1, and in these models androgen induction of HSD3B1 is abrogated via enzalutamide treatment. Androgen treatment increased flux from [3H]-dehydroepiandrosterone to androstenedione and other downstream metabolites. HSD3B1 expression was reduced 72 hours after castration in the VCaP xenograft mouse model, suggesting androgen receptor (AR) regulation of HSD3B1 also occurs in vivo. Overall, these data suggest that HSD3B1 is unexpectedly positively regulated by androgens and ARs. These data may have implications for the development of treatment strategies tailored to HSD3B1 genotype status.
Prostate cancer upregulates HSD3B1 in response to androgens, leading to increased DHEA metabolism by 3β-HSD1, suggesting that AR signaling directly regulates an increase in potent androgen synthesis.
Prostate cancer is a common and genetically heterogeneous disease that requires gonadal testosterone (T) for growth and progression (1). Advanced prostate cancer responds initially to androgen deprivation therapy (ADT; i.e., castration); however, metastatic cancer treated with ADT invariably results in the development of castration-resistant prostate cancer (CRPC) (2). 3βHSD1, encoded by HSD3B1, is the enzyme that converts dehydroepiandrosterone (DHEA) to androstenedione (AD), and regulates the rate limiting step in the conversion of DHEA to the potent androgens T and DHT (3). A genetic variant of HSD3B1—1245A>C; rs1047303—results in decreased proteasomal degradation of 3βHSD1 and in a strikingly dimorphic shift in DHEA metabolism, likely leading to increased DHT synthesis from extragonadal precursor steroids in metastatic tumors (4). In fact, in six separate cohorts, patients who inherit the HSD3B1 variant have worse outcomes when undergoing ADT for advanced prostate cancer (5–8). This is mechanistically attributable to the ability of tumors harboring the variant to more effectively use adrenal DHEA/DHEA sulfate for tumor androgen synthesis to fuel tumor growth when gonadal T is no longer available because of ADT (9). Conceptually, HSD3B1 variant status dictates tumor adrenal androgen dependence in the setting of ADT, with wild-type tumors having little reliance on adrenal androgens, heterozygous tumors having an intermediate dependence, and homozygous variant having the greatest dependence on adrenal androgens and, as such, the shortest response to ADT (9).
Many of the enzymes involved in androgen synthesis are transcriptionally upregulated in the metastatic setting (10–15). Steroid-5α-reductase (SRD5A) is an enzyme that converts AD to 5α-androstanedione and is an example of this phenomenon. SRD5A1 is upregulated in the metastatic CRPC setting and is directly regulated by androgens in vitro in metastatic prostate cancer models (13, 15, 16). These data suggest that androgen receptor (AR) stimulation can modulate androgen synthesis, perhaps even in the setting of CRPC (16, 17).
To date, the transcriptional regulation of HSD3B1 in prostatic tissue has not been fully elucidated. Initial studies found that interleukin-4 and -13 could upregulate HSD3B1 in several peripheral tissues, including normal prostate epithelium, immortalized keratinocytes, cervical cancer cells, colon cancer cells, normal mammary epithelial cells, and breast cancer cells (18). However, these cytokines were not found to induce HSD3B1 expression in prostate cancer (LNCaP or PC3) or choriocarcinoma (JAR and JEG-3) cell lines (19).
We sought to elucidate the transcriptional regulation of HSD3B1 in prostate cancer. We found that in multiple CRPC models, HSD3B1 is induced by the synthetic androgen R1881 and that this effect can be abrogated by enzalutamide. 3βHSD1 protein is also induced, and enzyme functionality was confirmed by measuring flux from DHEA to AD and other downstream metabolites via HPLC. Further, HSD3B1 mRNA and protein are suppressed in an in vivo VCaP xenograft model after castration. These data, although somewhat unexpected, strongly suggest that HSD3B1 is positively regulated by AR signaling in CRPC cell models.
Materials and Methods
Cell culture
LNCaP and CWR22Rv1 cells were purchased from American Type Culture Collection (ATCC) and cultured in RPMI 1640 without phenol red with 10% fetal bovine serum (FBS; Gemini Bio Products) and 1% penicillin/streptomycin (P/S). LAPC4 cells were generously provided by Dr. David Danielpour (Case Western Reserve University, Cleveland, OH) and cultured in Isocove’s modified Dulbecco’s medium with 10% FBS, 1% P/S, and 1% l-glutamine. VCaP cells were purchased from ATCC and cultured in DMEM with 10% FBS, 1% P/S, and 1% sodium pyruvate. HEK293T cells were purchased from ATCC and maintained in DMEM with 10% FBS and 1% P/S. All cells were cultured at 37°C in a 5% carbon dioxide humidified incubator and routinely tested for the presence of mycoplasma. For gene expression assays, cells were cultured in 10% charcoal stripped serum (CSS; Gemini Bio Products) for 48 hours before R1881 or enzalutamide treatment and maintained in CSS for the duration of the experiment.
Gene expression analysis
Total RNA was isolated from cells and tissues by using RNAzol RT (Sigma-Aldrich) and 1 μg of RNA was reverse transcribed to cDNA by using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR (qPCR) was then performed with an ABI 7500 Real-Time PCR machine (Applied Biosystems) by using iTaq Fast SYBR Green Supermix with ROX (Bio-Rad) in a final reaction volume of 20 µL. All qPCR assays were performed in triplicate with previously tested primer sets (4) as listed: PSA (forward: 5′-GCATGGGATGAAGTAAG-3′; reverse: 5′-CATCAAATCTGAGGGTTGTCTGGA-3′), FKBP5 (forward: 5′-AAAAGGCCACCTAGCTTTTT-3′; reverse: 5′-CCCCCTGGTGAACCATAATACA-3′), HSD3B1 (forward: 5′-CCATGTGGTTTGCTGTTACCAA-3′; reverse: 5′-TCAAACGACCCTCAAGTTAAAAGA-3′), and RPLP0 forward: 5′-CGAGGGCACCTGGAAAAC-3′; reverse: 5′-CACATTCCCCCGGATATGA-3′). Expression was quantitated by using the 2ΔΔCt method, normalizing to housekeeping gene RPLP0 and vehicle-treated cells. Actinomycin D (ActD; Sigma-Aldrich) was used at 5 mg to inhibit transcription in LNCaP cells treated with R1881. All gene expression studies were repeated at least once in independent experiments.
Western blot analysis
Total protein was isolated from cells by using ice-cold radioimmunoprecipitation assay buffer (Sigma-Aldrich) with protease inhibitors (Roche). A total of 50 to 75 µg of protein was separated on 12% Mini-PROTEAN TGX Precast Gels (Bio-Rad) and transferred to a nitrocellulose membrane (Millipore). Membranes were incubated with 3βHSD1 primary antibody provided by Dr. CE Gomez-Sanchez (University of Mississippi Medical Center, Jackson, MS; RRID: AB_2728753) (20) overnight at 4°C. The following day, membranes were washed in Tris-buffered saline with 0.1% Tween-20 for 45 minutes and incubated for 1 hour at room temperature with anti-mouse secondary antibody conjugated to peroxidase (GE Healthcare; catalog no. NA931; RRID: AB_772210). Membranes were washed again in Tris-buffered saline with 0.1% Tween-20 for a total of 45 minutes and then developed by using the chemiluminescent detection system Pico (Thermo Fisher Scientific) to detect bands with peroxidase activity. Primary and secondary antibodies were removed from the membrane by using Restore Western Blot Stripping Buffer (Thermo Fisher Scientific), and protein loading was subsequently measured by using an anti–glyceraldehyde 3-phosphate dehydrogenase antibody (Cell Signaling; catalog no. 5174; RRID: AB_10622025) and anti-rabbit secondary antibody (Thermo Fisher Scientific; catalog no. 31460; RRID: AB_228341) as described above.
Xenograft studies
All mouse studies were performed under an approved Cleveland Clinic Lerner Research Institute Institutional Animal Care and Use Committee protocol. All mice were NSG males purchased from Jackson Laboratory at 6 to 8 weeks of age. Approximately 107 VCaP cells in 50% matrigel/50% PBS were injected subcutaneously into the flank of each mouse. Tumors grew to approximately 200 to 300 mm3, at which time three mice were selected randomly for castration; all tumors were harvested 72 hours after castration. Approximately 50% of each xenograft tissue was fixed in 4% paraformaldehyde and embedded in paraffin, and ∼25% of the tissue was flash frozen and 25% was submerged in RNAlater (Thermo Fisher Scientific) for protein and gene expression analysis, respectively.
Cellular steroid metabolism
HPLC was used to determine DHEA metabolism in cells treated with R1881 as previously described (3, 4). Briefly, cells were cultured in fetal calf serum with R1881 or vehicle in a 10-cm dish and then seeded in a 12-well dish at 300,000 cells/well 1 day before media collection. [3H]-DHEA (100 nM, 300,000 to 600,000 cpm; PerkinElmer) was added to CSS cell media 10 days after initiation of R1881 or vehicle treatment, and media samples were collected at 8, 24, and 48 hours after addition of DHEA for HPLC analysis. Media samples were treated with β-glucuronidase (1000 units; Sigma-Aldrich) at 65°C for at least 2 hours, and deconjugated steroids were extracted with 1:1 ethyl acetate: isooctane and subsequently concentrated under nitrogen gas in glass tubes. Dried samples were dissolved in 50% methanol and injected on a Breeze 1525 system fitted with a model 717 plus autoinjector (Waters Corp.) and a Kinetix 100 × 2.1-mm, 2.6 µmol/L C18 reverse-phase column (Phenomenex) and methanol/water gradients at 30°C. Column effluent was evaluated using a β-RAM model 3 in-line radioactivity detector (IN/US Systems, Inc.) using Liquiscint scintillation mixture (National Diagnostics). Steroid metabolites were characterized by comparison with isotope-labeled standards of AD, T, DHEA, and DHT. All HPLC experiments were performed in triplicate, and results were reproduced in independent experiments.
Bioinformatics analysis of HSD3B1 regulation
Multiple studies have been conducted investigating genome wide binding of AR in the setting of androgen stimulation via chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) in prostate cancer models. Many of these studies have been made publically available via Cistrome Data Browser (http://cistrome.org/db/#/), a portal for mining large-throughput ChIP-seq datasets (21). We queried all available public datasets for those involving prostate cancer and AR. Each study retrieved was manually reviewed for AR stimulation with R1881 or DHT and appropriate ethanol control in desired cell models. Two representative studies were identified, one in LNCaP and one in VCaP, and were queried for HSD3B1 occupancy. Original data files were retrieved from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) via GEO accession number provided within the Cistrome Data Browser (LNCaP: GSE69043; VCaP: GSE55062). Data files were visualized using an integrated genomics viewer (http://software.broadinstitute.org/software/igv/) (22).
Results
HSD3B1 is transcriptionally upregulated in response to R1881
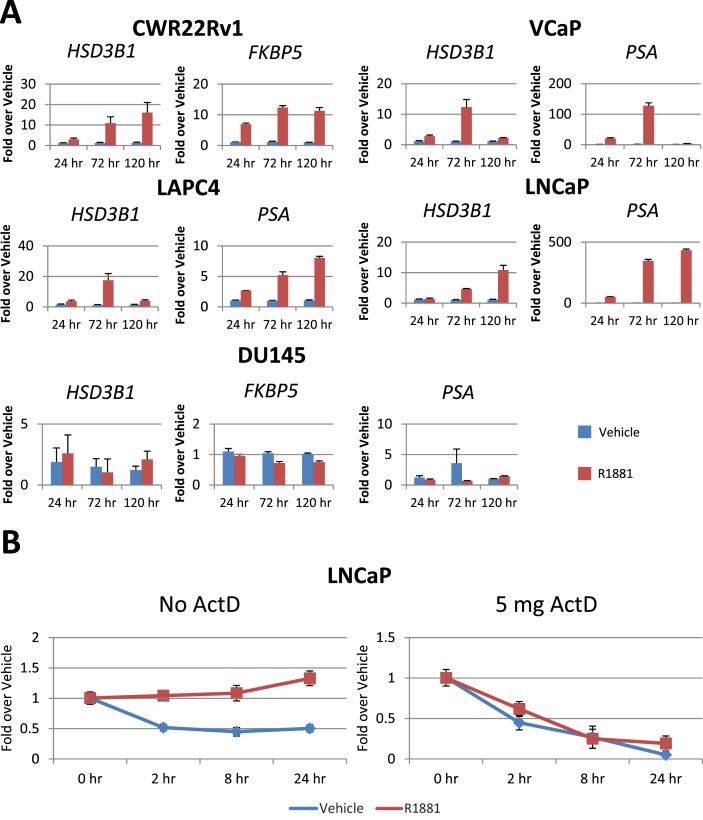
AR stimulation with R1881 increased HSD3B1 transcript relative to vehicle controls at 24, 72, and 120 hours after treatment in the AR-expressing cell lines LNCaP, VCaP, CWR22Rv1, and LAPC4 (Fig. 1A). DU145 cells, a model that does not express AR, was also treated with R1881 and no increase in HSD3B1 transcript was observed. To determine whether the effect of R1881 on HSD3B1 is attributable to increased transcript stability, LNCaP cells were treated with R1881 and subsequently treated with ActD for 2, 8, and 24 hours (Fig. 1B). HSD3B1 transcript loss was similar in vehicle- and R1881-treated cells, suggesting that increased transcript levels are attributable to transcriptional upregulation rather than increased transcript stability.
Figure 1.
HSD3B1 is induced by AR signaling through transcriptional upregulation. (A) Multiple CRPC models were incubated with 1 nM R1881 (red bars) or vehicle (70% ethanol, blue bars) in 10% CSS and media. Transcripts were normalized to RPLP0 and vehicle controls at each time point. Each experiment was independently repeated at least twice. Increases in HSD3B1 compared with vehicle demonstrate that AR signaling leads to upregulation of HSD3B1, suggesting HSD3B1 is an AR-regulated gene. (B) LNCaP cells were treated with 1 nM R1881 (red line) or vehicle (70% ethanol, blue line) followed by no ActD or 5 mg ActD for 2, 4, 8, or 24 hours. Data demonstrate that HSD3B1 requires intact transcriptional machinery to maintain increased HSD3B1 transcript levels, suggesting that increases in HSD3B1 transcript levels are the result of transcriptional upregulation rather than increased transcript stability. Error bars represent SD of triplicates.
AR stimulation increases functional 3βHSD1 protein
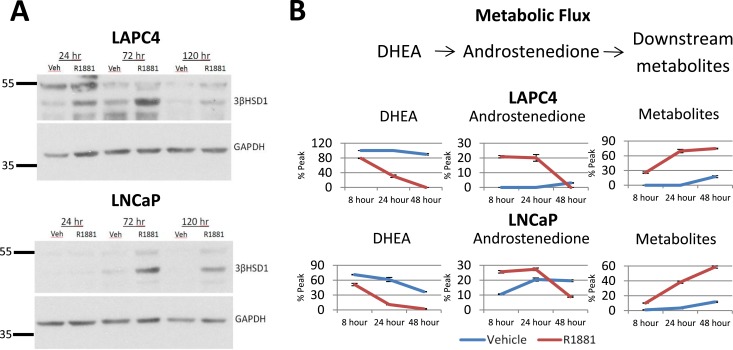
AR stimulation with R1881 led to an increase in 3βHSD1 protein at 24, 72, and 120 hours in both LAPC4 and LNCaP cell models (Fig. 2A). Increased protein levels were accompanied by an increase in metabolic flux from [3H]-DHEA to downstream metabolites in R1881-treated cells compared with vehicle, as detected by HPLC (Fig. 2B). Although the difference in DHEA metabolism between R1881-treated and vehicle-treated LNCaP cells was modest, this is expected based on the HSD3B1 variant genotype that confers an increase in baseline metabolism of DHEA in the LNCaP model. Taken together, these data suggest that the observed AR-induced HSD3B1 transcriptional upregulation results in increased 3βHSD1 protein levels and that this protein is functional, allowing conversion of DHEA to AD and other downstream metabolites.
Figure 2.
AR stimulation induces functional and enzymatically active 3βHSD1 protein. (A) Homozygous wild-type (LAPC4) and homozygous variant (LNCaP) 3βHSD1 protein increased upon AR stimulation (1 nM R1881) at 24, 72, and 120 hours compared with vehicle (70% ethanol). (B) Increased protein resulted in increased DHEA metabolism as measured by HPLC in cell media samples collected at 8, 24, and 48 hours after addition of [3H]-DHEA. R1881-stimulated cells are represented by red lines and vehicle-treated cells are represented by blue lines. Error bars represent SD of triplicates.
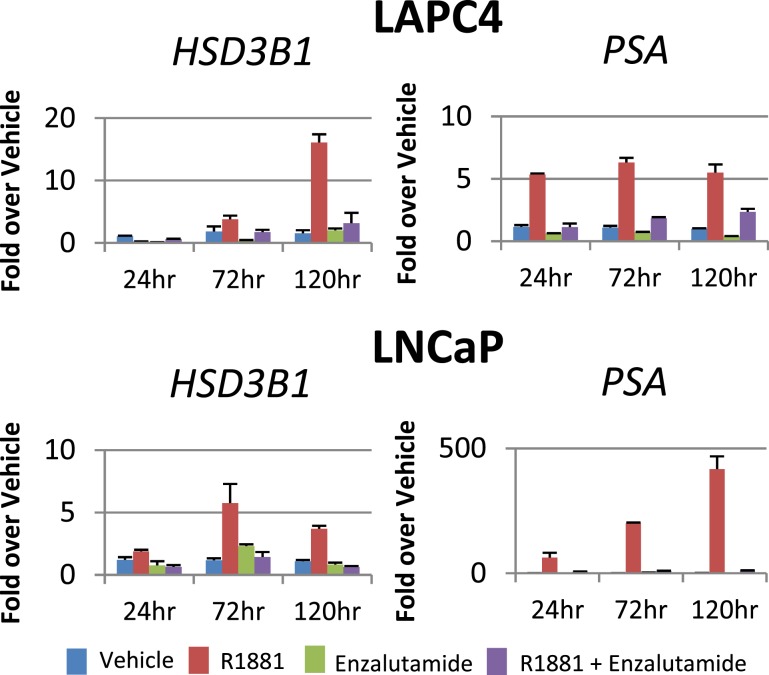
Enzalutamide treatment blocks induction of HSD3B1
To determine the AR-ligand–binding domain dependence of androgen-induced HSD3B1 transcript, we treated LAPC4 cells harboring wild-type HSD3B1 and LNCaP cells harboring variant HSD3B1 with R1881 in conjunction with the AR antagonist enzalutamide. Enzalutamide treatment abrogates the induction of HSD3B1 by R1881 as assessed by qPCR (Fig. 3). PSA expression data demonstrate that enzalutamide was appropriately dosed to block canonical AR signaling, and enzalutamide alone does not appear to affect HSD3B1 expression. These data suggest that expression of HSD3B1 requires the action of AR.
Figure 3.
Enzalutamide reverses HSD3B1 induction by R1881. LAPC4 and LNCaP cells were treated with vehicle (blue bar), R1881 (red bar), enzalutamide alone (green bar), or R1881 plus enzalutamide (purple bar) for 24, 72, and 120 hours. Enzalutamide (10 µM) treatment blocked HSD3B1 expression as well as canonical expression of PSA in the presence of R1881 (1 nM), suggesting that enzalutamide achieved an effective concentration and that AR activation is required for expression of HSD3B1. Error bars represent SD of triplicates.
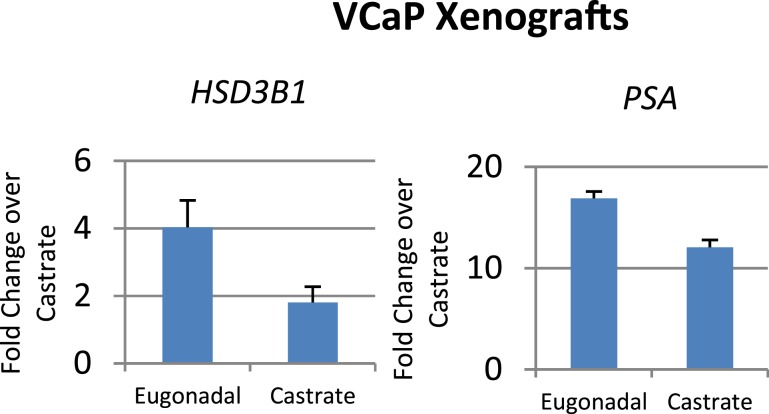
Castration suppresses HSD3B1 expression
To determine the in vivo relevance of our in vitro findings, VCaP xenograft tumors were grown in male NSG mice until tumors reached between 200 and 300 mm3, at which time half of the mice were randomly selected for castration. Three days after castration, all tumors were harvested and subsequently underwent RNA and protein isolation. A decrease in HSD3B1 transcript in tumors grown in castrated mice compared with tumors in eugonadal mice was found via qPCR (Fig. 4). These in vivo data suggest that HSD3B1 responds to changes in circulating androgens and confirms our in vitro findings, suggesting HSD3B1 is an androgen-regulated gene.
Figure 4.
Castration suppresses HSD3B1 transcript. Seven VCaP xenograft tumors were grown to approximately 200 to 300 mm3 in male NSG mice, at which time three were randomly selected for castration. Three days after castration, all tumors were harvested and RNA and protein isolated. HSD3B1 and PSA were normalized to RPLP0 and then to lowest expressing castrate sample. HSD3B1 was reduced upon castration, as was canonical androgen signaling, as demonstrated by an observable decrease in PSA expression.
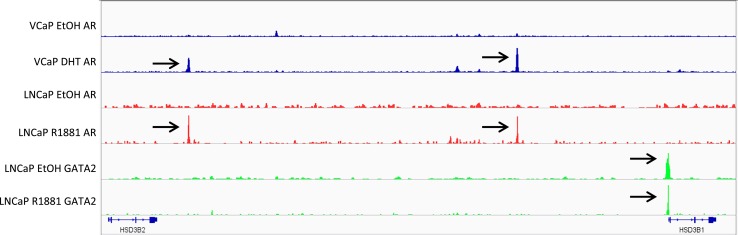
AR and GATA2 binding sites upstream of HSD3B1
To determine whether AR directly regulates HSD3B1 transcription, two publically available ChIP-seq datasets were queried for AR occupancy upstream of HSD3B1 (23, 24). We identified 2 AR binding sites and one potential GATA2 binding site (Fig. 5). Visualization in integrated genomics viewer revealed the GATA2 binding site resides less than 1 kb upstream of the HSD3B1 transcription start site (TSS), whereas both potential androgen regulatory element (ARE) sites are located further upstream at approximately 25 kb and 80 kb from the TSS, respectively. The same AR binding sites were commonly found in AR-stimulated VCaP and LNCaP models.
Figure 5.
AR and GATA2 binding sites upstream of HSD3B1. Publically available ChIP-seq datasets targeting AR and GATA2 in prostate cancer models reveal that GATA2 binds <1 kb upstream of the HSD3B1 transcriptional start site irrespective of androgen stimulation, whereas AR binds approximately 25 and 80 kb upstream in both LNCaP and VCaP models stimulated with androgens, but not with ethanol (EtOH) controls. Data were imported from the GEO (https://www.ncbi.nlm.nih.gov/geo/) (GEO accession nos.:/GSE69043 and GSE55062) and visualized with Integrated Genomics Viewer (http://software.broadinstitute.org/software/igv/). DHT indicates 10 nM DHT for 12 hours, and R1881 indicates treatment with 10 nM R1881 for 16 hours.
Discussion
We were surprised by our findings that HSD3B1 is transcriptionally upregulated in response to androgen treatment, leading to an elevation in functional 3βHSD1 protein and increased conversion from adrenal DHEA to potent downstream androgens. In fact, based on the general notion that gonadal T deprivation therapy appears to spur an increase in compensatory androgen synthesis, we had anticipated the exact opposite result. Namely, we fully expected that AR stimulation would instead suppress HSD3B1 transcript and potent androgen synthesis from extragonadal precursor steroids.
How can the difference between our observed and expected results be explained? First, it is possible that our data do not accurately reflect the physiologic state in vivo. We did observe suppression of HSD3B1 transcript after castration in our xenograft study, which is in line with our in vitro data. HSD3B1 expression analysis in our xenograft studies was carried out 72 hours after castration. Our in vitro studies of HSD3B1 regulation were conducted out to 120 hours of treatment. Longer durations of treatment with androgens or androgen withdrawal may yield results that differ from our observations. It is also possible that in vivo physiologic mechanisms regulate HSD3B1 transcript and downstream enzymatic activity that are not accounted for in our studies.
We are not aware of any studies that have previously characterized isolated 3β-HSD1 enzyme activity in response to AR stimulation in prostate cancer. Androgen stimulation and, alternatively, withdrawal perturb the expression of multiple transcripts that impinge on steroid metabolism (1, 15). Undoubtedly, the net results, from transcript expression to enzymatic activity across multiple catalytic reactions, are complex and temporally regulated. However, our data appear to establish a feed-forward regulatory mechanism of AR stimulation that spurs the additional generation of endogenous AR agonists, at least over the time frame tested in our experimental models.
Multiple studies investigating HSD3B1 regulation in placental tissue, where HSD3B1 is highly expressed, concluded that the zinc finger GATA family of transcription factors, specifically GATA2 (25, 26), regulates HSD3B1 expression. Similarly, our results indicate that GATA2 is bound to the HSD3B1 promoter in LNCaP, suggesting that prostate tissue also uses GATA2 to regulate HSD3B1. Interestingly, GATA2 acts as a cofactor with AR by enhancing AR binding to nearby ARE (24). This could account for the delayed expression and later attenuation of HSD3B1 transcript and 3βHSD1 protein, as our expression data and the distance from TSS to ARE suggest that HSD3B1 may not be regulated by AR in a canonical fashion. Further mechanistic studies are required to fully elucidate the activity of each ARE and how these interact with GATA2 in the regulation of HSD3B1.
3βHSD1 metabolic activity is regulated in major part by the common HSD3B1(1245C) germline variant, which encodes for a protein degradation–resistant enzyme (4), increases engagement with adrenal precursors for potent androgen synthesis. and predicts poor clinical outcomes after castration (5–8). Our data suggest that AR stimulation upregulates HSD3B1 transcript regardless of endogenous wild-type or variant expression.
What could these data mean for the clinical setting? It is possible that feed-forward regulation of AR by androgen-stimulation of HSD3B1 would allow the persistence of a basal level of signaling to propel tumors forward in the castrate state. Consistent with this scenario, a randomized phase II clinical trial of abiraterone (inhibitor of extragonadal androgen synthesis) vs enzalutamide (direct AR antagonist) yielded superior serum PSA declines with enzalutamide compared with abiraterone (27). This might speak to the potential clinical effects of directly blocking AR. However, there was no apparent difference in progression-free survival in this study. Furthermore, elevated levels of intratumoral androgens enabled by HSD3B1(1245C) germline variant inheritance may function to feedback and raise HSD3B1 transcript levels beyond that otherwise found in the wild-type HSD3B1 setting. The profound differences in clinical outcomes associated with HSD3B1 variant inheritance may therefore be attributable in part to transcript regulation. This might argue for testing the role of AR antagonists in blocking steroidogenesis-enabling elevated HSD3B1 transcript.
Acknowledgments
Financial Support: This work was supported by National Cancer Institute Grants R01CA168899, R01CA172382, and R01CA190289 and a Prostate Cancer Foundation Challenge Award (to N.S.) and a Howard Hughes Medical Institute Medical Fellowship Award (to D.H.).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ActD
actinomycin D
- AD
androstenedione
- ADT
androgen deprivation therapy
- AR
androgen receptor
- ARE
androgen regulatory element
- ATCC
American Type Culture Collection
- ChIP-seq
chromatin immunoprecipitation followed by next-generation sequencing
- CRPC
castration-resistant prostate cancer
- CSS
charcoal-stripped serum
- DHEA
dehydroepiandrosterone
- FBS
fetal bovine serum
- GEO
Gene Expression Omnibus
- P/S
penicillin/streptomycin
- qPCR
quantitative PCR
- SRD5A
steroid-5α-reductase
- T
testosterone
- TSS
transcription start site
References
- 1. Dai C, Heemers H, Sharifi N. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med. 2017;7(9):a030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sharifi N, Gulley JL, Dahut WL. Androgen deprivation therapy for prostate cancer. JAMA. 2005;294(2):238–244. [DOI] [PubMed] [Google Scholar]
- 3. Chang K-H, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, Sharifi N. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2011;108(33):13728–13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang K-H, Li R, Kuri B, Lotan Y, Roehrborn CG, Liu J, Vessella R, Nelson PS, Kapur P, Guo X, Mirzaei H, Auchus RJ, Sharifi N. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell. 2013;154(5):1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hearn JWD, AbuAli G, Reichard CA, Reddy CA, Magi-Galluzzi C, Chang K-H, Carlson R, Rangel L, Reagan K, Davis BJ, Karnes RJ, Kohli M, Tindall D, Klein EA, Sharifi N. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 2016;17(10):1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hearn JWD, Xie W, Nakabayashi M, Almassi N, Reichard CA, Pomerantz M, Kantoff PW, Sharifi N. Association of HSD3B1 genotype with response to androgen-deprivation therapy for biochemical recurrence after radiotherapy for localized prostate cancer. JAMA Oncol. 2018;4(4):558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Agarwal N, Hahn AW, Gill DM, Farnham JM, Poole AI, Cannon-Albright L. Independent validation of effect of HSD3B1 genotype on response to androgen-deprivation therapy in prostate cancer. JAMA Oncol. 2017;3(6):856–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shiota M, Fujimoto N, Imada K, Kashiwagi E, Takeuchi A, Inokuchi J, Tatsugami K, Kajioka S, Uchiumi T, Eto M. Independent validation of missense polymorphism in HSD3B1 in Japanese men treated with primary androgen-deprivation therapy for metastatic prostate cancer. J Clin Oncol. 2018;36(6_suppl):179. [Google Scholar]
- 9. Hettel D, Sharifi N. HSD3B1 status as a biomarker of androgen deprivation resistance and implications for prostate cancer. Nat Rev Urol. 2017;15(3):191–196. [DOI] [PubMed] [Google Scholar]
- 10. Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, Ryan C, Smith S, Scher H, Scardino P, Reuter V, Gerald WL. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164(1):217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamid ARAH, Pfeiffer MJ, Verhaegh GW, Schaafsma E, Brandt A, Sweep FCGJ, Sedelaar JPM, Schalken JA. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol Med. 2013;18(1):1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rasiah KK, Gardiner-Garden M, Padilla EJD, Möller G, Kench JG, Alles MC, Eggleton SA, Stricker PD, Adamski J, Sutherland RL, Henshall SM, Hayes VM. HSD17B4 overexpression, an independent biomarker of poor patient outcome in prostate cancer. Mol Cell Endocrinol. 2009;301(1-2):89–96. [DOI] [PubMed] [Google Scholar]
- 13. Mitsiades N, Sung CC, Schultz N, Danila DC, He B, Eedunuri VK, Fleisher M, Sander C, Sawyers CL, Scher HI. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res. 2012;72(23):6142–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pfeiffer MJ, Smit FP, Sedelaar JPM, Schalken JA. Steroidogenic enzymes and stem cell markers are upregulated during androgen deprivation in prostate cancer. Mol Med. 2011;17(7-8):657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, True LD, Nelson PS. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68(11):4447–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Audet-Walsh É, Yee T, Tam IS, Giguère V. Inverse regulation of DHT synthesis enzymes 5α-reductase types 1 and 2 by the androgen receptor in prostate cancer. Endocrinology. 2017;158(4):1015–1021. [DOI] [PubMed] [Google Scholar]
- 17. Powell K, Semaan L, Conley-LaComb MK, Asangani I, Wu Y-M, Ginsburg KB, Williams J, Squire JA, Maddipati KR, Cher ML, Chinni SR. ERG/AKR1C3/AR constitutes a feed-forward loop for AR signaling in prostate cancer cells. Clin Cancer Res. 2015;21(11):2569–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gingras S, Simard J. Induction of 3β-hydroxysteroid dehydrogenase/isomerase type 1 expression by interleukin-4 in human normal prostate epithelial cells, immortalized keratinocytes, colon, and cervix cancer cell lines. Endocrinology. 1999;140(10):4573–4584. [DOI] [PubMed] [Google Scholar]
- 19. Gingras S, Moriggl R, Groner B, Simard J. Induction of 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase type 1 gene transcription in human breast cancer cell lines and in normal mammary epithelial cells by interleukin-4 and interleukin-13. Mol Endocrinol. 1999;13(1):66–81. [DOI] [PubMed] [Google Scholar]
- 20. Gomez-Sanchez CE, Lewis M, Nanba K, Rainey WE, Kuppusamy M, Gomez-Sanchez EP. Development of monoclonal antibodies against the human 3β-hydroxysteroid dehydrogenase/isomerase isozymes. Steroids. 2017;127:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, Zhu M, Wu J, Shi X, Taing L, Liu T, Brown M, Meyer CA, Liu XS. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017;45(D1):D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu Y-M, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao JC, Fong K-W, Jin H-J, Yang YA, Kim J, Yu J. FOXA1 acts upstream of GATA2 and AR in hormonal regulation of gene expression. Oncogene. 2016;35(33):4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peng L, Huang Y, Jin F, Jiang S-W, Payne AH. Transcription enhancer factor-5 and a GATA-like protein determine placental-specific expression of the Type I human 3β-hydroxysteroid dehydrogenase gene, HSD3B1. Mol Endocrinol. 2004;18(8):2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lai T-C, Li H-F, Li Y-S, Hung P-Y, Shyu M-K, Hu M-C. Proximal GATA-binding sites are essential for human HSD3B1 gene transcription in the placenta. Sci Rep. 2017;7(1):4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chi KN, Annala M, Sunderland K, Khalaf D, Finch D, Oja CD, Vergidis J, Zulfiqar M, Beja K, Vandekerkhove G, Gleave M, Wyatt AW. A randomized phase II cross-over study of abiraterone + prednisone (ABI) vs enzalutamide (ENZ) for patients (pts) with metastatic, castration-resistant prostate cancer (mCRPC). J Clin Oncol. 2017;35(15_suppl):5002. [Google Scholar]