Abstract
Invariant Natural Killer T (iNKT) cells are innate lipid-reactive T cells that develop and differentiate in the thymus into iNKT1/2/17 subsets, akin to TH1/2/17 conventional CD4 T cell subsets. The factors driving the central priming of iNKT cells remain obscure, although strong/prolonged TCR signals appear to favor iNKT2 cell development. The Src homology domain-containing phosphatase 1 (Shp1) is a protein tyrosine phosphatase that has been identified as a negative regulator of TCR signaling. In this study, we found that mice with a T cell-specific deletion of Shp1 had normal iNKT cell numbers and peripheral distribution. However, iNKT cell differentiation was biased towards the iNKT2/17 subsets in the thymus, but not in peripheral tissues. Shp1-deficient iNKT cells were also functionally biased towards the production of TH2 cytokines, such as IL-4 and IL-13. Surprisingly, we found no evidence that Shp1 regulates the TCR and Slamf6 signaling cascades, which have been suggested to promote iNKT2 differentiation. Rather, Shp1 dampened iNKT cell proliferation in response to IL-2, IL-7 and IL-15, but not following TCR engagement. Our findings suggest that Shp1 controls iNKT cell effector differentiation independently of positive selection through the modulation of cytokine responsiveness.
INTRODUCTION
iNKT cells recognize self and foreign lipid antigens presented on the MHC Class Ib molecule CD1d and have been shown to play protective or deleterious functions in many diseases due to their capacity to rapidly secrete large amounts of cytokines and chemokines following antigen encounter(1). iNKT cell ontogeny occurs in the thymus and requires thymocyte-thymocyte interactions at the double positive (DP) stage, which provide signals mediated by the TCR(2) and by members of the signaling lymphocytic-activation molecule (SLAM) family, especially Slamf6 (Ly108) and Slamf1 (CD150) through their adaptor molecule SAP(3–5). iNKT cell development relies on strong or agonist TCR signals, similarly to other unconventional T cells such as Foxp3+ regulatory T (TREG) cells, γδ T cells, and CD8αα+ intraepithelial lymphocytes (IELs) (for review(6–10)). These stronger than normal TCR signals(11) impart iNKT cells with an effector/memory phenotype that is consistent with their innate effector functions, and largely governed by the expression of the transcription factor PLZF (promyelocytic leukemia zinc finger, Zbtb16)(12, 13).
iNKT cells appear to be “primed” in the thymus and functionally differentiate into discrete subsets that preferentially secrete TH1 (iNKT1), TH2 (iNKT2) and TH17 (iNKT17) cytokines(14). iNKT cell subsets can be identified by differential expression of PLZF as well as the other signature transcription factors T-bet, RORγt and to a lower extent GATA-3(8, 14). Although the factors controlling the differentiation of the various iNKT cell subsets are only poorly understood, it is suspected that TCR signal strength and duration plays a central role(14–17). In parallel, studies from multiple groups have shown that co-engagement of the TCR and Slamf6 enhances the expression of the early growth response (Egr)-2 and PLZF transcription factor in pre-selection double positive thymocytes (PSDPs)(18–20), which favors the iNKT2 effector fate(21). Several cell-intrinsic factors that impact TCR signaling and/or PLZF expression have been shown to influence iNKT cell selection or effector differentiation. These include several microRNAs(22, 23), the lipid phosphatase PTEN and other factors of the PI3K pathway(24), several components of the autophagy pathway such as mammalian target of rapamycin (mTOR)(25–27), the E protein transcription factor HEB and its negative regulators Id2 and Id3(28–30). As for extrinsic factors, certain cytokines such as IL-7 and IL-15 are necessary for iNKT cell homeostasis(31, 32), but their role in effector differentiation is unclear. Finally, the chemokine receptor CCR7 has been shown to drive iNKT cells from the thymic cortex into the medulla(33), but its role in iNKT cell maturation or effector differentiation has not been fully elucidated.
Tyrosine phosphorylation and dephosphorylation of target proteins by specific protein kinases and protein phosphatases is a central feature of signal transduction. The Src homology region 2 domain-containing phosphatase (Shp)-1 is a protein tyrosine phosphatase (encoded by the Ptpn6 gene) expressed in all hematopoietic cells, and plays important functions in T cell development and function(34). Shp1 is primarily considered to be a key negative regulator of TCR signaling(35), as well as many other immune receptors such as the B cell receptor(36), natural killer (NK) receptors(37, 38), chemokine and cytokine receptors(39, 40), SLAM receptors(20, 41), the death receptor FAS and integrins(37, 38). The role of Shp1 in signal transduction has been widely studied through the use of various strains of mice carrying partial or total loss-of-function mutations at the Ptpn6 locus (motheaten, motheaten viable and spin)(34, 42–44). These mice suffer from severe systemic inflammation and autoimmunity, which is a confounding and limiting factor. The recent development of cell-specific deletions of Shp1 has shed new light on its function in immune cells. For example, Shp1 deletion in B cells or dendritic cells (DCs) resulted in autoimmunity whereas Shp1 deletion in neutrophils led to cutaneous inflammation(45). T cell-specific deletion of Shp1 did not lead to autoimmunity or inflammation. Instead, these mice accumulated innate memory CD4 and CD8 T cells(46, 47). Recent work from Johnson et al. suggested that this phenotype was due to enhanced IL-4 sensitivity in T cells in the absence of Shp1(47). Overall, these studies argue that Shp1 has a minor(48), negligible(47) or even surprisingly positive(49) role in TCR signaling.
The agonistic nature of iNKT cell selection raised the question whether Shp1 controls the ontogeny and differentiation of these cells. One study by the Casorati and Dellabona groups used mice transgenic for the human CD1d molecule as well as heterozygous motheaten mice to suggest that Shp1 expression in iNKT cells prevents their hyperactivation in response to exogenous glycolipid antigens(50). To avoid extrinsic confounding factors, we characterized iNKT cell development and function using a T cell-specific Shp1 deletion (Shp1fl/fl CD4-cre mice). Although Shp1fl/fl CD4-cre mice had normal numbers of iNKT cells in all the tissues tested, they had a cell-intrinsic bias towards iNKT2 and iNKT17 cells in the thymus, but not in peripheral tissues. Shp-1-deficient iNKT cells from the thymus and spleen also had a functional bias towards a TH2 response upon activation in vitro and in vivo. Shp1-deficiency did not impact signaling downstream of the TCR or Slamf6 as previously proposed. Rather, Shp1 regulated iNKT cell proliferation in response to multiple cytokines. We propose a model whereby Shp1 regulates cytokine receptor signaling in iNKT cells to regulate their effector differentiation and functional responses.
MATERIALS AND METHODS
Mice and reagents
Mice were used between 6–8 weeks of age, unless otherwise specified. C57BL/6 (B6) wild-type (WT), CD4-creERT2 and RAG1−/− mice were purchased from Jackson Laboratories. Shp1fl/fl and Shp1fl/fl CD4-cre mice were generously provided by Dr. Benjamin Neel (Princess Margaret Cancer Center, Canada; UofT mice) and Dr. Toshiaki Kawakami (La Jolla Institute for Immunology, USA; LJI mice). As these mice have a mixed 129/C57BL/6 genetic background, littermate mice were analyzed with the exception of Fig. 1D, where CD45.1 WT non-littermate mice were used. CD1d−/− mice were generated and generously provided by Dr. Chyung-Ru Wang (Northwestern University, USA) (51). SAP−/− and CD5−/− mice were provided by Dr. Andre Veillette (Institut de Recherches Cliniques de Montréal, Canada) and Dr. Daniel Hawiger (Saint Louis University School of Medicine, St. Louis, MO, USA), respectively. All strains were housed at the Division of Comparative Medicine, University of Toronto animal facility and the La Jolla Institute for Immunology (LJI) vivarium, under specific-pathogen free conditions. All animal procedures were approved by the Faculty of Medicine and Pharmacy Animal Care Committee at the University of Toronto (Animal use protocols 20010135, 20010715, 20011113, 20011656), or by the LJI Institutional Animal Care and Use Committee. α-galactosylceramide (KRN7000, αGC) was purchased from Diagnocine. Antibodies used were purchased from eBiosciences, Biolegend or BD Biosciences. PBS57-loaded and unloaded biotinylated CD1d monomers were obtained from the NIH Tetramer Core Facility, and were tetramerized by addition of fluorochrome-conjugated streptavidin. For stimulation assays, mouse CD1d monomers were purified from the culture supernatant of transduced HEK293 cells obtained from the NIH Tetramer Core facility, using affinity chromatography.
Figure 1.
Shp1-deficient iNKT cells are biased towards iNKT2 and iNKT17 subsets. (A) Frequency (top row) and absolute numbers (bottom row) of iNKT cells in the indicated tissues of Shp1fl/fl and Shp1fl/fl CD4-cre mice. (B, C) iNKT cell subsets in the thymus (B) and spleen (C) of Shp1fl/fl (fl/fl) and Shp1fl/fl CD4-cre (cre) mice were assessed by PLZF and RORγt staining. (D) Frequency of iNKT cells in the thymus of single or mixed (mix) bone marrow chimeras. Rag1−/− mice were reconstituted with wild type CD45.1 and/or Shp1fl/fl CD4-cre CD45.2 bone marrow cells. (E, F) Frequency of iNKT2 and iNKT17 subsets in the thymus (E) and spleen (F) of 16-week-old Shp1fl/fl and Shp1fl/fl CD4-cre mice. Data shows representative dot plots, individual mice and the mean values +/− s.e.m. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student t test.
Bone marrow chimeras and adoptive transfer
Bone marrow was collected from femur and tibia of C57BL/6 and Shp1fl/fl CD4-cre mice, and injected i.v. either separately or in a 1:1 ratio into lethally irradiated (2 × 450 cGy) RAG1−/− recipient mice. Mice were analyzed 7 weeks post-transfer. For adoptive transfer experiments, magnetically-enriched thymic or splenic iNKT cells were injected i.v. into recipient mice. 5 days post-transfer, iNKT cells were identified using anti-TCRβ antibodies and PBS57-loaded CD1d tetramers by flow cytometry.
Flow cytometry and cell sorting
Cells were stained with Aqua Live/Dead (Life Technologies) in PBS for 30 min at room temperature (RT), and stained with antibodies and tetramers for 45 min in MACS buffer (PBS containing 0.5% fecal calf serum and 2 mM EDTA). For transcription factor staining, cells were fixed and permeabilized using the FoxP3/transcription factor buffer set (eBiosciences). For cytokine staining, cells were fixed and permeabilized using the Cytofix/Cytoperm buffer set (BD Biosciences). Cells were analyzed using the LSR Fortessa (BD Biosciences). For cell sorting, thymocytes and splenocytes were stained with APC conjugated PBS57 tetramers. iNKT cells were enriched from these preparations using an APC positive selection kit according to the manufacturer’s protocol (StemCell Technologies). Examples of the gating strategies used are depicted in Supplementary Fig. 1. For adoptive transfer experiments, iNKT cells were magnetically enriched using StemCell kits and identified in recipient mice by PBS57-loaded CD1d tetramer staining by flow cytometry using LSR Fortessa (BD Biosciences). iNKT cells were sorted as TCRβ+ CD1d-tetramer+ cells using a FACSAria (BD Biosciences).
In vitro and in vivo proliferation
Sorted iNKT cells were labelled with 5 ng/ml CFSE, washed and cultured for 4 days in 96-well plates in complete RPMI medium supplemented with 10 ng/ml of IL-2, IL-7 or IL-15. In parallel, 24-well plates were coated with 10 μg/ml of polyclonal anti-hamster antibody (Life Technologies) for 2 h at 37°C, washed, and incubated with the indicated concentrations of anti-CD3 antibody alone or together with anti-CD3 antibodies for 2 h at 37°C. Plates were washed and 3.3 × 106 CFSE-labelled splenocytes were added and cultured for 72 h. Cells were analyzed by flow cytometry. Shp1fl/fl and Shp1fl/fl CD4-cre mice received 3 consecutive daily intraperitoneal injections of 1 mg BrdU. BrdU incorporation by thymic and splenic iNKT cells and CD44high CD8 T cells was analyzed on day 4, using the APC BrdU Flow Kit (BD Biosciences), according to the manufacturer’s specifications.
Stimulation assays
Mice were injected with 0.5 μg αGC, and spleen and liver cells were collected after 90 min. Shp1fl/fl and Shp1fl/fl CD4-creERT2 received three consecutive intraperitoneal injections of 1 mg tamoxifen on days 0, 2 and 4. On day 11, mice were injected with 0.2 μg αGC, and spleen cells were collected after 90 min. For in vitro stimulations, 96-well flat-bottom plates were coated with 10 μg/ml of mouse CD1d monomers for 1 h at 37°C, washed and incubated with 5 μg/ml αGC in PBS containing 0.05% tyloxapol overnight. 5 × 105 thymocytes or splenocytes were stimulated by these CD1d/aGC complexes, or PMA and ionomycin, for 6 h at 37°C, with addition of Golgi stop after 2 h. In parallel, cells were stimulated with 100 ng/ml phorbol 12-myristate 13-acetate (PMA) and 5 μg/ml ionomycin. Cells were then analyzed by flow cytometry. To measure Nur77 and Egr2 upregulation, 96-well flat-bottom plates were coated with different concentrations of anti-CD3 antibody (clone 2C11) for 2 h at 37°C. 5 × 105 thymocytes or splenocytes were then added to each well and incubated for 2 h at 37°C. For co-culture experiments, bone marrow cells were cultured for 9 days in medium supplemented with 200 ng/ml recombinant FLT3L. 5 × 104 BMDCs and 5 × 104 sorted iNKT cells were co-cultured in 200 μl complete RPMI with isotype control, anti-CD25 or anti-CD127 antibodies (40 μg/ml) for 48 h. Cytokines were assessed using ProcartaPlex™ immunoassays (ThermoFisher Scientific) and a Luminex Magpix. In some experiments, IL-4 was assessed by ELISA using antibody pairs purchased from eBioscience (ThermoFisher Scientific), following standard procedures.
Egr2 and PLZF upregulation assay
Pre-selection double positive thymocytes (PSDPs) were enriched and stimulated as previously described(19). Flat-bottom 96-well plates were coated with 1 μg/ml anti-CD3 and 5 μg/ml of either anti-Slamf6, anti-CD28 or isotype control antibodies overnight. Plates were washed and seeded with 5 × 105 PSDPs for 30 min or 48 h to measure upregulation of Egr2 and PLZF, respectively. To measure downregulation of Egr2 expression, PSDPs were stimulated with anti-CD3 and anti-Slamf6 for 30 min, washed and seeded in complete RPMI medium for the indicated times.
Data Analysis
Flow cytometry data were analyzed using FlowJo (Tree Star). Statistical analysis was performed using GraphPad Prism. Statistical tests are indicated for each figure and were selected based on the normality test for each data set.
RESULTS
Cell intrinsic deletion of Shp1 biases iNKT cell effector differentiation
To investigate how Shp1 affects iNKT cell development and function, we took advantage of Shp1fl/fl CD4-cre mice in which Shp1 is deleted in αβ T cells at the DP stage of thymic development(47). Because iNKT cells are selected at the DP stage, the Ptpn6 locus is excised upon CD4 expression. We confirmed that the Ptpn6 gene was deleted in splenic iNKT cells from Shp1fl/fl CD4-cre mice (Supplementary Fig. 2A). Importantly, absence of Shp1 at the DP stage did not affect expression of CD1d or the SLAM family receptors SLAMF1 and SLAMF6, which are all essential for iNKT cell development and function (Supplementary Fig. 2B). Furthermore, CD1d expression levels on splenic CD11b+ and CD11c+ antigen-presenting cells were not affected in Shp1fl/fl CD4-cre mice (Supplementary Fig 2C, D). We found no difference in relative and absolute iNKT cell numbers between Shp1fl/fl and Shp1fl/fl CD4-cre mice in all tissues tested, as previously reported(52), with the exception of a significantly higher frequency in the inguinal lymph nodes of Shp1fl/fl CD4-cre mice (Fig. 1A). This indicated that iNKT cell selection and homing were largely unaltered. Upon or after positive selection, iNKT cells differentiate into IFN-γ-producing (iNKT1), IL-4-producing (iNKT2) or IL-17-producing (iNKT17) cells that can significantly alter the function of surrounding cells(14). These iNKT cell subsets can be identified as PLZFlo RORγt™ (iNKT1), PLZFhi RORγt™ (iNKT2) and PLZFint RORγt+ (iNKT17). We observed a stark increase in iNKT2 and iNKT17 cells, and a concomitant decrease in iNKT1 cells, in the thymus of Shp1fl/fl CD4-cre mice (Fig. 1B). Total iNKT cells in Shp1fl/fl CD4-cre mice expressed higher levels of the folate receptor 4 (FR4, or IZUMO1R) and PD-1, and lower levels of CD69 and CD44, consistent with a bias towards the iNKT2 phenotype (Supplementary Fig. 3A). iNKT1 cells found in Shp1fl/fl CD4-cre mice had a similar expression pattern (Supplementary Fig. 3B), suggesting that these cells are also more “iNKT2-like”. iNKT cell subsets were unaffected in the spleen of Shp1fl/fl CD4-cre mice (Fig. 1C), as well as other peripheral tissues, with the exception of modest iNKT17 cell alterations in inguinal and mesenteric lymph nodes of Shp1fl/fl CD4-cre mice (Supplementary Fig. 3C-E). In addition, iNKT cells subsets were unaffected in the thymus and spleen of Shp1fl/+ and Shp1fl/+ CD4-cre control mice (Supplementary Fig. 3F and not shown).
To test whether this phenotype was cell intrinsic, we performed single and competitive (mixed) bone marrow chimeras by reconstituting irradiated Rag1−/− recipient mice with CD45.1 wild type and/or CD45.2 Shp1fl/fl CD4-cre bone marrow cells (Fig. 1D, left). Thymic Shp1-deficient iNKT cells contained a significantly higher proportion of iNKT2 cells, and a lower proportion of iNKT1 cells, than Shp1-sufficient iNKT cells, in both single and mixed bone marrow chimeras (Fig. 1D). This result shows that Shp1 deficiency skews thymic iNKT cell effector differentiation towards the iNKT2 subset in a cell-intrinsic fashion. To test whether this phenotype was a transient developmental defect, we analyzed iNKT cell effector differentiation in 16-week old mice. Although the overall frequency of iNKT2 cells was much lower compared to 6-week-old mice(14), 16-week-old Shp1fl/fl CD4-cre mice nevertheless had an increased proportion of iNKT2 and iNKT17 cells in the thymus, but not the spleen (Fig. 1E). Taken together, these results demonstrate that Shp1 deficiency results in a cell intrinsic biased iNKT cell differentiation in favor of iNKT2 and iNKT17 cells in the thymus, but not the spleen.
Shp1-deficient iNKT cells have an increased TH2 response
We assessed the functional response of Shp1-deficient iNKT cells using several approaches. First, Shp1fl/fl and Shp1fl/fl CD4-cre mice were injected with αGC and we measured splenic iNKT cell production of IL-4 and IFN-γ by flow cytometry. Surprisingly, iNKT cells from Shp1fl/fl CD4-cre mice were not hyper-responsive, which is different from previous findings using motheaten mice(50). In contrast, Shp1-deficient iNKT cells produced lower amounts of IFN-γ, overall biasing their response towards a TH2 phenotype (Fig. 2A). Interestingly, the reduction in IFN-γ-producing cells was mainly the result of a decrease in IL-4+ IFN-γ+ double producer iNKT cells, and not in IL-4 or IFN-γ single producers (Fig. 2A). Next, we stimulated thymocytes and splenocytes from Shp1fl/fl and Shp1fl/fl CD4-cre mice in vitro with immobilized CD1d-αGC complexes or PMA/ionomycin for a short period of time, and assessed iNKT cell production of IFN-γ and IL-4 by flow cytometry. Thymic iNKT cells from Shp1fl/fl CD4-cre mice were overall less responsive and produced less IL-4 and less IFN-γ (Fig. 2B). Splenic iNKT cells from Shp1fl/fl CD4-cre mice produced slightly more IL-4 than Shp1fl/fl CD4-cre iNKT cells (Fig. 2C). Both thymic and splenic Shp1-deficient iNKT cells had elevated IL-4/IFN-γ ratios, demonstrating a bias towards a TH2 response upon stimulation with CD1d-αGC and to a lower extent with PMA/ionomycin, (Fig. 2B, C). In order to assess whether the biased TH2 response resulting from Shp1 deficiency was imprinted during iNKT cell development or induced upon activation, we crossed Shp1fl/fl mice with CD4-creERT2 mice, treated them with tamoxifen and injected αGC. In this setting, Shp1-deficient iNKT cells were hyporesponsive without the functional TH2 bias (Fig. 2D). Although Shp1-deficient iNKT cells newly generated in the thymus may have populated the spleen during the course of this experiment, this result suggested that the functional TH2 bias observed in Shp1fl/fl CD4-cre mice may be acquired during iNKT cell ontogeny.
Figure 2.
Shp1-deficient iNKT cells produce more TH2 cytokines. (A) Shp1fl/fl and Shp1fl/fl CD4-cre mice were injected i.v. with αGC (0.5 μg). Intracellular FACS analysis of IFN-γ and IL-4 in spleen iNKT cells 90 min post-injection. Data shows representative flow cytometry plots (left), as well as the corresponding frequency (individual mice and mean values +/− s.e.m.) of total, single and double producers of IFN-γ and/or IL-4 (right). (B, C) Thymocytes (B) and splenocytes (C) from Shp1fl/fl and Shp1fl/fl CD4-cre mice were stimulated with plate-bound CD1d-αGC, or PMA/ionomycin (P/I) for 6 hours. Intracellular FACS analysis of IFN-γ and IL-4 in iNKT cells. Data shows representative flow cytometry plots (left), as well as individual and mean values +/− s.e.m. of IL-4/IFN-γ ratios. (D) Shp1fl/fl and Shp1fl/fl CD4-creERT2 mice were treated with tamoxifen and subsequently injected i.v. with αGC (0.2 μg). Data shows individual and mean values +/− s.e.m. of IFN-γ+ or IL-4+ iNKT cells, as well as IL-4/IFN-γ ratios. (E) iNKT cells from Shp1fl/fl or Shp1fl/fl CD4-cre mice were sorted and cultured with BMDCs alone, BMDCs pre-loaded with αGC, or BMDCs in the presence of the indicated cytokine for 48 h. in the presence of indicated cytokines. IFN-γ, IL-4, IL-13, IL-17A and IL-22 production was assessed using a multiplex assay. (F) iNKT cells from Shp1fl/fl or Shp1fl/fl CD4-cre mice were sorted and cultured with BMDCs with isotype control, anti-CD25 or anti-CD127 antibodies (40 μg/ml) for 48 h. Data shows the mean +/− s.e.m. of triplicate values and is representative of 2 individual experiments. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student t test (A-D) or two-way ANOVA (E).
Finally, we sorted splenic iNKT cells from Shp1fl/fl and Shp1fl/fl CD4-cre mice, which contained similar effector subsets (Fig. 1), and cultured them with bone marrow-derived dendritic cells (BMDCs) with or without the addition of αGC (pre-loaded on BMDCs), IL-12, IL-25 or IL-23 for 48 h, which allowed us to detect a broader set of cytokines (Fig. 2E). Wild type iNKT cells produced IL-4 in response to BMDCs, but no other cytokine. Shp1-deficient iNKT cells produced significantly more IL-4 and IL-13 than Shp1-sufficient iNKT cells. That increased IL-4 and IL-13 signature remained the same regardless of the antigen or cytokine used. As expected, αGC strongly activated iNKT cells to produce all the cytokines tested. In contrast to short-term stimulations, Shp1-deficient iNKT cells produced significantly more IFN-γ, IL-13, IL17A and IL-22 than Shp1-sufficient iNKT cells when measured at 48 h, suggesting that Shp1 may regulate early and late stages of the response differently. IL-25 induced increased production of IL-4 and IL-13, but also other cytokines, in a similar way to that of αGC. IL-12 and IL-23 induced the production of IFN-γ and IL-17A/IL-22, respectively, with no difference between genotypes. We found that the enhanced steady-state TH2 response of Shp1-deficient iNKT cells required TCR stimulation as IL-4 production was abrogated in the presence of anti-CD1d antibodies (Fig. 2F). However, neutralizing CD25 or CD127, components of the IL-2 and IL-7 receptors, respectively, did not affect IL-4 levels (Fig. 2F), suggesting that potential homeostatic dysregulation during the 2-day culture period was not responsible for the increased TH2 response. Taken together, these data demonstrate that Shp1-deficient iNKT cells are functionally biased towards the production of TH2 cytokines.
Shp1 deficient iNKT cells induce memory Eomes+ CD8 T cell expansion in the thymus but not the spleen
Shp-1fl/fl CD4-cre mice have an expansion of memory CD4 and CD8 T cells defined by the high expression of CD44(47). Several studies have shown that iNKT cell-derived IL-4 drives the expansion of these Eomes+ innate memory CD8 T cells, which are typically also CD44high (14). As iNKT cells in Shp-1fl/fl CD4-cre mice produce higher amounts of IL-4 and IL-13, we investigated their contribution in the expansion of innate memory T cells. In agreement with a previous study, we found an increase of Eomes+ CD8 T cells in the spleen of Shp1fl/fl CD4-cre mice (Fig. 3A). The thymus of Shp1fl/fl CD4-cre mice also showed a 3-fold increase in the frequency of Eomes+ CD8 T cells (Fig. 3A), despite previous studies reporting no difference in CD44high cells the thymus(47). The frequency of CD44high CD8 T cells were unaffected in the thymus and spleen of Shp1fl/+ and Shp1fl/+ CD4-cre control mice (not shown). CD44high CD8 T cells from Shp1fl/fl CD4-cre mice incorporated more bromodeoxyuridine (BrdU) in vivo than their Shp1fl/fl counterpart, both in the thymus and in the spleen (Fig. 3B). To assess the role of iNKT cells, or other CD1d-restricted T cells, in this homeostatic expansion of innate memory CD8 T cells, we crossed Shp1fl/fl CD4-cre mice with CD1d−/− mice. Interestingly, deletion of CD1d reduced the frequency of Eomes+ CD8T cells back to wild type levels in the thymus, but not the spleen (Fig. 3B). This result suggests that although iNKT cell-derived IL-4 likely drives the homeostatic expansion of innate memory CD8 T cell in the thymus, a different source of IL-4 and/or CD8 T cell-intrinsic Shp1 deficiency(47) drive the expansion of Eomes+ CD8 T cells in the periphery.
Figure 3.
Shp1-deficient iNKT cells drive the expansion of innate memory CD8 T cells in the thymus. (A) Frequency of Eomes+ CD8 T cells in the thymus (left) and spleen (right) of Shp1fl/fl and Shp1fl/fl CD4-cre mice. (B) BrdU incorporation by CD44high CD8 T cells from the thymus (left) and spleen (right) of Shp1fl/fl and Shp1fl/fl CD4-cre mice. (C) Frequency of Eomes+ CD8 T cells in the thymus (top) and spleen (bottom) of Shp1fl/fl CD1d+/−, Shp1fl/fl CD1d−/−, Shp1fl/fl CD4-cre CD1d+/− and Shp1fl/fl CD4-cre CD1d−/− mice. Data represents individual experiments and mean values +/− s.e.m of 2 or 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student t test.
Shp1 deficiency does not regulate iNKT TCR signaling
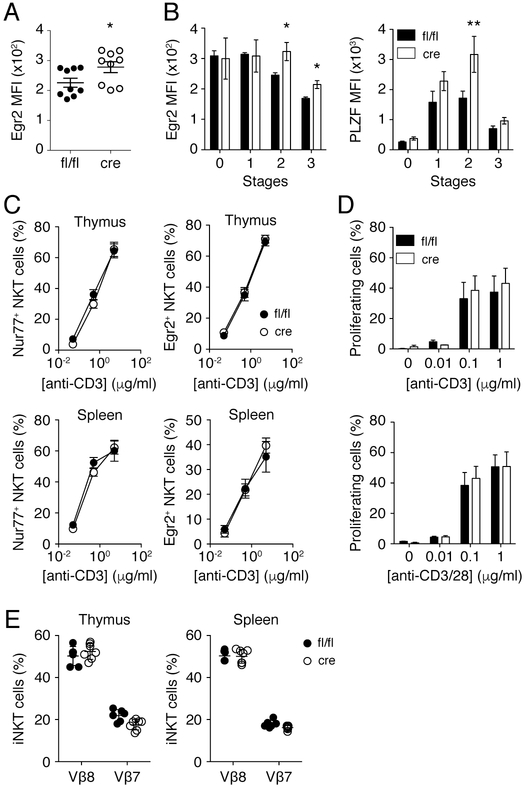
Multiple lines of evidence have suggested that strong TCR signaling favors iNKT2 differentiation(14, 15), specifically due to prolonged TCR interactions(15). The enhanced iNKT2 differentiation in the thymus of Shp1fl/fl CD4-cre mice is a priori consistent with a role for Shp1 in the dampening of TCR signaling(48). We found that Shp1-deficient thymic iNKT cells expressed higher levels of Egr2 (Fig. 4A), which is consistent with their higher expression of PLZF reported above. However, when we analyzed Egr2 and PLZF expression by the various iNKT cell sub-lineages (stages 0–3), we found that the newly selected CD24hi stage 0 iNKT cells from Shp1fl/fl and Shp1fl/fl CD4-cre mice expressed similar levels of Egr2 and PLZF (Fig. 4B), which suggest that these cells receive similar signals at the time of positive selection. Among the more “mature” iNKT cell sub-lineages, only stage 2–3 iNKT cells from Shp1fl/fl CD4-cre mice expressed higher levels of Egr2 and PLZF compared to iNKT cells found in control mice (Fig. 4B). This result suggests that iNKT cells from both strains receive similar signals at the time of positive selection, but that Shp1 may dampen these signals at later stages of iNKT cell development.
Figure 4.
Shp1 deficiency does not affect iNKT TCR signaling. (A) Expression of Egr2 in thymic iNKT cells from Shp1fl/fl (fl/fl) and Shp1fl/fl CD4-cre (cre) mice was determined by FACS. (B) Expression of Egr2 and PLZF in thymic stage 0 (CD24hi CD44™ NK1.1™), stage 1 (CD24™ CD44™ NK1.1™), stage 2 (CD24™ CD44+ NK1.1™) and stage 3 (CD24™ CD44+ NK1.1+) iNKT cells from Shp1fl/fl (fl/fl) and Shp1fl/fl CD4-cre (cre) mice. (C) Frequency of Nur77+ and Egr2+ thymic (top) or splenic (bottom) iNKT cells upon stimulation with different concentrations of anti-CD3 antibodies. (D) Splenocytes from Shp1fl/fl (fl/fl) and Shp1fl/fl CD4-cre (cre) mice were labelled with CellTrace™ CFSE and stimulated for 72 h with the indicated concentrations of anti-CD3 (top) or anti-CD3/CD28 (bottom) antibodies. Proliferation of iNKT cells (TCRβ+ PBS57-CD1d Tetramer+) was assessed by FACS. (E) Frequency of Vβ8+ and Vβ7+ iNKT cells in the thymus (left) and spleen (right) of Shp1fl/fl and Shp1fl/fl CD4-cre mice. Data represents individual mice and mean +/− s.e.m of 2-3 independent experiments (A, E) or the mean +/− s.e.m. of 2 to 3 independent experiments (n ≥ 4). *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student t test.
To determine whether Shp1 specifically regulated TCR signaling in iNKT cells, we first stimulated Shp1-sufficient and deficient iNKT cells in vitro with anti-CD3 antibodies and measured the expression of Egr2 and Nur77, both induced by strong TCR signals. Although thymic and splenic iNKT cells upregulated both Egr2 and Nur77 expression upon TCR stimulation, we found no difference between Shp1-sufficient and deficient iNKT cells (Fig. 4C). We obtained similar results following stimulation with immobilized CD1d-αGC complexes (not shown). Next, we analyzed iNKT cell proliferation and found that iNKT cells from Shp1fl/fl and Shp1fl/fl CD4-cre mice proliferated to the same extent after 3 days of culture with anti-CD3 antibodies alone or together with anti-CD28 antibodies (Fig. 4D). Finally, as it was suggested that Vβ7-containing iNKT TCRs preferentially recognize self-antigens(53), and that iNKT2s are enriched in Vβ7-containing TCRs(14), we assessed whether Shp-1 deficiency affected the TCRβ repertoire of iNKT cells. We found no difference in the frequency of Vβ8+ or Vβ7+ iNKT cells in the thymus or spleen of Shp1fl/fl and Shp1fl/fl CD4-cre mice (Fig. 4E).
We next analyzed the expression of CD5, which correlates with TCR strength in developing thymocytes(54, 55). iNKT cells from Shp1fl/fl and Shp1fl/fl CD4-cre mice expressed similar levels of CD5, as did CD4 and CD8 T cells (Supplementary Fig. 4A). However, splenic Shp1-deficient iNKT, CD4 and CD8 T cells had significantly higher CD5 expression than their Shp1-sufficient counterparts (Supplementary Fig. 4B). Peripheral CD5hi T cells have been shown to preferentially contribute to the effector/memory pool, which could be due to greater TCR sensitivity towards self and foreign peptide MHC complexes and/or enhanced sensitivity to the microenvironment(54, 56). To test whether the enhanced CD5 expression by peripheral iNKT cells was due to TCR-mediated signals, potentially regulated by Shp1, we adoptively transferred splenic iNKT cells from Vα14 transgenic Shp1fl/fl and Shp1fl/fl CD4-cre mice into either wild type or CD1d-deficient mice. CD5 levels on Shp1-deficient iNKT cells remained similarly high after transfer into CD1d-sufficient or deficient mice (Supplementary Fig. 4C), suggesting that this process was independent of peripheral TCR engagement with CD1d-self lipid complexes. Nevertheless, as CD5 has been shown to negatively regulate TCR signaling through the recruitment of Shp1, we analyzed the phenotype of iNKT cells in CD5−/− and CD5−/− Shp1fl/fl CD4-cre mice. We found that CD5-deficient iNKT cells were not biased towards the iNKT2 subset, and that only Shp1 deficiency led to the iNKT bias, regardless of the CD5 genotype (Supplementary Fig. 4D). In sum, these results strongly suggest that the phosphatase Shp1 does not negatively regulate the strength of TCR signaling during positive selection or following TCR-mediated activation of iNKT cells.
Shp1 deficiency does not affect Slamf6-mediated upregulation of Egr2 and PLZF
Aside from TCR-mediated signals, members of the signaling lymphocytic-activation molecule (SLAM) family, especially Slamf6 (Ly108) and Slamf1 (CD150), and their adaptor SAP provide complimentary signals that are essential for iNKT cell development(3–5). Slamf6 activation has been reported to enhance the TCR-mediated upregulation of both Egr2 and PLZF in preselection double positive (PSDP) thymocytes(19, 20), and favor iNKT2 cell development(21). In addition, several studies have suggested that Shp1 attenuates signaling of SLAM family receptors in NK and T cells(20, 41). To determine whether Shp1 regulates Slamf6 signaling within iNKT cells, we first measured PLZF expression in PSDP thymocytes upon crosslinking of CD3 and Slamf6. As previously described, we found that co-stimulation with anti-CD3 and anti-Slamf6 antibodies significantly increased the frequency of PLZF positive PSDPs compared to anti-CD3 or anti-CD3/28 (Fig. 5A). However, both Shp1-sufficient and Shp1-deficient PSDPs upregulated PLZF to the same extent (Fig. 5A), suggesting that Shp1 does not regulate SLAMF6 signaling. Egr2 is rapidly and transiently induced upon co-ligation of CD3 and Slamf6(19). Because phosphatases are often involved in the downregulation or termination of signaling cascades, we measured the Egr2 expression decay following stimulation. However, Shp1 deficiency did not affect the magnitude or duration of Egr2 expression in PSDPs (Fig 5B). Shp1 has been proposed to compete with SAP for binding to the Slamf6 receptor to modulate the development of iNKT cells and follicular helper T cells (20). Specifically, genetic deletion of Slamf6 in addition to SAP, partially restored the severe iNKT cell defect in SAP−/− mice(20). With this in mind, we crossed the Shp1fl/fl CD4-cre mice with Sh2d1a−/− (SAP−/−) mice. As expected, we found that iNKT cells were absent in SAP−/− Shp1fl/fl mice, but were not restored in SAP−/− Shp1fl/fl CD4-cre mice (Fig 5C). This shows that Shp1 does not compete with SAP for binding to Slamf6, as has been proposed(20), or that other phosphatases can compensate for the absence of Shp1. Taken together, these data suggest that Shp1 does not control iNKT cell effector differentiation through the regulation of the Slamf6 signaling cascade.
Figure 5.
Shp1 deletion does not affect Slamf6 mediated upregulation of Egr2 and PLZF. (A) Frequency of live PLZF+ PSDPs from Shp1fl/fl and Shp1fl/fl CD4-cre mice after 48 hours of stimulation with the indicated immobilized antibodies. (B) PSDPs were stimulated stimulation with anti-CD3 and anti-Slamf6 antibodies for 30 min., washed and analyzed for Egr2 expression at the indicated times following stimulus withdrawal. (C) Frequency of iNKT cells in the thymus of SAP−/− Shp1fl/fl, SAP−/− Shp1fl/fl CD4-cre and SAP+/− Shp1fl/fl CD4-cre mice Representative dot plots gated on live cells are shown. Data represents individual mice and mean +/− s.e.m. of 2-3 independent experiments.
Shp1 regulates iNKT cell cytokine-mediated proliferation
Although iNKT cells require certain cytokines such as IL-2, IL-7 and IL-15 for their survival and proliferation(31, 32, 57, 58), the role cytokines play in their effector differentiation has been largely overlooked. As several studies have suggested that Shp1 regulates cytokine and chemokine receptor signaling(39, 47, 59, 60), we compared the proliferative capacity of Shp1-sufficient and deficient iNKT cells in response to cytokines. Surprisingly, unlike TCR-mediated proliferation (Fig. 4D), we found that Shp1-deficient iNKT cells isolated from the thymus or the spleen had enhanced proliferation in response to IL-2, IL-7 and IL-15, compared to Shp1-sufficient iNKT cells (Fig. 6A). We analyzed the proliferation capacity iNKT cells at steady state using BrdU labelling in vivo. We found that iNKT cells from the thymus, but not the spleen, of Shp1fl/fl CD4-cre mice incorporated more BrdU than iNKT cells from Shp1fl/fl control mice (Fig. 6B), suggesting that their proliferation was enhanced in vivo. Because thymic and splenic iNKT cells from Shp1fl/fl and Shp1fl/fl CD4-cre mice express comparable levels of the IL-2Rα (CD25), IL-7Rα (CD127) and IL-2Rβ (CD122) chains (Fig. 6B), we hypothesize that Shp1 may negatively regulate signals downstream of these cytokine receptors(47). Overall, these data point to a role of Shp1 in maintaining cytokine signals in check to regulate iNKT cell homeostasis.
Figure 6.
Shp1 regulates cytokine-mediated iNKT cell proliferation. (A) Representative histogram plots (3 mice per group) of CFSE dilution profile of iNKT cells sorted from the thymus (left) or the spleen (right) of Shp1fl/fl and Shp1fl/fl CD4-cre mice cultured for 4 days in the presence of 10ng/ml of IL-2, IL-7 or IL-15. One experiment out of two is shown. (B) BrdU incorporation by iNKT cells from the thymus (left) and spleen (right) of Shp1fl/fl and Shp1fl/fl CD4-cre mice. (C) Expression of CD25, CD127 and CD122 on thymic (left) and splenic (right) iNKT cells from Shp1fl/fl and Shp1fl/fl CD4-cre mice. Data shows representative histogram plots as well as individual mice and the mean values +/− s.e.m of 2-3 independent experiments.
DISCUSSION
The making of an iNKT cell from developing DP thymocytes requires stronger than normal TCR signaling, as well as cooperative engagement of members of the signaling lymphocytic-activation molecule (SLAM) family of receptors, which culminate in the expression of PLZF, a master regulator of iNKT cell innate effector programs(8). Here, we found that the protein tyrosine phosphatase Shp1, traditionally viewed as a negative regulator of TCR signaling, regulates iNKT cell effector differentiation independently of TCR and Slamf6 signaling. Instead, Shp1 dampens iNKT cell proliferation in response to IL-2, IL-7 and IL-15, but not following TCR engagement, which suggests that certain cytokines may participate in iNKT cell effector programming.
Although Shp1 has been shown to regulate signaling downstream of various immune receptors(34), many initial studies have been confounded by the various degrees of autoimmunity/autoinflammation present in mice with constitutive total or partial Shp1 loss-of-function (motheaten, motheaten viable and spin)(42–44). Alternative approaches, including conditional Shp1 deletion, have so far failed to provide a consensus on Shp1 function in TCR signaling and T cell biology(46–48, 61). Conditional Shp1 deficiency enhanced iNKT2 differentiation in the thymus in a cell-intrinsic fashion, which was a priori consistent with increased TCR signaling in developing iNKT cells. However, we found no evidence that Shp1 dampens TCR signaling within iNKT cells. Shp1-deficient iNKT cells were not overtly autoreactive in vitro or in vivo, contrary to what could have been predicted from previous work(50). In fact, these cells were somewhat hyporesponsive under some stimulation conditions, which is reminiscent of Shp1-deficient NK cells(62). Shp1 deficiency also did not impact the ability of iNKT cells (or DPs) to upregulate expression of Nur77, Egr2 and PLZF, or proliferate following TCR engagement. In sum, although TCR interactions with self-lipid/CD1d complexes are crucial for iNKT cell positive selection(2), negative selection(63, 64) and effector differentiation(14, 15), this work suggests that Shp1 does not regulate TCR signaling in iNKT cells, and therefore controls their effector differentiation through TCR-independent mechanisms.
Homotypic Slamf6-Slamf6 interactions facilitated by DP-DP interactions in the cortex are also critical for iNKT cell development(5). In agreement with previous reports(19, 20), we found that Slamf6 co-engagement potentiates the TCR-mediated upregulation of Egr2 and PLZF in pre-selection DPs. Although Shp1 has been proposed to compete with SAP for interaction with Slamf6 to regulate Slamf6 signaling in iNKT cells and follicular helper T cells(20), we found no evidence of such regulation. Although Slamf1 (CD150) and Slamf3 (Ly-9 or CD229) have been shown to play a role in iNKT cell effector differentiation, co-engagement of these receptors failed to upregulate Egr2 and PLZF expression in pre-selection DPs in vitro (not shown). Given that mice with combined deficiency in multiple Slam family receptors have iNKT cell defects(65), it is possible that other Slam family receptors impact iNKT cell development and/or differentiation through the recruitment of Shp1.
We found that Shp1 regulates the proliferation of iNKT cells following exposure to IL-2, IL-7 and IL-15, without affecting cytokine receptor expression, which is in line with previous studies(39, 47, 59, 60). How Shp1 regulates cytokine-mediated proliferation, and how this increased proliferative capacity ties back to the in vivo bias towards iNKT2 differentiation remains unclear and will require further investigation. Generally, the role these cytokines play in iNKT cell homeostasis has been studied through the adoptive transfer of peripheral iNKT cells into various (often lymphopenic) recipient mice(31, 32). It is well-described that stage 3 iNKT cells (or iNKT1 cells) express the IL-15Rβ chain (CD122) and proliferate in response to IL-15(31, 32). Additionally, a recent study from Webster et al. suggested that IL-7 controls the homeostasis of IL-17-producing iNKT cells(58). To the best of our knowledge, there has been no clear demonstration of the role of cytokines, chemokines and their receptors in iNKT2 cell development, although IL-25(66) and IL-33(67) could be important contributors.
In agreement with previous studies(14, 28, 29, 68, 69), we found that the expansion of iNKT2s in Shp1fl/fl CD4-cre mice was accompanied by an increased prevalence of Eomes+ innate memory CD8+ T cells(47). Whereas the study from Johnson et al. reported an expansion of innate memory CD8+ T cells in the spleen but not the thymus, we found an expansion in both tissues. The discrepancy may lie in the use of CD44 vs. Eomes to identify these cells. This expansion of thymic Eomes+ CD8+ T cells was entirely abrogated in CD1d-deficient mice, suggesting that iNKT cells are a major source of IL-4 to drive the differentiation of these innate memory CD8+ T cells. Interestingly, the much larger expansion of peripheral Eomes+ CD8+ T cells was completely iNKT cell-independent, suggesting that other cells, such as CD4-dependent γδ T cells, are producing IL-4, and/or that the increased IL-4 sensitivity in CD8+ T cells is sufficient to promote a memory phenotype even in the absence of IL-4-producing iNKT cells(47).
In this study, we have shown that the protein tyrosine phosphatase Shp1 regulates the effector differentiation of iNKT cells after positive selection, and independently of the TCR and Slamf6 signaling cascades. We found that Shp1 dampens the cytokine-mediated proliferation of iNKT cells, suggesting that cytokines may play an underappreciated role in the functional programming of iNKT cells.
Supplementary Material
KEY POINTS.
Shp1-deficient iNKT cells are biased towards the iNKT2/17 subsets.
Shp1 deficiency does not alter TCR or Slam signaling.
Shp1 dampens iNKT cell proliferation in response to cytokines.
ACKNOWLEDGEMENTS
We express our gratitude to Dr. Benjamin Neel for providing Shp1fl/fl CD4-cre mice, and to Drs. Heather Melichar and Christophe Paget for the critical review of the manuscript. The authors have no conflict of interest.
This work was supported by a Canadian Institutes of Health Research (CIHR) grant (MOP – 114911), and a Canada Foundation for Innovation Physical Infrastructure Grant (29186) to T.M., and NIH Grants R01 AI1922 (M.K.) and T32 AR064194 (M.Z.). M.C.T was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Doctoral Research Award. T.M. is supported by a Canada Research Chair in NKT cell Immunobiology.
ABBREVIATIONS
- BMDC
bone marrow-derived dendritic cell
- DC
dendritic cell
- iNKT
invariant Natural Killer T cell
- DP
double positive
- PSDP
pre-selection double positive
- TCR
T cell receptor
- NK
Natural Killer
- αGC
α-galactosylceramide
REFERENCES
- 1.Brennan PJ, Brigl M, and Brenner MB. 2013. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol 13: 101–117. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A 1995. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med 182: 2091–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquier B, Yin L, Fondanèche M-C, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, and Latour S. 2005. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J. Exp. Med 201: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols KE, Hom J, Gong S-Y, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, and Stein PL. 2005. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med 11: 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, and Bendelac A. 2007. Homotypic Interactions Mediated by Slamf1 and Slamf6 Receptors Control NKT Cell Lineage Development. Immunity 27: 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Josefowicz SZ, Lu L-F, and Rudensky AY. 2012. Regulatory T Cells: Mechanisms of Differentiation and Function. Annu. Rev. Immunol 30: 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stritesky GL, Jameson SC, and Hogquist KA. 2012. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol 30: 95–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gapin L 2016. Science Direct Development of invariant natural killer T cells. Curr. Opin. Immunol 39: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogquist KA, and Jameson SC. 2014. The self-obsession of T cells: how TCR signaling thresholds affect fate “decisions” and effector function. Nat. Immunol 15: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarin P, Chen ELY, In TSH, Anderson MK, and Zúñiga-Pflücker JC. 2015. Gamma delta T-cell differentiation and effector function programming, TCR signal strength, when and how much? Cellular Immunology 296: 70–75. [DOI] [PubMed] [Google Scholar]
- 11.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, and Hogquist KA. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. Journal of Experimental Medicine 208: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim H-J, Im JS, Pandolfi PP, and Sant’angelo DB. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol 9: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A. 2008. The Transcription Factor PLZF Directs the Effector Program of the NKT Cell Lineage. Immunity 29: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol 14: 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz Tleugabulova M, Escalante NK, Deng S, Fieve S, Ereno-Orbea J, Savage PB, Julien JP, and Mallevaey T. 2016. Discrete TCR Binding Kinetics Control Invariant NKT Cell Selection and Central Priming. The Journal of Immunology 197: 3959–3969. [DOI] [PubMed] [Google Scholar]
- 16.Zhao M, Svensson MND, Venken K, Chawla A, Liang S, Engel I, Mydel P, Day J, Elewaut D, Bottini N, and Kronenberg M. 2018. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat Comms 9: 2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, Dragone LL, Lefferts A, Halluszczak C, Riemondy K, Hesselberth JR, Rao A, O’Connor BP, Marrack P, Scott-Browne J, and Gapin L. 2018. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat Comms 9: 2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiler MP, Mathew R, Liszewski MK, Spooner C, Barr K, Meng F, Singh H, and Bendelac A. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol 13: 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta M, Kraus ZJ, Gomez-Rodriguez J, Hwang S-H, Cannons JL, Cheng J, Lee S-Y, Wiest DL, Wakeland EK, and Schwartzberg PL. 2013. A Role for Ly108 in the Induction of Promyelocytic Zinc Finger Transcription Factor in Developing Thymocytes. The Journal of Immunology 190: 2121–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, and Crotty S. 2012. The Receptor Ly108 Functions as a SAP Adaptor-Dependent On-Off Switch for T Cell Help to B Cells and NKT Cell Development. Immunity 36: 986–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel M-L, Lenoir C, Massot B, Diem S, Pasquier B, Sawa S, Rignault-Bricard R, Lehuen A, Eberl G, Veillette A, Leite de Moraes M, and Latour S. 2016. SLAM-associated protein favors the development of iNKT2 over iNKT17 cells. Eur. J. Immunol 46: 2162–2174. [DOI] [PubMed] [Google Scholar]
- 22.Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM, Kimura MY, Sharrow SO, Guinter TI, Feigenbaum L, and Singer A. 2015. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat. Immunol 16: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zietara N, Lyszkiewicz M, Witzlau K, Naumann R, Hurwitz R, Langemeier J, Bohne J, Sandrock I, Ballmaier M, Weiss S, Prinz I, and Krueger A. 2013. Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc. Natl. Acad. Sci. U.S.A 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishimoto H, Ohteki T, Yajima N, Kawahara K, Natsui M, Kawarasaki S, Hamada K, Horie Y, Kubo Y, Arase S, Taniguchi M, Vanhaesebroeck B, Mak TW, Nakano T, Koyasu S, Sasaki T, and Suzuki A. 2007. The Pten/PI3K pathway governs the homeostasis of Valpha14iNKT cells. Blood 109: 3316–3324. [DOI] [PubMed] [Google Scholar]
- 25.Wei J, Yang K, and Chi H. 2014. Cutting edge: Discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. The Journal of Immunology 193: 4297–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Tschumi BO, Corgnac S, Rüegg MA, Hall MN, Mach J-P, Romero P, and Donda A. 2014. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. The Journal of Immunology 193: 1759–1765. [DOI] [PubMed] [Google Scholar]
- 27.Prevot N, Pyaram K, Bischoff E, Sen JM, Powell JD, and Chang C-H. 2015. Mammalian target of rapamycin complex 2 regulates invariant NKT cell development and function independent of promyelocytic leukemia zinc-finger. The Journal of Immunology 194: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Cruz LM, Stradner MH, Yang CY, and Goldrath AW. 2014. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. The Journal of Immunology 192: 2227–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verykokakis M, Krishnamoorthy V, Iavarone A, Lasorella A, Sigvardsson M, and Kee BL. 2013. Essential Functions for ID Proteins at Multiple Checkpoints in Invariant NKT Cell Development. The Journal of Immunology 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu T, Wang H, Simmons A, Bajaña S, Zhao Y, Kovats S, Sun X-H, and Alberola-Ila J. 2013. Increased level of E protein activity during invariant NKT development promotes differentiation of invariant NKT2 and invariant NKT17 subsets. The Journal of Immunology 191: 5065–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, and Kronenberg M. 2002. Homeostasis of Vα14i NKT cells. Nat. Immunol 3: 966–974. [DOI] [PubMed] [Google Scholar]
- 32.Ranson T, Vosshenrich CAJ, Corcuff E, Richard O, Laloux V, Lehuen A, and Di Santo JP. 2003. IL-15 availability conditions homeostasis of peripheral natural killer T cells. Proc. Natl. Acad. Sci. U.S.A 100: 2663–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowan JE, McCarthy NI, Parnell SM, White AJ, Bacon A, Serge A, Irla M, Lane PJL, Jenkinson EJ, Jenkinson WE, and Anderson G. 2014. Differential requirement for CCR4 and CCR7 during the development of innate and adaptive αβT cells in the adult thymus. The Journal of Immunology 193: 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenz U 2009. SHP-1 and SHP-2 in T cells: two phosphatases functioning at many levels. Immunological reviews 228: 342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plas DR, Johnson R, Pingel JT, Matthews RJ, Dalton M, Roy G, Chan AC, and Thomas ML. 1996. Direct regulation of ZAP-70 by SHP-1 in T cell antigen receptor signaling. Science 272: 1173–1176. [DOI] [PubMed] [Google Scholar]
- 36.Cyster JG, and Goodnow CC. 1995. Protein tyrosine phosphatase 1C negatively regulates antigen receptor signaling in B lymphocytes and determines thresholds for negative selection. Immunity 2: 13–24. [DOI] [PubMed] [Google Scholar]
- 37.Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet JP, and Long EO. 1996. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity 4: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura MC, Niemi EC, Fisher MJ, Shultz LD, Seaman WE, and Ryan JC. 1997. Mouse Ly-49A interrupts early signaling events in natural killer cell cytotoxicity and functionally associates with the SHP-1 tyrosine phosphatase. J. Exp. Med 185: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minoo P, Zadeh MM, Rottapel R, Lebrun J-J, and Ali S. 2004. A novel SHP-1/Grb2-dependent mechanism of negative regulation of cytokine-receptor signaling: contribution of SHP-1 C-terminal tyrosines in cytokine signaling. Blood 103: 1398–1407. [DOI] [PubMed] [Google Scholar]
- 40.Xiao W, Hong H, Kawakami Y, Kato Y, Wu D, Yasudo H, Kimura A, Kubagawa H, Bertoli LF, Davis RS, Chau LA, Madrenas J, Hsia CC, Xenocostas A, Kipps TJ, Hennighausen L, Iwama A, Nakauchi H, and Kawakami T. 2009. Tumor suppression by phospholipase C-beta3 via SHP-1-mediated dephosphorylation of Stat5. Cancer Cell 16: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu N, Zhong M-C, Roncagalli R, Pérez-Quintero L-A, Guo H, Zhang Z, Lenoir C, Dong Z, Latour S, and Veillette A. 2016. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat. Immunol 17: 387–396. [DOI] [PubMed] [Google Scholar]
- 42.Shultz LD, Schweitzer PA, Rajan TV, Yi T, Ihle JN, Matthews RJ, Thomas ML, and Beier DR. 1993. Mutations at the murine motheaten locus are within the hematopoietic cell protein-tyrosine phosphatase (Hcph) gene. Cell 73: 1445–1454. [DOI] [PubMed] [Google Scholar]
- 43.Tsui HW, Siminovitch KA, de Souza L, and Tsui FW. 1993. Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat. Genet 4: 124–129. [DOI] [PubMed] [Google Scholar]
- 44.Croker BA, Lawson BR, Rutschmann S, Berger M, Eidenschenk C, Blasius AL, Moresco EMY, Sovath S, Cengia L, Shultz LD, Theofilopoulos AN, Pettersson S, and Beutler BA. 2008. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc. Natl. Acad. Sci. U.S.A 105: 15028–15033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pao LI, Lam K-P, Henderson JM, Kutok JL, Alimzhanov M, Nitschke L, Thomas ML, Neel BG, and Rajewsky K. 2007. B Cell-Specific Deletion of Protein-Tyrosine Phosphatase Shp1 Promotes B-1a Cell Development and Causes Systemic Autoimmunity. Immunity 27: 35–48. [DOI] [PubMed] [Google Scholar]
- 46.Fowler CC, Pao LI, Blattman JN, and Greenberg PD. 2010. SHP-1 in T Cells Limits the Production of CD8 Effector Cells without Impacting the Formation of Long-Lived Central Memory Cells. J. Immunol 185: 3256–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson DJ, Pao LI, Dhanji S, Murakami K, Ohashi PS, and Neel BG. 2013. Shp1 regulates T cell homeostasis by limiting IL-4 signals. Journal of Experimental Medicine 210: 1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez RJ, Morris AB, Neeld DK, and Evavold BD. 2016. Targeted loss of SHP1 in murine thymocytes dampens TCR signaling late in selection. Eur. J. Immunol 46: 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa E, Kosako H, Yasuda T, Ohmuraya M, Araki K, Kurosaki T, Saito T, and Yamasaki S. 2016. Protein kinase D regulates positive selection of CD4+ thymocytes through phosphorylation of SHP-1. Nat Comms 7: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napolitano A, Pittoni P, Beaudoin L, Lehuen A, Voehringer D, MacDonald HR, Dellabona P, and Casorati G. 2013. Functional Education of Invariant NKT Cells by Dendritic Cell Tuning of SHP-1. The Journal of Immunology 190: 3299–3308. [DOI] [PubMed] [Google Scholar]
- 51.Chen YH, Chiu NM, Mandal M, Wang N, and Wang CR. 1997. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity 6: 459–467. [DOI] [PubMed] [Google Scholar]
- 52.Miah SMS, Jayasuriya CT, Salter AI, Reilly EC, Fugère C, Yang W, Chen Q, and Brossay L. 2017. Ptpn11 Deletion in CD4+ Cells Does Not Affect T Cell Development and Functions but Causes Cartilage Tumors in a T Cell-Independent Manner. Front Immunol 8: 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schumann J, Mycko MP, Dellabona P, Casorati G, and MacDonald HR. 2006. Cutting Edge: Influence of the TCR V Domain on the Selection of Semi-Invariant NKT Cells by Endogenous Ligands. The Journal of Immunology 176: 2064–2068. [DOI] [PubMed] [Google Scholar]
- 54.Hogquist KA, and Jameson SC. 2014. The self-obsession of T cells: how TCR signaling thresholds affect fate “decisions” and effector function. Nat. Immunol 15: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, and Love PE. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med 188: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fulton RB, Hamilton SE, Xing Y, Best JA, Goldrath AW, Hogquist KA, and Jameson SC. 2014. The TCR’s sensitivity to self peptide–MHC dictates the ability of naive CD8+ T cells to respond to foreign antigens. Nat. Immunol 16: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iyoda T, Ushida M, Kimura Y, Minamino K, Hayuka A, Yokohata S, Ehara H, and Inaba K. 2010. Invariant NKT cell anergy is induced by a strong TCR-mediated signal plus co-stimulation. International Immunology 22: 905–913. [DOI] [PubMed] [Google Scholar]
- 58.Webster KE, Kim H-O, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, Brink R, Belz GT, Cho JH, Godfrey DI, and Sprent J. 2014. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol 7. [DOI] [PubMed] [Google Scholar]
- 59.Jiao H, Berrada K, Yang W, Tabrizi M, Platanias LC, and Yi T. 1996. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol. Cell. Biol 16: 6985–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klingmüller U, Lorenz U, Cantley LC, Neel BG, and Lodish HF. 1995. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell 80: 729–738. [DOI] [PubMed] [Google Scholar]
- 61.Fu G, Casas J, Rigaud S, Rybakin V, Lambolez F, Brzostek J, Hoerter JAH, Paster W, Acuto O, Cheroutre H, Sauer K, and Gascoigne NRJ. 2013. Themis sets the signal threshold for positive and negative selection in T-cell development. Nature 504: 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viant C, Fenis A, Chicanne G, Payrastre B, Ugolini S, and Vivier E. 2014. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Comms 5: 5108. [DOI] [PubMed] [Google Scholar]
- 63.Chun T, Page MJ, Gapin L, Matsuda JL, Xu H, Nguyen H, Kang H-S, Stanic AK, Joyce S, Koltun WA, Chorney MJ, Kronenberg M, and Wang C-R. 2003. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J. Exp. Med 197: 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bedel R, Berry R, Mallevaey T, Matsuda JL, Zhang J, Godfrey DI, Rossjohn J, Kappler JW, Marrack P, and Gapin L. 2014. Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc. Natl. Acad. Sci. U.S.A 111: E119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu JK, Crampton JC, Locci M, and Crotty S. 2016. CRISPR-Mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ Triple Gene Disruption Reveals NKT Cell Defects but Not T Follicular Helper Cell Defects. PloS one 11: e0156074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, Iwamura C, Nakajima H, Nakayama T, and Taniguchi M. 2008. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. Journal of Experimental Medicine 205: 2727–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bourgeois E, Van LP, Samson M, Diem S, Barra A, Roga S, Gombert J-M, Schneider E, Dy M, Gourdy P, Girard J-P, and Herbelin A. 2009. The pro-Th2 cytokine IL-33 directly interacts with invariant NKT and NK cells to induce IFN-gamma production. Eur. J. Immunol 39: 1046–1055. [DOI] [PubMed] [Google Scholar]
- 68.Weinreich MA, Odumade OA, Jameson SC, and al E. 2010. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai D, Zhu J, Wang T, Hu-Li J, Terabe M, Berzofsky JA, Clayberger C, and Krensky AM. 2011. KLF13 sustains thymic memory-like CD8(+) T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. Journal of Experimental Medicine 208: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.