Abstract
INTRODUCTION:
The main goal of this work is to investigate the feasibility of estimating an anatomical index that can be used as an Alzheimer’s disease (AD) risk factor in the Women’s Health Initiative Magnetic Resonance Imaging Study (WHIMS-MRI) using MRI data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), a well-characterized imaging database of AD patients and cognitively normal subjects. We called this index AD Pattern Similarity (AD-PS) scores. To demonstrate the construct validity of the scores, we investigated their associations with several AD risk factors. The ADNI and WHIMS imaging databases were collected with different goals, populations and data acquisition protocols: it is important to demonstrate that the approach to estimating AD-PS scores can bridge these differences.
METHODS:
MRI data from both studies were processed using high-dimensional warping methods. High-dimensional classifiers were then estimated using the ADNI MRI data. Next, the classifiers were applied to baseline and follow-up WHIMS-MRI GM data to generate the GM AD-PS scores. To study the validity of the scores we investigated associations between GM AD-PS scores at baseline (Scan 1) and their longitudinal changes (Scan2 –Scan 1) with: 1) age, cognitive scores, white matter small vessel ischemic disease (WM SVID) volume at baseline and 2) age, cognitive scores, WM SVID volume longitudinal changes respectively. In addition, we investigated their associations with time until classification of independently adjudicated status in WHIMS-MRI.
RESULTS:
Higher GM AD-PS scores from WHIMS-MRI baseline data were associated with older age, lower cognitive scores, and higher WM SVID volume. Longitudinal changes in GM AD-PS scores (Scan2 – Scan 1) were also associated with age and changes in WM SVID volumes and cognitive test scores. Increases in the GM AD-PS scores predicted decreases in cognitive scores and increases in WM SVID volume. GM AD-PS scores and their longitudinal changes also were associated with time until classification of cognitive impairment. Finally, receiver operating characteristic curves showed that baseline GM AD-PS scores of cognitively normal participants carried information about future cognitive status determined during follow-up.
DISCUSSION:
We applied a high-dimensional machine learning approach to estimate a novel AD risk factor for WHIMS-MRI study participants using ADNI data. The GM AD-PS scores showed strong associations with incident cognitive impairment and cross-sectional and longitudinal associations with age, cognitive function, cognitive status and WM SVID volume lending support to the ongoing validation of the GM AD-PS score.
Keywords: Alzheimer’s disease, MRI, AD-PS, machine learning, High-Dimensional
INTRODUCTION
Machine learning is becoming an increasingly popular approach in biomedical research related to high-dimensional data. In Alzheimer’s disease (AD) research the challenge of early detection is of paramount importance, since pathological processes develop many years before the cognitive impairment is observed. Early detection of AD might help in selection of patients for clinical trials and improve the efficacy of clinical or behavioral interventions.
Massive amounts of data from different sources are being used to develop early detection models (Weiner, et al., 2013). One of the more common types of neuroimaging data is structural MRI. Brain MRI is used to identify neuropathologies such as brain atrophy. Traditional approaches to study the impact of AD on brain structure are based on specific regions of interest (ROI) or voxel-based morphometry; these are univariate and cannot reveal complex spatial patterns of atrophy related to AD. Machine learning approaches are well-suited to address these challenges because they can capture complex patterns hidden in the data and build powerful prediction models based on high-dimensional data.
Many recent studies have sought to derive machine learning models based on structural MRI to predict AD or MCI-AD conversion. Much of this work has used the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset, which was developed with highly standardized protocols to distinguish individuals with and without AD. For these methods to be adopted in practice, it is important to demonstrate their utility with different protocols for identifying dementia and MRI outcomes. These efforts could enable the AD research community to more fully use large amounts of imaging data already collected.
A few groups have developed machine learning models using one imaging database, and then applied them to other databases using different populations and imaging protocols. Kloppel and colleagues showed the feasibility of automatic classification of AD using whole brain MRI scans collected in different centers and support vector machines (SVM)(Kloppel, et al., 2008). Davatzikos and colleagues used the ADNI dataset to train a SVM classifier used to estimate an index of AD anatomical risk called Spatial Pattern of Abnormality for Recognition of Early Alzheimer’s disease (SPARE-AD). They investigated the association of this index with cognitive decline using longitudinal structural MRI data from cognitively normal participants and those with mild cognitive impairment (MCI) in the Baltimore Longitudinal Aging Study (BLSA)(Davatzikos, et al., 2009). Associations between the SPARE-AD index, plasma analytes, and AD risk factors have been investigated using other cohorts (Habes, et al., 2016a,Habes, et al., 2016b,Toledo, et al., 2013).
This study extends our previous research on AD pattern similarity (AD-PS) scores developed using structural MRI from ADNI (Casanova, et al., 2013). Our main goal here is to create a metric based on MRI to be used by Women Health Initiative investigators, as an AD risk factor reflecting the presence of AD-related spatial patterns in the brain. This study is part of an ongoing effort to investigate the feasibility and constructive validity of estimating a novel AD risk factor based on elastic net regularization and WHIMS-MRI data. To do this we computed AD-PS scores for the Women’s Health Initiative Memory MRI study cohort, and then we evaluated whether these scores are associated with classifications of cognitive impairment (normal, MCI, and probable dementia), incident cognitive impairment, age, performance on a test of cognitive function, and white matter hyperintensity volume and other risk factors.
MATERIALS AND METHODS
WHIMS-MRI
The Women’s Health Initiative Memory Study (WHIMS) investigated the effects of postmenopausal hormone therapy on the risk of dementia and changes in cognitive function in women aged 65–79 at enrollment (1996–1998) into the WHI randomized placebo-controlled hormone therapy clinical trials (Espeland, et al., 2004,Shumaker, et al., 1998). The WHIMS-MRI study enrolled WHIMS participants from 14 of 39 sites (Jaramillo, et al., 2007,Resnick, et al., 2009) from January 2005 through April 2006, an average of 8 years after they had enrolled in the WHIMS trial and 1–3 years after study medications ended (Coker, et al; 2014). WHIMS-MRI participants who continued WHIMS follow-up were invited to join the WHIMS-MRI2 study; a second MRI scan was obtained an average of 4.7 years after the first one. WHIMS-MRI2 exclusion criteria included absolute contraindications and health-related factors and were identical to those for WHIMS-MRI (Coker, et al., 2014,Jaramillo, et al., 2007).
We analyzed baseline and follow-up images from 1,365 and 712 participants, respectively, who met WHIMS-MRI reading criteria. This study was conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent for both WHIMS-MRI studies.
MRI Data Acquisition and Processing
MRI scans were performed using a standardized protocol developed by the MRI Quality Control Center in the Department of Radiology of the University of Pennsylvania (Coker, et al., 2009,Resnick, et al., 2009). Briefly, the scans were obtained with a 22 cm field of view and a matrix of 256×256 in 1.5T scanners. Included were oblique axial spin density/T2-weighted spin echo (TR:3200 ms, TE=30/120 ms, slice thickness=3 mm), fluid-attenuated inversion recovery (FLAIR) T2-weighted spin echo (TR=8000 ms, TI=2000 ms, TE=100 ms, slice thickness=3 mm), and oblique axial three-dimensional T1-weighted gradient echo (flip angle=30 degrees, TR=21 ms, TE=8 ms, slice thickness=1.5 mm) images from the vertex to the skull base parallel to the anterior commissure-posterior commissure (AC-PC) plane. Total brain white matter hyperintensity volumes were computed based on a machine learning methodology previously described, validated and deployed in several imaging studies (Lao, et al., 2008,Launer, et al., 2011).
ADNI
ADNI database
The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations. Its primary goal was to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment could be combined to measure the progression of mild cognitive impairment (MCI) and early AD. From over 50 sites across the U.S. and Canada, in its first phase ADNI (ADNI-1) recruited 819 participants, aged 55 to 90 years, including 229 cognitively normal (CN) individuals to be followed for 3 years, 398 people with MCI to be followed for 3 years, and 192 people with early AD to be followed for 2 years. For up-to-date information about the cohort, see www.adni-info.org.
MRI Data
In this work, we used ADNI baseline structural MRI data from 359 Caucasians (Casanova, et al., 2013) (See Table S1). Of those, 188 were CN (86 women) and 171 had AD (76 women) (Petersen, et al., 2010). Baseline 1.5T T1-weighted MRI scans were collected using 3D MPRAGE sequences, as described in the ADNI acquisition protocol (Hua, et al., 2008,Jack, et al., 2008). The ADNI protocol acquires 2 repeated scans of structural MRI data at each visit that are rated for image quality and artifacts by ADNI investigators. To enhance standardization across sites and platforms, the best dataset undergoes additional pre-processing, including corrections for gradient non-linearity and intensity non-uniformity.
ADNI and WHIMS-MRI Image Processing
The open source package Advanced Normalization Tools (ANTs) was used to process the MRI images. Its capabilities include diffeomorphic transformations for image warping that preserve topology. These are based on symmetric normalization algorithms (Avants, et al., 2008,Avants, et al., 2011a), which are among the top-performing image warping approaches in large comparative studies (Klein, et al., 2009,Murphy, et al., 2011). A more detailed description of the ANTs pipeline components is in Tustison et al. (Tustison, et al., 2014). All the necessary preprocessing steps consist of previously published algorithms for bias correction (Tustison, et al., 2010), brain extraction, n-tissue segmentation (Avants, et al., 2011b), template construction (Avants, et al., 2010), and image normalization (Avants, et al., 2011a). Further details of the MRI image processing and its implementation can be found in the appendix.
Cognitive Assessment
Participants underwent annual cognitive assessments during their in-person clinic visits to evaluate global cognitive function in the first phase of WHIMS (years 1–10), as described previously (Shumaker, et al., 1998). Briefly, the Modified Mini-Mental State Exam (3MS) (Teng and Chui, 1987) was administered to all participants. Age and education specific cut points determined which participants were referred for an extensive clinical evaluation, which involved additional cognitive testing and questionnaires, a physical exam by an experienced physician and proxy interviews (Shumaker, et al., 1998). All data were submitted to a central adjudication panel of expert clinicians who classified each case as no impairment, MCI using Petersen’s criteria (Petersen, et al., 1999,Petersen, et al., 2001) or probable dementia probable dementia(American Psychiatric Association, 1994).
Beginning in 2007, WHI clinic visits ceased and WHIMS transitioned to a telephone-based cognitive assessment battery (Rapp, et al., 2012), questionnaires, proxy interviews and central adjudication of cognitive impairment as in the initial phase of WHIMS. The telephone battery included the modified Telephone Interview for Cognitive Status (TICS-m), (Brandt, et al., 1988,Plassman, et al., 1994,Welsh, et al., 1993), a measure of global cognitive function and tests of memory, verbal fluency, attention, working memory and executive function; questionnaires assessing depression and sleep; and the Dementia Questionnaire (DQ) (Kawas, et al., 1994), a standardized interview assessing cognitive and behavioral changes and medical events related to dementia and MCI administered to a proxy informant. The DQ was administered when TICS-m scores fell below 31 points. All data were forwarded for central adjudication. Following central adjudication of dementia, each adjudicator was required to subtype the classification using DSM-IV diagnostic criteria for Alzheimer’s, Vascular and Other dementias. Disagreements were resolved in regular consensus conferences.
Estimation of the GM AD-PS scores in the WHIMS-MRI cohort
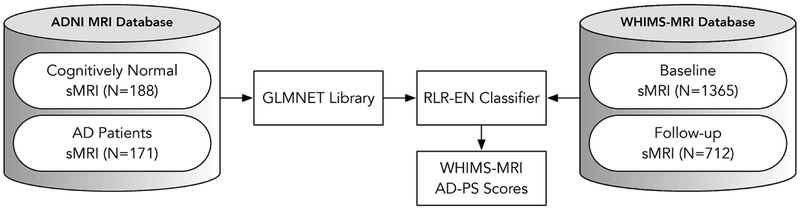
Once ADNI and WHIMS-MRI were aligned into a template (see appendix A) , we used high-dimensional machine learning methods to estimate GM AD-PS scores in the WHIMS-MRI cohort. The general approach followed is depicted in Figure 1. Details of the machine learning algorithms were published previously (R. Casanova, et al., 2012,Casanova, et al., 2013) and also can be found in appendix B. An elastic net regularized logistic regression (EN-RLR) classifier was estimated using the GM probability maps (described above) from CN and AD participants in ADNI. The weights estimated after solving the optimization problem associated with the EN-RLR classifier (Casanova, et al., 2013,Friedman, et al., 2009) are used to estimate the conditional probabilities. These were computed as the median values of 5 repetitions of the computations, to account for variability due to random partitioning of cross-validation that occurred during model estimation (Casanova, et al., 2013). The scores for WHIMS-MRI participants were estimated by providing the corresponding baseline and follow-up GM probability maps to the classifiers. Discriminative maps (voxels with coefficients different from zero) pinpointing the GM areas involved in discrimination of AD patients from CN participants were generated. For simplicity from now on we will refer to the GM AD-PS scores as AD-PS scores.
Figure 1 -.
General approach to estimate Alzheimer’s disease pattern similarity (AD-PS) scores for the WHIMS-MRI cohort once the images from both datasets have been processed.
Analyses
The cross-sectional relationships of AD-PS scores of baseline WHIMS-MRI participants and three risk factors for cognitive impairment (age at MRI, most proximal cognitive function test score, and abnormal white matter volumes) were assessed with linear regression with covariate adjustment for age, education, race and hormone therapy assignment. Relationships between longitudinal changes in AD-PS scores and changes in age and WM SVID volumes were assessed with linear models.
When the initial MRIs were collected, the clinic-based cognitive assessment protocol of WHIMS was ongoing. When repeat MRIs were collected in a subset of women, global cognitive function assessment had transitioned to telephone-based TICS. To obtain a measure of relative change in global cognitive function over time, we standardized 3MS (administered in person) and TICS scores by subtracting them from the cohort-wide mean and dividing this by the cohort-wide standard deviation. We used the difference between the standardized 3MS and TICS scores as a measure of relative change, using the scores most proximal to the initial and repeat MRI. Linear models were then used to assess relationships between changes in AD-PS scores and changes in standardized cognitive function scores. Survival analyses, using proportional hazards regression, were performed to investigate associations of the AD-PS scores with time to diagnosis of cognitive impairment(Cox DR and D., 1984) as MCI, probable dementia, and MCI/probable dementia adjusted for age, education, HT treatment assignment and race/ethnicity, using the 27 cases of probable dementia and 33 cases of MCI that were observed among the 1365 women between their initial MRI through January 2010 (when follow-up MRIs began). Associations that changes in AD-PS scores between the initial and follow-up MRI (in 712 women) had with subsequent cognitive impairment was based on 54 cases of probable dementia and 37 cases of MCI that occurred during later follow-up. Analyses were adjusted for age, education, race and HT treatment assignment.
Boxplots, receiver operating characteristic curves (ROC), and area under the curve (AUC) estimates were generated for baseline, follow-up, and longitudinal changes in AD-PS scores to evaluate their sensitivity to cognitive status. To generate confidence intervals for AUC we used the R package pROC(Robin, et al., 2011). When generating these plots for the baseline AD-PS scores, we used only participants who were CN at baseline MRI and with date of last cognitive assessment in 2010 or afterwards (4.5 years on average) or whose cognitive impairment was diagnosed between baseline MRI and January 1st 2010 (Total N = 834). Of those, 733 remained stable in that time period as CN, 33 converted to MCI and 28 to probable dementia, including 12 cases adjudicated as AD. For follow up AD-PS scores and change plots, we also used participants who were CN at baseline MRI, who had both scans and with date of last cognitive assessment in 2010 or afterwards or whose cognitive impairment was diagnosed at any point during the follow up (N = 571). Of those, 455 remained stable as CN until 2009, 53 converted to MCI and 63 to probable dementia including 6 cases adjudicated as AD.
RESULTS
Women who had only a baseline MRI scan in WHIMS-MRI were older, more likely to be minority, hypertensive and diabetic, and had lower 3MS scores compared with women who had both baseline and follow-up MRI scans (p<0.05) (Table 1). Baseline AD-PS scores were significantly higher for the 653 women who only had baseline MRIs compared with the 712 who had baseline and repeat MRIs (mean ± SD: 0.39 ± 0.25 versus 0.28 ± 0.20; p=0.0002). This is consistent with a previous report by WHIMS-MRI researchers indicating that women who had both MRI scans had greater hippocampal and brain volumes than those who had only the baseline scan (Coker, et al., 2014). For participants who had both scans, follow-up scores were significantly higher than baseline scores when adjusted by age, education, race and hormone therapy treatment (0.44 ± 0.26; p < 0.0001).
Table 1 -.
Baseline characteristics of WHIMS-MRI participants
| Variable | Mean (SD) or Frequency (%) | |||
|---|---|---|---|---|
| Had Baseline MRI (n=1365) | Had Baseline MRI Only (n=653) | Had Baseline and Follow-up MRI (n=712) | P-value* | |
| Age | 70.53 (3.63) | 71.02 (3.76) | 70.08 (3.47) | p<0.0001 |
| Body mass index | 28.23 (5.42) | 28.28 (5.79) | 28.18 (5.08) | p=0.7388 |
| Other | 40 (2.93%) | 21 (3.22%) | 19 (2.67%) | |
| > High school | 985 (72.32%) | 486 (74.43%) | 499 (70.38%) | |
| Current | 57 (4.21%) | 28 (4.31%) | 29 (4.11%) | |
| Cardiovascular disease ever | 86 (6.30%) | 49 (7.50%) | 37 (5.20%) | p=0.1503 |
| Hypertension ever | 650 (47.62%) | 333 (51.00%) | 317 (44.52%) | p=0.0254 |
| Prior use of hormone therapy | 449 (32.89%) | 201 (30.78%) | 248 (34.83%) | p=0.1116 |
| Diabetes treated ever (oral therapy or injected insulin) | 73 (5.35%) | 47 (7.20%) | 26 (3.65%) | p=0.0036 |
| Baseline score on Mini-Mental Examination | 96.10 (3.45) | 95.68 (3.88) | 96.50 (2.96) | p<0.0001 |
Comparing those with a baseline MRI only versus those who had both baseline and follow-up MRIs.
AD-PS scores were significantly associated with age at baseline MRI, 3MS scores, and WM SVID volume after adjusting for age, education, race and hormone therapy treatment assignment (p < 0.001) (Table 2). In participants who had baseline and follow-up MRIs, changes in AD-PS scores were also significantly related to changes in age, cognitive function, and abnormal white matter volumes. At baseline, women assigned to hormone therapy had slightly, but not significantly, higher mean (standard error) AD-PS scores compared to the placebo group (0.337 ± 0.008 versus 0.320 ± 0.008; p=0.14). There was no difference in mean changes in AD-PS scores over time between intervention groups.
Table 2 -.
Cross-sectional and longitudinal relationships that risk factors for cognitive impairment have with AD-PS scores and their longitudinal changes, with covariate adjustment for age, education, race and hormone therapy assignment.
| Risk Factor | Cross-sectional Relationships Between Risk Factors and AD-PS at Time 1 | Relationships Between Changes in Risk Factors and Changes in AD-PS From Time 1 to Time 2 | ||
|---|---|---|---|---|
| Slope* (SE) | p-value | Slope** (SE) |
p-value | |
| Age, yrs | 0.027 (0.002) |
t= 17.17 <.001 |
0.034 (0.001) |
t= 29.55 <.001 |
| Cognitive Function, SD | −0.036 (0.004) |
t= −8.81 <0.001 |
−0.019 (0.008) |
t= −2.41 0.02 |
| WM SVID volume, SDs | 0.024 (0.004) |
t= 4.22 <0.001 |
0.149 (0.008) |
t= 18.36 <.001 |
Unit AD-PS score per year (for age) or per Time 1 standard deviation (for cognitive function and WM SVID volume)
Unit change in AD-PS score between Time 1 and Time 2 per year (for age) or per standard deviation (for cognitive function and WM SVID volume)
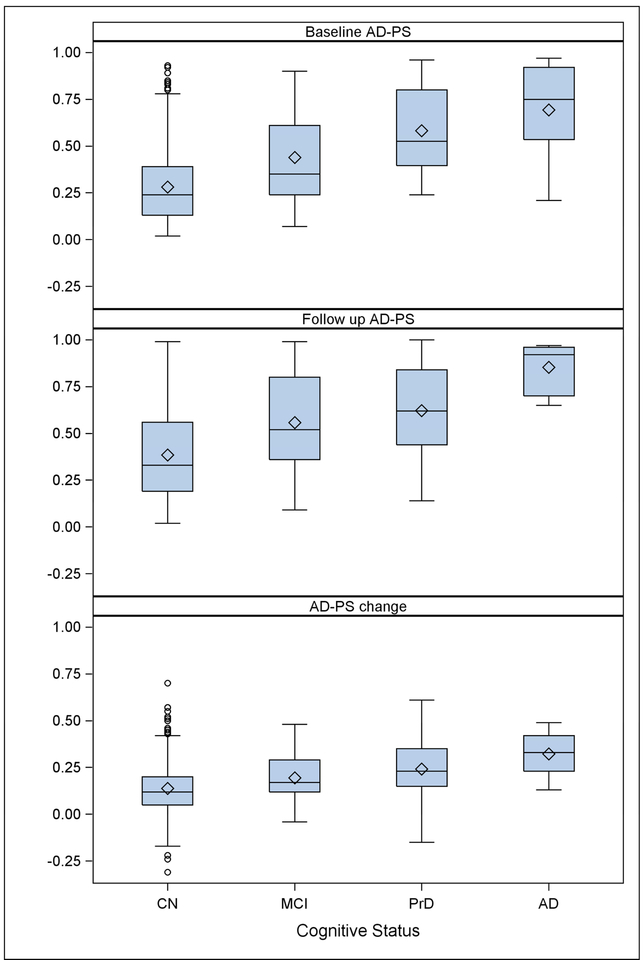
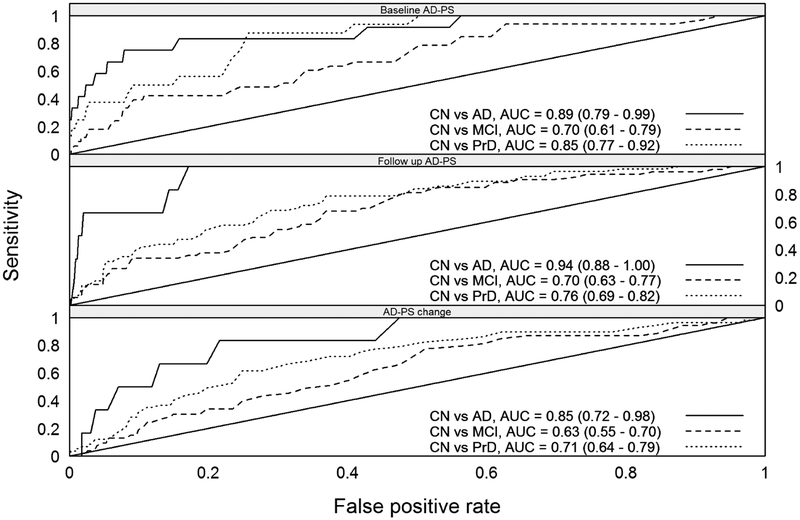
Figure 2 shows the values of the AD-PS scores at Scan 1, Scan 2 and their longitudinal change for a subset of WHIMS-MRI participants CN at baseline and with cognitive follow-up until 2010 or afterwards, or whose cognitive impairment was diagnosed during follow-up. In all cases the scores and their longitudinal change increased with the severity of the cognitive impairment. Additional plots of the scores can be found in Figures S2–S3 in supplementary materials. ROC curves evaluating discrimination of the cognitive impaired groups’ AD-PS scores relative to the CN are presented in Figure 3. Each panel evaluates discrimination of the data from the corresponding panel of Figure 2. The level of discrimination according to the estimated AUC increases as the severity of the cognitive impairment increases.
Figure 2 -.
The upper panel shows the baseline (Scan 1) AD-PS scores for only participants who were cognitively normal at baseline MRI and with date of last cognitive assessment in 2010 (4.5 years in average) or afterwards. The abbreviation PrD stands for probable dementia. Of those 733 remained stable as CN in that time period, 33 converted to MCI and 28 converted to probable dementia probable dementiaincluding 12 cases adjudicated as AD. For the follow up AD-PS scores and change plots we also used only participants who were CN at baseline MRI with date of last cognitive assessment in 2010 or afterwards whose cognitive impairment was diagnosed at any point during the follow up (N = 571). Of those 455 remained stable as CN until 2009, 53 converted to MCI and 57 converted to Probable dementia including 6 cases adjudicated as AD during follow up.
Figure 3 -.
The ROC curves evaluating discrimination of the AD-PS scores for each impaired cognitive group with respect to the CN group. Each panel in this Figure shows discrimination of the data from the corresponding panel in Figure 2. The abbreviation PrD stands for probable dementia.
Baseline AD-PS scores and change sin AD-PS scores over time were both significantly (p<0.0001) associated with the incidence of cognitive impairment (Table 3). For baseline (Scan 1) scores, hazard ratios per 0.1 greater score were 1.84, 1.29 and 1.50 for probable dementia, MCI and MCI/probable dementia respectively. For the longitudinal changes in scores, hazard ratios per 0.1 increases of the AD-PS scores were 1.63, 1.38 and 1.51 for probable dementia, MCI and MCI/probable dementia respectively.
Table 3 -.
Survival analyses investigating associations of AD-PS baseline scores and their longitudinal change with date of cognitive impairment diagnosis.
| Category | Hazard ratio per unit = 0.1 | 95% Confidence Interval | p-values | |
|---|---|---|---|---|
| Baseline AD-PS scores | MCI | 1.29 | 1.10 – 1.52 | 0.002 |
| Probable dementia | 1.84 | 1.55 – 2.18 | <.0001 | |
| MCI+probable dementia | 1.51 | 1.35 – 1.69 | <.0001 | |
| AD-PS Change (FUP-BS) | MCI | 1.38 | 1.09 – 1.74 | 0.0069 |
| PrD | 1.63 | 1.36 – 1.95 | <.0001 | |
| MCI+probable dementia | 1.51 | 1.31 – 1.75 | <.0001 |
For baseline analyses 27, 33 and 60 cases of probable dementia, MCI and MCI/probable dementia were available while 1132, 1099 and 1099 were censored respectively. For longitudinal change analyses (which spans the time from the second MRI until the end of follow-up) 54, 37 and 90 cases of probable dementia, MCI and MCI/probable dementia were available while 485, 448, and 448 were censored respectively. Analyses were adjusted by age, education and race and HT assignment.
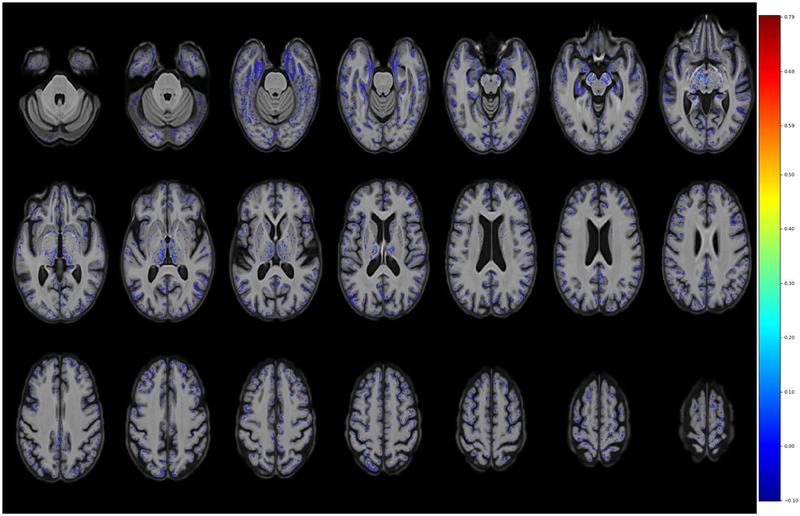
Figure 4 shows a discriminative map indicating the areas involved in discrimination of AD from CN, derived from the elastic net regularization constraints. In addition, in the supplementary materials, Figure S3 shows a mask to indicate the location identified by the elastic net algorithm as relevant. The map includes many regions thought to be involved in AD (e.g. amygdala, hippocampus, parahippocampal gyrus, thalamus, bilateral inferior temporal lobe areas)(Braak and Braak, 1991,Mu and Gage, 2011,Poulin, et al., 2011,Zanchi, et al., 2017), but others less reported as well (e.g. midbrain)(Lee, et al., 2015). This part of the midbrain corresponds to substantia nigra which is formed by dopaminergic neurons. Very likely few voxel-based machine learning studies (or none) have included this area in the parameter space which is part of the GM segmentation provided by ANTS. There are some groups that promote the idea of early involvement of the brainstem in AD (Simic, et al., 2009). There is also a growing literature that suggests midbrain and dopamine involvement in AD (D’Amelio and Nistico, 2018,D’Amelio, et al., 2018,Koch, et al., 2014,Martorana and Koch, 2014,Nobili, et al., 2017,Uematsu, et al., 2018). However, it should be kept in mind that possibility that the maps are affected by noise coming from errors in the segmentation, normalization process or other causes.
Figure 4 -.
Discriminative maps indicating the areas involved in discrimination of AD from CN derived from the elastic net regularization constraints are shown in neurological convention. Some of the pinpointed areas are amygdala, hippocampus, parahippocampal gyrus, thalamus, inferior temporal lobe areas and midbrain (all bilateral).
DISCUSSION
WHIMS is a cohort where AD biomarkers (e.g. amyloid PET, blood or cerebrospinal fluid biomarkers, etc.) are not available however MRI scans are available for a relatively large number of participants. The long term goal of this work is to provide the WHIMS-MRI database with a metric (AD-PS scores) based on MRI that can be used by WHIMS investigators as an anatomical AD risk factor to address research questions. This is part of an ongoing work aiming at validating the AD-PS scores in the WHIMS-MRI cohort. Here we develop further our previous work (Casanova, et al., 2013) by estimating the AD-PS scores in a second cohort, the WHIMS-MRI study. The scores characterize how similar an individual’s patterns of atrophy are to those of AD patients. Previously in ADNI we found the scores to be associated with age, cognitive status, time to conversion from MCI to AD and physical function as assessed by the functional assessment questionnaire (Casanova, et al., 2013). In addition, a recent report of our group (Espeland, et al., 2018) found the AD-PS scores to be associated with trajectories of cognitive function tests scores in WHIMS-MRI. Women were grouped into five clusters of trajectories using a latent class approach(Jones BL and DS., 2007). AD-PS scores varied significantly among clusters of trajectories with relationships that were more strong and consistent than those for other traditional risk factors (education, diabetes, and APOE-e4 genotype).
In this work we further investigated the constructive validity of the WHIMS-MRI AD-PS scores founding that the WHIMS-MRI AD-PS scores had significant cross-sectional and longitudinal relationships with known risk factors and cognitive status. After adjustment, the AD-PS scores were associated with age, cognitive scores, and WM SVID volumes at baseline. At follow-up, greater age, poorer cognitive test scores, and greater WM SVID volume were associated with higher AD-PS scores. The scores were correlated with age, as expected. However, distinguishing between effects attributable to normal aging versus AD could be challenging. In our case the strong association of the scores with incident impairment and the observation that including AD-PS scores in our survival analyses rendered non-significant the associations of age and other covariates suggest that the AD-PS scores are a stronger risk factor than age. There is growing evidence (Brickman, et al., 2009,Prins, et al., 2004) that increased burden of WM small vessel disease increases the risk of AD. Furthermore, several studies have reported associations of GM tissue atrophy with WM lesion burden in older adults. Bilello et al. investigated the correlation of brain atrophy and white matter lesions with cognitive decline in CN, MCI and AD subjects finding that both were associated with cognitive decline(Bilello, et al., 2015). Three City Study researchers, using data from 1792 adults free of dementia, reported negative associations between WM lesion volumes with GM and hippocampal volumes(Godin, et al., 2009). In a cohort of 740 cognitive normal older adults, GM and WM lesions were found to be inversely correlated (Raji, et al., 2012). Crane et al. reported association between increased WM lesion burden and GM volume losses in several brain areas, including the hippocampus (Crane, et al., 2015). Recent work by Habes and colleagues supports the hypothesis that white matter hyperintensities also contribute to brain atrophy patterns in regions related to Alzheimer’s disease (Habes, et al., 2016a). In this context our work provides further support to this hypothesis based on a large cohort of older women with available baseline and follow-up MRI data. However, we should notice that we did not take any measure to mask out the WM hyperintensities from the images which could be a source of confound in our analyses. Higher baseline AD-PS scores predicted which CN women would transition to poorer cognition. Changes of the AD-PS scores in time (Scan 2 – Scan 1) were also associated with changes in cognitive scores: increases in the AD-PS scores predicted decreases in cognitive scores and increases in WM SVID volume. The scores were also strongly associated with incident cognitive impairment. An increase of 0.1 units in AD-PS scores between MRI scans increased the hazard for cognitive impairment (combined group of MCI/probable dementia cases) was associated to a hazard ratio of 1.51 (1.31–1.75) (see Table 3). There are a few other studies that investigated value of MRI measures to investigate the progression of CN individuals to MCI and AD. Csernansky et al. used hippocampal volume and shape to predict conversion to cognitive impairment in a cohort of 49 individuals followed for 4.9 years (Csernansky, et al., 2005). They reported both measures predicted time to conversion. Toledo et al. investigated in ADNI the risk of conversion from CN to MCI/AD based on combinations of biomarkers, using 326 subjects with 7 years of follow-up. They reported hazard ratios of 3.11 (1.84–5.25) and 1.46 (1.12–1.92) for hippocampal volume and SPARE-AD respectively.(Toledo, et al., 2014). Albert et al. followed a cohort of 224 individuals cognitive normal individuals for an average of 11 years (Albert, et al., 2018). The volumes of the right hippocampus and entorhinal cortex thickness derived from MRI scans were associated with MCI incidence with HRs of 0.728 (0.552 – 0.961) and 0.668 (0.492 – 0.905), respectively.
In addition, other unadjusted results (Table 1) support further the constructive validity of the scores. Women who had only a baseline MRI scan in WHIMS-MRI were older, more likely to be minority, hypertensive and diabetic, and had lower 3MS scores compared with women who had both baseline and follow-up MRI scans. Baseline AD-PS scores were significantly higher for the 653 women who only had baseline MRIs compared with the 712 who had baseline and repeat MRIs. For participants who had both scans, follow-up scores were significantly higher than baseline scores when adjusted by age, education, race and hormone therapy treatment.
We did not attempt accurate classification according to cognitive status in WHIMS-MRI. There are several reasons why classification of participants may not align with the adjudication of probable dementia implemented in WHIMS-MRI. It presents an unbalanced classification problem. The overwhelming majority of WHIMS-MRI participants met study criteria for CN. This poses technical challenges because the overall accuracy is dominated by the intra-class accuracy of the majority group. A relatively large fraction of WHIMS-MRI women meeting study criteria for CN are miss-classified as AD, very likely because they present AD-like patterns but have not progressed sufficiently to meet study adjudication criteria based on cognitive testing. Also only women performing below a fixed threshold of the 3MS scores underwent adjudication process. Women above the threshold were presumed to be CN and not thoroughly tested which in addition could lead to the categorization as CN of MCI cases further confounding classification. Here instead we tackled successfully a less complex problem. Rather than an accurate classifier of cognitive status, in essence we derived a metric (AD-PS scores) in WHIMS-MRI that acts as a “detector” of AD-like patterns in the brain of WHIMS participants. Though we have shown significant statistical associations of the scores with cognitive status in WHIMS-MRI (among several other factors) these statistical associations across groups do not necessarily translate in accurate classification per class.
We used the Advanced Normalization Tools software to put both databases into a common coordinate system. This software warped all images into a customized ADNI template based on a subset of subjects. Many other strategies are possible, but testing all of them is a very time- and storage space-consuming exercise, which was well beyond our resources.
The AD-PS scores can potentially be used as an anatomical AD risk factor in research, since they seem to capture subtle spatial patterns of AD-related tissue atrophy. They could also help to select subjects for clinical trials of AD prevention or treatment (Weiner, et al., 2013) or as a criterion to select participants more or less likely to progress to greater cognitive impairment. These strategies could increase statistical power while saving resources.
To the best of our knowledge, only the SPARE-AD index (Davatzikos, et al., 2009) originally computed for the ADNI dataset has been estimated for one cohort with longitudinal data, BLSA. The SPARE-AD index was associated with age and could discriminate which CN subjects were more likely to convert to MCI or remain stable as CN. The approach presented here differs from that behind the SPARE-AD in several respects. Our classification method is not based on a support vector machine model, but on elastic net regularized logistic regression. We determined cognitive classifications directly in the voxel space (via GM probability maps) taking advantage of the sparsity properties of the elastic net regularization and the speed of the coordinate descent (Friedman, et al., 2009). We used a different image warping approach based on high-dimensional diffeomorphic transformation and cross-database alignment strategy. Finally, here we investigated the feasibility of using ADNI MRI data to estimate the AD-PS scores in WHIMS-MRI. ADNI is a clinical dataset designed for AD research, whereas WHIMS-MRI was a clinical trial focused on hormone therapy, with data collected across multiple sites with different MRI protocols. Our work further demonstrates the value of cross-database machine learning analyses aimed at AD applications, pioneered by others (Davatzikos, et al., 2009,Kloppel, et al., 2008). It also demonstrates the feasibility of performing cross-database high-dimensional machine learning analyses at the voxel level using the elastic net regularization approach.
Our study has several limitations. Although WHIMS-MRI includes only women, we used data from both women and men in ADNI to train our classifiers: this provided a larger sample size that would be expect to increase prediction performance. In addition, the classifiers were trained using MRI data from Caucasians ADNI participants. While the WHIMS-MRI cohort is overwhelmingly composed by Caucasian women (91% at baseline and 93% at follow-up) there are still some participants of other race/ethnicities for whom generalization may be unwarranted until further validation. In WHIMS-MRI relatively few women had adjudicated MCI or dementia, and many women did not have a second MRI. These rates are in line with the full WHIMS cohort (Espeland, et al., 2015). This may be attributable to a healthy volunteer effect, i.e. the women who enrolled in WHIMS were healthier than more general cohorts. As we noted, the WHIMS study shifted to a telephone assessment of cognitive function, so not all data were collected in person.
CONCLUSIONS
In this work we applied a high-dimensional machine learning approach to estimate the AD-PS scores based on GM for WHIMS-MRI study participants, using ADNI imaging data. To investigate the value of the scores as an AD anatomical risk factor we studied their associations with several AD risk factors using WHIMS-MRI data. The scores of AD risk showed associations with incident cognitive impairment, age, cognitive function, cognitive status and WM SVID volume. Our work lends additional support to the utility of this machine learning-derived score. Future work will aim at refinement of the machine learning methodology and its applications to other available imaging databases.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by NIH grant R21AG051113 (Casanova and Chen). Also RC, MAE, KMH and SRR receive funding from the Wake Forest Alzheimer’s Disease Core Center (P30AG049638-01A1). The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. We acknowledge the following WHI investigators: Program Office: (National Heart, Lung, and Blood Institute, Bethesda, Maryland) Jacques Rossouw, Shari Ludlam, Joan McGowan, Leslie Ford, and Nancy Geller. Clinical Coordinating Center: (Fred Hutchinson Cancer Research Center, Seattle, WA) Garnet Anderson, Ross Prentice, Andrea LaCroix, and Charles Kooperberg. Investigators and Academic Centers: (Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) JoAnn E. Manson; (MedStar Health Research Institute/Howard University, Washington, DC) Barbara V. Howard; (Stanford Prevention Research Center, Stanford, CA) Marcia L. Stefanick; (The Ohio State University, Columbus, OH) Rebecca Jackson; (University of Arizona, Tucson/Phoenix, AZ) Cynthia A. Thomson; (University at Buffalo, Buffalo, NY) Jean Wactawski-Wende; (University of Florida, Gainesville/Jacksonville, FL) Marian Limacher; (University of Iowa, Iowa City/Davenport, LA) Jennifer Robinson; (University of Pittsburgh, Pittsburgh, PA) Lewis Kuller; (Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker; (University of Nevada, Reno, NV) Robert Brunner; (University of Minnesota, Minneapolis, MN) Karen L. Margolis. Women’s Health Initiative Memory Study: (Wake Forest University School of Medicine, Winston-Salem, NC) Mark Espeland. We also acknowledge the editorial assistance of Karen Klein, MA, in the Wake Forest Clinical and Translational Science Institute (UL1 TR001420; PI: McClain).
Data collection and sharing for this project was also funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Appendix A
Group template generation
Images from participants in both studies were aligned into a group template created using ADNI MRI images of female participants. Images of 50 CN women (mean age 76.8) from the ADNI study were selected at random to generate a group template. The template generation process consists of four main stages: (1) group template assembly, (2) brain extraction, (3) brain tissue labeling, and (4) generation of the prior probability images.
Group template assembly
uses the antsMultivariateTemplateConstruction.sh ANTs script to build an anatomical template based on the input images (Avants, et al., 2011a). This script first creates an initial group template based on the average of the input images. The input images are then registered and warped to the initial group template. These warped images are then averaged again, creating a second group template image. The original input images are again registered and warped to the refined group template, and the cycle repeats for 16 iterations. The averaged image from the final iteration is the anatomical (T1 space), non-segmented group template.
Brain extraction
uses the antsBrainExtraction.sh script, which strips the skull from the group template image, producing a usable, skull-stripped group template. This procedure registers a brain extraction template (Tustison N.J, et al., 2017,Tustison, et al., 2014) to the group template. The brain extraction template includes a brain extraction mask, which is warped to the group template. The mask is then applied to the group template to produce a skull-stripped version of the group template.
Brain tissue segmentation
uses a multi-atlas segmentation technique (Wang, et al., 2013) via the antsJointLabelFusion.sh ANTs script. This script uses a set of 15 independent anatomical images (“atlases”), each of which is paired with an expert-segmented label image that identifies the tissue type of each voxel in the anatomical image (Tustison, et al., 2014). The atlas images are warped to the group template and the resulting deformations are applied to the label images, which are then combined using the joint label fusion technique referenced above. The atlas images used for brain template segmentation include six tissue types: cerebrospinal fluid (CSF), cortical gray matter, white matter, subcortical gray matter, brain stem, and cerebellum. The group template segmentation is thus labeled with the same tissue types.
Prior probability generation:
Prior probability images depict the prior spatial probability of each voxel being of a particular tissue type. To obtain these from the segmented group template image, we combined two approaches. First, a set of prior probability images for each tissue type is obtained via the antsJointLabelFusion.sh script, which propagates labels from a set of expert-labeled atlases onto the skull-stripped template. Second, the antsCorticalThickness.sh script is used to produce a prior image for the CSF. This additional CSF prior probability image is subtracted from the other tissue type prior probability images to correct for a bias in differentiating between CSF and grey matter, wherein the latter is favored over the former (Tustison, et al., 2014).
The result is a set of prior probability images for the aforementioned tissue types. Several additional prior images are created from combinations of these six, by adding single-tissue prior probability images together using the ANTs ImageMath program. In this report, we use a GM prior probability image that combines cortical gray matter and deep gray matter structures.
Processing of the individual images
Once the ADNI group template was generated, all MRI images from both studies were segmented and warped into the ADNI template. We briefly describe the main steps. The antsCorticalThickness.sh script registers input images to the group template. This script consists of six stages: brain extraction; template registration; tissue segmentation; an optional, improved template registration; cortical thickness estimation; and quality control and summary measurements. To obtain a registration of input images to the group template image, only the first four stages are needed. These stages produce a number of output images and transform files. The output images are all in the space of the original input image; they include a brain extraction mask, an N4 bias field-corrected version of the input image, and segmented posterior probability images for the six tissue types previously described. The transforms include affine transformation matrices and high-dimensional deformation fields for transforming an image from the input subject’s space to the group template space and vice-versa. The output images are warped to the group template space via the antsApplyTransforms program. Finally, we produce a Jacobian determinant image and a log-Jacobian image using the CreateJacobianDeterminantImage program on the high-dimensional deformation field. The GM posterior probability maps from all participants were used as input for the machine learning algorithms that generate the AD-PS scores. The images were thresholded using a mask derived from the group template GM prior probability map retaining voxels above 0.4. This generated a set of 441,678 features. The images were not modulated nor smoothed. Intracranial volume is estimated from the brain extraction mask in the space of the original input image. This mask is a binary image in which voxels corresponding to brain tissue are assigned a value of one and all other voxels are assigned a value of zero. Thus, the sum of all of the mask’s voxels gives the number of voxels corresponding to brain tissue. Multiplying the number of brain tissue voxels by the image’s voxel volume gives the total volume of brain tissue.
Implementation details
For these analyses, we used a cluster of over 1500 CPUs available at Wake Forest School of Medicine. ANTs provides support for several parallelization mechanisms (e.g. Sun Grid Engine, pexec, Apple Xgrid, and PBS/Torque); our cluster uses the SLURM resource scheduler, which was not supported by ANTs. However, because of the open source philosophy of the ANTs developers, we could adapt their code to support parallelization based on SLURM. These enhancements were contributed back to the core ANTs development team by way of a GitHub pull request, which was merged into the main code base (https://github.com/stnava/ANTs/pull/230). SLURM support is now available to the ANTs user community.
Appendix B
Machine learning methodology
The RLR method used here is based on the implementation provided by the GLMNET library (Friedman, et al., 2010), which uses a very efficient optimization technique called coordinate-wise descent technique (Friedman, et al., 2007). The general form of the optimization problem solved by the library is of the form:
| (1) |
| (2) |
| (3) |
where xi ∈ Rp is the ith sample or feature vector containing the ith participant MRI data, p is the number of variables (voxels) entering the analysis, yi ∈ {0,1} is the ith label (0 for cognitively normal participants, 1 for participants with Alzheimer’s disease),β0,β ∈ Rp are the parameters of the model, and λ is the regularization parameter. The regularization scheme described by Eq.(1) contains two terms: a loss term C(β0,β,x,y) and a penalty term P called elastic net penalty, which is given by Eq.(3). The regularization parameter λ establishes a trade-off between the two terms and it is determined from the data using cross-validation combined with grid search. Our software implementation is based on MATLAB, where the GLMNET library is called using a freely available MATLAB wrapper developed by Hui Jiang (http://www-stat.stanford.edu/~tibs/glmnet-matlab/).
2.6 Optimization of regularization parameters
To estimate the optimal values of the regularization parameters, we combined a three-way split of the data (training-validation-testing) with 10-fold cross-validations (CV) and grid search. This was done to avoid upward bias in the metrics of performance estimates (Casanova, et al., 2011,Ryali, et al., 2011,Zhang, et al., 2011). We implemented an external K1-fold CV where at each step we leave one fold for testing and use the remaining K1-1 folds for training and validation. These last two procedures are implemented by using a nested K2-fold CV. We divided the K1-1 folds into K2 folds and we left one fold for validation and K2-1 folds for training combined with a grid search to determine the optimal parameters. The grid we used in our analyses was λ = 0.5,1,5,10,11,12…98,99,100,200,500,1000. For the sMRI data, we fixed in advance one of the regularization parameters (α = 0.001) and optimized the second. We have observed in practice working with high-dimensional imaging data that this choice works well, avoiding the heavier computational burden related to the optimization of both parameters while still producing a sparse solution (R Casanova, et al., 2012). At each grid point, the classifier is trained and its performance is assessed using the fold left for validation by estimating the classification accuracy. We select the regularization parameters that produce maximum average accuracy across the K2 folds of the internal CV procedure. The classifier is then retrained using the data in the K1-1 folds left for training and validation and the selected optimal regularization parameters. The classifier’s generalization capability is then evaluated by computing the classification accuracy, sensitivity and specificity using the fold originally left for testing in the external CV. This is repeated K1 times and the average classification accuracy is reported. In our analyses we used K1= 10 and K2= 10.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- Albert M, Zhu Y, Moghekar A, Mori S, Miller MI, Soldan A, Pettigrew C, Selnes O, Li S, Wang MC 2018. Predicting progression from normal cognition to mild cognitive impairment for individuals at 5 years. Brain : a journal of neurology. doi: 10.1093/brain/awx365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association; 1994. Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition: DSM-IV. American Psychiatric Association, Washington, DC. [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis 12(1), 26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC 2011a. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54(3), 2033–44. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC 2011b. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9(4), 381–400. doi: 10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC 2010. The optimal template effect in hippocampus studies of diseased populations. NeuroImage 49(3), 2457–66. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello M, Doshi J, Nabavizadeh SA, Toledo JB, Erus G, Xie SX, Trojanowski JQ, Han X, Davatzikos C 2015. Correlating Cognitive Decline with White Matter Lesion and Brain Atrophy Magnetic Resonance Imaging Measurements in Alzheimer’s Disease. Journal of Alzheimer’s disease : JAD 48(4), 987–94. doi: 10.3233/JAD-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E 1991. Alzheimer’s disease affects limbic nuclei of the thalamus. Acta neuropathologica 81(3), 261–8. [DOI] [PubMed] [Google Scholar]
- Brandt J, Spencer M, Folstein M 1988. The Telephone Interview for Cognitive Status. Neuropsychiatry, Neuropsychology, and Behavioral Neurology 1, 111–7. [Google Scholar]
- Brickman AM, Muraskin J, Zimmerman ME 2009. Structural neuroimaging in Altheimer’s disease: do white matter hyperintensities matter? Dialogues in clinical neuroscience 11(2), 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova R, Hsu F-C, Espeland MA 2012. Classification of structural MRI images in Alzheimer’s disease from the perspective of ill-posed problems. PLOS One 7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova R, Hsu FC, Espeland MA, Alzheimer’s Disease Neuroimaging, I. 2012. Classification of structural MRI images in Alzheimer’s disease from the perspective of ill-posed problems. PloS one 7(10), e44877. doi: 10.1371/journal.pone.0044877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova R, Hsu FC, Sink KM, Rapp SR, Williamson JD, Resnick SM, Espeland MA, Alzheimer’s Disease Neuroimaging, I. 2013. Alzheimer’s disease risk assessment using large-scale machine learning methods. PloS one 8(11), e77949. doi: 10.1371/journal.pone.0077949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova R, Whitlow CT, Wagner B, Williamson J, Shumaker SA, Maldjian JA, Espeland MA 2011. High dimensional classification of structural MRI Alzheimer’s disease data based on large scale regularization. Frontiers of Neuroscience in Neuroinformatics 5:22 Epub 2011 Oct 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker LH, Espeland MA, Hogan PE, Resnick SM, Bryan RN, Robinson JG, Goveas JS, Davatzikos C, Kuller LH, Williamson JD, Bushnell CD, Shumaker SA, Group W-MS 2014. Change in brain and lesion volumes after CEE therapies: the WHIMS-MRI studies. Neurology 82(5), 427–34. doi: 10.1212/WNL.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker LH, Hogan PE, Bryan NR, Kuller LH, Margolis KL, Bettermann K, Wallace RB, Lao Z, Freeman R, Stefanick ML, Shumaker SA 2009. Postmenopausal hormone therapy and subclinical cerebrovascular disease: the WHIMS-MRI Study. Neurology 72(2), 125–34. doi: 10.1212/01.wnl.0000339036.88842.9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, D. O. 1984. Analysis of Survival Data. Chapman and Hall, London. [Google Scholar]
- Crane DE, Black SE, Ganda A, Mikulis DJ, Nestor SM, Donahue MJ, MacIntosh BJ 2015. Gray matter blood flow and volume are reduced in association with white matter hyperintensity lesion burden: a cross-sectional MRI study. Frontiers in aging neuroscience 7, 131. doi: 10.3389/fnagi.2015.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC 2005. Preclinical detection of Alzheimer’s disease: hippocampal shape and volume predict dementia onset in the elderly. NeuroImage 25(3), 783–92. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- D’Amelio M, Nistico R 2018. Unlocking the secrets of dopamine in Alzheimer’s Disease. Pharmacological research 128, 399. doi: 10.1016/j.phrs.2017.06.018. [DOI] [PubMed] [Google Scholar]
- D’Amelio M, Puglisi-Allegra S, Mercuri N 2018. The role of dopaminergic midbrain in Alzheimer’s disease: Translating basic science into clinical practice. Pharmacological research. doi: 10.1016/j.phrs.2018.01.016. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Xu F, An Y, Fan Y, Resnick SM 2009. Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain 132(Pt 8), 2026–35. doi: 10.1093/brain/awp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeland MA, Brinton RD, Hugenschmidt C, Manson JE, Craft S, Yaffe K, Weitlauf J, Vaughan L, Johnson KC, Padula CB, Jackson RD, Resnick SM, Group WS 2015. Impact of Type 2 Diabetes and Postmenopausal Hormone Therapy on Incidence of Cognitive Impairment in Older Women. Diabetes care 38(12), 2316–24. doi: 10.2337/dc15-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeland MA, Chen JC, Weitlauf J, Hayden KM, Rapp SR, Resnick SM, Garcia L, Cannell B, Baker LD, Sachs BC, Tindle HA, Wallace R, Casanova R, Women’s Health Initiative Memory Study Magnetic Resonance Imaging Study, G. 2018. Trajectories of Relative Performance with 2 Measures of Global Cognitive Function. Journal of the American Geriatrics Society. doi: 10.1111/jgs.15431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeland MA, Rapp SR, Shumaker SA, Brunner R, Manson JE, Sherwin BB, Hsia J, Margolis KL, Hogan PE, Wallace R, Dailey M, Freeman R, Hays J, Women’s Health Initiative Memory, S. 2004. Conjugated equine estrogens and global cognitive function in postmenopausal women: Women’s Health Initiative Memory Study. Jama 291(24), 2959–68. doi: 10.1001/jama.291.24.2959. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Hofling H, Tibshirani R 2007. PATHWISE COORDINATE OPTIMIZATION. The Annals of Applied Statistics 1(2), 302–32. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R 2009. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software, 1–24. [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R 2010. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software 33(1), 1–22. [PMC free article] [PubMed] [Google Scholar]
- Godin O, Maillard P, Crivello F, Alperovitch A, Mazoyer B, Tzourio C, Dufouil C 2009. Association of white-matter lesions with brain atrophy markers: the three-city Dijon MRI study. Cerebrovascular diseases 28(2), 177–84. doi: 10.1159/000226117. [DOI] [PubMed] [Google Scholar]
- Habes M, Erus G, Toledo JB, Zhang T, Bryan N, Launer LJ, Rosseel Y, Janowitz D, Doshi J, Van der Auwera S, von Sarnowski B, Hegenscheid K, Hosten N, Homuth G, Volzke H, Schminke U, Hoffmann W, Grabe HJ, Davatzikos C 2016a. White matter hyperintensities and imaging patterns of brain ageing in the general population. Brain : a journal of neurology 139(Pt 4), 1164–79. doi: 10.1093/brain/aww008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habes M, Janowitz D, Erus G, Toledo JB, Resnick SM, Doshi J, Van der Auwera S, Wittfeld K, Hegenscheid K, Hosten N, Biffar R, Homuth G, Volzke H, Grabe HJ, Hoffmann W, Davatzikos C 2016b. Advanced brain aging: relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Translational psychiatry 6, e775. doi: 10.1038/tp.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR Jr., Weiner MW, Thompson PM, Alzheimer’s Disease Neuroimaging, I. 2008. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. NeuroImage 43(3), 458–69. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr., Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW 2008. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of magnetic resonance imaging : JMRI 27(4), 685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo SA, Felton D, Andrews L, Desiderio L, Hallarn RK, Jackson SD, Coker LH, Robinson JG, Ockene JK, Espeland MA, Women’s Health Initiative Memory Study Research, G. 2007. Enrollment in a brain magnetic resonance study: results from the Women’s Health Initiative Memory Study Magnetic Resonance Imaging Study (WHIMS-MRI). Acad Radiol 14(5), 603–12. doi: 10.1016/j.acra.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BL, DS. N 2007. Advances in group-based trajectory modeling and a SAS procedure for estimating them. Sociological Meth Res 35, 542–74. [Google Scholar]
- Kawas C, Segal J, Stewart WF, Corrada M, Thal LJ 1994. A validation study of the Dementia Questionnaire. Arch Neurol 51(9), 901–6. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV 2009. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 46(3), 786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR Jr., Ashburner J, Frackowiak RS 2008. Automatic classification of MR scans in Alzheimer’s disease. Brain 131(Pt 3), 681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G, Di Lorenzo F, Bonni S, Giacobbe V, Bozzali M, Caltagirone C, Martorana A 2014. Dopaminergic modulation of cortical plasticity in Alzheimer’s disease patients. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 39(11), 2654–61. doi: 10.1038/npp.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Z, Shen D, Liu D, Jawad AF, Melhem ER, Launer LJ, Bryan RN, Davatzikos C 2008. Computer-assisted segmentation of white matter lesions in 3D MR images using support vector machine. Academic radiology 15(3), 300–13. doi: 10.1016/j.acra.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launer LJ, Miller ME, Williamson JD, Lazar RM, Gerstein HC, Murray AM, Sullivan M, Horowitz KR, Ding J, Marcovina S, Lovato LC, Lovato J, Margolis KL, O’Connor P, Lipkin EW, Hirsch J, Coker L, Maldjian J, Sunshine JL, Truwit C, Davatzikos C, Bryan RN, investigators AM 2011. Effects of intensive glucose lowering on brain structure and function in people with type 2 diabetes (ACCORD MIND): a randomised open-label substudy. The Lancet Neurology 10(11), 969–77. doi: 10.1016/S1474-4422(11)70188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ryan J, Andreescu C, Aizenstein H, Lim HK 2015. Brainstem morphological changes in Alzheimer’s disease. Neuroreport 26(7), 411–5. doi: 10.1097/WNR.0000000000000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorana A, Koch G 2014. “Is dopamine involved in Alzheimer’s disease?”. Frontiers in aging neuroscience 6, 252. doi: 10.3389/fnagi.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Gage FH 2011. Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Molecular neurodegeneration 6, 85. doi: 10.1186/1750-1326-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V, Vercauteren T, Ayache N, Commowick O, Malandain G, Glocker B, Paragios N, Navab N, Gorbunova V, Sporring J, de Bruijne M, Han X, Heinrich MP, Schnabel JA, Jenkinson M, Lorenz C, Modat M, McClelland JR, Ourselin S, Muenzing SE, Viergever MA, De Nigris D, Collins DL, Arbel T, Peroni M, Li R, Sharp GC, Schmidt-Richberg A, Ehrhardt J, Werner R, Smeets D, Loeckx D, Song G, Tustison N, Avants B, Gee JC, Staring M, Klein S, Stoel BC, Urschler M, Werlberger M, Vandemeulebroucke J, Rit S, Sarrut D, Pluim JP 2011. Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE transactions on medical imaging 30(11), 1901–20. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- Nobili A, Latagliata EC, Viscomi MT, Cavallucci V, Cutuli D, Giacovazzo G, Krashia P, Rizzo FR, Marino R, Federici M, De Bartolo P, Aversa D, Dell’Acqua MC, Cordella A, Sancandi M, Keller F, Petrosini L, Puglisi-Allegra S, Mercuri NB, Coccurello R, Berretta N, D’Amelio M 2017. Dopamine neuronal loss contributes to memory and reward dysfunction in a model of Alzheimer’s disease. Nature communications 8, 14727. doi: 10.1038/ncomms14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR Jr., Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW 2010. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 74(3), 201–9. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E 1999. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol 56(3), 303–8. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Stevens JC, Ganguli M, Tangalos EG, Cummings JL, DeKosky ST 2001. Practice parameter: early detection of dementia: mild cognitive impairment (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology 56(9), 1133–42. [DOI] [PubMed] [Google Scholar]
- Plassman BL, Newman TT, Welsh KA, Helms M, Breitner JCS 1994. Properties of the Telephone Interview for Cognitive Status. Application in epidemiological and longitudinal studies. Neuropsychiatry, Neuropsychology, and Behavioral Neurology 7, 235–41. [Google Scholar]
- Poulin SP, Dautoff R, Morris JC, Barrett LF, Dickerson BC, Alzheimer’s Disease Neuroimaging, I. 2011. Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry research 194(1), 7–13. doi: 10.1016/j.pscychresns.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins ND, van Dijk EJ, den Heijer T, Vermeer SE, Koudstaal PJ, Oudkerk M, Hofman A, Breteler MM 2004. Cerebral white matter lesions and the risk of dementia. Arch Neurol 61(10), 1531–4. doi: 10.1001/archneur.61.10.1531. [DOI] [PubMed] [Google Scholar]
- Raji CA, Lopez OL, Kuller LH, Carmichael OT, Longstreth WT Jr., Gach HM, Boardman J, Bernick CB, Thompson PM, Becker JT 2012. White matter lesions and brain gray matter volume in cognitively normal elders. Neurobiology of aging 33(4), 834 e7–16. doi: 10.1016/j.neurobiolaging.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp SR, Legault C, Espeland MA, Resnick SM, Hogan PE, Coker LH, Dailey M, Shumaker SA, Group CATS 2012. Validation of a cognitive assessment battery administered over the telephone. J Am Geriatr Soc 60(9), 1616–23. doi: 10.1111/j.1532-5415.2012.04111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Espeland MA, Jaramillo SA, Hirsch C, Stefanick ML, Murray AM, Ockene J, Davatzikos C 2009. Postmenopausal hormone therapy and regional brain volumes: the WHIMS-MRI Study. Neurology 72(2), 135–42. doi: 10.1212/01.wnl.0000339037.76336.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics 12, 77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryali S, Supekar K, Abrams DA, Menon V 2011. Sparse logistic regression for whole-brain classification of fMRI data. Neuroimage 51(2), 752–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker SA, Reboussin BA, Espeland MA, Rapp SR, McBee WL, Dailey M, Bowen D, Terrell T, Jones BN 1998. The Women’s Health Initiative Memory Study (WHIMS): a trial of the effect of estrogen therapy in preventing and slowing the progression of dementia. Controlled clinical trials 19(6), 604–21. [DOI] [PubMed] [Google Scholar]
- Simic G, Stanic G, Mladinov M, Jovanov-Milosevic N, Kostovic I, Hof PR 2009. Does Alzheimer’s disease begin in the brainstem? Neuropathology and applied neurobiology 35(6), 532–54. doi: 10.1111/j.1365-2990.2009.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng EL, Chui HC 1987. The Modified Mini-Mental State (3MS) examination. J Clin Psychiatry 48(8), 314–8. [PubMed] [Google Scholar]
- Toledo JB, Da X, Bhatt P, Wolk DA, Arnold SE, Shaw LM, Trojanowski JQ, Davatzikos C, Alzheimer’s Disease Neuroimaging, I. 2013. Relationship between plasma analytes and SPARE-AD defined brain atrophy patterns in ADNI. PloS one 8(2), e55531. doi: 10.1371/journal.pone.0055531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Weiner MW, Wolk DA, Da X, Chen K, Arnold SE, Jagust W, Jack C, Reiman EM, Davatzikos C, Shaw LM, Trojanowski JQ, Alzheimer’s Disease Neuroimaging, I. 2014. Neuronal injury biomarkers and prognosis in ADNI subjects with normal cognition. Acta neuropathologica communications 2, 26. doi: 10.1186/2051-5960-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Holbrook AJ, Avants BB, Roberts JM, Cook PA, Stone JA, Gillen ML,Y 2017. The ANTs Longitudinal Cortical Thickness Pipeline 13th ADPD Conference, Viena. [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC 2010. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging 29(6), 1310–20. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Cook PA, Klein A, Song G, Das SR, Duda JT, Kandel BM, van Strien N, Stone JR, Gee JC, Avants BB 2014. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. NeuroImage 99, 166–79. doi: 10.1016/j.neuroimage.2014.05.044. [DOI] [PubMed] [Google Scholar]
- Uematsu M, Nakamura A, Ebashi M, Hirokawa K, Takahashi R, Uchihara T 2018. Brainstem tau pathology in Alzheimer’s disease is characterized by increase of three repeat tau and independent of amyloid beta. Acta neuropathologica communications 6(1), 1. doi: 10.1186/s40478-017-0501-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA 2013. Multi-Atlas Segmentation with Joint Label Fusion. IEEE transactions on pattern analysis and machine intelligence 35(3), 611–23. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Saykin AJ, Schmidt ME, Shaw L, Shen L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ, Alzheimer’s Disease Neuroimaging, I. 2013. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 9(5), e111–94. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh KA, Breitner J, Magruder-Habib KM 1993. Detection of dementia in the elderly using the telephone interview for cognitive status. Neuropsychiatry, Neuropsychology, & Behavioral Neurology 6, 103–10. [Google Scholar]
- Zanchi D, Giannakopoulos P, Borgwardt S, Rodriguez C, Haller S 2017. Hippocampal and Amygdala Gray Matter Loss in Elderly Controls with Subtle Cognitive Decline. Frontiers in aging neuroscience 9, 50. doi: 10.3389/fnagi.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D, Alzheimer’s Disease Neuroimaging, I. 2011. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage 55(3), 856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.