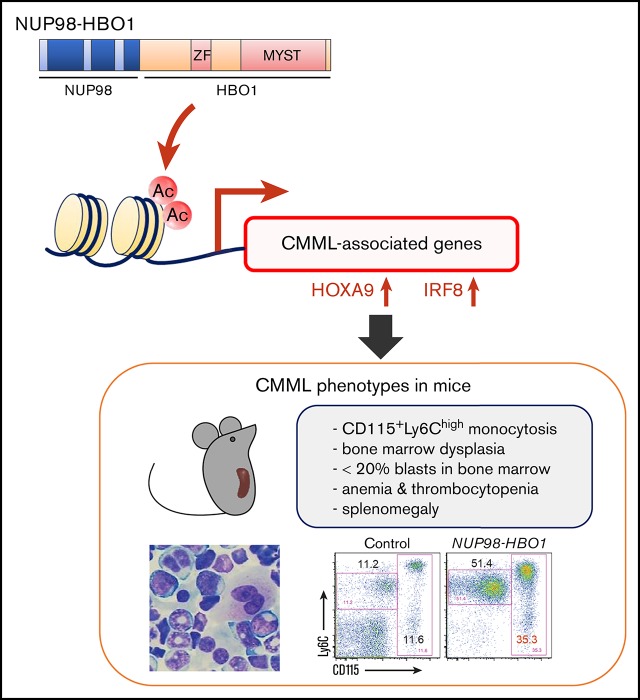
Key Points
NUP98-HBO1 induces clinically relevant diversity of CMML phenotypes, such as increased classical monocytes, myelodysplasia, and cachexia.
CMML-specific transcriptional signature is induced through aberrant histone acetylation.
Abstract
Chronic myelomonocytic leukemia (CMML) constitutes a hematopoietic stem cell (HSC) disorder characterized by prominent monocytosis and myelodysplasia. Although genome sequencing has revealed the CMML mutation profile, the mechanism of disease development remains unclear. Here we show that aberrant histone acetylation by nucleoporin-98 (NUP98)-HBO1, a newly identified fusion in a patient with CMML, is sufficient to generate clinically relevant CMML pathogenesis. Overexpression of NUP98-HBO1 in murine HSC/progenitors (HSC/Ps) induced diverse CMML phenotypes, such as severe leukocytosis, increased CD115+ Ly6Chigh monocytes (an equivalent subpopulation to human classical CD14+ CD16− monocytes), macrocytic anemia, thrombocytopenia, megakaryocyte-lineage dysplasia, splenomegaly, and cachexia. A NUP98-HBO1–mediated transcriptional signature in human CD34+ cells was specifically activated in HSC/Ps from a CMML patient cohort. Besides critical determinants of monocytic cell fate choice in HSC/Ps, an oncogenic HOXA9 signature was significantly activated by NUP98-HBO1 fusion through aberrant histone acetylation. Increased HOXA9 gene expression level with disease progression was confirmed in our CMML cohort. Genetic disruption of NUP98-HBO1 histone acetyltransferase activity abrogated its leukemogenic potential and disease development in human cells and a mouse model. Furthermore, treatment of azacytidine was effective in our CMML mice. The recapitulation of CMML clinical phenotypes and gene expression profile by the HBO1 fusion suggests our new model as a useful platform for elucidating the central downstream mediators underlying diverse CMML-related mutations and testing multiple compounds, providing novel therapeutic potential.
Visual Abstract
Introduction
Aging of the global population increases the incidence of myeloid malignancies, which generally occur in elderly individuals, such as myelodysplastic syndromes (MDS) and myeloproliferative neoplasms. Among these, chronic myelomonocytic leukemia (CMML) constitutes a discrete clonal hematopoietic malignancy that shares clinical features with MDS and myeloproliferative neoplasms1-4 but is characterized by an absolute increase in peripheral blood (PB) monocytes and myelodysplasia.1-4 Systemic symptoms, such as weight loss and cachexia, are also prominent compared with other myeloid disorders.2 However, despite the distinct clinical features of CMML, the molecular pathogenesis of disease development has remained elusive.
To understand the molecular basis of CMML, recent work in the field has focused on genome sequencing and revealed the mutation status of patients with CMML.3 Although many recurrent gene mutations have been identified in almost all patients with CMML,3 the mutational profile is similar to that of related disorders, such as MDS. Mouse modeling of each mutation frequently found in patients did not display bona fide CMML phenotypes.5-7 Thus, the precise mechanisms of how these mutations give rise to the characteristic CMML phenotypes are largely unknown. Moreover, under the current circumstance in which limited preclinical models are available for dissecting CMML pathobiology and testing new treatment options,8,9 no curative options are available for the majority of the patients with CMML.4
Aberrant acetylation of histones has been reported in various cancers, and its contribution to tumorigenesis has been demonstrated. Histone acetyltransferases (HATs), which target lysine residues on nucleosomal histones, function as transcriptional activators and regulators. Among HATs, the Moz, Ybf2/Sas3, Sas2, and Tip60 (MYST) family is composed of evolutionarily conserved enzymes that are assembled into multi-subunit protein complexes.10 HBO1 (also known as MYST2 and KAT7) is a HAT belonging to a MYST family that includes TIP60, MOZ/MORF, and MOF in humans. MYST HATs play critical roles in gene-specific transcriptional regulation, DNA damage response and repair, as well as DNA replication.10-13 Moreover, MYST family members, except for HBO1, have been shown to exhibit oncogenic potential,10 and their critical roles in leukemia development are well documented.14-17 Aberrant expression of HBO1 has also been reported in some cancers.18 However, less is known regarding the role of HBO1 HAT activity in leukemogenesis.
Recently, we identified a new nucleoporin-98 (NUP98)-HBO1 fusion containing an intact MYST domain in a patient with CMML. HBO1 is the first NUP98 fusion partner encoding HAT. Many NUP98 fusion proteins are suspected to act as aberrant transcription factors. Given the critical role of the HBO1 MYST domain in regulating histone acetylation status, we hypothesized that the NUP98-HBO1 fusion could induce aberrant histone acetylation and sequential dysregulation of target genes, leading to CMML development. Thus, using a mouse model system and human cells, we evaluated the pathobiologic impact of the NUP98-HBO1 fusion on disease development in the present study.
Materials and methods
Patients
We examined a patient with CMML, whose diagnosis was based on morphologic, immunophenotypic, and genetic studies according to the 2017 version of World Health Organization classification. Mononuclear cells of patients were isolated from bone marrow (BM) or PB samples. For the validation of HOXA9 messenger RNA (mRNA) expression, we examined CD34+ BM cells obtained from healthy donors (HDs; n = 4) or from patients with MDS with single lineage dysplasia/multilineage dysplasia (n = 3), MDS with excess blast (EB; n = 6), CMML1 (n = 5), and CMML2 (n = 3). The study was approved by the institutional review board at Juntendo University School of Medicine and Bunkyo Gakuin University. Patients provided written informed consent for the study, according to the Declaration of Helsinki. Chromosomal analysis and double-color split fluorescence in situ hybridization (FISH) assay using NUP98 (mapped 11q15) and D11Z1 (chromosome 11 centromere) probes were performed in accordance with standard techniques (SRL Inc).
Mice
C57BL/6 mice (7-week-old, female) were purchased from Tokyo Laboratory Animals Science (Tokyo, Japan). All animal studies were conducted according to an approved institutional animal care protocol and guidelines of the Tokyo University of Pharmacy and Life Sciences.
Mouse BMT
Plat-E packaging cells were transfected with the retroviral constructs, and mouse bone marrow transplantation (BMT) was performed as described previously.19 C57BL/6 donor mice were euthanized under anesthesia with cervical dislocation 3 days after intraperitoneal administration of 150 mg/kg 5-fluorouracil. BM mononuclear cells were isolated from the femurs and tibias of the donor mice immediately after euthanasia. After stimulation with 50 ng/mL mouse stem cell factor, mouse FMS-like tyrosine kinase 3 (FLT3) ligand (FLT3-L), mouse interleukin 6 (IL-6), and mouse thrombopoietin, the cells were transduced with the retrovirus for 60 hours using RetroNectin-coated dishes. Then, 3 to 5 × 105 of the nonsorted cells were injected into sublethally irradiated wild-type recipient mice.
Flow cytometry
Flow cytometric analysis was performed using a fluorescence-activated cell sorter (FACS) Canto (Becton Dickinson [BD]). Murine PB and BM cells were immunostained using the following anti-mouse antibodies: Gr-1 (RB6-8C5), CD115 (AFS98), c-Kit (2B8), and Ly6C (HK1.4) (Biolegend). Phosphate-buffered saline containing 2% fetal bovine serum (FBS) and 2 mM EDTA was used as a FACS buffer. Data were analyzed using FlowJo software (Tree Star). Human cells were immunostained using the following antibodies: CD34 (YB5.B8), CD117 (104D2), CD13 (WM15), CD33 (WM53), CD11b (ICRF44), CD14 (M5E2), CD56 (B159) (BD), and CD16 (B73.1) (Biolegend). For cell-cycle analysis, the cells were stained using an allophycocyanin BrdU Flow Kit (BD).
Microarray
FACS-sorted CB CD34+ cells transduced with NUP98-HBO1 fusion or empty vector were subjected to RNA extraction and hybridization on the CodeLink Human Whole Genome Bioarray. Total RNA was extracted from the CB CD34+ cells using TRIzol (Invitrogen). The data have been deposited in Gene Expression Omnibus (GSE112909).
Gene set enrichment analysis (GSEA)
To generate the NUP98-HBO1–induced gene set, 434 genes demonstrating over twofold upregulation in the NUP98-HBO1–transduced human CB CD34+ cells compared with the control cells were used. The NUP98-HBO1–induced gene set was then tested for enrichment in human cohorts of CMML and MDS using GSEA (v3.0).20 The following parameters were used: 1000 gene_set permutations, Signal2Noise ranking metric, and descending order real mode for gene sorting.
Genes identified by Olsson et al26(Fig1b,2d) were used to generate the gene sets of determinants for monocytic or granulocytic cell fate in hematopoietic stem cell/progenitors (HSC/Ps). GSEA analysis was run on Log2 expression data of human CB CD34+ cells transduced with NUP98-HBO1 or empty control vector. The following parameters were used; 1000 gene_set permutations, Diff_of_Classes ranking metric, and descending order real mode for gene sorting.
Retrovirus transduction into human CD34+ primary cells and cell culture
The Plat-GP packaging cell line was transfected with pMXs and pCMV.VSV-G plasmids as described previously.21 Human CD34+ cells were purified from cord blood (CB) cells with the CD34 MicroBead Kit using an autoMACS system (Miltenyi Biotec). At 3 days after transduction, GFP+ cells were sorted by FACS Aria (BD). Cells were cultured continuously in Iscove modified Dulbecco medium (IMDM) containing 20% FBS; 100 ng/mL each of human FLT3 (hFLT3) ligand (hFLT3-L), human stem cell factor (hSCF), and human thrombopoietin; and 2 mM l-glutamine. For long-term growth, IMDM was supplemented with the following: 20% FBS; 100 ng/mL each of hFLT3-L, hSCF, and human thrombopoietin; 20 ng/mL each of human interleukin-6 (hIL-6) and hIL-3 (Peprotech); and 2 mM l-glutamine. Cells were counted weekly using Trypan blue dye exclusion and analyzed by flow cytometry and morphology analyses.
CFU replating assays
Sorted cells (1 × 104) were resuspended in MethoCult H4034 medium (StemCell Technologies) containing hSCF, human granulocyte-colony stimulating factor, human granulocyte/macrophage-colony stimulating factor, hIL-3, and human erythropoietin. After 14 days in culture, colonies were counted. Cells were then suspended in methylcellulose medium, and 104 cells were plated again for the colony-forming unit (CFU) replating assay. Remaining cells were used for cell number counting and cytospin centrifugation for morphological and flow cytometry analyses.
Treatment with 5-Aza
FACS-sorted NUP98-HBO1 cells from the diseased mice were used for in vitro and in vivo treatment. In in vitro cell proliferation assay, 1 × 104 cells were cultured in IMDM containing 20% fetal calf serum, 50 ng/mL mouse stem cell factor, 10 ng/mL mouse IL-6, and IL-3. Freshly prepared 5-Azacytidine (5-Aza; Sigma) was added every other day. For in vivo treatment, cells from the NUP98-HBO1 clone obtained from the diseased mice were injected into sublethally irradiated wild-type recipient mice. From day 14, phosphate-buffered saline or 2.5 to 5.0 mg/kg 5-Aza were injected intraperitoneally once every other day for a total of 5 injections. For second-round treatment, 5-Aza was injected intraperitoneally once every other day for a total of 5 injections from day 50.
Statistical analysis
For comparison of 2 independent samples, normally distributed variables were compared using the Student t test and nonnormally distributed variables by the Mann-Whitney U test. For multiple pairwise comparisons, the data were analyzed by 1-way analysis of variance followed by Dunnett’s multiple comparison test, or differences between individual groups were estimated using the Steel-Dwass test. The data in Figure 5D were assessed with the Shirley-Williams test. Survival curves were estimated by Kaplan-Meier analysis and compared using the log-rank test. P < .05 was considered statistically significant.
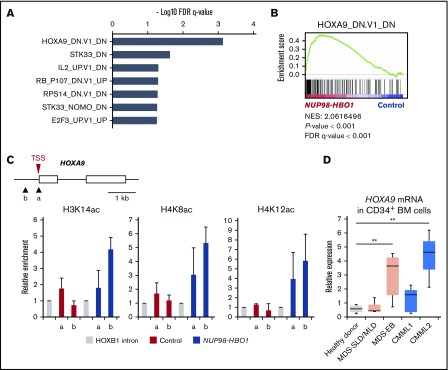
Figure 5.
Upregulation of oncogenic HOXA9 signature by NUP98-HBO1–mediated aberrant histone acetylation. (A-B) GSEA of MSigDB C6-oncogenic signatures. The top-ranked signatures (A) and representative plot of ranked HOXA9-induced gene signature (B) are shown. (C) Acetylation levels of H3K14, H4K8, and H4K12 of the HOXA9 gene promoter. ChIP-qPCR was performed using CB CD34+ cells. In the schematic of HOXA9 promoter region, arrowheads (a and b) indicate the location of the primers. (D) HOXA9 mRNA expression in CD34+ BM cells obtained from the indicated patients and HDs. MDS with single lineage dysplasia/multilineage dysplasia, n = 3; MDS with EB, n = 6; CMML1, n = 5; CMML2, n = 3; HD, n = 4. Data are presented as mean ± SD. **P < .01. TSS, transcription start site.
Results
Identification of a novel NUP98-HBO1 fusion in a patient with CMML
A 69-year-old woman with a history of gastric cancer infiltration of the pancreas was referred to our hematology department for leukocytosis (36.19 × 109/L). She had received postoperative chemotherapy, including fluorouracil, cisplatin, docetaxel, and gemcitabine, for 3 years. In PB, myeloblasts were detected (14%), and monocytes were increased to 37%. Neither eosinophilia nor basophilia was observed. Macrocytic anemia and thrombocytopenia were also observed (supplemental Table 1). BM aspirate showed hypercellular marrow with 15% myeloperoxidase-negative myeloblasts without Auer rods. Multilineage dysplasia was observed (Figure 1A). A cytogenetic study of the BM cells revealed a clonal chromosomal abnormality, 46,XX, t(11;17)(p11.5;q21) in 20 of 20 metaphases (Figure 1B). Double-color split FISH assay using NUP98 (mapped to 11q15) and D11Z1 (chromosome 11 centromere) probes indicated the presence of NUP98 rearrangements (Figure 1C). No material was available for further mutational analysis. The patient had a diagnosis of CMML but was not able to be treated with intensive chemotherapy because of multiple metastasis of gastric cancer to, for example, the liver and lung. Despite cytoreduced chemotherapy, she died of uncontrollable leukocytosis in 3 months.
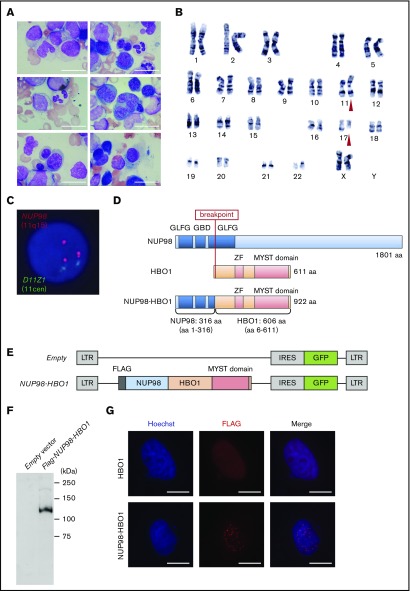
Figure 1.
Identification of a novel HBO1-fusion in a patient with CMML. (A) BM smear from the patient with CMML. Scale bar, 20 µm (original magnification ×1000; Wright-Giemsa stain). (B) G-banding karyotyping analysis of BM cells from the patient with CMML. Red triangles indicate the location of chromosomal translocation. (C) FISH analysis of BM cells from the patient with CMML. (D) Schematic of the CMML patient-derived NUP98-HBO1 fusion. (E) Schematic of the retrovirus vectors. (F) Expression of the NUP98-HBO1 fusion product in 293T cells. (G) Localization of the NUP98-HBO1 fusion protein. HeLa cells were transfected with the indicated vectors 24 hours before immunofluorescence staining. Scale bar, 20 µm (original magnification ×1000; immunofluorescence stain).
As no other cytogenetic abnormality was identified in this patient, we hypothesized that the NUP98 rearrangement on t(11;17)(p11.5;q21) could be responsible for the CMML development. Thus, we attempted to identify the fusion partner gene of NUP98. Using panhandle polymerase chain reaction (PCR) methods, we identified an in-frame fusion of exon 8 of NUP98 and exon 2 of HBO1, resulting in an NUP98-HBO1 fusion gene (Figure 1D). In the fusion protein, a relatively short N-terminus of NUP98, including Gly-Leu-Phe-Gly repeats, was fused to an almost full-length HBO1 protein containing an intact MYST domain. Expression of this fusion gene in the patient cells was confirmed by reverse transcription PCR (RT-PCR; supplemental Figure 1). We cloned the coding region of the full-length NUP98-HBO1 complementary DNA produced by the patient’s cells and generated expression vectors (Figure 1E-F). Consistent with the localization pattern of other NUP98 fusion proteins in previous reports,22 the NUP98-HBO1 fusion protein exhibited a punctate pattern in the nucleus (Figure 1G), which is distinct from the localization pattern of the HBO1 protein and other nuclear components, such as PML bodies (supplemental Figure 2).
Patient-derived novel NUP98-HBO1 fusion generates a variety of CMML phenotypes in mice
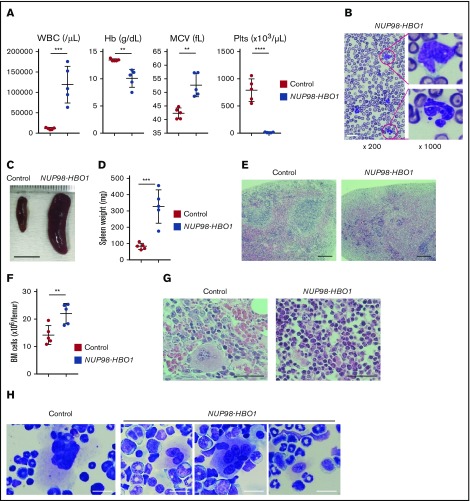
To determine whether NUP98-HBO1 could cause CMML pathogenesis, we retrovirally introduced the NUP98-HBO1 complementary DNA into murine BM cells obtained from wild-type C57BL/6 mice and performed an in vivo BMT assay. The NUP98-HBO1–transduced cells were transplanted into sublethally irradiated wild-type recipient mice. Empty vector-transduced cells served as controls. After transplantation, the NUP98-HBO1 BMT mice developed severe leukocytosis, macrocytic anemia, and thrombocytopenia within 1 month (Figure 2A). Both monocytes and neutrophils were dominantly increased in PB from the NUP98-HBO1 BMT mice (Figure 2B). Consistent with observations in the majority of CMML cases, abnormally lobated nuclei were prominent in PB neutrophils from the NUP98-HBO1 BMT mice (Figure 2B). The NUP98-HBO1 BMT mice showed progressive weight loss and splenomegaly, which also comprise common clinical features of CMML (Figure 2C-D; supplemental Figure 3). Normal follicular structure of spleen was disrupted in the NUP98-HBO1 BMT mice (Figure 2E). We next assessed the cellularity and morphology of the BM from the moribund NUP98-HBO1 BMT mice. Total BM cell number was significantly increased in the NUP98-HBO1 BMT mice compared with the control mice (Figure 2F-G). Consistent with the phenotypes of nearly 80% of patients with CMML, micro-megakaryocytes and megakaryocytes with abnormally lobated nuclei were found in BM from the NUP98-HBO1 BMT mice (Figure 2H). All the NUP98-HBO1 BMT mice died within 2 to 6 months after BMT without transforming into acute leukemia. These data suggest the sufficient role of NUP98-HBO1 to generate clinically relevant CMML phenotypes.
Figure 2.
Clinically relevant CMML phenotypes in the patient-derived NUP98-HBO1 BMT mice. (A) White blood cell (WBC) counts, hemoglobin (Hb), mean corpuscular volume (MCV), and platelet (Plts) counts in PB from the control mice transplanted with empty vector transduced cell (Control) and NUP98-HBO1 BMT mice (NUP98-HBO1) at 8 to 10 weeks after transplantation (n = 5 per group). Data are presented as mean ± standard deviation (SD). (B) Diff-Quick (Modified Giemsa) staining of PB smear from the NUP98-HBO1 BMT mice. Scale bar, 20 µm. (C-D) Representative images (C; scale bar, 10 mm) and weight (D) of the spleen from the control and NUP98-HBO1 BMT mice. Data are presented as mean ± SD. (E) Spleen sections from the control and NUP98-HBO1 BMT mice. Scale bar, 200 µm (original magnification ×40; hematoxylin and eosin stain). (F) BM cellularity in the indicated mice. (G) BM sections from the control and NUP98-HBO1 BMT mice. Scale bar, 40 µm (original magnification ×200; hematoxylin and eosin stain). (H) BM cells from the control and NUP98-HBO1 BMT mice. Scale bar, 20 µm (original magnification ×1000; Diff-Quick stain). **P < .01, ***P < .001, ****P < .0001.
NUP98-HBO1 BMT mice faithfully recapitulate the characteristic monocytosis of CMML
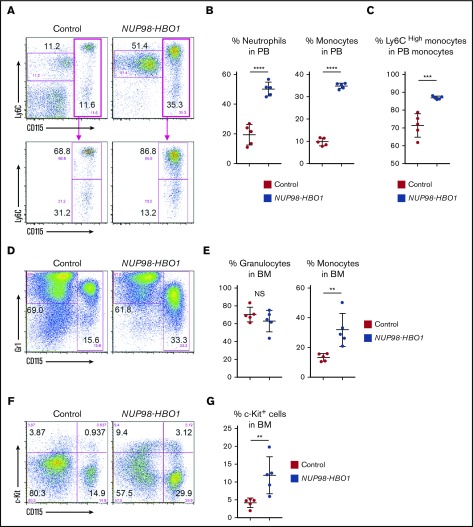
To further dissect the NUP98-HBO1–induced hematological phenotypes, we analyzed PB and BM cells using flow cytometry. Consistent with the morphological assessment, both monocytes and neutrophils were significantly increased in PB from the NUP98-HBO1 BMT mice compared with control mice (Figure 3A-B). Accumulating clinical evidence has indicated a variety of characteristic phenotypes of CMML. Recently it has been reported that an increase in classical CD14+ CD16− monocytes serves as a useful diagnostic signature of CMML, exhibiting both high sensitivity and specificity.23 Notably, we found significantly increased CD115+ Ly6Chigh monocytes, an equivalent subpopulation to human classical CD14+ CD16− monocytes, in the NUP98-HBO1 BMT mice (Figure 3A,C). Monocytes were also significantly increased in BM from the moribund NUP98-HBO1 BMT mice compared with control mice, while the frequency of granulocytes did not change (Figure 3D-E). Although immature c-Kit+ cells, including monocytic lineage cells, were increased in BM from the moribund NUP98-HBO1 BMT mice, their frequency in the BM was <20% (Figure 3F-G). Taken together, these results indicate that NUP98-HBO1 induces a clinically relevant monocytic feature of CMML.
Figure 3.
Increased classical monocytes in NUP98-HBO1 BMT mice. (A) Flow cytometric analysis of CD115/Ly6C expression in PB WBCs from the control and NUP98-HBO1 BMT mice at 8 to 10 weeks after transplantation. (B) Percentage of monocytes (CD115+) and neutrophils (CD115− Ly6Cdim) in PB WBCs from the control and NUP98-HBO1 BMT mice. (C) Percentage of inflammatory monocytes (CD115+ Ly6Chigh) in PB monocytes from the control and NUP98-HBO1 BMT mice. (D) Flow cytometric analysis of Gr1/CD115 expression in BM cells from the control at 8 to 10 weeks after BMT and moribund NUP98-HBO1 BMT mice. (E) Percentage of granulocytes (CD115− Gr1high) and monocytes (CD115+) in BM from the control and NUP98-HBO1 BMT mice. (F) Flow cytometric analysis of c-Kit/CD115 expression in BM cells from the control and NUP98-HBO1 BMT mice. (G) Percentage of c-Kit+ immature cells in BM from the control and NUP98-HBO1 BMT mice. Throughout, n = 5 per group; data are presented as mean ± SD. **P < .01, ***P < .001, ****P < .0001. NS, not significant.
NUP98-HBO1 induces a CMML-specific gene signature through histone acetylation
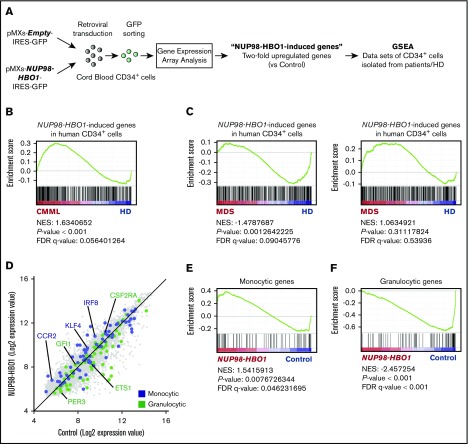
To clarify the effect of the NUP98-HBO1 fusion oncoprotein on downstream gene regulation, we next performed gene expression array analysis of NUP98-HBO1–transduced human CB CD34+ cells (Figure 4A). The expression pattern of cell surface makers of NUP98-HBO1–transduced CB CD34+ cells are shown in supplemental Figure 4. Expansion of CD14+ CD16− population and decrease in CD14+ CD16+ population were observed (supplemental Figure 5). We next generated an NUP98-HBO1–induced gene set using the 434 genes exhibiting twofold upregulation in the NUP98-HBO1–transduced human CB CD34+ cells compared with control cells (Figure 4A). We then tested these genes for enrichment in a human CMML cohort (E-MTAB-1044) using GSEA. This cohort contains patients with CMML (n = 15) and HDs (n = 4). GSEA revealed that expression of the NUP98-HBO1–induced genes is significantly upregulated in the CMML cohort (Figure 4B; supplemental Figure 6). Given that CMML shares some common phenotypes, such as BM dysplasia, with MDS, we also tested these genes in human MDS cohorts (GSE43399 and GSE19429)24,25 (Figure 4C). One cohort contains BM CD34+ cells from patients with MDS (n = 26) and HD (n = 4) (GSE43399). The other cohort contains BM CD34+ cells from patients with MDS (n = 183) and HD (n = 17) (GSE19429). However, these genes were not significantly upregulated in the MDS cohorts (Figure 4C), suggesting that the NUP98-HBO1 fusion oncoprotein could induce a CMML-associated gene signature.
Figure 4.
CMML-specific gene signature in NUP98-HBO1–transduced human CD34+cells. (A) Schematic diagram of the experimental strategy for gene expression array analysis. (B) GSEA plot showing the ranked NUP98-HBO1–induced genes in CB CD34+ cells. NUP98-HBO1–induced genes were tested in the data set of PB CD34+ cells from patients with CMML (n = 15) and HDs (n = 4) (E-MTAB-1044). (C) GSEA plot showing the ranked NUP98-HBO1–induced genes in CB CD34+ cells. NUP98-HBO1–induced genes were tested in the data set of BM CD34+ cells from patients with MDS (n = 26) and HD (n = 4) (GSE43399) or MDS (n = 183) and HD (n = 17) (GSE19429). (D) Scatter plot of NUP98-HBO1–induced genes in CB CD34+ cells. (E-F) Ranked monocytic gene signature (E) and granulocytic gene signature (F) were tested in the data set of NUP98-HBO1–induced genes in CB CD34+ cells.
As the mice transplanted with NUP98-HBO1–transduced BM HSC/Ps developed prominent monocytosis, we hypothesized that NUP98-HBO1 could activate genes that are responsible for monocytic lineage cell fate choice in HSC/Ps. Recently, a single-cell-RNA-Sequencing–based study has identified critical genes for monocytic or granulocytic lineage commitment in murine HSC/Ps.26 Thus, we tested these monocytic or granulocytic genes for enrichment in the NUP98-HBO1–transduced CB CD34+ cells. GSEA revealed that genes that are critical for monocytic cell fate choice, such as IRF8, KLF4, and CCR2, are significantly enriched in NUP98-HBO1–transduced CB CD34+ cells compared with control cells (Figure 4D-E). In contrast, genes that are responsible for granulocytic cell fate choice were significantly downregulated in NUP98-HBO1–transduced CB CD34+ cells compared with control cells (Figure 4D-F). These results support the phenotype of strong monocytic bias in the NUP98-HBO1 BMT mice.
Dysregulation of histone acetylation induces an oncogenic HOXA9 signature in NUP98-HBO1–transduced human CD34+ cells
To dissect the NUP98-HBO1–induced oncogenic gene signature, we further tested gene sets of oncogenic signatures that are dysregulated in cancer (MSigDB)20 for enrichment in the NUP98-HBO1–transduced CB CD34+ cells (Figure 5A). Among the several gene sets that were significantly upregulated in NUP98-HBO1–transduced cells, the HOXA9-induced gene signature27 comprised the most significantly enriched gene set (Figure 5A-B). As histone acetylation constitutes a critical mechanism of MYST family member–mediated tumorigenesis, we hypothesized that the NUP98-HBO1–derived oncogenic signatures are regulated by histone acetylation. Whereas MOZ and MORF HATs are considered to be responsible for histone 3 (H3) acetylation, HBO1 HAT has been reported to primarily acetylate histone 4 (H4),28-30 although it can also acetylate H3 at K14 (H3K14).31-33 To test our hypothesis, we focused on HOXA9 and performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) assays using the NUP98-HBO1–transduced CB CD34+ cells. ChIP-qPCR revealed a significant increase in the acetylation levels of H4K8, H4K12, and H3K14 at the promoter of HOXA9 in the NUP98-HBO1–transduced CB CD34+ cells (Figure 5C). We note that the expression level of HOXA9 mRNA was significantly upregulated in BM CD34+ cells from patients with CMML2, as well as MDS-EB, compared with that in patients with low-risk MDS and HDs (Figure 5D). Although there is no statistical significance, the mean expression level of HOXA9 mRNA in the CMML1 samples was 2.22-fold higher than that in the HD samples (Figure 5D). This indicates that the activation of a HOXA9 signature may constitute a common event in patients with CMML and also play a critical role for disease progression. Taken together, these results suggest that NUP98/HBO1 activates oncogenic signatures through acetylation of H4 and H3, resulting in CMML development.
Histone acetyl transferase activity is critical for NUP98-HBO1–mediated clonal advantage and cell proliferation
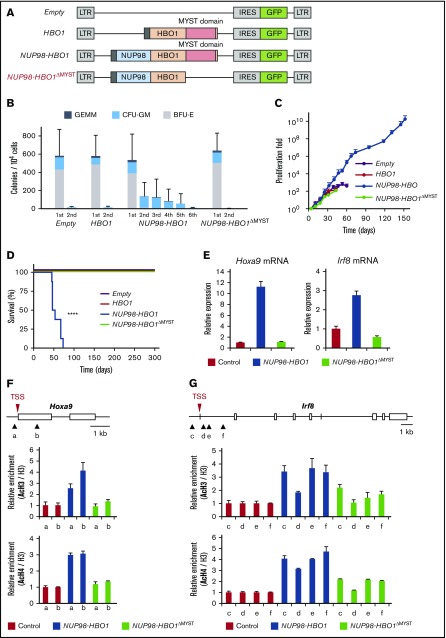
To clarify the essential role of histone acetylation in NUP98-HBO1–mediated CMML pathogenesis, we generated a deletion mutant of the MYST domain (NUP98-HBO1ΔMYST) (Figure 6A; supplemental Figure 7). In serial replating CFU assays, empty vector– or wild-type HBO1-transduced CB CD34+ cells stopped growing after the second round of replating (Figure 6B). In contrast, the NUP98-HBO1–transduced CB CD34+ cells could be replated up to 6 times (Figure 6B), indicating the increased self-renewal potential in NUP98-HBO1–transduced cells. Notably, deletion of the MYST domain completely attenuated the replating potential in NUP98-HBO1–transduced cells (Figure 6B). Using long-term liquid culture assay, we also assessed the impact of MYST domain deletion on the proliferation ability of the NUP98-HBO1–transduced cells. The NUP98-HBO1–transduced CB CD34+ cells continued growing over 1010-fold for 150 days (Figure 6C; supplemental Figure 8). However, the NUP98-HBO1ΔMYST–transduced cells lost their proliferation advantage (Figure 6C). Taken together, these results suggest that the MYST domain is critical for NUP98-HBO1–mediated clonal advantage and cell expansion.
Figure 6.
Critical role of the MYST domain in NUP98-HBO1–driven leukemogenesis in vitro and CMML development in vivo. (A) Schematic of the retrovirus vectors. (B) CFU serial replating assay using CB CD34+ cells transduced with the indicated vectors. Data are presented as mean ± SD from triplicates. (C) Cell proliferation in the liquid culture of the CB CD34+ cells transduced with the indicated vectors. Data are presented as mean ± SD from triplicates. (D) Survival of empty-control (n = 8), HBO1 (n = 6), NUP98-HBO1 (n = 8), and NUP98-HBO1ΔMYST–BMT mice (n = 4). (E) Hoxa9 mRNA and Irf8 mRNA expression in murine BM HSC/Ps transduced with the indicated vectors. Data are presented as mean ± SD from triplicates. (F-G) Acetylation levels of H3 and H4 at the indicated locus of the Hoxa9 (F) and Irf8 gene (G). ChIP-qPCR was performed using BM HSC/Ps transduced with the indicated vectors. In the schematic of HOXA9 promoter region, arrowheads (a to f) indicate the location of the primers. AcH3, acetyl-histone 3; AcH4, acetyl-histone 4; BFU-E, burst forming unit-erythroid; CFU-GM, colony-forming unit granulocyte/monocyte; GEMM, colony-forming unit–granulocyte, erythrocyte, macrophage, megakaryocyte.
Elimination of the HAT activity of NUP98-HBO1 blocks CMML development in vivo
To evaluate the significance of HAT activity in NUP98-HBO1–mediated disease development in vivo, we performed a BMT assay using NUP98-HBO1ΔMYST–transduced cells. The NUP98-HBO1–BMT mice developed a lethal CMML phenotype, whereas mice that received transplants of empty or wild-type HBO1 vectors remained healthy over 12 months (Figure 6D). Notably, deletion of the MYST domain completely attenuated CMML development (Figure 6D). As hypermethylation of DNA is one of the characteristic features of CMML, we also assessed the NUP98-HBO1–mediated DNA methylation status. However, we found no prominent alteration of DNA methylation status in NUP98-HBO1–transduced murine BM cells (supplemental Figure 9). Taken together, these results suggest that dysregulated histone acetylation is critical for generating NUP98-HBO1–mediated CMML pathogenesis.
Given that the gene signatures of HOXA9 and monocytic cell fate choice were activated in NUP98-HBO1–transduced human CB CD34+ cells, we confirmed the expression of Hoxa9 and Irf8, a critical cell fate determinant toward the monocytic lineage, in our CMML model. The expression level of Hoxa9 and Irf8 transcripts in the NUP98-HBO1–transduced murine BM cells was significantly upregulated compared with that in control cells (Figure 6E). In contrast, significant reduction in the expression level of Hoxa9 and Irf8 transcripts was observed in the NUP98-HBO1ΔMYST–transduced cells compared with that in the NUP98-HBO1–transduced cells (Figure 6E). MYST domain abrogation significantly attenuated the acetylation levels of H4 and H3 at the promoters of Hoxa9 and Irf8 (Figure 6F-G). These results suggest that dysregulated HAT activity is critical for activating oncogenic signatures and monocytic genes in NUP98-HBO1 HSC/Ps, leading to CMML development.
NUP98-HBO1 clones are sensitive to the clinically used therapeutic option for patients with CMML
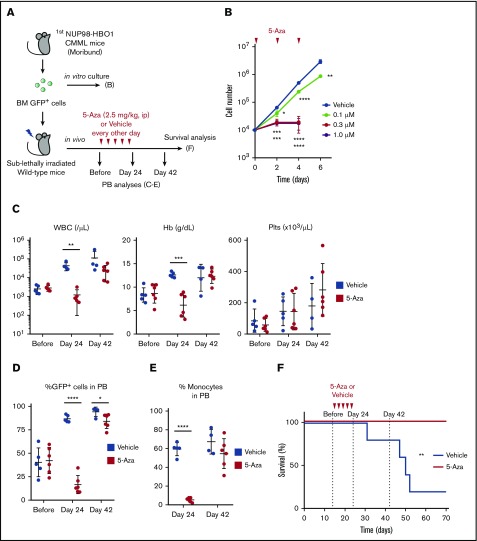
Hypomethylating agents (HMAs) such as 5-Aza and 5-aza-2-deoxycytidine have been used for the treatment of patients with MDS and CMML.4,34,35 In fact, DNA hypomethylation is induced by these HMAs. However, the mechanisms of how these agents work and the targets of these agents remain largely unclear.34,35 Nonetheless, these HMAs have been widely used in clinical practice for a decade. Given that the NUP98-HBO1 fusion recapitulates clinically relevant CMML phenotypes and transcriptional signature, we tested whether 5-Aza might exhibit therapeutic efficacy in our NUP98-HBO1–induced CMML model (Figure 7A). To evaluate the growth inhibitory effect of 5-Aza in vitro, we sorted NUP98-HBO1 clones from the diseased mice and treated them with 5-Aza (Figure 7A-B). Administration of 5-Aza significantly inhibited the proliferation of NUP98-HBO1 clones even at low concentration, although DNA methylation status and the expression levels of Hoxa9 and Irf8 transcripts did not change (Figure 7B; supplemental Figure 10A-B). To determine the therapeutic efficacy of 5-Aza in vivo, we next transplanted the NUP98-HBO1 clones obtained from the diseased mice into sublethally irradiated wild-type recipient mice (Figure 7A). The secondary recipient mice also developed the CMML phenotype without transformation toward an acute leukemic phase, indicating that the NUP98-HBO1–induced CMML phenotype is transplantable. In response to the 5-Aza treatment, tumor burden was temporarily but significantly decreased with limited hematological adverse effects (Figure 7C-D). Monocytosis was also temporarily improved (Figure 7E). Although the NUP98-HBO1 clones were not eradicated in the responding NUP98-HBO1 CMML mice, low-dose 5-Aza treatment significantly prolonged their survival in vivo (Figure 7F; supplemental Figure 11A-B). These results suggest that NUP98-HBO1–induced CMML cells are sensitive to HMA.
Figure 7.
Efficacy of the current therapeutic option in the NUP98-HBO1–derived CMML model. (A) Schematic of in vitro and in vivo treatment. (B) Cell proliferation in the liquid culture of the NUP98-HBO1–transduced murine BM cells. 5-Aza was added every other day to maintain the indicated concentration. Data are presented as mean ± SD from triplicates. (C-D) WBC counts, Hb, and Plts counts in PB (C) and PB chimerism (D) of the NUP98-HBO1 BMT mice with or without 5-Aza treatment (Vehicle, n = 5; 5-Aza, n = 6). Data are presented as mean ± SD. (E) Percentage of monocytes (CD11b+ CD115+) in PB from the NUP98-HBO1 BMT mice with or without 5-Aza treatment (Vehicle, n = 5; 5-Aza, n = 6). Data are presented as mean ± SD. (F) Survival of NUP98-HBO1 BMT mice with or without 5-Aza treatment (Vehicle, n = 5; 5-Aza, n = 6). *P < .05, **P < .01, ***P < .001, ****P < .0001. ip, intraperitoneally.
Discussion
Despite efforts to identify the mutational profile of patients with CMML, the incidence of mutations does not correspond directly to disease phenotypes. Here, we identified a new chimeric gene, NUP98-HBO1, in a patient with CMML. Using the patient-derived NUP98-HBO1 fusion, we established a mouse model that faithfully recapitulates clinically relevant phenotypic features of CMML. We also confirmed that NUP98-HBO1 induces a CMML-specific gene signature in human CD34+ cells. Notably, a clinically used therapeutic option for patients with CMML was effective in our mouse model at a low dose. These findings indicate the usefulness of this novel CMML model as a platform for dissecting disease pathogenesis and a potent preclinical tool for testing a variety of therapeutic options, thus potentially benefiting the field at both basic research and clinical application levels.
Oncogenic signature analysis highlighted HOXA9-induced genes as the most significantly enriched signature in our CMML model, whereas HOXA9 expression was repressed in patients with early-stage MDS.36 Notably, the expression level of the HOXA9 transcript was increased in BM CD34+ cells from patients with CMML exhibiting disease progression. Given the important roles of HOXA9 in regulating HSC proliferation and repopulation abilities,37 upregulation of HOXA9 and its downstream signaling may explain the NUP98-HBO1–mediated clonal expansion advantage. However, upregulation of HOXA9 signaling alone cannot explain the diverse CMML phenotypes observed in the NUP98-HBO1 BMT mice.14,38 CMML arises from HSCs with cytogenetic and molecular defects.3 As persistent monocytosis constitutes the most characteristic feature of CMML, we hypothesized that NUP98-HBO1 may also regulate genes involved in cell fate choice toward the monocytic lineage at the level of HSC/Ps. Consistent with this, many genes related to monocytic cell fate choice, such as IRF8 and KLF4,26 were upregulated in NUP98-HBO1–transduced human CD34+ cells. This likely underlies the potent monocytic bias in NUP98-HBO1–mediated pathogenesis. In contrast, the majority of genes related to granulocytic cell fate choice were downregulated in the NUP98-HBO1–transduced cells. However, several genes, such as GFI1 and CSF2RA, were upregulated. As GFI1 is a critical cell fate determinant toward granulocytes,26 the upregulation of GFI1 may explain the increase of neutrophils in PB from the NUP98-HBO1–mediated CMML model. Hypersensitivity to granulocyte-macrophage colony-stimulating factor is one of the hallmarks of CMML.39 Notably, CSF2RA encodes the α−subunit protein of the granulocyte-macrophage colony-stimulating factor receptor.
In particular, we note that the NUP98-HBO1–induced genes in human CD34+ cells were significantly enriched in the HSC/Ps from patients with CMML, but not those with MDS. The NUP98-HBO1 fusion generates a clinically relevant diversity of CMML phenotypes without any additional gene mutations. These results afford the possibility that genes dysregulated by NUP98-HBO1 comprise the common pathobiologic mediators of CMML development, which could link the diversity of gene mutations with the specific clinical features for CMML. Further investigation of the overlap between the genes induced by NUP98-HBO1 and the gene signatures driven by CMML-associated mutations will reveal the common underlying molecular mechanisms for this intractable HSC disorder.
Furthermore, low-dose 5-Aza treatment was effective in our CMML model. Unlike MDS, the optimal drug dosage, timing, and duration of HMA administration for patients with CMML have not been well established. Thus, our results may provide a clue for establishing an optimal regimen of HMA treatment of patients with CMML. Aberrant DNA hypermethylation profile is a well-known feature in MDS and CMML and is associated with poor outcome.34,40 DNA hypomethylation and sequential alteration of gene signature are indeed induced by HMAs, and the sensitivity of MDS and CMML clones to HMAs is evident.34,41 Thus, it has been proposed that DNA demethylation is a central mechanism underlying clinical antitumor effect of HMAs. However, the precise mechanisms of how these agents clinically work are still in debate.34,35,42 Besides the effect of DNA demethylation, several other mechanisms, such as viral mimicry, have been reported.35,42 In our current study, although 5-Aza treatment significantly inhibited the proliferation of NUP98-HBO1 clones in vitro and prolonged the survival of the NUP98-HBO1–BMT mice in vivo, we did not find obvious alteration in the global DNA methylation status after 5-Aza treatment. These results indicate the possibility of site-specific hypomethylation by 5-Aza. It is also likely that other mechanisms cooperated with DNA demethylation to induce antitumor effect in the NUP98-HBO1–BMT mice. Considering that our CMML model faithfully mimics the diversity of CMML phenotypes as well as the specific gene signature observed in patients with CMML, our model has potential for revealing the precise molecular mechanisms of HMAs in the pathogenesis of myeloid malignancies.
In summary, our findings suggest a novel histone acetylation-mediated mechanism of CMML development. Specifically, by using a patient-derived NUP98-HBO1 fusion, we established a clinically relevant CMML mouse model that faithfully mimics a variety of phenotypes and gene expression profiles observed in the patients with CMML. Our current mouse model may serve as a useful preclinical tool for dissecting the complex mechanisms of CMML pathogenesis and systemic symptoms observed in the patients with CMML and other cancers, as well as for testing a variety of preclinical options, potentially leading to novel therapeutic targets and strategies.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
This study was supported in part by the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grants 24591398, 25461422, 15K09460 and 16K09831) (H.H. and Y. Harada), and the Grant for Joint Research Project of the Institute of Medical Science, the University of Tokyo (H.H.).
Footnotes
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE112909).
Authorship
Contribution: Y. Hayashi and Y. Harada designed the research, performed experiments, and wrote the manuscript; Y.D., N. Kato, and H.M. performed experiments and prepared the manuscript; Y.K., S.N., J.I., S.H., and Y.M. performed experiments; N.S. provided patients’ samples and clinical information; J.K., A. Ito, I.K., A. Iwama, N. Komatsu, and T.K. contributed helpful discussion; and H.H. conceived and designed the research, collected and interpreted the data, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hironori Harada, Laboratory of Oncology, Tokyo University of Pharmacy and Life Sciences, 1432-1 Horinouchi, Hachioji, Tokyo 192-0392, Japan; e-mail: hharada@toyaku.ac.jp.
References
- 1.Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia. 2013;27(7):1441-1450. [DOI] [PubMed] [Google Scholar]
- 2.Patnaik MM, Parikh SA, Hanson CA, Tefferi A. Chronic myelomonocytic leukaemia: a concise clinical and pathophysiological review. Br J Haematol. 2014;165(3):273-286. [DOI] [PubMed] [Google Scholar]
- 3.Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6(2):e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solary E, Itzykson R. How I treat chronic myelomonocytic leukemia. Blood. 2017;130(2):126-136. [DOI] [PubMed] [Google Scholar]
- 5.Moran-Crusio K, Reavie L, Shih A, et al. . Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue D, Kitaura J, Togami K, et al. . Myelodysplastic syndromes are induced by histone methylation–altering ASXL1 mutations. J Clin Invest. 2013;123(11):4627-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Ilagan JO, Liang Y, et al. . SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakata Y, Ueda T, Nagamachi A, et al. . Acquired expression of Cbl Q367P in mice induces dysplastic myelopoiesis mimicking chronic myelomonocytic leukemia. Blood. 2017;129(15):2148-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunimoto H, Meydan C, Nazir A, et al. . Cooperative epigenetic remodeling by TET2 loss and NRAS mutation drives myeloid transformation and MEK inhibitor sensitivity. Cancer Cell. 2018;33(1):44-59.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avvakumov N, Côté J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26(37):5395-5407. [DOI] [PubMed] [Google Scholar]
- 11.Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22(19):2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avvakumov N, Lalonde ME, Saksouk N, et al. . Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol. 2012;32(3):689-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsunuma R, Niida H, Ohhata T, et al. . UV damage-induced phosphorylation of HBO1 triggers CRL4DDB2-mediated degradation to regulate cell proliferation [published correction appears in Mol Cell Biol. 2018;38(7):e00572-17]. Mol Cell Biol. 2015;36(3):394-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow J, Stanton VP Jr, Andresen JM, et al. . The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14(1):33-41. [DOI] [PubMed] [Google Scholar]
- 15.Kitabayashi I, Aikawa Y, Nguyen LA, Yokoyama A, Ohki M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 2001;20(24):7184-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panagopoulos I, Fioretos T, Isaksson M, et al. . Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13). Hum Mol Genet. 2001;10(4):395-404. [DOI] [PubMed] [Google Scholar]
- 17.Deguchi K, Ayton PM, Carapeti M, et al. . MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3(3):259-271. [DOI] [PubMed] [Google Scholar]
- 18.Iizuka M, Takahashi Y, Mizzen CA, et al. . Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436(1-2):108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe-Okochi N, Kitaura J, Ono R, et al. . AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111(8):4297-4308. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada Y, Inoue D, Ding Y, et al. . RUNX1/AML1 mutant collaborates with BMI1 overexpression in the development of human and murine myelodysplastic syndromes. Blood. 2013;121(17):3434-3446. [DOI] [PubMed] [Google Scholar]
- 22.Fahrenkrog B, Martinelli V, Nilles N, et al. . Expression of leukemia-associated Nup98 fusion proteins generates an aberrant nuclear envelope phenotype. PLoS One. 2016;11(3):e0152321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selimoglu-Buet D, Wagner-Ballon O, Saada V, et al. ; Francophone Myelodysplasia Group. Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood. 2015;125(23):3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellagatti A, Cazzola M, Giagounidis A, et al. . Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24(4):756-764. [DOI] [PubMed] [Google Scholar]
- 25.Heinrichs S, Conover LF, Bueso-Ramos CE, et al. . MYBL2 is a sub-haploinsufficient tumor suppressor gene in myeloid malignancy. eLife. 2013;2:e00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson A, Venkatasubramanian M, Chaudhri VK, et al. . Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537(7622):698-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faber J, Krivtsov AV, Stubbs MC, et al. . HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyon Y, Cayrou C, Ullah M, et al. . ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21(1):51-64. [DOI] [PubMed] [Google Scholar]
- 29.Foy RL, Song IY, Chitalia VC, et al. . Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283(43):28817-28826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell. 2010;37(1):57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishima Y, Miyagi S, Saraya A, et al. . The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011;118(9):2443-2453. [DOI] [PubMed] [Google Scholar]
- 32.Mishima Y, Wang C, Miyagi S, et al. . Histone acetylation mediated by Brd1 is crucial for Cd8 gene activation during early thymocyte development. Nat Commun. 2014;5(1):5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Y, Vlassis A, Roques C, et al. . BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016;35(2):176-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Manero G, Fenaux P. Hypomethylating agents and other novel strategies in myelodysplastic syndromes. J Clin Oncol. 2011;29(5):516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff F, Leisch M, Greil R, Risch A, Pleyer L. The double-edged sword of (re)expression of genes by hypomethylating agents: from viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell Commun Signal. 2017;15(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sashida G, Harada H, Matsui H, et al. . Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun. 2014;5(1):4177. [DOI] [PubMed] [Google Scholar]
- 37.Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14(3):281-291. [DOI] [PubMed] [Google Scholar]
- 38.Abdel-Wahab O, Adli M, LaFave LM, et al. . ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padron E, Painter JS, Kunigal S, et al. . GM-CSF-dependent pSTAT5 sensitivity is a feature with therapeutic potential in chronic myelomonocytic leukemia. Blood. 2013;121(25):5068-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen L, Kantarjian H, Guo Y, et al. . DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28(4):605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figueroa ME, Skrabanek L, Li Y, et al. . MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114(16):3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roulois D, Loo Yau H, Singhania R, et al. . DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.