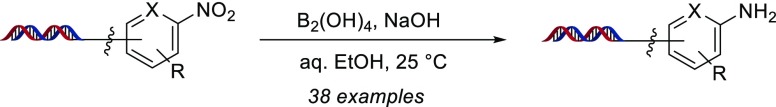Abstract
A hypodiboric acid system for the reduction of nitro groups on DNA–chemical conjugates has been developed. This transformation provided good to excellent yields of the reduced amine product for a variety of functionalized aromatic, heterocyclic, and aliphatic nitro compounds. DNA tolerance to reaction conditions, extension to decigram scale reductions, successful use in a DNA-encoded chemical library synthesis, and subsequent target selection are also described.
DNA-encoded chemical library (DECL) screens are an economic and efficient method for hit discovery.1 Recent advances in DNA high-throughput sequencing and DNA synthesis have enabled routine screens of large DECL collections,2 and successful reports of DECL-based target campaigns have spurred wide interest in the platform.1,3 However, solution-phase synthesis of DECLs is limited by constraints imposed by DNA-integrity and DNA-solubility concerns.4 Adaptation of common chemical reactions to a mild, generally applicable, and partially aqueous condition is needed to expand the repertoire of both DNA-compatible chemical transformations5 as well as available DECL chemical space.6
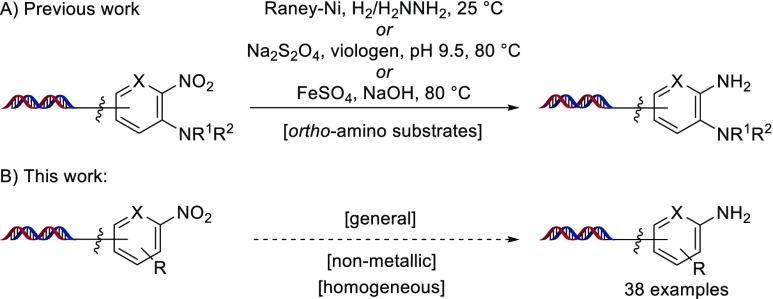
Nitro-substituted, bifunctional compounds represent a versatile set of building blocks for the synthesis of DECLs—as an incorporation of the nitro functional group for select drug–target classes,7 as a substituent to facilitate other reactions under DNA-compatible conditions (e.g., nucleophilic aromatic substitution8), but most importantly as a widely commercially available set of masked, bifunctional anilines that can be unveiled and screened directly or further functionalized in a subsequent build cycle. As a routine transformation in drug synthesis, numerous methods9 have been developed for nitro reduction with metal reagents, such as iron10a or zinc,10b as well as nonmetallic reagents, such as sodium dithionite10c and trichlorosilane.10d However, in the context of DECL synthesis there are limited disclosed methods. Satz and co-workers have utilized a Raney-nickel system,5a we have described a sodium dithionite protocol,11 and recently, Ding et al. have developed an iron sulfate condition12 for use in the on-DNA synthesis of benzimidazoles through an SNAr, o-amino nitro reduction, and aldehyde condensation sequence (Figure 1A). However, to the best of our knowledge, all of these conditions have only been widely demonstrated in the reduction of o-aminonitroarenes, and our attempts to develop dithionite as a general reducing agent for nitroarenes was not successful.13 Seeking a general condition for the synthesis of nonbenzimidazole DECLs, we initiated studies to develop a mild and general nitro reduction for application to a wide variety of on-DNA nitro substrates (Figure 1, B).
Figure 1.
On-DNA nitro reductions: (A) conditions from previous work using Raney-Ni, sodium dithionite, or iron sulfate for the reduction of ortho-amino nitroarenes; (B) this work.
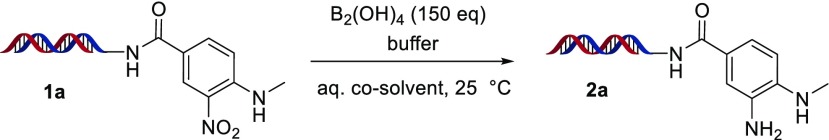
We were intrigued by recent reports which utilized diboron reagents for nitro reduction, with bis(pinacolato)diboron in basic, alcoholic solvent at 110 °C14a or hypodiboric acid in neutral water at 80 °C,14b conditions that might be tolerated by DNA. We initiated our studies on substrate-functionalized “headpiece” DNA commonly used in DECL synthesis, which features a distal amino group for compound attachments covalently connected to two complementary DNA strands via a PEG-type bifurcated linker (our “headpiece” DNA is 17-bp dsDNA with a 3′ overhang; see the Supporting Information for the full structure). As our DECL reaction optimization and applications were conducted on nanomole amounts of DNA at micromolar concentrations, reaction assessments were conducted by LC–MS.15 Preliminary on-DNA nitro reduction experiments utilizing bis(pinacolato)diboron failed, likely due to aqueous temperature limitations and poor reagent solubility.
However, upon switching to water-soluble hypodiboric acid,16 a small amount of 2a was observed for model nitroarene 1a at ambient temperature in aqueous alcoholic solvent (Table 1, entry 1). Encouraged by this result, we began a pH screen (Table 1, entries 2–5) which revealed a clear trend toward enhanced conversions under increasingly basic conditions. This is consistent with a proposed mechanism for diboron-mediated nitro reduction, in which reduction is promoted by coordination of an alkoxide to boron.14 We next set out to determine cosolvent effects, utilizing several common, water-miscible organic solvents. Use of acetonitrile, dimethylacetamide, dimethylformamide, and pure water provided lowered conversion to 2a, and although DMSO provided the aniline in excellent conversion, high amounts of DMSO may impair isolation of DNA (Table 1, entries 5–10). Ultimately, use of aqueous ethanol with B2(OH)4 and NaOH cleanly provided 2a in >95% conversion after only 2 h (Table 1, entry 11).
Table 1. Optimization of Nitro Reduction Conditions.
| entry | buffer/basea | cosolventb | 2ac (%) |
|---|---|---|---|
| 1 | pH 8.2 borate | MIPOd | 11 |
| 2 | pH 5.5 phosphate | MIPO | <5 |
| 3 | pH 10.6 borate | MIPO | 28 |
| 4 | pH 12 phosphate | MIPO | 26 |
| 5 | NaOH | MIPO | 81 |
| 6 | NaOH | CH3CN | 80 |
| 7 | NaOH | none | 78 |
| 8 | NaOH | DMAe | 68 |
| 9 | NaOH | DMFf | 40 |
| 10 | NaOH | DMSOg | 93 |
| 11 | NaOH | CH3CH2OH | >95 |
500 equiv used.
30% (v/v) cosolvent used.
Conversion determined by LC–MS.
1-Methoxy-2-isopropanol.
Dimethylacetamide.
Dimethylformamide.
Dimethylsulfoxide.
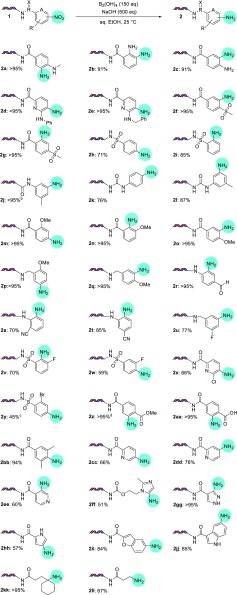
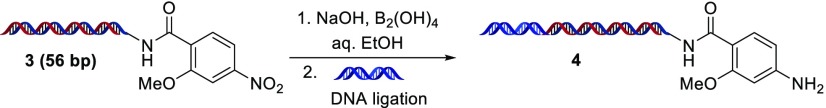
With this established condition in hand, we explored the scope of this reaction with a range of nitroarene substrates (Scheme 1). Based on our previous success, we first applied these conditions against o-amino nitroarenes. Gratifyingly, o-amino anilines 2a–e were cleanly formed, suggesting this protocol could be useful for benzimidazole DECL processes. To examine electronic effects, we investigated the formation of electronic deficient sulfonyl-substituted anilines 2f–i, neutral anilines 2j–l, and electron-rich methoxy-substituted anilines 2m–q. All provided the desired anilines in good to excellent yields, although electron-rich nitro substrates were reduced at slower rates. Notably, the urea linker used in 2k and 2l was tolerated, as ureas may be cleaved under some basic conditions. Substrates with functional groups susceptible to off-target reduction, such as aldehyde 1r and nitriles 1s and 1t, underwent nitro reduction without significant formation of reduced side products. Halogenated nitroarenes are known to be problematic substrates due to the formation of dehalogenated byproducts.17 Although formation of halogenated 2u–y was realized, the desired anilines were formed in lowered yields, and significant dehalogenation was observed for brominated 1y. Potential coordination of a nearby group did not inhibit reduction (methyl ester 2z and free acid 2aa), and steric effects appeared minimal (ortho-disubstituted 2bb). Extension of these conditions to nitro-substituted heterocycles was successful, with pyridyl substrates 1cc–ee, azoles 1ff and 1gg, and electron-rich heterocycles 1hh–jj all providing the desired amine product in moderate to excellent yields. Finally, application to aliphatic nitro substrates 1kk and 1ll furnished the expected amines. Some on-DNA transformations exhibit DNA length related effects, particularly when optimized on short DNA substrates.18 To simulate a late-stage substrate in a DECL synthesis, we prepared substrate 3 from nitroarene 1m, which features a 56-bp dsDNA tag. Application of the reduction conditions efficiently provided the desired aniline, no evidence of DNA decomposition was detected by LC–MS or gel electrophoresis, and the product underwent efficient ligation with an additional DNA tag to form 4 (Scheme 2; see the Supporting Information for full details).
Scheme 1. Substrate Scope of the Nitro Reduction.
The conversions of nitro substrates 1 to amino products 2 are shown. Conversion determined by LC–MS.
On a 1,4-cyclohexanedicarboxylic acid linker.
Significant debromination also observed.
Product observed as the carboxylic acid.
Scheme 2. DNA Length, Stability, and Ligation Test.
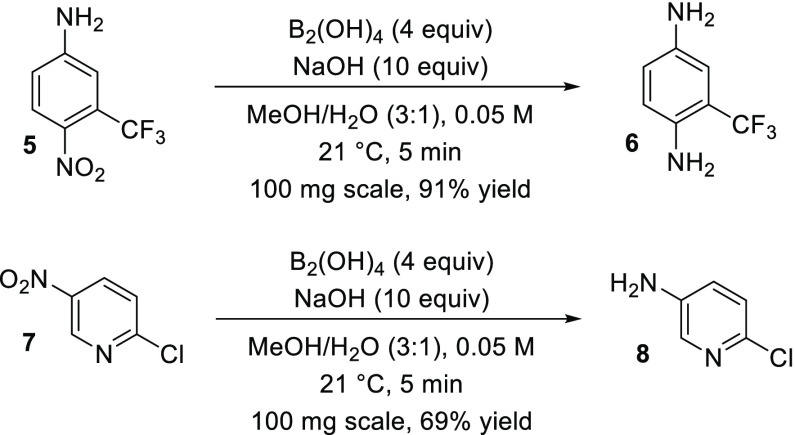
As this reaction proceeded at significantly lower temperatures than previously reported,14 we sought to further validate formation of the presumed on-DNA aniline product. Using similar conditions modified for decigram scale, nitroarene substrates 5 and 7 both provided the desired amine products in good or better isolated yield within 5 min (Scheme 3). Thus, further optimization of this system may be useful for applications in which acidic/hydrogenation conditions are problematic or for which metal contamination must be minimized.19
Scheme 3. Nitro Reduction with B2(OH)4 on Decigram Scale.
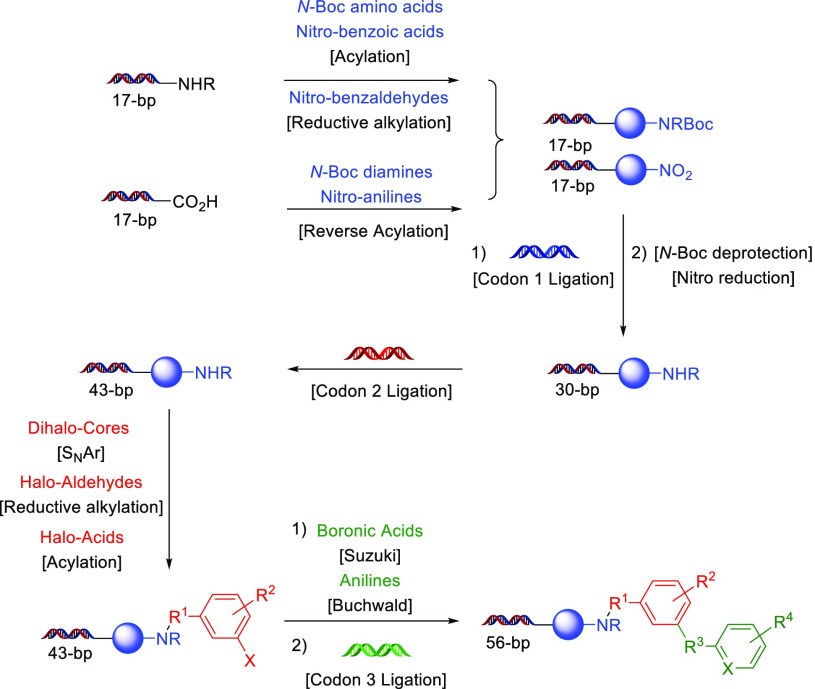
Having demonstrated the wide substrate scope of this reaction, we next sought to use the nitro reduction within a three-cycle, split-and-pool type DECL synthesis. Within this DECL synthesis, each cycle consists of an initial split of DNA “headpiece”-derived materials into hundreds of wells, the attachment and/or transformation of a unique building block, ligation of a unique encoding pair (“codon”) of DNA oligonucleotides, and a final pooling. Thus, a three-cycle DECL featuring hundreds of unique building blocks per cycle will produce a collection of millions of small-molecule structures each connected to a unique, identifying DNA sequence. Within the first cycle, all transformations may be directly observed by LC–MS, but later cycles have complex spectra due to pooling and require other analysis methods.20 After DECL synthesis is completed, adapted samples of the DECL may be amplified and sequenced to allow analysis of the population of DNA sequences within the sample. Altogether, evaluation of post-transformation cycle 1 LC–MS traces, analysis of intra-DECL sequence populations, comparison of the sequencing fidelity/amplification efficiency to other in-house DECLs of identical DNA architecture,21 and the discovery of structurally confirmed DECL hits are definitive tests to determine a new transformation’s DNA tolerance, susceptibility to DNA base-specific effects, and overall suitability for practical DECL applications.
Starting from a DNA headpiece functionalized with linkers featuring either amino or carboxyl termini that had been split into hundreds of individual wells, N-Boc-protected amino acids and nitro-functionalized benzoic acids were connected through acylation, nitro-functionalized benzaldehydes were added through reductive alkylation, and N-Boc protected diamines and nitro-functionalized anilines were attached through a “reverse” acylation reaction (Scheme 4). Within each well, unique encoding DNA tags (codon 1) were then ligated, followed by deprotection of N-Boc carbamates or reduction of a nitro group to the corresponding amine. Before pooling, each well was assessed by LC–MS to determine if the building block coupled, if DNA ligation was completed, if the amino group was fully formed, and if the final cycle 1 product was made in high purity. As the deprotections were performed after the codon 1 tag ligation, the nitro reduction conditions were applied to hundreds of DNA-linked substrates that each contained a unique 30-bp dsDNA tag. Although some variability in amidation efficiencies was observed (e.g., for very electron-deficient nitro-anilines), in general, coupled nitro substrates cleanly reduced to the amine with our hypodiboric acid conditions (>80% conversion). However, consistent with previous rate observations, some substrates with electron-donating groups para to the nitro group had moderate conversions (∼50%). Upon pooling and precipitation, post-cycle 1 DNA recovery was excellent and similar to other in-house DECL productions. After this material was split into hundreds of wells, well-specific ligation of cycle 2 codons and amine substrate functionalization by either nucleophilic substitution of heteroaromatic dihalides, acylation of carboxyl-functionalized aromatic halides, or reductive amination of aldehyde-functionalized aromatic halides was performed. After pooling, this material underwent a third cycle of splitting, functionalization through palladium-catalyzed cross-coupling of boronic acids and anilines, and ligation of cycle 3 codons to ultimately provide a DECL of ∼75 million compounds (full details are available in the Supporting Information).
Scheme 4. Use of Nitro Reduction in a DECL Synthesis.
To probe for potential sequence-specific or well-specific effects, a sample of the library was elaborated with DNA tags to enable a “naïve” sequencing to assess the distribution of codons within the unselected DECL. In DECL productions unbiased by reaction-induced DNA damage, incomplete codon ligations, or varying DNA recovery, cycle 1 codon populations are expected to follow a Gaussian distribution.22 Furthermore, as only a subset of the DECL proceeded through a nitro reduction pathway, comparison of codon populations encoding substrates that underwent the nitro reduction versus the N-Boc deprotection would serve as an intra-DECL control for potential nonspecific DNA damage. Gratifyingly, naïve sample amplification efficiency and the sequence fidelity of amplicons were within expected ranges,21 no sequence-dependent population effects were detected, codon 1 distributions were approximately Gaussian, and the normalized populations of cycle 1 codons for each deprotection pathway were nearly equivalent (Figure 2).22
Figure 2.
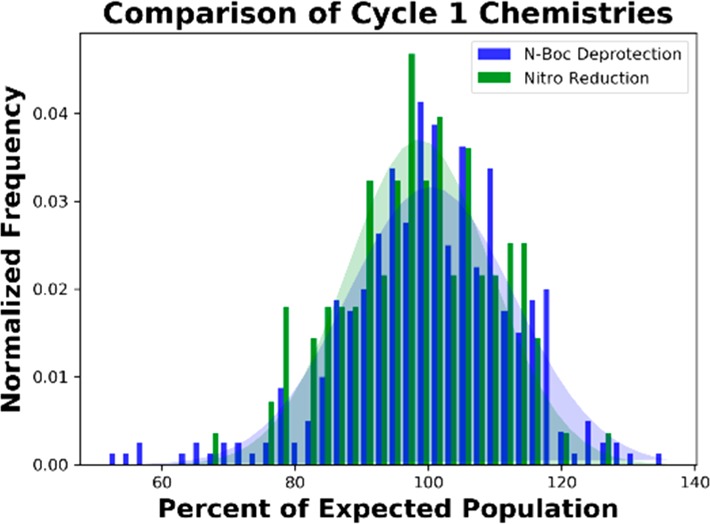
Normalized, observed cycle 1 codon populations from the unselected DECL split by an associated chemical deprotection method (N-Boc deprotection or nitro reduction).22
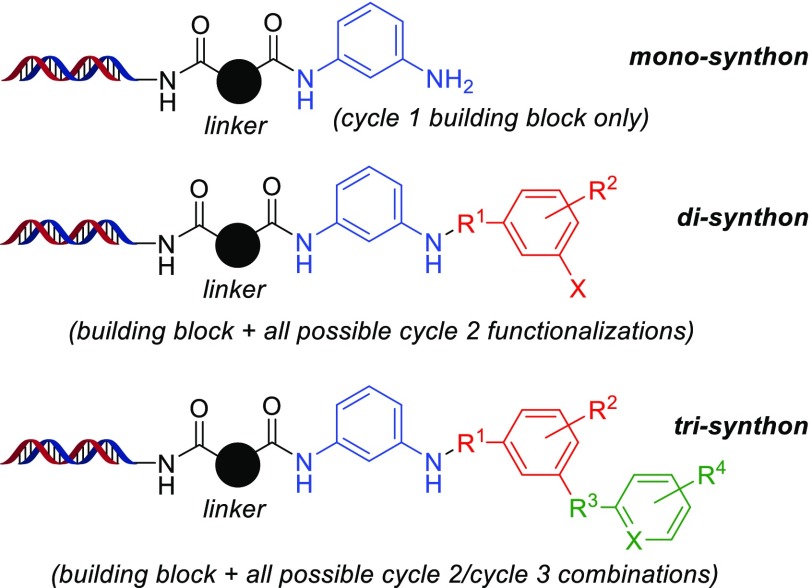
Finally, this DECL was utilized in a selection23 against the kinase PLK1, an important regulator of cell division in eukaryotic cells.24 After a three-round selection of the DECL against recombinant His-PLK1, amplification, and sequencing, cheminformatics analysis25 and hit resynthesis revealed a compound series which inhibited PLK1 kinase activity at low nanomolar concentrations. This series contained 1,3-diaminobenzene26 which had been produced in cycle 1 through two independent pathways: the N-Boc deprotection of acylated 3-(Boc-amino)aniline and the reduction of acylated 3-nitroaniline. Enrichment of DNA encoding this building block by each pathway was comparable at the mono-, di-, and trisynthon27 levels (Figure 3), providing additional evidence that the nitro reduction provided the expected amine product and did not induce pathway-specific effects on later DECL synthetic transformations (see the Supporting Information for selection details and n-synthon analysis).
Figure 3.
Structures of the mono-, di-, and trisynthons of the PLK1 hit series with 1,3-diaminobenzene. Components from cycle 1 are black/blue, cycle 2 are red, and cycle 3 are green.
In summary, we have developed a DNA-compatible nitro reduction for application to aromatic and aliphatic nitro groups that maintains DNA integrity and ligation efficiency. The utility of this methodology was demonstrated in the synthesis of a DECL which was applied to a kinase target. This methodology should complement existing nitro reduction approaches and be useful in future DECL productions.
Acknowledgments
The authors acknowledge Kevin MacKenzie (Baylor College of Medicine) for NMR spectroscopic assistance and Pranavanand Nyshadham (Baylor College of Medicine) and Phil Rosner (Baylor College of Medicine) for mass spectrometry assistance. This work was supported by the Welch Foundation (Grant Q-0042), a Core Facility Support Award (RP160805) from the Cancer Prevention Research Institute of Texas (CPRIT), Grant No. OPP1160866 from The Bill and Melinda Gates Foundation, and National Institutes of Health Grant No. P01HD087157 from The Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.orglett.9b00497.
Details of experimental procedures, DNA structures, DECL construction, and selection experiments (PDF)
Author Contributions
† H.-C.D. and N.S. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- For reviews on DNA-encoded chemical libraries, see:; a Neri D. Twenty-five Years of DNA-Encoded Chemical Libraries. ChemBioChem 2017, 18, 827–828. 10.1002/cbic.201700130. [DOI] [PubMed] [Google Scholar]; b Lerner R. A.; Brenner S. DNA-Encoded Compound Libraries as Open Source: A Powerful Pathway to New Drugs. Angew. Chem., Int. Ed. 2017, 56, 1164–1165. 10.1002/anie.201612143. [DOI] [PubMed] [Google Scholar]; c Franzini R. M.; Neri D.; Scheuermann J. DNA-Encoded Chemical Libraries: Advancing beyond Conventional Small-Molecule Libraries. Acc. Chem. Res. 2014, 47, 1247–1255. 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]; d Neri D.; Lerner R. A. DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu. Rev. Biochem. 2018, 87, 479–502. 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Goodnow R. A. Jr.; Dumelin C. E.; Keefe A. D. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat. Rev. Drug Discovery 2017, 16, 131–147. 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- a Arico-Muendel C. C. From haystack to needle: finding value with DNA encoded library technology at GSK. MedChemComm 2016, 7, 1898–1909. 10.1039/C6MD00341A. [DOI] [Google Scholar]; b Haro R.; Román J. P.; de Blas J.; Jessop T. C.; Castañón J. Design and Development of a Technology Platform for DNA-Encoded Library Production and Affinity Selection. SLAS Discovery 2018, 23, 387–396. 10.1177/2472555217752091. [DOI] [PubMed] [Google Scholar]
- For selected examples of successful DECL-based screening campaigns, see:; a Salamon H.; Škopić M. K.; Jung K.; Bugain O.; Brunschweiger A. Chemical Biology Probes from Advanced DNA-encoded Libraries. ACS Chem. Biol. 2016, 11, 296–307. 10.1021/acschembio.5b00981. [DOI] [PubMed] [Google Scholar]; b Harris P. A.; King B. W.; Bandyopadhyay D.; Berger S. B.; Campobasso N.; Capriotti C. A.; Cox J. A.; Dare L.; Dong X.; Finger J. N.; Grady L. C.; Hoffman S. J.; Jeong J. U.; Kang J.; Kasparcova V.; Lakdawala A. S.; Lehr R.; McNulty D. E.; Nagilla R.; Ouellette M. T.; Pao C. S.; Rendina AlR.; Schaeffer M. C.; Summerfield J. D.; Swift B. A.; Totoritis R. D.; Ward P.; Zhang A.; Zhang D.; Marquis R. W.; Bertin J.; Gough P. J. DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors. J. Med. Chem. 2016, 59, 2163–2178. 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]; c Usanov D. L.; Chan A. I.; Maianti J. P.; Liu D. R. Second-generation DNA-templated macrocycle libraries for the discovery of bioactive small molecules. Nat. Chem. 2018, 10, 704–714. 10.1038/s41557-018-0033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Johannes J. W.; Bates S.; Beigie C.; Belmonte M.; Breen J.; Cao S.; Centrella P. A.; Clark M. A.; Cuozzo J. W.; Dumelin C. E.; Ferguson A. D.; Habeshian S.; Hargreaves D.; Joubran C.; Kazmirski S.; Keefe A. D.; Lamb M. L.; Lan H.; Li Y.; Ma H.; Mlynarski S.; Packer M. J.; Rawlins P. B.; Robbins D. W.; Shen H.; Sigel E. A.; Soutter H. H.; Su N.; Troast D. M.; Wang H.; Wickson K. F.; Wu C.; Zhang Y.; Zhao Q.; Zheng X.; Hird A. W. Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors. ACS Med. Chem. Lett. 2017, 8, 239–244. 10.1021/acsmedchemlett.6b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For a discussion of DNA stability concerns, see:; a Malone M. L.; Paegel B. M. What is a “DNA-Compatible” Reaction. ACS Comb. Sci. 2016, 18, 182–187. 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Halpin D. R.; Lee J. A.; Wrenn S. J.; Harbury P. B. DNA Display III. Solid-Phase Organic Synthesis on Unprotected DNA. PLoS Biol. 2004, 2, 1031–1038. 10.1371/journal.pbio.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For select examples of DECL-compatible reactions, see:; a Satz A. L.; Cai J.; Chen Y.; Goodnow R.; Gruber F.; Kowalczyk A.; Petersen A.; Naderi-Oboodi G.; Orzechowski L.; Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjugate Chem. 2015, 26, 1623–1632. 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]; b Kölmel D. K.; Loach R. P.; Knauber T.; Flanagan M. E. Employing Photoredox Catalysis for DNA-Encoded Chemistry: Decarboxylative Alkylation of α-Amino Acids. ChemMedChem 2018, 13, 2159–2165. 10.1002/cmdc.201800492. [DOI] [PubMed] [Google Scholar]; c Li J.-Y.; Huang H. Development of DNA-Compatible Suzuki-Miyaura Reaction in Aqueous Media. Bioconjugate Chem. 2018, 29, 3841–3846. 10.1021/acs.bioconjchem.8b00676. [DOI] [PubMed] [Google Scholar]; d Ruff Y.; Berst F. Efficient copper-catalyzed amination of DNA-conjugated aryl iodides under mild aqueous conditions. MedChemComm 2018, 9, 1188–1193. 10.1039/C8MD00185E. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Wang J.; Lundberg H.; Asai S.; Martín-Acosta P.; Chen J. S.; Brown S.; Farrell W.; Dushin R. G.; O’Donnell C. J.; Ratnayake A. S.; Richardson P.; Liu Z.; Qin T.; Blackmond D. G.; Baran P. S. Kinetically guided radical-based synthesis of C(sp3)–C(sp3) linkages on DNA. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E6404–E6410. 10.1073/pnas.1806900115. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Ding Y.; Clark M. A. Robust Suzuki–Miyaura Cross-Coupling on DNA-Linked Substrates. ACS Comb. Sci. 2015, 17, 1–4. 10.1021/co5001037. [DOI] [PubMed] [Google Scholar]; g Li Y.; Gabriele E.; Samain F.; Favalli N.; Sladojevich F.; Scheuermann J.; Neri D. Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb. Sci. 2016, 18, 438–443. 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Some types of DECLs can be built without semiaqueous conditions, see:; a MacConnell A. B.; McEnaney P. J.; Cavett V. J.; Paegel B. M. DNA-Encoded Solid-Phase Synthesis: Encoding Language Design and Complex Oligomer Library Synthesis. ACS Comb. Sci. 2015, 17, 518–534. 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Škopić M. K.; Salamon H.; Bugain O.; Jung K.; Gohla A.; Doetsch L. J.; dos Santos D.; Bhat A.; Wagner B.; Brunschweiger A. Acid- and Au(I)-mediated synthesis of hexathymidine-DNA-heterocycle chimeras, an efficient entry to DNA-encoded libraries inspired by drug structures. Chem. Sci. 2017, 8, 3356–3361. 10.1039/C7SC00455A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepali K.; Lee H.-Y.; Liou J.-P. Nitro-Group-Containing Drugs. J. Med. Chem. 2018, 10.1021/acs.jmedchem.8b00147. [DOI] [PubMed] [Google Scholar]
- Blaziak K.; Danikiewicz W.; Mąkosza M. How Does Nucleophilic Aromatic Substitution Really Proceed in Nitroarenes? Computational Prediction and Experimental Verification. J. Am. Chem. Soc. 2016, 138, 7276–7281. 10.1021/jacs.5b13365. [DOI] [PubMed] [Google Scholar]
- For a recent review of nitro reduction conditions, see:Orlandi M.; Brenna D.; Harms R.; Jost S.; Benaglia M. Recent Developments in the Reduction of Aromatic and Aliphatic Nitro Compounds to Amines. Org. Process Res. Dev. 2018, 22, 430–445. 10.1021/acs.oprd.6b00205. [DOI] [Google Scholar]
- a Gamble A. B.; Garner J.; Gordon C.; O’Conner S. J. J.; Keller P. A. Aryl Nitro Reduction with Iron Powder or Stannous Chloride under Ultrasonic Irradiation. Synth. Commun. 2007, 37, 2777–2786. 10.1080/00397910701481195. [DOI] [Google Scholar]; b Kumar P.; Rai K. L. Reduction of aromatic nitro compound to amines using zinc and aqueous chelating ethers: Mild and efficient method for zinc activation. Chemical Papers 2012, 66, 772–778. 10.2478/s11696-012-0195-6. [DOI] [Google Scholar]; c Park K. K.; Oh C. H.; Joung W. K. Sodium dithionite reduction of nitroarenes using viologen as an electron phase-transfer catalyst. Tetrahedron Lett. 1993, 34, 7445–7446. 10.1016/S0040-4039(00)60148-X. [DOI] [Google Scholar]; d Orlandi M.; Tosi F.; Bonsignore M.; Benaglia M. Metal-Free Reduction of Aromatic and Aliphatic Nitro Compounds to Amines: A HSiCl3- Mediated Reaction of Wide General Applicability. Org. Lett. 2015, 17, 3941–3943. 10.1021/acs.orglett.5b01698. [DOI] [PubMed] [Google Scholar]
- Du H. C.; Huang H. DNA-Compatible Nitro Reduction and Synthesis of Benzimidazoles. Bioconjugate Chem. 2017, 28, 2575–2580. 10.1021/acs.bioconjchem.7b00416. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Chai J.; Centrella P. A.; Gondo C.; DeLorey J. L.; Clark M. A. Development and Synthesis of DNA-Encoded Benzimidazole Library. ACS Comb. Sci. 2018, 20, 251–255. 10.1021/acscombsci.8b00009. [DOI] [PubMed] [Google Scholar]
- Du H.-C. Unpublished results. Generally low yields and byproducts were observed.
- a Lu H.; Geng Z.; Li J.; Zou D.; Wu Y.; Wu Y. Metal-Free Reduction of Aromatic Nitro Compounds to Aromatic Amines with B2pin2 in Isopropanol. Org. Lett. 2016, 18, 2774–2776. 10.1021/acs.orglett.6b01274. [DOI] [PubMed] [Google Scholar]; b Chen D.; Zhou Y.; Zhou H.; Liu S.; Liu Q.; Zhang K.; Uozumi Y. Metal-free Reduction of Nitro Aromatics to Amines with B2(OH)4/H2O. Synlett 2018, 29, 1765–1768. 10.1055/s-0037-1610086. [DOI] [Google Scholar]
- Although the masses of our headpiece DNA substrates are large (>12 kDa), DNA may be ionized with multiple charges leading to complex m/z patterns in the 500–2000 range. With automated m/z deconvolution software, small mass changes even as low as 1–2 Da can be detected.
- Hypodiboric acid has been utilized in other reductions; see:; a Cummings S. P.; Le T.-N.; Fernandez G. E.; Quiambao L. G.; Stokes B. J. Tetrahydroxydiboron-Mediated Palladium-Catalyzed Transfer Hydrogenation and Deuteriation of Alkenes and Alkynes Using Water as the Stoichiometric H or D Atom Donor. J. Am. Chem. Soc. 2016, 138, 6107–6110. 10.1021/jacs.6b02132. [DOI] [PubMed] [Google Scholar]; b Londregan A. T.; Piotrowski D. W.; Xiao J. Rapid and Selective in situ Reduction of Pyridine-N-oxides with Tetrahydroxydiboron. Synlett 2013, 24, 2695–2700. 10.1055/s-0033-1340010. [DOI] [Google Scholar]
- Kasparian A. J.; Savarin C.; Allgeier A. M.; Walker S. D. Selective Catalytic Hydrogenation of Nitro Groups in the Presence of Activated Heteroaryl Halides. J. Org. Chem. 2011, 76, 9841–9844. 10.1021/jo2015664. [DOI] [PubMed] [Google Scholar]
- We suspect this arises from intermolecular DNA interactions and/or changes in DNA solubility in mixed aqueous–organic solutions.
- Miyamoto H.; Sakumoto C.; Takekoshi E.; Maeda Y.; Hiramoto N.; Itoh T.; Kato Y. Effective Method To Remove Metal Elements from Pharmaceutical Intermediates with Polychelated Resin Scavenger. Org. Process Res. Dev. 2015, 19, 1054–1061. 10.1021/acs.oprd.5b00161. [DOI] [Google Scholar]; See also references cited therein.
- Although major mass shifts (e.g., codon ligation) can be followed by LC–MS or gel electrophoresis, we often follow postpool DECL synthetic steps by monitoring a spiked-in DNA substrate that can be chromatographically separated from the DECL on LC–MS; see the Supporting Information for more details.
- As our DECLs are built with chemically modified, intramolecularly joined segments of dsDNA, amplification does not proceed at theoretical rates. Furthermore, amplification efficiency is significantly affected by the design of adapters added to enable amplification/sequencing/decoding (e.g., degenerate segments, primer overhangs, etc.). Separately, DNA sequence fidelity is affected by the chosen method of DNA sequencing and sample preparation. However, analysis of hundreds of naïve/selection experiments using dozens of DECLs that have been produced/sequenced with identical overall DNA architectures/methods have provided an experimental baseline to which can compare the performance of new DECLs.
- Assuming that equal concentrations of cycle 1 codons are present in the library sample, the observed counts for cycle 1 codons after DNA sequencing should follow a normal distribution centered at the average count. DNA sequencing of the naïve DECL resulted in a number of decoded molecules equal to 21674.6 times the diversity of the cycle 1 encoding region. Consequently, the observed average count for all c1 codons was 21674.6, and the count distribution of c1 codons roughly followed a normal distribution with mean 21674.6. In Figure 2, counts for c1 codons are normalized by the mean count to produce percentage population fractions.
- DECL affinity “selections” are experiments designed to enrich small-molecule binders of a biological target within a pool of DNA-encoded compounds, and the structures of enriched binders are then determined by DNA sequencing and bio- and cheminformatic analysis. For a discussion of the principles and practice of DECL selection, see:Chan A. I.; McGregor L. M.; Liu D. R. Novel selection methods for DNA-encoded chemical libraries. Curr. Opin. Chem. Biol. 2015, 26, 55–61. 10.1016/j.cbpa.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes G.; Alharbi I.; Braga L. G.; Elowe S. Playing polo during mitosis: PLK1 takes the lead. Oncogene 2017, 36, 4819–4827. 10.1038/onc.2017.113. [DOI] [PubMed] [Google Scholar]
- For a discussion of our enrichment analysis method, see:Faver J. C.; Riehle K.; Lancia D. R.; Milbank J. B. J.; Kollmann C. S.; Simmons N.; Yu Z.; Matzuk M. M. Quantitative Comparison of Enrichment from DNA-Encoded Chemical Library Selections. ACS Comb. Sci. 2019, 21, 75–82. 10.1021/acscombsci.8b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1,3-Diaminobenzene is the deprotected structure of the cycle 1 building block; the full hit structure is not shown.
- The term “synthon” refers to each building block/encoding DNA component and may be analyzed by itself (“mono-synthon”) or in combination with components from other cycles (i.e., “di-synthon” or “tri-synthon” within a three-cycle DECL).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.