Abstract
Both zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1) are intimately involved in many aspects of early intestinal mucosal repair after acute injury, but the exact mechanisms that control their cellular abundances remain largely unknown. The present study shows that microRNA-222 (miR-222) interacts with the mRNAs encoding ZBP1 and PLCγ1 and regulates ZBP1 and PLCγ1 expression in intestinal epithelial cells (IECs). The biotinylated miR-222 bound specifically to the ZBP1 and PLCγ1 mRNAs in IECs. Ectopically expressed miR-222 precursor destabilized the ZBP1 and PLCγ1 mRNAs and consequently lowered the levels of cellular ZBP1 and PLCγ1 proteins. Conversely, decreasing the levels of cellular miR-222 by transfection with its antagonism increased the stability of the ZBP1 and PLCγ1 mRNAs and increased the levels of ZBP1 and PLCγ1 proteins. Overexpression of miR-222 also inhibited cell migration over the wounded area, which was partially abolished by overexpressing ZBP1 and PLCγ1. Furthermore, prevention of the increased levels of ZBP1 and PLCγ1 in the miR-222-silenced cells by transfection with specific small interfering RNAs targeting ZBP1 or PLCγ1 mRNA inhibited cell migration after wounding. These findings indicate that induced miR-222 represses expression of ZBP1 and PLCγ1 at the posttranscriptional level, thus inhibiting IEC migration during intestinal epithelial restitution after wounding.
Keywords: cell migration, early rapid repair, HCT 116 cells, microRNAs, mRNA stability, mucosal injury, posttranscriptional regulation
INTRODUCTION
Zipcode binding protein-1 (ZBP1) is a member of the highly conserved family of RNA-binding proteins (RBPs), which consist of three mammalian paralogs (ZBP-1–3) (14, 15, 55). ZBP1 facilitates intestinal morphogenesis and also regulates normal growth and development of the intestinal mucosa (17, 30). ZBP1 modulates cytoskeletal organization, cellular adhesion, and cell migration via distinct cellular mechanisms (9, 10, 14). For example, ZBP1 affects the speed of tumor cell migration by modulating MAPK4 and ACTB expression (20); it also promotes cell migration by directing actin monomers to the site of active protrusion, lamellipodia formation, and intrinsic polarization (19, 44). Phospholipase C-γ1 (PLCγ1) is an important regulatory enzyme that catalyzes hydrolysis of the phospholipid phosphatidylinositol-4,5-bisphosphate (PIP2) to generate diacylglycerol (DAG) and inositol-1,4,5-trisphosphate (IP3), both of which are implicated in the regulation of a variety of cellular processes including cell motility (22, 34, 35, 40). Inhibition of PLCγ1 activity represses cell migration and reduces cell invasiveness in breast, prostate, and glioblastoma multiform cancer cell lines (26, 49). We have demonstrated that activation of PLCγ1 stimulates intestinal epithelial cell (IEC) migration over the wounded area by activating Ca2+ signaling, whereas PLCγ1 silencing represses intestinal epithelial restitution by downing regulating Ca2+ influx (34, 35, 48). However, the exact mechanisms by which the cellular abundances of ZBP1 and PLCγ1 are regulated after acute injury remain largely unknown.
In mammalian cells, regulation of the mRNA stability and translation is a crucial step in the control of gene expression (25, 50, 51). MicroRNAs (miRNAs) bind to specific mRNAs and decrease the stability and translation of target mRNAs, thus inhibiting gene expression (12, 47). Recently, miRNAs have emerged as master regulators of gut epithelial homeostasis (1, 27, 47). We have shown that several miRNAs, including miR-222, miR-29b, miR-195, miR-503, and miR-675, are highly expressed in the intestinal epithelium and their expression levels are rapidly affected in response to stressful environments such as food starvation and acute mucosal injury (3, 6, 52, 53, 57, 58). miR-222 is highly conserved among different species and is clustered in tandem with miR-221 on the X chromosome (11). miR-222 is shown to play an important role in distinct cellular functions (2, 5, 43). We have reported that miR-222 and RBP CUG-binding protein 1 (CUGBP1) synergistically inhibit translation of cyclin dependent kinase 4 (CDK4) (52) and that intestinal epithelial tissue-specific transgenic expression of miR-222 disrupts the mucosal regeneration by targeting multiple genes including Wnt-receptor Frizzled-7 (6).
An en mass search for miR-222 targets identified the ZBP1 and PLCγ1 mRNAs as putative miR-222 targets and computationally detected several miR-222 binding sites in their transcripts. Given our long-term interest in understanding the mechanisms underlying gut mucosal repair and homeostasis, we studied the influence of miR-222 on the stability of ZBP1 and PLCγ1 mRNAs. We set out to determine if miR-222 binds to the ZBP1 and PLCγ1 mRNAs and further elucidate the functional consequences of these interactions. miR-222 was found to directly interact with the ZBP1 and PLCγ1 mRNAs and enhance their degradation. Moreover, ectopically expressed miR-222 inhibited cell migration after wounding, and this inhibition was prevented by overexpressing ZBP1 and PLCγ1. These results indicate that miR-222 downregulates expression of ZBP1 and PLCγ1 posttranscriptionally and that miR-222-mediated repression of ZBP1 and PLCγ1 expression plays an important role in the regulation of IEC migration after acute injury, thus affecting the integrity of the intestinal epithelium.
MATERIALS AND METHODS
Chemicals and cell culture.
Disposable culture ware was purchased from Corning Glass Works (Corning, NY). Tissue culture media, LipofectAMINE 2000, and fetal bovine serum (FBS) were obtained from Invitrogen (Carlsbad, CA), and biochemicals were obtained from Sigma (St. Louis, MO). The affinity-purified rabbit polyclonal antibodies against ZBP1 (cat. no. 8482), PCNA (cat. no. 13110), and PLCγ1 (cat. no. 5690) were purchased from Cell Signaling Technologies (Danvers, MA), and the antibody against GAPDH (cat. no. sc-25778) was from Santa Cruz Biotechnology (Santa Cruz, CA). The secondary antibodies conjugated to horseradish peroxidase (goat anti-rabbit IgG-HRP, cat. no. sc-2004; goat anti-mouse IgG HRP, cat. no. sc-2005) were purchased from Santa Cruz Biotechnology. Pre-miR miRNA precursor and anti-miR miRNA inhibitor of miR-222 were purchased from Ambion (Austin, TX). Biotin-labeled miRNA-222 was custom made by Dharmacon (Lafayette, CO). Human ZBP1 and PLCγ1 cDNAs and siRNAs were purchased from OriGene Technologies (Rockville, MD). The HCT-116 cells (a line of human colorectal carcinoma cells) were purchased from the American Type Culture Collection (cat. no. CCL-247; ATCC). Stock cells were maintained in T-150 flasks in McCoy’s 5A modified medium supplemented with 10% heat-inactivated FBS. Flasks were incubated at 37°C in a humidified atmosphere of 95%-5% CO2, and the first 10 passages were used in the experiments as described previously (7, 8, 38). The differentiated rat IECs (line of IEC-Cdx2L1) were maintained as described in our previous studies (34, 35, 38, 48).
RT and quantitative real-time PCR analyses.
Total RNA was isolated using RNeasy Mini Kit (Qiagen) and used in reverse transcription (RT) and PCR amplification reactions as described previously (48, 53, 56). The levels of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) PCR product were assessed to monitor the evenness of RNA input in RT–PCR samples. Quantitative real-time PCR (qPCR) analysis was performed using 7500-Fast Real-Time PCR Systems with specific primers, probes, and software (Applied Biosystems). For miRNA studies, the levels of miR-222 were quantified by qPCR using a TaqMan MicroRNA assay (Applied Biosystems); levels of small nuclear RNA (snRNA) U6 were measured as an endogenous control.
Measurement of mRNA stability.
The stability of mRNAs was examined by measuring the mRNA half-life as described previously (4). Forty-eight hours after the transfections, cells were incubated with actinomycin D (Sigma-Aldrich) at the concentration of 5 μg/ml to the medium. Total cellular RNA was isolated at the indicated times after exposure to actinomycin D, and the remaining levels of ZBP1, PLCγ1, or GAPDH mRNAs were measured by qRT-PCR analysis. Nonlinear regression analysis was used to calculate mRNA half-life.
Western immunoblotting analysis.
Whole cell lysates were prepared using 2% SDS, sonicated, and centrifuged (12,000 rpm) at 4°C for 15 min as described previously (8, 38). Briefly, the supernatants were boiled for 5 min and size fractionated by SDS-PAGE (10% acrylamide). After proteins were transferred onto nitrocellulose membranes, the membranes were incubated for 1 h in 5% nonfat dry milk in 1× TBS-T buffer (0.1% Tween-20). Immunologic evaluation was then performed overnight at 4°C in 5% nonfat dry milk/TBS-T buffer containing a specific antibody against ZBP1 or PLCγ1. The membranes were subsequently washed with 1× TBS-T and incubated with the secondary antibodies conjugated with horseradish peroxidase for 1 h at room temperature. The immunocomplexes on the membranes were reacted for 1 min with Chemiluminescence Reagent (NEL-100 DuPont NEN). All antibodies utilized in this study were thoroughly validated for species specificity. Antibody dilutions used for ZBP1 (first Ab 1:1,000; second AB 1:2,000); PLCγ1 (first Ab 1:2,000; second Ab 1:2,000); PCNA (first Ab 1:1,000; second Ab 1:3,000); and GAPDH (first Ab 1:1,000; second Ab 1:1,000). Relative levels of protein expression was analyzed by using Bio-Rad Chemidoc and XRS system equipped with Image laboratory software (version 4.1), and we have utilized “Quantity tool” to determine the band intensity volume and the values were normalized with internal loading control GAPDH.
Biotin-labeled miR-222 pull-down assays.
Binding of miR-222 to target mRNAs was examined by RNA pull-down assays using biotin labeled miR-222 as described previously (6, 53, 56, 58). Briefly, cells were transfected with biotin-labeled miR-222, and whole cell lysates were collected 24 h after the transfection. The cell lysates were mixed with streptavidin-Dynabeads and incubated at 4°C with rotation overnight. After the beads were washed thoroughly, the bead-bound RNA was isolated and subjected to RT-PCR followed by qPCR analysis. Input RNA was extracted and served as a control.
Measurement of cell migration.
Migration assays were carried out as described in our earlier publications (7, 34, 37, 38). HCT116 cells were plated at 6.25 × 104/cm2 in Mccoy’s 5A medium containing FBS on 60-mm dishes thinly coated with Matrigel according to the manufacturer’s instructions (BD Biosciences, Bedford, MA) and were incubated as described for stock cultures. Cells were fed on day 2, followed by appropriate cotransfection assays; cell migration was assayed 48 h after transfection. To initiate migration, the cell layer was scratched with a single edge razor blade cut to ~27 mm in length. The scratch was made over the diameter of the dish and extended over an area 7- to 10-mm wide. The migrating cells in six contiguous 0.1-mm squares were counted at ×100 magnification beginning at the scratch line and extending as far out as the cells had migrated. All experiments were carried out independently three to five times, and the results are reported as number of migrating cells per millimeter of scratch.
Statistical analysis.
All data for migration experiments are expressed as means ± SE from six dishes in each experiment and independently repeated three times (n = 3). qRT-PCR and immunoblotting analyses were repeated three times (n = 3). The significance of the difference between means was determined by one-way ANOVA with Dunnett’s post hoc test (GraphPad Instat Prism 5, San Diego, CA). The level of significance was determined using the Duncan’s multiple-range test (18), and values of P < 0.05 were considered statistically significant. For nonparametric analysis rank comparison, the Kruskal-Wallis test was conducted.
RESULTS
The mRNAs encoding ZBP1 and PLCγ1 are novel targets of miR-222.
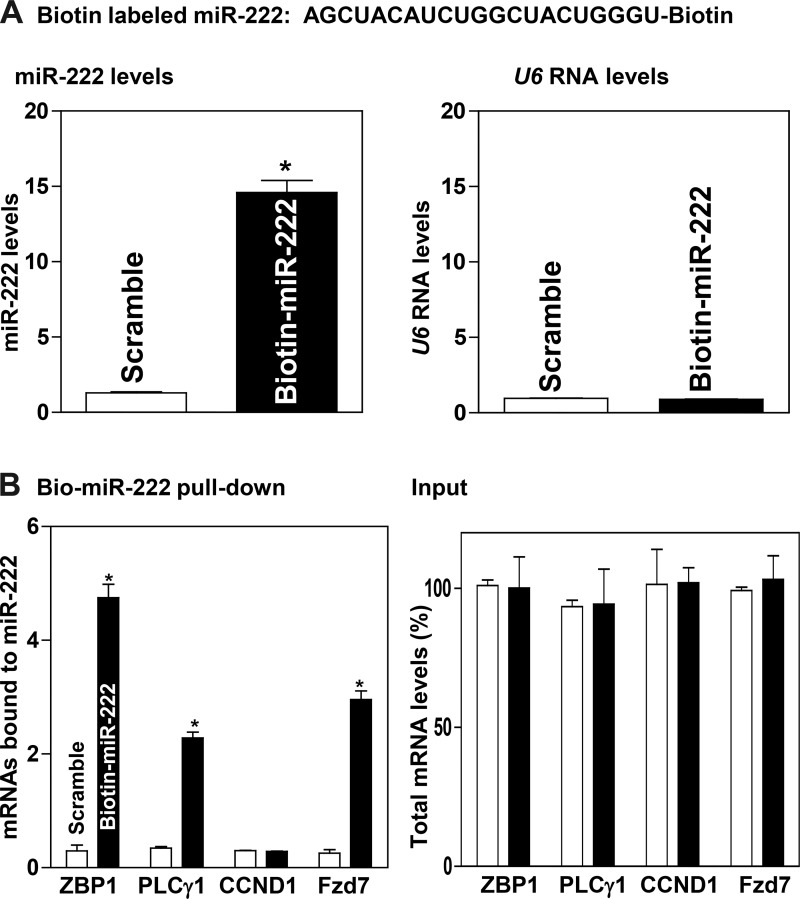
To determine the role of miR-222 in the regulation of ZBP1 and PLCγ1 expression in IECs, we analyzed the association of miR-222 with the ZBP1 and PLCγ1 mRNAs by RNA pulldown assays using biotin-labeled miR-222. Twenty-four hours after HCT 116 cells were transfected with biotin-labeled miR-222, the levels of miR-222 increased significantly (Fig. 1A, left), although levels of the housekeeping noncoding RNA U6 did not (Fig. 1A, right). The ZBP1 and PLCγ1 mRNAs were enriched in the materials pulled down by biotin-miR-222 but not in materials from cells transfected with control scrambled RNA (Fig. 1B, left). The enrichment of the Fzd7 mRNA was also examined and served as a positive control, because the Fzd7 mRNA was known as a target of miR-222 (6). The associations of miR-222 with the ZBP1 and PLCγ1 mRNAs were specific, since increasing the levels of biotin-miR-222 did not increase its interaction with the mRNA encoding claudin-1 (CCND1). In addition, the transfection with biotin-labeled miR-222 for 24 h did not alter the steady-state levels (input) of ZBP1, PLCγ1, CCND1, and Fzd7 mRNAs (Fig. 1B, right). These results indicate that the ZBP1 and PLCγ1 mRNAs are the targets of miR-222 in IECs.
Fig. 1.
Interaction of microRNA-222 (miR-222) with the zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1) mRNAs. A: miR-222 and U6 RNA levels in HCT-116 cells transfected with biotinylated miR-222 for 24 h. Values are means ± SE from 3 independent experiments (n = 4). *P < 0.01 compared with cells transfected with control scramble oligomer as analyzed by one-way ANOVA followed by Duncan’s test. B: levels of ZBP1, PLCγ1, claudin-1 (CCND1), and FZD7 mRNAs in the materials pulled down by biotin-miR-222 (left) and total input mRNAs (right) in cells described in A. Fzd7 served as a positive control.
Induced miR-222 represses ZBP1 and PLCγ1 expression by destabilizing their mRNAs.
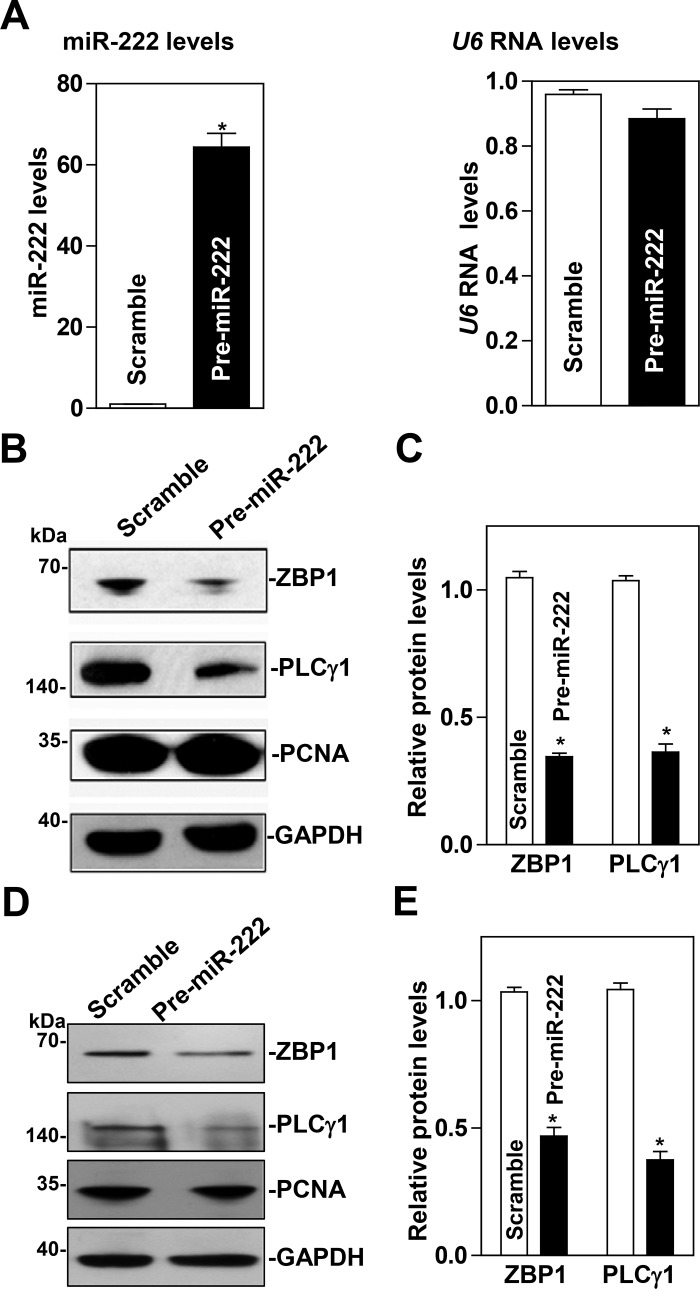
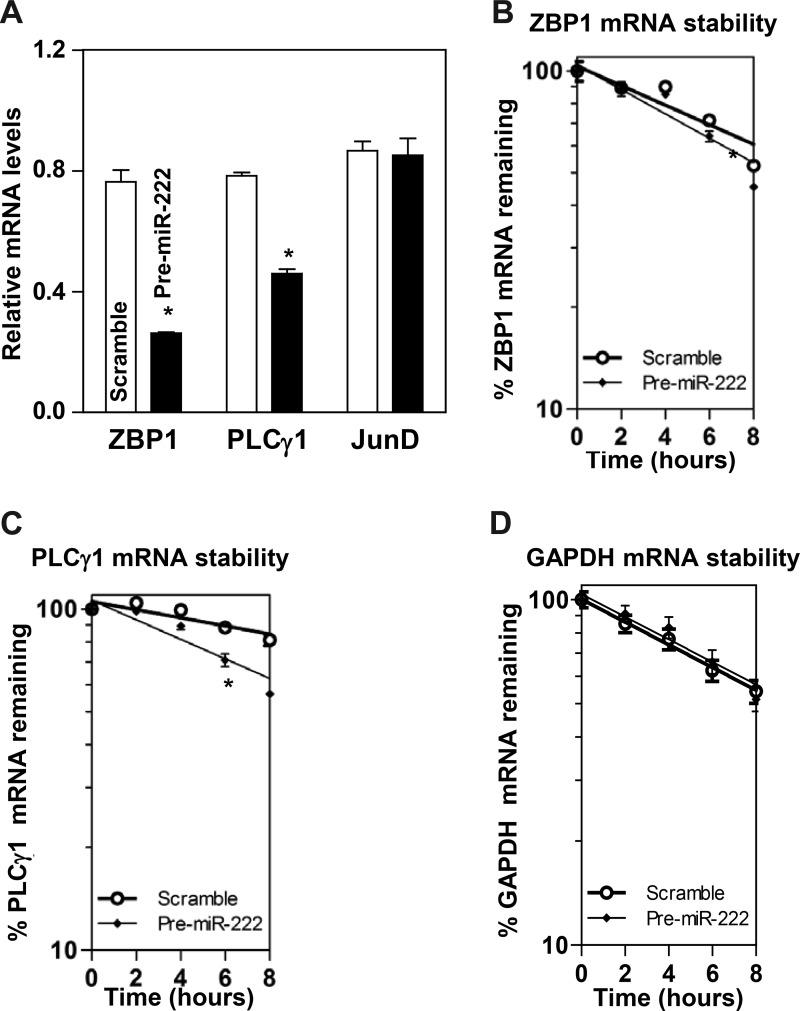
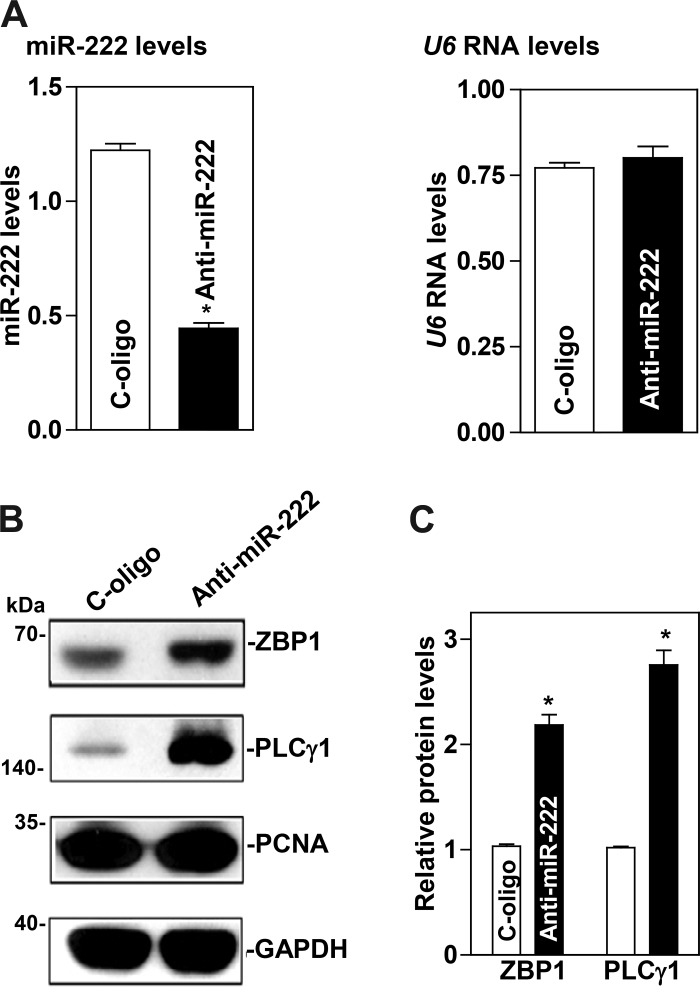
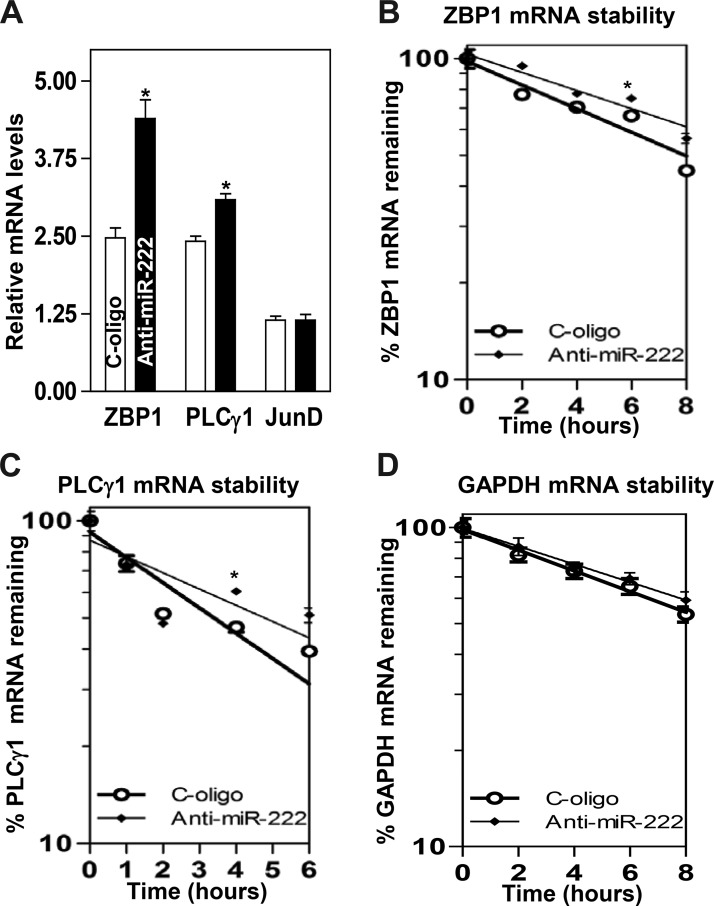
To examine the functional consequences of the miR-222 association with the ZBP1 and PLCγ1 mRNAs, we first investigated whether increasing the levels of miR-222 through transfection with miR-222 precursor (pre-miR-222) repressed ZBP1 and PLCγ1 expression. As shown in Fig. 2A, the levels of miR-222 increased dramatically 48 h after transfection with pre-miR-222. Ectopically expressed miR-222 decreased the abundance of cellular ZBP1 and PLCγ1 proteins (Fig. 2, B and C). Similar experiments were also conducted in another IECs, differentiated IEC-Cdx2L1 cells, and showed that miR-222 overexpression also inhibited expression of both ZBP1 and PLCγ1 (Fig. 2, D and E). To determine whether miR-222 inhibited ZBP1 and PLCγ1 expression by decreasing their mRNAs, we examined the mRNA levels of ZBP1 and PLCγ1 in cells overexpressing miR-222. The levels ZBP1 and PLCγ1 mRNAs decreased significantly in cells transfected with pre-miR-222 compared with those observed in cells transfected with the scrambled oligomer (Fig. 3A). Moreover, miR-222 overexpression enhanced decay of the ZBP1 and PLCγ1 mRNAs, as indicated by a rapid degradation after treatment with actinomycin D (Fig. 3, B and C). However, the stability of GAPDH mRNA was not altered by miR-222 overexpression (Fig. 3D). The decay rates of ZBP1 and PLCγ1 mRNA in the cells overexpressing miR-222 were enhanced by ~35 and ~55%, respectively, although PLCγ1 mRNA decay in control cells was quicker than that of the ZBP1 mRNA. Finally, we examined the influence of decreasing the level of miR-222 by transfecting the corresponding antisense oligomer (anti-miR-222) on ZBP1 and PLCγ1 expression. The levels of miR-222 decreased remarkably 48 h after transfection with anti-miR-222 without effect on U6 RNA levels (Fig. 4A). Decreased levels of miR-222 by anti-miR-222 stimulated expression of ZBP1 and PLCγ1, as shown by an increase in the levels of ZBP1 and PLCγ1 proteins (Fig. 4, B and C). Consistently, miR-222 silencing increased the levels of ZBP1 and PLCγ1 mRNAs (Fig. 5A) by increasing their stability, since half-lives of the ZBP1 and PLCγ1 mRNAs in miR-222-silenced cells were increased by ~40% and ~45%, respectively (Fig. 5, B and C). There were massive variations between the levels of ZBP1 and PLCγ1 mRNAs and levels of their proteins after miR-222 silencing in this study. Although the exact reasons causing this difference remain unknown, these results suggest that miR-222 silencing might also affect the translational efficiency of ZBP1 and PLCγ1 mRNAs. Taken together, our results indicate that miR-222 inhibits expression of ZBP1 and PLCγ1 at least partially by enhancing their mRNA degradation.
Fig. 2.
Ectopically expressed microRNA-222 (miR-222) represses the expression of zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1). A: levels of miR-222 and U6 RNA 48 h after transfection with pre-miR-222 as measured by quantitative PCR analysis. Values are means ± SE from independent experiments (n = 4). *P < 0.05, compared with cells transfected with control scrambled oligomer analyzed by one-way ANOVA followed by Duncan’s test. B: immunoblots of ZBP1, PLCγ1, and PCNA proteins in HCT-116 cells described in A. Whole cell lysates were harvested and prepared for Western blotting; equal loading was monitored by assessing GAPDH levels. C: quantitative analysis of ZBP1 and PLCγ1 immunoblotting signals as measured by densitometry using Bio-Rad-XRS system equipped with Image laboratory software (version 4.1) and used “Quantity tool” to determine the band intensity volume. The values were normalized with internal loading control GAPDH. Values are means ± SE of data from 3 independent experiments (n = 3). D and E: changes in ZBP1, PLCγ1, and PCNA proteins in IEC-Cdx2L1 cells 48 h after transfection with pre-miR-222. Values are means ± SE (n = 3). Statistical test: means are compared with the scramble (cells exposed to pre-miR-222) by nonparametric comparison (*P < 0.0495, Kruskal-Wallis test).
Fig. 3.
microRNA-222 (miR-222) overexpression destabilizes the zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1) mRNAs. A: levels of the ZBP1 and PLCγ1 mRNAs in cells transfected with pre-miR-222 for 48 h. JunD served as a negative control. Values are the means ± SE from independent experiments (n = 3). *P < 0.01, compared with cells transfected with control scramble oligomer as analyzed by one-way ANOVA followed by Duncan’s test. B–D: half-lives of the ZBP1, PLCγ1, and GAPDH mRNA in cells described in A. Total cellular RNA was isolated at indicated times after administration of actinomycin D (5 μg/ml), and the levels of ZBP1, PLCγ1, and GAPDH mRNAs were measured by quantitative PCR analysis. GAPDH mRNA served as a control. *P < 0.05, compared with cells transfected with C-oligo as analyzed by one-way ANOVA followed by Duncan’s test.
Fig. 4.
microRNA-222 (miR-222) silencing enhances the expression of zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1). A: levels of miR-222 and U6 RNA 48 h after transfection with anti-miR-222. Values are the means ± SE from 3 independent experiments (n = 4). *P < 0.05, compared with cells transfected with control oligomer (C-oligo). B: immunoblots of ZBP1, PLCγ, and PCNA proteins in cells described in A. C: quantitative analysis of ZBP1 and PLCγ1 immunoblotting signals by densitometry using Bio-Rad-XRS system equipped with Image laboratory software (version 4.1) and used “Quantity tool” to determine the band intensity volume. The values were normalized with internal loading control GAPDH. Values are means ± SE of data from 3 independent experiments (n = 3). Statistical test: means are compared with the scramble (cells exposed to Anti-miR-222) by nonparametric comparison (*P < 0.0495, Kruskal-Wallis test).
Fig. 5.
microRNA-222 (miR-222) silencing increases the stability of the zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1) mRNAs. A: levels of the ZBP1 and PLCγ1 mRNAs in cells transfected with anti-miR-222 for 48 h. JunD served as a negative control. Values are the means ± SE from independent experiments (n = 4). *P < 0.01, compared with cells transfected with C-oligo. B–D: half-lives of the ZBP1, PLCγ1, and GAPDH mRNA in cells described in A. Total cellular RNA was isolated at indicated times after administration of actinomycin D (5 μg/ml), and the levels of ZBP1, PLCγ1, and GAPDH mRNAs were measured by quantitative PCR analysis. GAPDH mRNA served as a control. *P < 0.05, compared with cells transfected with C-oligo as analyzed by one-way ANOVA followed by Duncan’s test.
Inhibition of ZBP1 and PLCγ1 by miR-222 downregulates intestinal epithelial restitution after wounding.
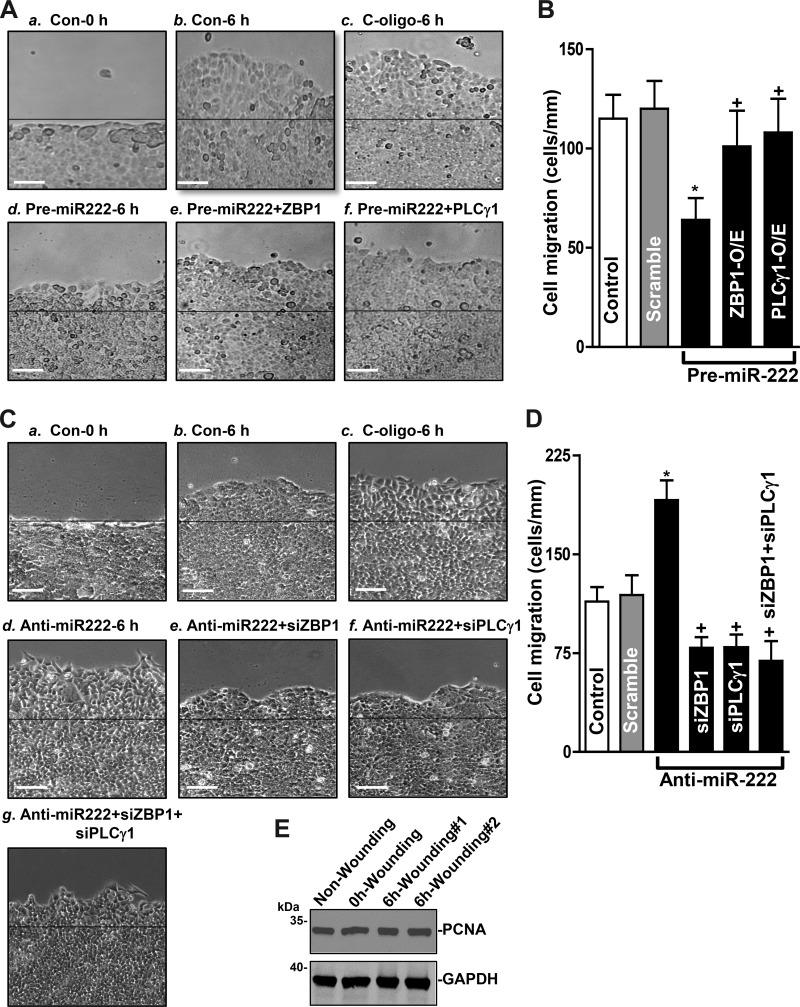
To determine if the role of miR-222-mediated regulation of ZBP1 and PLCγ1 expression in intestinal epithelial repair after wounding, an in vitro model that mimics proliferation-independent rapid epithelial restitution (7, 38, 56) was used in this study. HCT 116 cells were initially grown for 2 days and then cotransfected with pre-miR-222 and the vector expressing ZBP1 or PLCγ1. Consistent with our previous studies (7, 35), increasing the levels of miR-222 by transfection with pre-miR-222 inhibited cell migration after wounding. Interestingly, this miR-222-induced inhibition of cell migration was almost completely prevented by overexpressing ZBP1 or PLCγ1 (Fig. 6A). As shown, the numbers of migrating cells over the denuded area in cells cotransfected with pre-miR-222 and ZBP1 or PLCγ1 expression vector were indistinguishable from those observed in cells transfected with control scramble (Fig. 6B). Moreover, miR-222 silencing increased the number of cells over the wounded area and this stimulation of cell migration in miR-222-silenced cells was lost when ZBP1 and/or PLCγ1 expression was inhibited by transfection with small interference RNAs targeting ZBP1 (siZBP1) or PLCγ1 (siPLCγ1) (Fig. 6, C and D). On the other hand, there was no additional inhibitory effect on cell migration when the cells were cotransfected with siZBP1 and siPLCγ1 in miR-222 silenced cells. To determine the involvement of cell proliferation in this process, we examined changes in the levels of cell proliferation marker PCNA after wounding. Our results clearly showed that there were no significant changes in PCNA abundance 6 h after wounding (Fig. 6E); indicating that cell proliferation does not play a role in this rapid epithelial restitution. In addition, transfection with siZBP1 or siPLCγ1 did not affect cell viability as measured by trypan blue staining. These results indicate that miR-222-regulated expression of ZBP1 and PLCγ1 contributes to the control of intestinal epithelial restitution after wounding.
Fig. 6.
microRNA-222 (miR-222)-regulated expression of zipcode binding protein-1 (ZBP1) and phospholipase C-γ1 (PLCγ1) modulates rapid epithelial restitution after wounding. A: images of cell migration: a, 0 h after wounding in control cells (Con-0 h); b, 6 h after wounding in control cells (Con-6 h); c, 6 h after wounding in cells transfected with scramble oligo (C-oligo-6 h); d, 6 h after wounding in cells transfected with pre-miR-222 alone for 48 h (Pre-miR-222–6 h); e, 6 h after wounding in cells cotransfected with pre-miR-222 and the expression vector encoding ZBP1 (Pre-miR222 + ZBP1); f, 6 h after wounding in cells cotransfected with pre-miR-222 and the expression vector encoding PLCγ1 (Pre-miR222+ PLCγ1). Scale bar, 100 μm; magnification, ×100. B: summarized data showing rates of cell migration 6 h after wounding in cells described in A. Values are the means ± SE of data from 6 dishes and repeated 4 times independently (n = 4). *,+P < 0.05, compared with cells transfected with scramble and cells transfected with pre-miR-222, respectively as analyzed by one-way ANOVA followed by Duncan’s test. C: images of cell migration: a: 0 h after wounding in control cells; b: 6 h after wounding in control cells; c, 6 h after wounding in cells transfected with scramble oligo; d: 6 h after wounding in cells transfected with anti-miR-222 alone for 48 h; e: 6 h after wounding in cells cotransfected with anti-miR-222 and siZBP1 (Anti-miR222 + siZBP1); f: 6 h after wounding in cells cotransfected with anti-miR-222 and siPLCγ1 (Anti-miR222 + siPLCγ1); and g: 6 h after wounding in cells cotransfected with anti-miR-222, siZBP1 and siPLCγ1 (anti-miR222 + siZBP1 + siPLCγ1). Scale bar, 100 μm; magnification, ×100. D: summarized data showing rates of cell migration in cells described in C. Values are the means ± SE of data from 6 dishes and repeated four times independently (n = 4). *,+P < 0.05, compared with cells transfected with scramble and cells transfected with Anti-miR-222, respectively as analyzed by one-way ANOVA followed by Duncan’s test. E: immunoblot of PCNA protein in nonwounding and 0 h and 6 h after wounding (#1 and #2). Whole cell lysates were harvested and prepared for Western blotting; equal loading was monitored by assessing GAPDH levels.
DISCUSSION
miR-222 is encoded in tandem from a gene cluster containing identical seed sequences, and it is highly conserved in vertebrates (11). Our previous studies show that miR-222 acts as a negative regulator of intestinal epithelium homeostasis and that transgenic expression of miR-222 in IECs represses intestinal mucosal regeneration and disrupts the epithelial barrier function in mice (6). Mechanistically, miR-222 was found to repress CDK4 translation (52) and downregulate Wnt signaling activity by inhibiting the expression of Wnt-receptor Frizzled-7 posttranscriptionally (6). In the present study, we provide new evidence that miR-222 represses expression of ZBP1 and PLCγ1 by destabilizing their mRNAs. Our findings also show that miR-222-mediated regulation of ZBP1 and PLCγ1 expression plays an important role in the regulation of intestinal epithelial restitution after wounding, thus advancing our understanding of the biological functions of miR-222 in the intestinal epithelium.
ZBP1 and PLCγ1 interact with their different partner proteins and are implicated in many aspects of cellular functions (33, 34, 41, 48). Several miRNAs such as miR-98, miR-140, and miR-491 are shown to regulate ZBP1 expression at the posttranscriptional level, thereby affecting cell adhesion and motility (13, 21, 45). Similarly, miR-218, miR-429, and miR-132 are associated with and decrease the stability and translation of the mRNA encoding PLCγ1 (16, 23, 46). In this study, we identify miR-222 as a repressor of both ZBP1 and PLCγ1 by directly interacting with their mRNAs in IECs and provide additional new evidence showing the mechanism underlying the control of cellular ZBP1 and PLCγ1 levels. As shown, miR-222 associates with both ZBP1 and PLCγ1 mRNAs as measured by RNA pulldown assays using biotin-labeled miR-222, whereas ectopically expressed miR-222 enhances degradation of these two transcripts. In support of these findings, there are several computationally predicted binding sites of miR-222 in the 3′-untranslated regions (3′-UTR) of the ZBP1 and PLCγ1 mRNAs. Several studies have demonstrated that miR-222 exerts its regulatory actions through interaction with the 3′-UTRs of target transcripts in most cases (24, 31, 47, 54), although it can also bind occasionally to the coding region (52).
The data obtained in our study also indicates that miR-222-modulated expression of ZBP1 and PLCγ1 is of biological significance and plays a critical role in the regulation of intestinal epithelial restitution after wounding. The epithelium of the intestinal mucosa is a rapidly self-renewing tissue, and its homeostasis is preserved through strict regulation of cell proliferation, migration, differentiation, and apoptosis (8, 32, 38). Increased levels of ZBP1 and PLCγ1 in the population of miR-222-silenced cells were associated with an induction in IEC migration over the wounded area, which was abolished by silencing ZBP1 or PLCγ1. In contrast, decreasing the levels of ZBP1 and PLCγ1 by miR-222 overexpression inhibited cell migration, but cell migration in pre-miR-222-transfected cells was restored to near normal level by overexpressing ZBP1 or PLCγ1. Several studies from our group and others have revealed that intestinal epithelial restitution is tightly controlled by many factors including STIM1 (36), TRPC1 (7), caveolin-1 (37), Rac1 (35), Wnt signaling (28), and Cdc42 (29). On the other hand, the activity of these signaling pathways is also highly regulated by numerous intracellular and extracellular factors including RBPs (4, 47), miRNAs (3, 53, 56, 57), and cellular polyamines (34, 36, 42, 48, 52). Our current studies advance our knowledge and provide additional evidence showing that cellular levels of ZBP1 and PLCγ1 in IECs are negatively regulated by miR-222 at the posttranscriptional level.
In summary, these results indicate that miR-222 downregulates ZBP1 and PLCγ1 expression through a direct interaction with the ZBP1 and PLCγ1 mRNAs and that miR-222-regulated expression of ZBP1 and PLCγ1 plays an important role in the control of intestinal epithelial restitution after wounding. Ectopically expressed miR-222 decreases the levels of cellular ZBP1 and PLCγ1 primarily by destabilizing their mRNAs, whereas miR-222 silencing increases cellular abundances of ZBP1 and PLCγ1 by increasing the stability of ZBP1 and PLCγ1 mRNAs. Since ZBP1 and PLCγ1 are necessary for intestinal epithelial restitution after wounding and their cellular levels are tightly regulated by miR-222, the control of ZBP1 and PLCγ1 expression by miR-222 is essential for maintaining intestinal epithelial homeostasis and integrity under biological and pathologic conditions.
GRANTS
This work was supported by Merit Review Awards from the US Department of Veterans Affairs (J. N. Rao and J. Y. Wang) and National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-57819, DK-61972, and DK-68491 (J. Y. Wang). L.-P. Jiang was supported by the China Scholarship Council (no. 2011843319).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by authors.
AUTHOR CONTRIBUTIONS
L.-P.J. and J.N.R. conceived and designed research; L.-P.J., S.R.W., H.K.C., and S.B. performed experiments; L.-P.J., H.K.C., J.-Y.W., and J.N.R. analyzed data; L.-P.J. and J.N.R. prepared figures; L.-P.J. drafted manuscript; H.K.C., J.-Y.W., and J.N.R. interpreted results of experiments; J.-Y.W. and J.N.R. edited and revised manuscript; J.-Y.W. and J.N.R. approved final version of manuscript.
ACKNOWLEDGMENTS
Present address of L.-P. Jiang: Department of Parasitology, Xiangya School of Medicine, Central South University, Changsha, Hunan 410013, PR China.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature 431: 350–355, 2004. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Cai K, Shen F, Cui JH, Yu Y, Pan HQ. Expression of miR-221 in colon cancer correlates with prognosis. Int J Clin Exp Med 8: 2794–2798, 2015. [PMC free article] [PubMed] [Google Scholar]
- 3.Cao S, Xiao L, Rao JN, Zou T, Liu L, Zhang D, Turner DJ, Gorospe M, Wang JY. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-β/Smad2 signaling and intestinal epithelial homeostasis. Mol Biol Cell 25: 1234–1243, 2014. doi: 10.1091/mbc.e13-09-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem J 446: 113–123, 2012. doi: 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistiakov DA, Sobenin IA, Orekhov AN, Bobryshev YV. Human miR-221/222 in physiological and atherosclerotic vascular remodeling. BioMed Res Int 2015: 354517, 2015. doi: 10.1155/2015/354517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung HK, Chen Y, Rao JN, Liu L, Xiao L, Turner DJ, Yang P, Gorospe M, Wang JY. Transgenic expression of miR-222 disrupts intestinal epithelial regeneration by targeting multiple genes including Frizzled-7. Mol Med 21: 676–687, 2015. doi: 10.2119/molmed.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung HK, Rathor N, Wang SR, Wang JY, Rao JN. RhoA enhances store-operated Ca2+ entry and intestinal epithelial restitution by interacting with TRPC1 after wounding. Am J Physiol Gastrointest Liver Physiol 309: G759–G767, 2015. doi: 10.1152/ajpgi.00185.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung HK, Wang SR, Xiao L, Rathor N, Turner DJ, Yang P, Gorospe M, Rao JN, Wang JY. α4 coordinates small intestinal epithelium homeostasis by regulating stability of HuR. Mol Cell Biol 38: e00631-e17, 2018. doi: 10.1128/MCB.00631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degrauwe N, Suvà ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev 30: 2459–2474, 2016. doi: 10.1101/gad.287540.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87, 2003. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 282: 23716–23724, 2007. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 12.Gardiner AS, Twiss JL, Perrone-Bizzozero NI. Competing interactions of RNA-binding proteins, microRNAs, and their targets control neuronal development and function. Biomolecules 5: 2903–2918, 2015. doi: 10.3390/biom5042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong F, Ren P, Zhang Y, Jiang J, Zhang H. MicroRNAs-491-5p suppresses cell proliferation and invasion by inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res 8: 485–495, 2016. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Gu W, Pan F, Singer RH. Blocking β-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J Cell Sci 122: 1895–1905, 2009. doi: 10.1242/jcs.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Katz Z, Wu B, Park HY, Li D, Lin S, Wells AL, Singer RH. Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J Cell Sci 125: 81–91, 2012. doi: 10.1242/jcs.086132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan H, Wei G, Wu J, Fang D, Liao Z, Xiao H, Li M, Li Y. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab 98: E1334–E1344, 2013. doi: 10.1210/jc.2013-1053. [DOI] [PubMed] [Google Scholar]
- 17.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4464, 2004. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harter JL. Critical values for Duncan’s new multiple range test. Biometrics 16: 671–685, 1960. doi: 10.2307/2527770. [DOI] [Google Scholar]
- 19.Hsieh YT, Chou MM, Chen HC, Tseng JJ. IMP1 promotes choriocarcinoma cell migration and invasion through the novel effectors RSK2 and PPME1. Gynecol Oncol 131: 182–190, 2013. doi: 10.1016/j.ygyno.2013.07.106. [DOI] [PubMed] [Google Scholar]
- 20.Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515, 2005. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 21.Jiang T, Li M, Li Q, Guo Z, Sun X, Zhang X, Liu Y, Yao W, Xiao P. MicroRNA-98–5p inhibits cell proliferation and induces cell apoptosis in hepatocellular carcinoma via targeting IGF2BP1. Oncol Res 25: 1117–1127, 2017. doi: 10.3727/096504016X14821952695683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones NP, Peak J, Brader S, Eccles SA, Katan M. PLCγ1 is essential for early events in integrin signalling required for cell motility. J Cell Sci 118: 2695–2706, 2005. doi: 10.1242/jcs.02374. [DOI] [PubMed] [Google Scholar]
- 23.Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, Kunugi H, Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 165: 1301–1311, 2010. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 24.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020, 2010. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 25.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543, 2007. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 26.Lattanzio R, Piantelli M, Falasca M. Role of phospholipase C in cell invasion and metastasis. Adv Biol Regul 53: 309–318, 2013. doi: 10.1016/j.jbior.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Park EJ, Kiyono H. MicroRNA-orchestrated pathophysiologic control in gut homeostasis and inflammation. BMB Rep 49: 263–269, 2016. doi: 10.5483/BMBRep.2016.49.5.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Christodoulou-Vafeiadou E, Rao JN, Zou T, Xiao L, Chung HK, Yang H, Gorospe M, Kontoyiannis D, Wang JY. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell 25: 3308–3318, 2014. doi: 10.1091/mbc.e14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Zhuang R, Xiao L, Chung HK, Luo J, Turner DJ, Rao JN, Gorospe M, Wang JY. HuR enhances early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol 37: e00574-e16, 2017. doi: 10.1128/MCB.00574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manieri NA, Drylewicz MR, Miyoshi H, Stappenbeck TS. Igf2bp1 is required for full induction of Ptgs2 mRNA in colonic mesenchymal stem cells in mice. Gastroenterology 143: 110–21.e10, 2012. doi: 10.1053/j.gastro.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuzaki J, Suzuki H. Role of microRNAs-221/222 in digestive systems. J Clin Med 4: 1566–1577, 2015. doi: 10.3390/jcm4081566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nusrat A, Delp C, Madara JL. Intestinal epithelial restitution. Characterization of a cell culture model and mapping of cytoskeletal elements in migrating cells. J Clin Invest 89: 1501–1511, 1992. doi: 10.1172/JCI115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, Chao JA. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26: 43–53, 2012. doi: 10.1101/gad.177428.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao JN, Liu L, Zou T, Marasa BS, Boneva D, Wang SR, Malone DL, Turner DJ, Wang JY. Polyamines are required for phospholipase C-γ1 expression promoting intestinal epithelial restitution after wounding. Am J Physiol Gastrointest Liver Physiol 292: G335–G343, 2007. doi: 10.1152/ajpgi.00282.2006. [DOI] [PubMed] [Google Scholar]
- 35.Rao JN, Liu SV, Zou T, Liu L, Xiao L, Zhang X, Bellavance E, Yuan JX, Wang JY. Rac1 promotes intestinal epithelial restitution by increasing Ca2+ influx through interaction with phospholipase C-(γ)1 after wounding. Am J Physiol Cell Physiol 295: C1499–C1509, 2008. doi: 10.1152/ajpcell.00232.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao JN, Rathor N, Zhuang R, Zou T, Liu L, Xiao L, Turner DJ, Wang JY. Polyamines regulate intestinal epithelial restitution through TRPC1-mediated Ca2+ signaling by differentially modulating STIM1 and STIM2. Am J Physiol Cell Physiol 303: C308–C317, 2012. doi: 10.1152/ajpcell.00120.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathor N, Chung HK, Wang SR, Wang JY, Turner DJ, Rao JN. Caveolin-1 enhances rapid mucosal restitution by activating TRPC1-mediated Ca2+ signaling. Physiol Rep 2: e12193, 2014. doi: 10.14814/phy2.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rathor N, Chung HK, Wang SR, Qian M, Turner DJ, Wang JY, Rao JN. β-PIX plays an important role in regulation of intestinal epithelial restitution by interacting with GIT1 and Rac1 after wounding. Am J Physiol Gastrointest Liver Physiol 314: G399–G407, 2018. doi: 10.1152/ajpgi.00296.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebucci M, Sermeus A, Leonard E, Delaive E, Dieu M, Fransolet M, Arnould T, Michiels C. miRNA-196b inhibits cell proliferation and induces apoptosis in HepG2 cells by targeting IGF2BP1. Mol Cancer 14: 79, 2015. doi: 10.1186/s12943-015-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem 70: 281–312, 2001. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local β-actin synthesis and growth cone turning. J Neurosci 30: 9349–9358, 2010. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seiler N, Raul F. Polyamines and the intestinal tract. Crit Rev Clin Lab Sci 44: 365–411, 2007. doi: 10.1080/10408360701250016. [DOI] [PubMed] [Google Scholar]
- 43.Song J, Ouyang Y, Che J, Li X, Zhao Y, Yang K, Zhao X, Chen Y, Fan C, Yuan W. Potential value of miR-221/222 as diagnostic, prognostic, and therapeutic biomarkers for diseases. Front Immunol 8: 56, 2017. doi: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stöhr N, Köhn M, Lederer M, Glass M, Reinke C, Singer RH, Hüttelmaier S. IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes Dev 26: 176–189, 2012. doi: 10.1101/gad.177642.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y, Xiong J, Hu J, Wei X, Zhang X, Rao L. MicroRNA-140-5p targets insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) to suppress cervical cancer growth and metastasis. Oncotarget 7: 68397–68411, 2016. doi: 10.18632/oncotarget.11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlmann S, Zhang JD, Schwäger A, Mannsperger H, Riazalhosseini Y, Burmester S, Ward A, Korf U, Wiemann S, Sahin O. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29: 4297–4306, 2010. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 47.Wang JY, Xiao L, Wang JY. Posttranscriptional regulation of intestinal epithelial integrity by noncoding RNAs. Wiley Interdiscip Rev RNA 8: e1399, 2017. doi: 10.1002/wrna.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang PY, Wang SR, Xiao L, Chen J, Wang JY, Rao JN. c-Jun enhances intestinal epithelial restitution after wounding by increasing phospholipase C-γ1 transcription. Am J Physiol Cell Physiol 312: C367–C375, 2017. doi: 10.1152/ajpcell.00330.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waugh MG. Chromosomal instability and phosphoinositide pathway gene signatures in glioblastoma multiforme. Mol Neurobiol 53: 621–630, 2016. doi: 10.1007/s12035-014-9034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2: 237–246, 2001. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 51.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene 500: 10–21, 2012. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell 22: 3055–3069, 2011. doi: 10.1091/mbc.e11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, Gorospe M, Wang JY. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology 154: 599–611, 2018. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu G, Yang F, Ding CL, Wang J, Zhao P, Wang W, Ren H. MiR-221 accentuates IFN′s anti-HCV effect by downregulating SOCS1 and SOCS3. Virology 462-463: 343–350, 2014. doi: 10.1016/j.virol.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287: 49–54, 2002. doi: 10.1016/S0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- 56.Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res 41: 7905–7919, 2013. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou T, Rao JN, Liu L, Xiao L, Chung HK, Li Y, Chen G, Gorospe M, Wang JY. JunD enhances miR-29b levels transcriptionally and posttranscriptionally to inhibit proliferation of intestinal epithelial cells. Am J Physiol Cell Physiol 308: C813–C824, 2015. doi: 10.1152/ajpcell.00027.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, Xu Y, Gorospe M, Wang JY. H19 long noncoding RNA regulates intestinal epithelial barrier functio n via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol 36: 1332–1341, 2016. doi: 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]