 Biofilm formation by mycobacteria can lead to enhanced antibiotic tolerance.
Biofilm formation by mycobacteria can lead to enhanced antibiotic tolerance.
Abstract
Biofilm formation by mycobacteria can lead to enhanced antibiotic tolerance. Herein, we report on the identification of a series of 2-aminobenzimidazole (2-ABI) derivatives that potently inhibit biofilm formation by Mycobacterium smegmatis.
Bacterial biofilms are defined as surface attached communities of bacteria encased in an extracellular matrix of biomolecules (typically polysaccharides).1 Within a biofilm state, bacteria are upwards of 1000-fold more tolerant to antibiotics and microbicides in comparison to their free-floating (planktonic) brethren.2 Biofilms typically underpin chronic bacterial infections, such as those found in Cystic Fibrosis patients and in infected indwelling medical devices (catheters, stents, etc.).3,4 To date, there is no approved therapeutic to directly target bacteria within a biofilm state.
Mycobacterium tuberculosis is the causative agent of tuberculosis (TB), a disease that was responsible for 1.6 million deaths worldwide in 2017 (1.3 million among HIV-negative people and 300 000 deaths among HIV-positive people), while it is estimated that 10 million people developed TB disease in 2017.5 Current treatment regimens are centered on implementing a combination therapy of isoniazid, rifampin, pyrazinamide and ethambutol for two months followed by isoniazid and rifampin for four months.6 The deleterious side effects of such aggressive antibiotic therapy can lead to poor patient compliance, disease relapse, and emergence of multi-drug resistance.7
This lengthy antibiotic regimen is employed to eradicate reservoirs of persistent TB within the infected individual. Although the origin of these reservoirs has not been definitively established, recent studies have shown that M. tuberculosis has the ability to form biofilm-like structures when grown in vitro on lysed cellular debris in the absence of detergent (to mimic lung lesions encountered in patients suffering from an active TB infection).8–10 Importantly, these communities are significantly more drug tolerant than M. tuberculosis grown under standard culture conditions. Therefore, the discovery of small molecules that inhibit the formation of these biofilm-like communities could form the basis for the discovery of future therapeutics that can be combined with current antibiotic treatments to shorten treatment time and ultimately improve patient compliance.
As M. tuberculosis is a slow growing and pathogenic organism requiring BSL3 containment, M. smegmatis is typically employed as a screening surrogate due to increased growth rate and safety profile.11 These two bacteria share over 2000 genetic homologs,12 with anti-TB antibiotics discovered through screening initially with M. smegmatis.13 We have previously shown that a standard crystal violet-reporter assay can be employed to identify molecules that disrupt M. smegmatis biofilm formation and that these compounds are active at reversing biofilm-like growth of M. tuberculosis grown in vitro on cellular debris and in the process restore antibiotic susceptibility.14
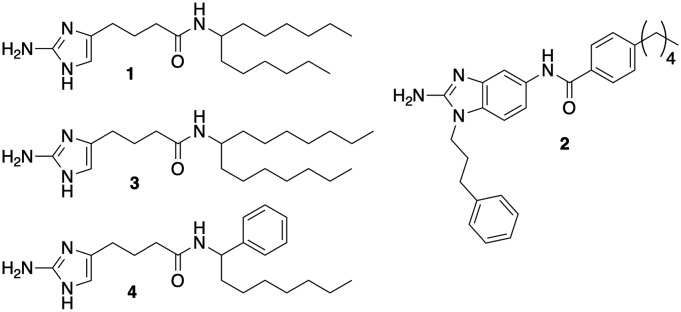
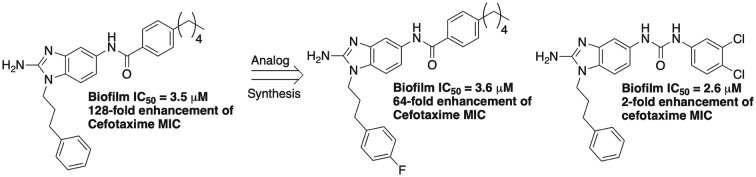
One of the lead compounds from this initial study was 2-aminoimidazole 1 (Fig. 1). Interestingly, this compound also demonstrated the ability to sensitize M. tuberculosis to the effect of β-lactam antibiotics by a mechanism independent of directly inhibiting β-lactamase activity. We subsequently performed a comprehensive screen of our in-house small molecule library, and identified 2-aminobenzimidazole (2-ABI) 2 as also having the ability to potently sensitize M. smegmatis to cefotaxime as well as other β-lactam antibiotics.15 Against multi-drug resistant M. tuberculosis strains, 2-ABI 2 was able to suppress the minimum inhibitory concentration (MIC) of β-lactams upwards of 4096-fold. A subsequent structure–activity relationship (SAR) study demonstrated a tight SAR window, with a majority of modifications to 2-ABI 2 resulting in either significantly reduced or abrogated activity. A follow up SAR study on compound 1 revealed that one could decouple β-lactam potentiation from biofilm inhibition and identify additional lead compounds that were either more potent towards biofilm disruption or β-lactam potentiation (compounds 3 and 4 respectively).16 Interestingly, compound 2 had also been discovered to inhibit M. smegmatis biofilm formation but was not pursued due to its inability to disperse bacterial biofilms. Given that the SAR study on compound 1 demonstrated that one could optimize anti-biofilm activity despite loss of β-lactam potentiation, we hypothesized that additional analogs of 2 either from our initial library studies or from an expanded library would have equal or greater activity than compound 2 in the context of inhibiting M. smegmatis biofilms. Herein we detail our studies into the antibiofilm activities of compound 2 and its derivatives against M. smegmatis.
Fig. 1. Structures of 2-Aminoimidazoles 1, 3, 4, and 2-ABI 2.
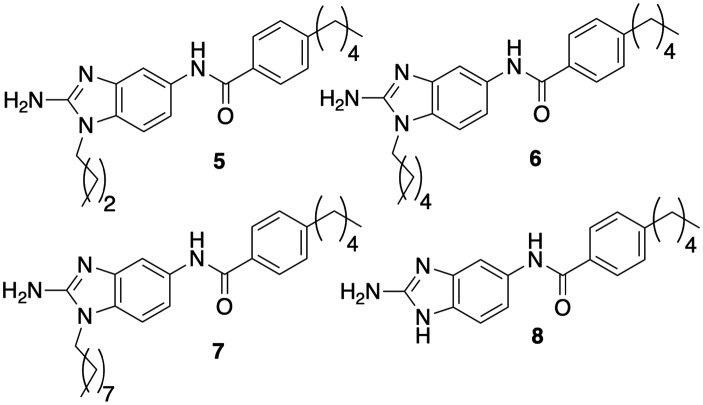
Compound 2 had initially returned an IC50 value (concentration required to inhibit 50% biofilm formation) of 8.0 μM from library stock using a crystal violet reporter assay.14 Upon resynthesis and reevaluation, compound 2, however, returned an IC50 of 3.5 μM. In comparison, compound 1 consistently returned IC50s of 5.3 μM. Compound 2 was also relatively non-toxic itself to M. smegmatis, registering a minimum inhibitory concentration (MIC) of 60 μM. We initiated analog screening by assaying previously synthesized alkyl derivatives at N-117 (Fig. 2, activity summarized in Table 1). The three alkyl derivatives (5, 6, and 7) were all significantly less active than 2, returning IC50's between 22.4–25.6 μM but were also essentially non-toxic to planktonic bacteria (MICs ≥ 200 μM). Complete removal of a substituent, giving the base 2-ABI derivative 8, returned activity (13.6 μM) superior to the alkly derivatives but still inferior to 2.
Fig. 2. Alkyl and unsubstituted 2-ABI derivatives.
Table 1. Biofilm inhibition as a function of N-1 substitution (μM).
| Compound | MIC | IC50 |
| 2 | 60 | 3.5 ± 0.9 |
| 5 | >200 | 22.4 ± 2.6 |
| 6 | >200 | 25.6 ± 3.3 |
| 7 | 200 | 23.2 ± 4.5 |
| 8 | 150 | 13.6 ± 2.3 |
| 9 | 100 | 12.2 ± 0.9 |
| 10 | 100 | 5.4 ± 0.8 |
| 11 | 100 | 4.3 ± 0.8 |
| 12 | 120 | 11.3 ± 1.8 |
| 13 | 200 | 12.5 ± 1.3 |
| 14 | 120 | 8.8 ± 2.1 |
| 15 | 100 | 4.3 ± 0.8 |
| 16 | >200 | 12.5 ± 1.4 |
| 17 | 120 | 3.6 ± 1.0 |
| 18 | 120 | 6.4 ± 1.5 |
| 19 | 100 | 6.2 ± 0.9 |
| 20 | 100 | 9.2 ± 2.5 |
| 21 | 120 | 10.5 ± 1.1 |
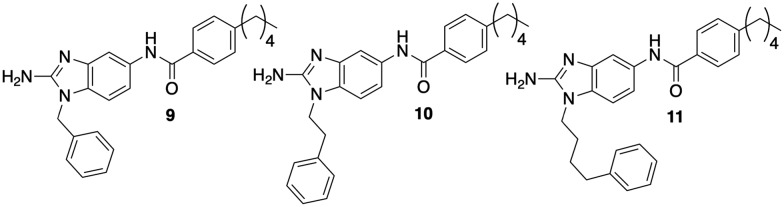
Next, we explored the impact linker length between the 2-ABI and phenyl group had on activity (Fig. 3). Modulating the linker length between the core 2-ABI and the appended aryl group also impacted activity, with the one, two, and four carbon linkers (9, 10, and 11) all exhibiting less activity than compound 2. All three compounds were less toxic with MICs 100–120 μM.
Fig. 3. Varying the linker length between the 2-ABI and phenyl substituent.
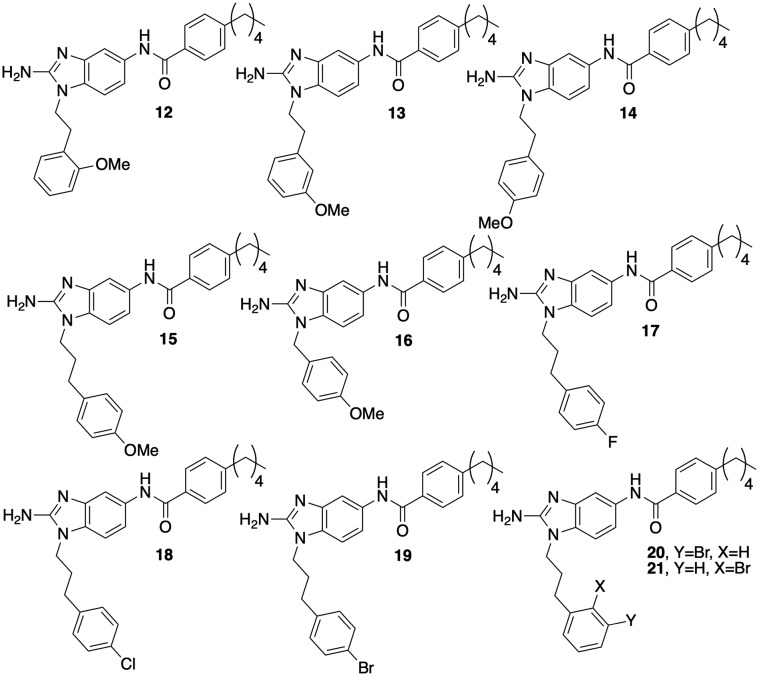
In the initial library, we had substituted the phenyl derivative with a 2-carbon linker (10) with methoxy groups at the ortho, meta, and para positions (12–14, Fig. 4). When assayed for biofilm inhibition, all three compounds were active, with compound 14 returning an essentially equipotent IC50 (8.8 μM) as compound 10 (8.5 μM). All three compounds were less toxic than 2, registering MICs of 120–200 μM. To test whether substitution tolerance was a general trend, we synthesized compound 15 and tested it along with compound 16. Compounds 15 and 16 returned IC50 values of 4.3 and 12.5 μM respectively, which again is comparable in activity to their respective phenyl derivatives, compound 2 (3.5 μM) and compound 12 (11.3 μM). We further tested the tolerance at the appended phenyl ring by synthesizing the corresponding p-fluoro, chloro, and bromo derivatives (17, 18, and 19) and assaying them for antibiofilm activity against M. smegmatis. The p-fluoro derivative, compound 17, was most active and equipotent to the lead with an IC50 of 3.6 μM. Both the chloro and bromo analogs were ca. 2-fold less active. Finally, we further tested the impact position had upon activity by synthesizing compounds 20 and 21. Upon testing, both compounds were less active with IC50's of 9.2 and 10.5 μM respectively. All new compounds, as well as 16, were less toxic than 2, returning MICs of ≥100 μM.
Fig. 4. Analogs to probe tolerance of substitution on the phenyl ring.
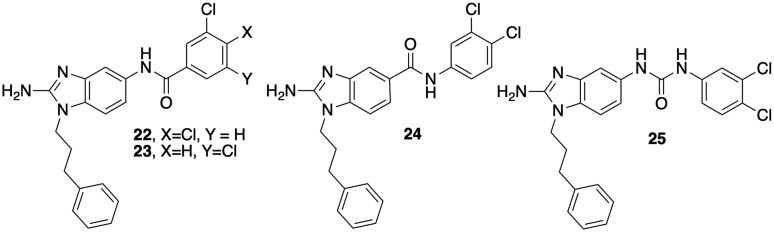
In a previous report, we had shown that introduction of a dichlorinated halogenation pattern at the tail aryl ring of the 2-AI scaffold can deliver anti-biofilm agents in the context of Salmonella biofilms.18 To explore whether this observation translates to the current studies, we assayed compounds 22 and 23 (Fig. 5) for activity (Table 2). Both compounds were less active than 2, with IC50's of 8.8 and 9.4 μM respectively. Finally, we tested the impact of reversing the amide (24) as well as replacement of the amide with a urea (25). Reversing the amide had no impact on activity (24, 8.8 μM); however replacement with a urea delivered a derivative (25, IC50 2.6 μM) that was essentially equivalent to the original lead 2. Again, all new derivatives were less toxic than 2, with 22–24 registering MICs of 200 μM each and 25 returning an MIC of 100 μM.
Fig. 5. Modifications to the 2-ABI tail.
Table 2. Biofilm inhibition for dichloro-substituted 2-ABIs (μM).
| Compound | MIC | IC50 |
| 22 | 200 | 8.8 ± 1.8 |
| 23 | 200 | 9.4 ± 2.5 |
| 24 | 200 | 8.6 ± 1.4 |
| 25 | 100 | 2.6 ± 1.1 |
As many members of our original library had anti-biofilm activity in the absence of the ability to potentiate β-lactam activity, we elected to evaluate whether this trend continued for a select number of the more potent derivatives synthesized in this study. To this end, we studied the ability of compounds 15, 17–19, and 25 to potentiate the activity of ampicillin or cefotaxime against M. smegmatis (Table 3). Lead compound 2, at 20 μM, reduced the ampicillin MIC 16-fold (256 to 16 μg mL–1) and the cefotaxime MIC 128-fold (128 to 1 μg mL–1). At 20 μM, all five compounds were less active than 2. Compounds 15, and 17–19 reduced the ampicillin MIC four-fold, while compound 25 was negligibly active (two-fold reduction). In combination with cefotaxime, compound 25 was again essentially inactive (two-fold reduction), while compounds 15 and 19 showed modest activity (16-fold reduction). The p-chloro derivative 18 was also moderately active (eight-fold reduction). Interestingly, compound 17, which shows the least structural deviation from 2, maintains activity and reduces the cefotaxime MIC 64-fold.
Table 3. Antibiotic potentiation with the most potent biofilm inhibitors.
| Compound | Ampicillin | Fold reduction | Cefotaxime | Fold reduction |
| — | 256 | — | 128 | — |
| 2 | 16 a | 16 | 1 | 128 |
| 15 | 64 | 4 | 8 | 16 |
| 17 | 64 | 4 | 2 | 64 |
| 18 | 64 | 4 | 16 | 8 |
| 19 | 64 | 4 | 8 | 16 |
| 25 | 128 | 2 | 64 | 2 |
aCompounds were dosed at 20 μM. All values are in μg mL–1.
In conclusion, screening of 21 additional 2-ABIs produced 12 additional compounds with sub 10 μM IC50 values. The original lead 2-ABI 2, as well as 17 and 25 were found to be the most potent, returning IC50's < 4.0 μM. Interestingly, all analogs in this study were less toxic to the bacterium than 2 as assessed by the MIC assay. Follow up studies of colony count analysis of bacterial culture treated with either 17 or 25 at their IC50 values demonstrated no difference between treated and untreated samples, further establishing that these compounds are non-toxic to bacteria. As we observed in SAR studies with compound 1, anti-biofilm and β-lactam potentiation appear to be de-coupled, as compounds 17 and 25 are equipotent with 2 yet show reduced or abrogated potentiation capability. On-going efforts are being made to identify novel 2-ABIs with suitable physiochemical properties that will allow evaluation of activity in vivo as well as mechanistic studies to determine how these molecules inhibit biofilm formation.
The authors would like to thank the National Institutes of Health (AI106733) for support as well as C. Norris for insight.
Conflicts of interest
Dr. Melander is co-founder of Agile Sciences, a biotechnology company that seeks to commercialize both anti-biofilm and antibiotic adjuvant small molecules.
Supplementary Material
Footnotes
†Electronic supplementary information (ESI) available: New compound characterization, MIC and biofilm assay screens. See DOI: 10.1039/c9md00025a
References
- Donlan R. M., Costerton J. W. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. B., Givskov M. Int. J. Med. Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Davies D. Nat. Rev. Drug Discovery. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Singh P. K., Schaefer A. L., Parsek M. R., Moninger T. O., Welsh M. J., Greenberg E. P. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Geneva: World Health Organization, 2018, Licence: CC BY-NC-SA 3.0 IGO.
- Frieden T. R., Sterling T., Pablos-Mendez A., Kilburn J. O., Cauthen G. M., Dooley S. W. N. Engl. J. Med. 1993;328:521–526. doi: 10.1056/NEJM199302253280801. [DOI] [PubMed] [Google Scholar]
- Awofeso N. Bull. W. H. O. 2008;86:B–D. doi: 10.2471/BLT.07.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackart D. F., Hascall-Dove L., Caceres S. M., Kirk N. M., Podell B. K., Melander C., Orme I. M., Leid J. G., Nick J. A., Basaraba R. J. Pathog. Dis. 2014;70:359–369. doi: 10.1111/2049-632X.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S., Richards J. P., Ojha A. K. Expert Rev. Anti-infect. Ther. 2012;10:1055–1066. doi: 10.1586/eri.12.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaraba R. J., Ojha A. K. Microbiol. Spectrum. 2017;5(3):TBTB2-0024-2016. doi: 10.1128/microbiolspec.TBTB2-0024-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A., Grant S. S., Kawate T., Iwase N., Shimizu M., Wivagg C., Silvis M., Kazyanskaya E., Aquadro J., Golas A., Fitzgerald M., Dai H., Zhang L., Hung D. T. ACS Chem. Biol. 2012;7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ge F., Tan Y., Zhang G., Li W. Int. J. Mol. Sci. 2016;17:689. doi: 10.3390/ijms17050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes L., Ebsworth-Mojica K., DiDone L., Li S. G., Freundlich J. S., Connell N., Dunman P. M., Krysan D. J. PLoS One. 2015;10:e0129234. doi: 10.1371/journal.pone.0129234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackart D. F., Lindsey E. A., Podell B. K., Melander R. J., Basaraba R. J., Melander C. Pathog. Dis. 2014;70:370–378. doi: 10.1111/2049-632X.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. V., Blackledge M. S., Lindsey E. A., Minrovic B. M., Ackart D. F., Jeon A. B., Obregon-Henao A., Melander R. J., Basaraba R. J., Melander C. Angew. Chem., Int. Ed. 2017;56:3940–3944. doi: 10.1002/anie.201612006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. E., Nguyen C. M., Basaraba R. J., Melander C. Chem. Biol. Drug Des. 2018;92:1403–1408. doi: 10.1111/cbdd.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey E. A., Brackett C. M., Mullikin T., Alcaraz C., Melander C. MedChemComm. 2012;3:1462–1465. doi: 10.1039/C2MD20244A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins W. M., Nguyen T. Vu, Hahn N. A., Baker J. T., Kuo L. G., Kaur D., Melander R. J., Gunn J. S., Melander C. MedChemComm. 2018;9:1547–1552. doi: 10.1039/c8md00298c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.