Abstract
The Eph-ephrin signaling pathway mediates developmental processes and the proper functioning of the adult human body. This distinctive bidirectional signaling pathway includes a canonical downstream signal cascade inside the Eph-bearing cells, as well as a reverse signaling in the ephrin-bearing cells. The signaling is terminated by ADAM metalloproteinase cleavage, internalization, and degradation of the Eph/ephrin complexes. Consequently, the Eph-ephrin-ADAM signaling cascade has emerged as a key target with immense therapeutic potential particularly in the context of cancer. An interesting twist was brought forth by the emergence of ephrins as the entry receptors for the pathological Henipaviruses, which has spurred new studies to target the viral entry. The availability of high-resolution structures of the multi-modular Eph receptors in complexes with ephrins and other binding partners, such as peptides, small molecule inhibitors and antibodies, offers a wealth of information for the structure-guided development of therapeutic intervention. Furthermore, genomic data mining of Eph mutants involved in cancer provides information for targeted drug development. In this review we summarize the distinct avenues for targeting the Eph-ephrin signaling pathway, including its termination by ADAM proteinases. We highlight the latest developments in Eph-related pharmacology in the context of Eph-ephrin-ADAM-based antibodies and small molecules. Finally, the future prospects of genomics- and proteomics-based medicine are discussed.
Keywords: Eph receptor, ADAM proteinase, antibody drug, Eph-specific peptide, small molecule inhibitor
Graphical abstract
Background
Eph receptors, the largest family of Receptor Tyrosine Kinases (RTKs), are key mediators of both developmental processes and the proper function of the adult body (Taylor et al., 2017). Consequently, in recent years they have emerged as some of the most interesting targets for drug development (Boyd et al., 2014; Lamminmaki et al., 2015). This excitement is based on several unique features of the Eph receptors. First, they are activated by the binding of their membrane-attached ligands, the ephrins, which initiates a distinctive ‘bidirectional’ signaling, where the information is transmitted downstream in both the receptor- and the ligand-bearing cells (Egea and Klein, 2007; Janes et al., 2012). An interesting new study, though, suggests that a direct cell contact might not always be necessary for signaling as Eph-containing extracellular vesicles (exosomes) were able to induce reverse signaling in ephrin-bearing cells (Gong et al., 2016). Second, the Eph receptors are multi-domain proteins, offering an ample choice of specific target regions: the extracellular ligand-binding domain, the cysteine-rich domain, the two fibronectin domains, and the cytoplasmic kinase domain (Himanen et al., 2007) (Figure 1). Even the very C-terminal SAM domain could offer an attractive drug target, since it operates as a docking surface for several cytoplasmic down-stream signaling proteins (Wang et al., 2016). Targeting SAM for drug discovery (Mercurio and Leone, 2016; Mercurio et al., 2016) should gain more interest in the future taking into account the fact that Eph’s have an unusual property to be able to signal even in the absence of cytoplasmic tyrosine phosphorylation (Truitt and Freywald, 2011). Importantly, high-resolution structures of all the domains, either as isolated molecules or in complex with ligands and other binding partners, including peptides, and small molecular inhibitors/modifiers, have been determined and are available (Barton et al., 2004; Noberini et al., 2012b). This offers an unprecedented platform for structure-based development of therapeutic modalities. Particularly, the well-studied ligand-binding domain, and its hydrophobic ligand-binding surface cavity, has proven to be a treasure trove for the identification and structure-based improvement of small-molecule binders that can not only inhibit ligand binding, but also elicit agonistic or antagonistic features on the receptor. The activation mechanism of the receptors is also well-studied (Lisabeth et al., 2013). This includes both the initial recognition and initiation of the signaling, and its termination, which has brought about very exciting recent development in the field of ADAM metalloproteinase inhibition (see below) (Atapattu et al., 2014; Atapattu et al., 2012). The vast amount of genomic data on Eph mutants involved in cancer offers yet another avenue for targeted drug development approach (Cerami et al., 2012). Future expansion of these sequence databases will show whether similar parallels can be drawn between Eph mutations and other diseases where they are involved, such as Alzheimer’s disease, amyotrophic lateral sclerosis, diabetes, and inflammation. Finally, the unexpected emergence of ephrins as entry receptors for some pathological viruses, has initiated a series of interesting studies aimed to inhibit or prevent the proliferation of these deadly microbes (Steffen et al., 2012). We will summarize here the latest studies on the field of Eph-related pharmacology, concentrating on Eph/ephrin/ADAM-based antibodies and small molecules, as well as Henipavirus targeting, and will discuss the future of genomics/proteomics-based medicine.
Figure 1:
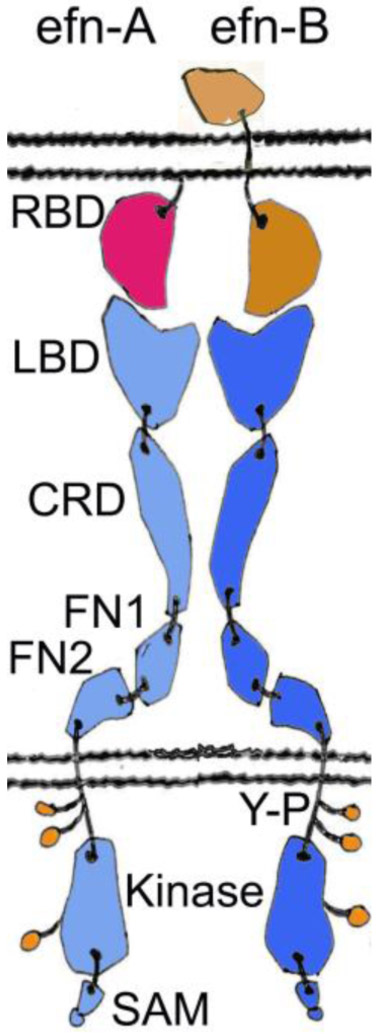
Schematic representation of the Eph receptors and their ephrin (efn) ligands. Ephrin-A is shown in red, ephrin-B in light brown, EphA receptor in cyan, and EphB receptor in blue. The A class ligands are attached to the cell membrane by a glycosylphosphatidylinositol (GPI) linkage and bind nearly exclusively the A class receptors. On the other hand, the B ligands that bind almost exclusively the B class receptors, contain a transmembrane stretch and a small cytoplasmic domain. The overall structures of the EphA and the EphB receptors are very similar. The ectodomain forms a fairly rigid, rod-like structure that undergoes minimal conformational change upon activation. Ligand binding causes a clustering of the Eph receptors, leading to the opening of an activation loop, and a subsequent tyrosine phosphorylation (shown in green) of the kinase domain. SAM domain offers a docking surface for downstream signaling and is reported to be involved in receptor oligomerization (Shi et al., 2017). RBD, Receptor-Binding Domain; LBD, Ligand-Binding Domain; CRD, Cysteine-Rich Domain; FN, Fibronectin III domain; Y-P, Phospho-Tyrosine; SAM, Sterile Alpha Motif.
Targeting the Eph/ephrin complex for drug development
Many different approaches have been employed in efforts to target the Eph/ephrin signaling system; antibodies, peptides and soluble fragments of the receptors/ligands, small molecule inhibitors of protein-protein interactions (PPIs), kinase inhibitors, and siRNAs (Lodola et al., 2017); (Charmsaz et al., 2017); (Chen et al., 2017); (Barquilla and Pasquale, 2015); (Charmsaz and Boyd, 2017) (Salgia et al., 2018). Although few have entered clinical trials, the relative infancy of the field and the enormous potential of various ‘druggable’ domains, should guarantee the continuation of these efforts. Kinase inhibitors, specific for the Eph receptors, as well as siRNA molecules that downregulate the expression levels of the Eph’s, have been described elsewhere (Amero et al., 2016; Biao-xue et al., 2011; Boyd et al., 2014; Dong et al., 2015; Duan et al., 2017; Heinzlmeir et al., 2017; Kung et al., 2016; Unzue et al., 2016; Zang et al., 2016) and will not be discussed further.
A). Peptides and small molecules
The first Eph-specific peptides were identified by phage display (Koolpe et al., 2002). Since then, many more have been characterized and studied, some modeled from the 15-amino acid Eph-binding ephrin loop, some through rational de novo design, with most peptides targeting EphA2, -A4, -B2, or -B4. A recent review summarizes the development of various peptides and their modifications (Riedl and Pasquale, 2015). Peptides offer several clear advantages: i) they can bind with considerably high affinity; ii) they can easily be modified to have different sizes in order to fit in the Eph surface cavity; iii) peptides usually have low toxicity and iv) low immunogenicity; v) the tissue penetration properties of peptides are often very efficient; vi) production costs are low. However, peptides also possess some distinct disadvantages, such as i) they are known to target only the LBD-cavity of the Eph receptors; ii) generally speaking, they have poor pharmacokinetics and bioavailability; iii) the stability of peptides in the plasma is notoriously low.
Considering the overall structural similarity of Eph receptors, it’s quite surprising that several peptides show remarkable specificity towards only one or two receptors. This might be a result of the flexible loops of the Eph receptors surrounding the hydrophobic ligand-binding cavity that differ quite considerably between otherwise structurally similar receptors (Himanen, 2012). For example, a peptide identified by phage display binds selectively only to EphA2, causing receptor activation as measured by tyrosine phosphorylation (Koolpe et al., 2002). Another interesting feature of the EphA2-specific peptides is that they are agonists, unlike most other peptides. The use of either agonist or antagonist peptides might lead in the future to a better understanding of Eph activities in pathology (Riedl and Pasquale, 2015).
The binding affinities of Eph-binding peptides, originally in the μM level, were increased to sub-μM levels after further screening and/or optimization steps following the determination of crystal structures of Eph/peptide complexes, and later some Kd values have been reported in the nM range (Xiong et al., 2011). An especially interesting study reports a cyclic, EphA4-specific peptide, with an IC50 value of about 30 nM and the ability to inhibit ephrin-induced EphA4 phosphorylation and neuronal growth cone collapse (Lamberto et al., 2014). Other studies have further indicated how cyclization improves not only affinity but also stability (Craik et al., 2013). For example, a cyclic form of an EphB4-specific TNYL-RAW peptide has been conjugated to doxorubicin-loaded nanoparticles and shown to selectively deliver the chemotherapeutic agent to EphB4-positive xenografts and to cause complete regression of most tumor cells (You et al., 2012). Another homing peptide, the EphA2-specific YSA, was recently used to target EphA2-positive breast cancer cells with doxorubicin-conjugated mesoporous silica nanoparticles (Liu et al., 2018). Other ways to improve peptide stability and, hence, resistance to plasma proteases are N-terminal modifications, PEGylation, conjugation to nanoparticles, fusion to Fc constructs, and complexation with streptavidin (Riedl and Pasquale, 2015). It’s worth keeping in mind, though, that the nM cyclic peptide (Lamberto et al., 2014), as well as all characterized peptides so far, target only the ligand-binding cavity on the receptor surface. It remains to be seen whether it’s possible to identify and develop peptides targeting other interfaces of the Eph ectodomain. Indeed, it would be a significantly difficult task to develop a peptide able to specifically recognize, bind to, and interfere with a protein-protein interface with a surface area around 2000 Å or larger.
Taking into consideration their substantial selectivity and binding affinity, it’s no surprise that several peptides have been tested for their potential in medical, especially neurological applications (Riedl and Pasquale, 2015). For example, EphA4 antagonistic peptides have been studied in neuroprotection and neural repair (Fabes et al., 2007; Galimberti et al., 2010; Murai et al., 2003; Olson et al., 2016). An EphA4-specific peptide (KYL) can regulate the viability of neural stem cells (DeVeale et al., 2014). Since EphA4 has been reported to have a role in Alzheimer’s disease, blocking it by the KYL-peptide can reverse the pathological effects of amyloid-beta oligomers in a cell-based assay for Alzheimer’s (Vargas et al., 2014). In addition, the KYL-peptide enhances the recovery of injured axons in a rat model for spinal cord injury (Fabes et al., 2007) and delays disease onset in a rat model for ALS (amyotrophic lateral sclerosis) (Robberecht and Philips, 2013). Another EphA4-specific peptide has been shown to impair the formation of longterm fear memory in rats (Dines and Lamprecht, 2014). Two peptides (KYL and TYY) might also have influence for breast cancer stem cell state and capillary-like tube formation in HUVE cells (Han et al., 2013; Riedl and Pasquale, 2015). Recently, another family of EphA4-specific molecules were designed and characterized (Wu et al., 2017), which selectively bind to the EphA4-LBD with nanomolar affinity and exhibit efficacy in an amyotrophic lateral sclerosis mouse model. Furthermore, an EphB2-specific SNEW-peptide and an EphB4-specific TNYL-RAW-peptide are reportedly active in inhibiting the pathological forms of angiogenesis e.g. retinal vascular disease (Barquilla and Pasquale, 2015). Remarkably, Azurin, a bacterial cupredoxin with structural similarity to ephrin, has a C-terminal peptide fragment (residues 88-113) that is claimed to inhibit prostate cancer cell DU145 phosphorylation and cell growth (Chaudhari et al., 2007). Another peptide fragment (residues 108-122) of the same protein, conjugated to a radiosensitizer nicotinic acid, has a nM affinity to Eph and the ability to increase the efficacy of Lewis lung carcinoma radiotherapy by several folds in artificial metastasis and solid tumor engraftment models (Micewicz et al., 2011). Although these peptide fragments show no specificity for a single Eph receptor (they bind to all Eph receptors tested, EphA2, -B2, and -B4), the 15-meric peptide (108-122), which contains unusual and D-amino acids, has an improved stability in human serum.
Soluble recombinant proteins have also been shown to inhibit Eph/ephrin interactions, thus showing a potential for therapeutic development (Barquilla and Pasquale, 2015; Boyd et al., 2014; Brantley et al., 2002; Dobrzanski et al., 2004). Hence, a soluble human ectodomain of EphB4 has been stabilized by human serum albumin (HSA; (Shi et al., 2012)) and has entered clinical trials for treating patients with advanced or metastatic solid tumors. Another soluble fragment of an Eph receptor ectodomain (sEphA7) has been demonstrated as an effective antitumor agent in xenografted human lymphoma cells (Oricchio et al., 2011). This effect can be enhanced by fusing the sEphA7 to an anti-CD20 antibody (rituximab). Furthermore, an Fc-fused ectodomain of EphA4 has been effectively used in promoting recovery after spinal cord injury in rats (Spanevello et al., 2013). Deletion of three glycosylation sited in EphA4 dramatically improves its pharmacokinetic characteristics (Pegg et al., 2017). The ability of ephrin-A5 to bind EphA2, -A3, and -B2 was used to target simultaneously all three receptors with pseudomonas exotoxin A on receptor-positive glioblastoma tumors (Ferluga et al., 2016). The ephrin-A5-Fc-based cytotoxin specifically killed glioblastoma cells with an IC50 of 10−11 M.
In addition to direct application, peptides can be conjugated with other molecules for selective delivery of imaging or therapeutic agents to Eph-expressing cells (Patel et al., 2016; Riedl and Pasquale, 2015; Salem et al., 2018a). For example, a dimeric form of EphA2-targeting YSA peptide, when conjugated to paclitaxel, is effective in targeting circulating tumor cells and decreasing tumor size in prostate and pancreatic cancer xenograft models and in inhibiting metastases in breast cancer models (Barile et al., 2014; Salem et al., 2018b; Wu et al., 2015b) . Apart from targeting, these peptides can make the conjugated agents more soluble and bioavailable. Similar chemical approaches have also been used for the development of antibody- and small molecule-drug conjugates (Cazzamalli et al., 2018). Overall, just as with Antibody-Drug-Conjugates (ADCs) where numerous pharmaceuticals are in clinical trials, Peptide-Drug Conjugates (PDCs) have reached a point in development where engineering success is expected to lead to therapeutic achievements. Not only the design of new conjugation strategies, but also improvement of the stability, cell penetration, safety, and efficacy of existing reagents, should facilitate advancement to the clinic (Fosgerau and Hoffmann, 2015; Giorgio et al., 2016; Skotland et al., 2015).
Protein-Protein Interactions (PPI) are at the center of perhaps all biological processes. Consequently, PPI inhibitors have become a hot but challenging field in the modern drug discovery (Jin et al., 2014; Nero et al., 2014; Wu et al., 2015a; Xu et al., 2017; Xu et al., 2016). Although some scientists consider the PPI’s ‘un-druggable’, the special structural features of the Eph/ephrin interactions has prompted a plethora of studies for developing pharmaceutically relevant small molecule inhibitors targeting the Eph/ephrin signaling (Noberini et al., 2011) (Noberini et al., 2012b). Indeed, the long ephrin loop penetrating into the hydrophobic Eph surface cavity offers plenty of specific chemical interactions to try to mimic, such as polar and hydrophobic interactions, and a salt bridge between Arg103 (EphA2) and Glu119 (ephrin-A1) (Himanen et al., 2009). To this date, both natural and synthetic small chemical compounds have been discovered, all targeting, not surprisingly, the ephrin-binding cavity of Eph’s (Tognolini and Lodola, 2015). Structural studies of Eph receptors in complex with various bound molecules and compounds (Figure 2) have assisted in discovering new compounds with high selectivity and desirable binding characteristics.
Figure 2.
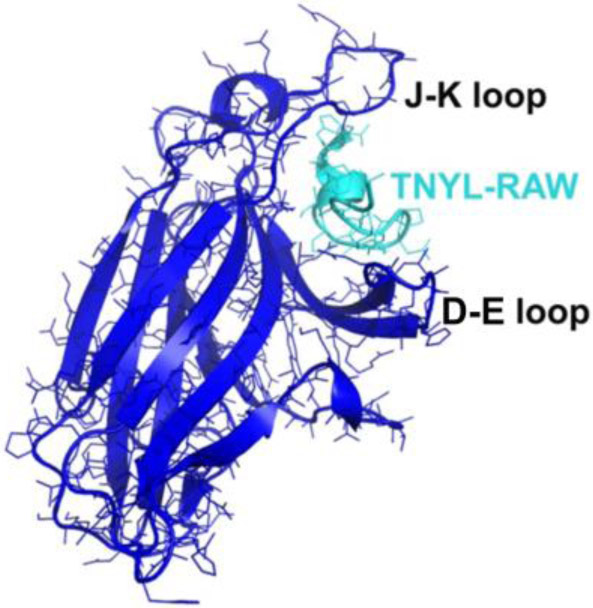
Ligand-Binding Domain (LBD) of the EphB4 receptor (in blue) in complex with an antagonistic peptide, TNYL-RAW (in cyan) (Chrencik et al., 2006). LBD has a jellyroll folding topology with 13 antiparallel B-sheets connected by several loops of varying lengths. Binding of TNYL-RAW makes the fairly flexible D-E and J-K loops more structured and they can be visualized in the electron density map.
Of the natural compounds, several tea polyphenols identified in a library screen have been shown to inhibit the EphA4/ephrin-A5 interactions at the μM level, interfere with the receptor phosphorylation, and reduce tube formation in HUVE cells (Mohamed et al., 2011; Noberini et al., 2012a; Tang et al., 2007). In addition, protocatechuic acid and pyrogallol have been shown to inhibit ligand-induced EphA2 phosphorylation in PC3 cells (Tognolini et al., 2012). However, the polyphenols show low degree of selectivity and poor stability. Another natural compound, rhynchophylline, an alkaloid in a Chinese medicinal herb, was claimed to be effective in a mouse model for Alzheimer’s disease (Fu et al., 2014), but these observations were later put under scrutiny, since only about 25% reduction in the EphA4 phosphorylation after rhynchophylline exposure could be recorded (Tognolini et al., 2014).
Benzoic acid derivatives were the first synthetic compounds found in High Throughput Screens (HTS) and were shown to inhibit binding of EphA2 and EphA4 to their biological ligands (Noberini et al., 2008; Takano et al., 2015). Once again, however, stability problems and aspecific binding characteristics have all but put further investigations of these compounds on hold (Tognolini et al., 2014). One of the more interesting chemicals to interfere with Eph/ephrin binding so far is lithocholic acid and its derivatives. Originally identified through an ELISA-based chemical screening as an EphA2-binding compound (Giorgio et al., 2011), lithocholic acid has since gone through an impressive cycle of structure- and modeling-based modifications in order to generate new analogues with improved affinity and other binding characteristics (Giorgio et al., 2018; Giorgio et al., 2016; Tognolini and Lodola, 2015). Analysis of a synthetic set of amino acid derivatives of lithocholic acid has led to the identification and characterization of a compound, UniPR129 (an L-homo-Trp conjugate of lithocholic acid) and its derivatives (UniPR139, UniPR502, UniPR1331), with enhanced potency, measured using both biochemical and cell-based functional assays (Hassan-Mohamed et al., 2014). UniPR129 can effectively inhibit EphA2 phosphorylation, PC3 cell retraction, and possess anti-angiogenic properties as measured by using HUVEC cultures. The UniPR compounds were shown to inhibit ephrin-induced EphA2 activation in a dose-dependent manner, and to exhibit anti-angiogenic activities (Giorgio et al., 2018). In addition, UniPR139 shows improved bioavailability and can be detected in the plasma with a t1/2 of 44 minutes. UniPR1331 was shown to be an anti-angiogenic agent in endothelial cells and orally bioavailable in a mouse model (Castelli et al., 2015). Moreover, UniPR1331 has been reported to possess anti-tumor activities in xenograft and orthotopic models of glioblastoma multiforme (Festuccia et al., 2018).
Other interesting new chemical inhibitors include stilbene carboxylic acid and cholenic acid derivatives (Pala et al., 2014), as well as doxazosin and its derivatives (Petty et al., 2018; Petty et al., 2012). Doxazosin is an EphA2 agonist that has very intriguing features, including inhibition of Akt and ERK activities in EphA2-expressing cells, ability to block the migration of glioma, prostate, and breast cancer cells, and capability to increase survival in prostate cancer-treated mice.
B). Antibodies
The use of antibodies (Ab) is the most common approach in developing new therapies and Eph receptors are no exception. Several members of the family (particularly EphA2, EphA3, EphB2, and EphB4) have been targeted with antibodies, mostly to configure new ways to treat cancers (Boyd et al., 2014; Charmsaz et al., 2017). The uncharacteristic dual role of Eph’s in cancer makes their targeting even more attractive: while there are numerous studies showing the correlation between Eph overexpression and poor cancer outcome, substantial evidence has also been presented to show how the same receptor, in certain cancer types and stages, can act as a tumor suppressor (Barquilla and Pasquale, 2015; Husa et al., 2016). Consequently, both agonistic and antagonistic (monoclonal) Ab’s have been developed for therapeutic use. For example, anti-EphA2 Ab’s have been shown to possess antitumor activities in ovarian, breast, and lung cancer xenografts (Gokmen-Polar et al., 2011; Landen et al., 2006; Zhuang et al., 2010). Anti-mouse EphA2 mAb (IF7) delays the course of MLL-AF9 leukemia when conjugated to 177lutetium (Charmsaz et al., 2015).
Interestingly, afucosylation of the carbohydrate chain in the Fc domain of another anti-EphA2 mAb (DS-8895a) has been shown to increase the Ab-dependent cellular toxicity (ADCC) and inhibit tumor growth in xenograft mouse models for breast and gastric cancers (Hasegawa et al., 2016). An antagonistic EphB2 Ab, on the other hand, was shown to inhibit the growth of colon carcinoma xenografts only as a conjugate with monomethyl auristatin E, an antimitotic agent (Mao et al., 2004). anti-EphB4 mAb’s have also been shown to inhibit angiogenesis and tumor growth in xenografted mice (Krasnoperov et al., 2010). Because EphB4 is highly expressed in ~30% of acute myeloid leukemia (AML) samples, it can be targeted with one of the antibodies (mAb131) (Merchant etal., 2017). By a conjugation with Cy5.5, these Ab’s can also be used for non-invasive near-infrared fluorescence imaging where they can potentially not only predict the individual immunotherapy responders, but also monitor their therapeutic response (Li et al., 2013).
At least four anti-EphA Ab’s have entered the clinic. One of them, the first Eph-related in-human clinical trial utilized an Ab-auristatin conjugate, but the trial had to be terminated because of unexpected side effects (Annunziata et al., 2013). The other one, developed from an agonistic anti-EphA3 mAb (Vearing et al., 2005), is currently undergoing a clinical trial to treat various hematological cancers (Janes et al., 2014; Swords et al., 2016). The original Ab has been shown to function through damaging the tumor stroma and microvasculature and changing the cell morphology (Vail et al., 2014; Vearing et al., 2005), and hence, the efficacy of the current Ab in clinical trials might depend on its ability to induce tumor apoptosis, cytotoxicity, and vasculature disruption. The current status of these clinical trials is summarized in (Lodola et al., 2017).
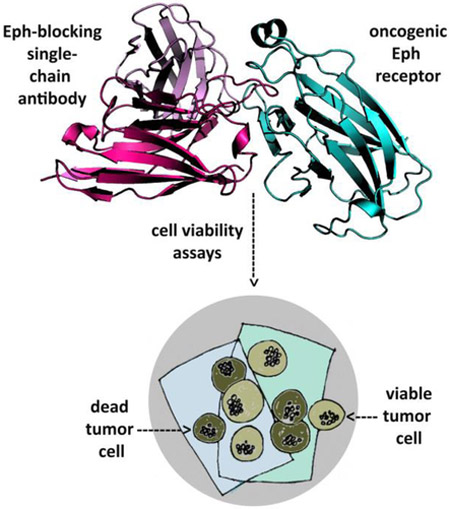
In addition to the ‘classical’ Ab production system, new technologies for producing nanobodies (Fridy et al., 2014) or single-chain (scFv) Ab’s (Hagemeyer et al., 2009; Lamminmaki et al., 2015) are expected to gain ground in the near future. These techniques can considerably improve the generation of Ab’s against Eph’s, which are usually poorly immunogenic due to their extremely high sequence similaritybetween species. Synthetic, phage display-based recombinant Ab libraries, on the other hand, can fairly quickly produce many human antibodies against freely-chosen antigens. These scFv antibodies not only have similar binding affinities and specificities as the conventional Ab’s, but their production from bacterial clones, readily expressed in a simple expression system, can potentially enhance their use as therapeutics. Indeed, scFv’s have been used e.g. as an anti-EphA2/anti-CD3 bispecific construct to kill EphA2-expressing tumor cells in vitro and in mice (Hammond et al., 2007). Another anti-EphA2 scFv Ab was shown to destroy prostate cancer cells, after being converted to a full-length IgG (Ha et al., 2014). Recently, yet another anti-EphA2 scFv fragment was obtained from a large human synthetic Ab library by a selection against the ligand-binding domain of the receptor. This scFv is capable of inducing apoptosis in lymphoma and leukemia cells, and structural studies showed that the Ab heavy chain is almost entirely responsible for binding to EphA2 (Figure 3) (Goldgur et al., 2014).
Figure 3.
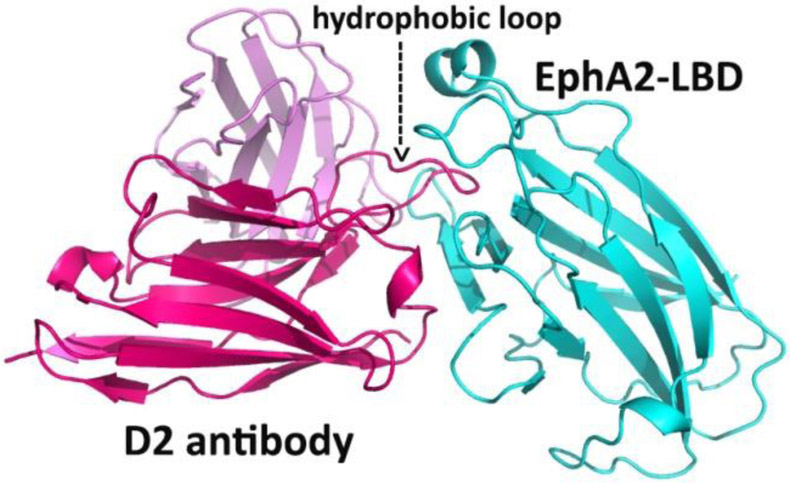
Crystal structure of an anti-EphA2 scFv Ab in complex with the ligand-binding domain of EphA2. The scFv variable light chain is pink, the variable heavy chain is red, and the EphA2 receptor is cyan. A long loop of the D2 heavy chain penetrates a hydrophobic surface cavity of the EphA2 in a way very similar to Ephrin binding to Eph.
The studies summarized above have clearly demonstrated the great potential of using fragments or full-length Ab’s as therapeutic agents. A special advantage in using Ab’s is their unlimited capacity to be raised against any surface-exposed epitope. Moreover, it is fairly straightforward to develop bispecific Ab’s, consisting of two scFv’s (or Fab’s) with a flexible linker between, each targeting a different epitope. Such Ab’s can offer a bridge between antigens expressed on a tumor and immune cells, thus promoting the recognition of tumor cells by the immune system, e.g. by the redirection of cytotoxic T cells (Taki et al., 2015).
In a rather rare study (Abengozar et al., 2012), the ephrin-B2 ligand, instead of the corresponding receptor, was used as a target for antibodies that were shown to suppress endothelial cell migration and tube formation. This highlights the potential of developing anti-angiogenic antibodies, although the wide expression of both EphB4 and ephrin-B2 in the normal human vasculature could nullify their use as therapeutic agents. An anti-ephrin-A4 mAb conjugated to the DNA-damaging agent calicheamicin has been shown to cause sustained tumor regressions in triple-negative breast and ovarian cancer in vivo (Damelin et al., 2015) and is now in a clinical trial.
It is interesting to note that the ephrin-specific antibodies might find applications beyond their original intentions because ephrins have been found to be the entry receptors for Hendra (HeV) and Nipah (NiV) viruses (Eaton et al., 2006; Eaton et al., 2004; Wang et al., 2013). Henipa belong to the family of deadly and zoonotic paramyxo viruses having natural reservoirs in fruit bats (flying foxes) in the genus Pteropus (Halpin et al., 2011). Both NiV and HeV have a unique and very broad host infection tropism, and can cause disease in animals and humans, resulting in a systemic and often fatal respiratory and/or neurological disease. The most severe pathology is seen in the respiratory system and central nervous system (CNS), with a predominance of an acute encephalitis syndrome, ulceration, thrombosis, and microinfarction (Ong and Wong, 2015). Therefore, they are recognized as important biosecurity agents of concern to human populations and economically important livestock, particularly horses and pigs (Broder, 2010; 2013; Field et al., 2012). The outbreaks of henipaviruses have very high rates of fatality in people (75-100%) (Anonymous, 2013; Homaira et al., 2010; Rahman et al., 2011).
Recent biochemical and structural studies have shed light on the initial receptor recognition and cell entry mechanisms of henipaviruses (Field, 2016) (Weatherman et al., 2017). In short, HeV and NiV attach and infect cells through the coordinated functions of the receptor-binding G-protein and the fusion-mediating F-protein on the viral envelope (Bossart et al., 2013; Dutch, 2010). After ephrin engagement, there’s a conformational or other alteration in the G-protein that either releases the F-protein towards a fusion-active state or induces conformational changes in the F-protein, eventually leading to membrane fusion (Dutta et al., 2016; Steffen et al., 2012).
The head domain structures of both NiV and HeV G-protein have been determined, both alone and in complex with their host cell ephrin receptors (Bowden et al., 2008a; Bowden et al., 2008b; Colgrave et al., 2011; Steffen et al., 2012; Xu et al., 2012b; Xu et al., 2008). Because ephrins have about 40% sequence identity, it’s understandable that all available ephrin/G-protein structures are very similar (Bowden et al., 2008a; Xu et al., 2012a; Xu et al., 2008). In all of the complexes the binding centers around a hydrophobic area on top of the G-protein and the so- called G-H loop of the ephrin. The core of this hydrophobic interface is surrounded by more polar area, stabilized mostly by hydrogen bonds (Figure 4) (Lee et al., 2015; Porotto et al., 2007). The high-resolution structural studies offer a road-map for designing special organic molecules, peptides, or protein fragments for inhibiting the ephrin/G-protein interaction, thus paving a way for new-generation antiviral agents.
Figure 4.
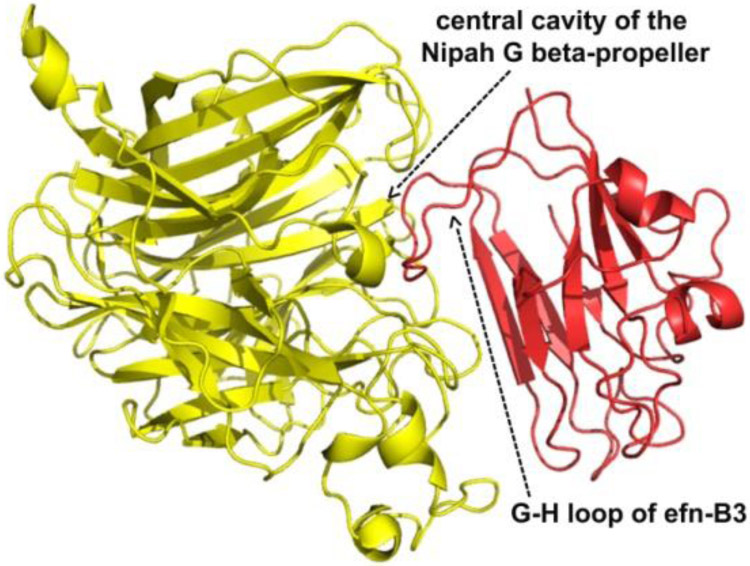
Crystal structure of the Nipah virus G protein in complex with ephrin-B3. G protein is shown in yellow and Ephrin in red. The main binding interfaces appear around a long G-H loop of Ephrin and the central cavity of the G protein beta-propeller. The same surface elements of Ephrin are involved in both Eph and Nipah G binding.
Although Henipa virus epidemics are fairly rare, their exceptionally broad species and cellular tropism as well as remarkably high morbidity and mortality rates (sometimes approaching 100%), have raised concerns among health professionals. Hence, henipavirus therapeutic efforts have gained ground in recent years. Considering the structural information available on the henipavirus glycoproteins and their ephrin complexes, it is clear that the G and the F glycoproteins are the major antigenic targets of the virus-neutralizing antibody response in infected animals and are presently the most important targets of antiviral strategies aimed at blocking infection (Broder et al., 2012). In addition, ephrin-specific Ab’s should in the future be studied for their anti-viral activities. Despite the lack of licensed treatment against henipa viruses in humans, a post-infection treatment by a human monoclonal antibody (mAb) has been shown to be effective against virus infection. The cross-reactivity of this mAb (m102.4) can now be explained by the crystal structure of a complex between the henipa G protein and the neutralizing m102.4, isolated from a recombinant naïve human phage-display Fab library (Zhu et al., 2008). Importantly, the epitope of m102.4 maps to the ephrin-binding site of the virus. All African green monkeys and ferrets treated with the antibody survived viral infection, while the untreated animals succumbed to severe systemic disease (Bossart et al., 2011; Bossart et al., 2009). Moreover, three human individuals, potentially infected by HeV, have received m102.4 on a compassionate basis and none have developed detectable HeV infection (Broder et al., 2013). A monoclonal antibody (m102.3), identical to m102.4 except for a few amino acid changes in its light chain distant from the CDRs, has been crystallized in complex with HeV G (Xu et al., 2013a). If this mAb can effectively serve as ‘ephrin’, it suggests that the neutralization activity is based on an effective binding to the virus and triggering an irreversible fusion cascade. When this occurs without attachment to a cell, it effectively defuses and neutralizes the virus.
C). Regulation of Eph/ephrin signaling by ADAM-proteases
Upon cell-cell contact, the Eph receptors and their ephrin ligands form heterotetramers that are further assembled into large signaling clusters between the interacting cells (Wimmer-Kleikamp et al., 2004). The resulting multivalent interaction needs to be broken to terminate signaling and ensure cell-cell repulsion. It has been now established that ADAM10 and other members of the ADAM family play a major role in the regulation of signaling by Eph and ephrins (Figure 5). An elegant study (Hattori et al., 2000) has demonstrated a general mechanism, according to which ligand- and receptor-expressing cell interaction is followed by ADAM 10 activation and the cleavage of Eph-bound ephrins from the cell surface, allowing ligand-receptor complex to be internalized and degraded inside the receptor-expressing cell. Ephrin-A2 was shown to be cleaved by ADAM10 and the cleavage was proposed to occur in cis (within the same cell membrane harboring both the ADAM proteinase and the substrate). Ephrin-A2 was found to be associated with ADAM10 via non-catalytic regions and ephrin cleavage was facilitated by binding to its receptor, EphA3.
Figure 5.
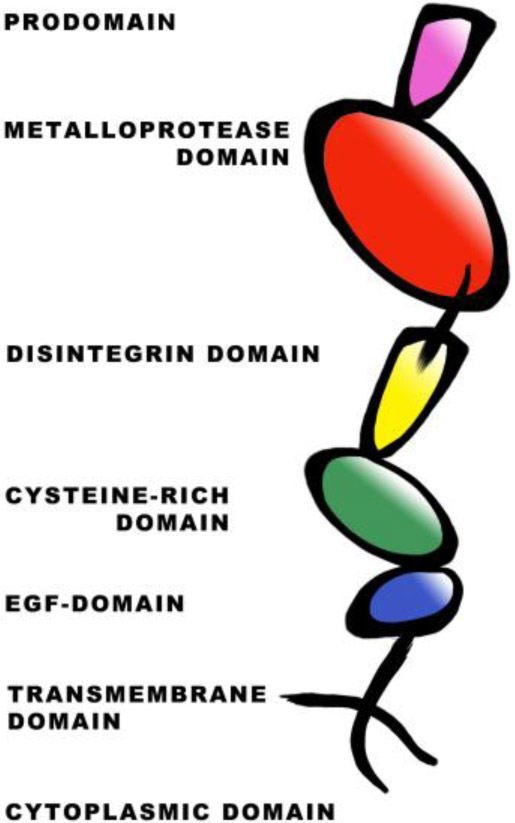
Domain organization of transmembrane ADAM metalloproteases. ADAMs contain a prodomain that is cleaved upon maturation. The metalloprotease (MP) harbors the active site. The disintegrin (D) and cysteine–rich domains are primarily responsible for substrate-recognition. The epidermal growth factor (EGF)-like domain is absent in ADAM10 and ADAM17. The EGF domain is followed by the transmembrane domain and a cytoplasmic tail.
The mechanistic insight into the recognition and cleavage of Eph-associated ephrins from the opposing cell has been illustrated by the structure-function studies of ADAM10 in the context of EphA3/ephrin-A5 signaling (Janes et al., 2005). Crystal structure of the ADAM10 disintegrin (D) and cysteine-rich (C) domains, along with functional studies, defined a substrate-recognition module that associates with the EphA3/ephrin-A5 complex. Prior to cell-cell contact, ADAM10 is constitutively associated with EphA3 to ensure its close proximity to potential substrates. Cell-cell contact mediated by the high-affinity EphA3/ephrin-A5 complex creates a new molecular recognition motif to which ADAM10 binds via its C-domain. The ADAM D-domain, which forms a continuous rigid structure with the C-domain, positions the metalloprotease (MP) domain to cleave the stem–region of ephrin-A5 from the opposing cell (in trans). Consequently, the molecular tethers between opposing cells surfaces are disrupted and the EphA3/ephrin-A5 complex is internalized. In addition to ephrin-As, ADAM10 also binds and cleaves ephrin-B2 during Xenopus embryogenesis, an interaction mediated by flotillin-1 (Ji et al., 2014).
Other ADAM family members are also known to play a role in Eph signaling. For instance, ADAM12 binds to EphA1 and enhances shedding of ephrin-A1 in trans in response to transforming growth factor β1 in primary tumors. This results in lung hyperpermeability leading to lung metastasis (Ieguchi et al., 2014). Likewise, ephrin-B1 and ephrin-B2 are known to be cleaved by ADAM13 during cranial/trunk neural crest migration in cells and embryos (Wei et al., 2010). Shedding by ADAM10 provides an essential switch (Hattori et al., 2000; Janes et al., 2005) regulating the function of Eph receptors (Boyd and Lackmann, 2001; Pasquale, 2005; Poliakov et al., 2004) by cleaving their ephrin ligands (Janes et al., 2005). Studies have elucidated the involvement of ADAM10 in EGFR signaling triggered by EphA2 and ephrin-A1-mediated cell-cell contact, a process that controls tissue homeostasis and wound healing in epithelial barrier tissues like cornea (Kaplan et al., 2018). Recent studies have also unfolded the importance of ADAM10 as a major sheddase of ephrin-B2 in fibroblasts. It has been shown that the transforming growth factor (TGF) β1 elevates the expression of ADAM10, which in turns sheds ephrin-B2 in myofibroblasts. Inhibition of ADAM10 by the small molecule inhibitor GI254023X decreases the shedding of ephrin-B2 and prevents lung fibrosis in mice (Lagares et al., 2017). This raises the possibility of targeting ephrin-B2, its receptors EphB3 and EphB4, as well as ADAM10 as therapeutic targets in the treatment of fibrosis.
Other proteases such as matrix metalloproteinases MMP-2 and MMP-9 are known to cleave the EphB2 receptor and this cleavage is induced by interaction with its ligand ephrin-B2 (Lin et al., 2008). Also, the proteolytically active complex comprising of tissue factor/serine protease factor VIIa, (TF/FVIIa) cleaves the EphB2 receptor and this causes EphB2 dependent repulsion by its ligand ephrin-B1 (Eriksson et al., 2014). Consequently, the molecular tethers between the receptor EphB2 and its ligand ephrin-B1 or ephrin-B2 are severed, causing cell-cell repulsion and signal termination. Likewise, the dynamics of EphB2/NMDA receptor interaction, critical for stress-related plasticity in the amygdala, is regulated by a serine protease neuropsin. In mouse models, it has been elegantly demonstrated that stress causes neuropsin to cleave EphB2, resulting in the dissociation of EphB2 from the NR1 subunit of the NMDA. This is essential for membrane turnover of EphB2 (Attwood et al., 2011).
A dysregulation of ectodomain shedding by the ADAM proteases can lead to neurodegeneration, autoimmune and inflammatory diseases, cardiovascular diseases, and cancer (Drag and Salvesen, 2010; Turk, 2006). For instance, ADAM10-mediated shedding of cadherins (Solanas et al., 2011) and ephrins (Janes et al., 2005) regulate tumor development and metastasis (Adams, 2003; Brantley-Sieders and Chen, 2004; Nievergall et al., 2012; Wimmer-Kleikamp and Lackmann, 2005). To that end, significant efforts have been made to develop ADAM metalloprotease inhibitors, targeting the protease catalytic site. However, until now, all clinical trials using broad-spectrum metalloprotease inhibitors have failed (DasGupta et al., 2009; Moss and Lambert, 2002; Saftig and Reiss, 2011). The lack of efficacy and specificity of the inhibitors is due to the fact that (i) the MP domain of the ADAMs is similar to that of the matrix metalloproteinases (MMP) and (ii) the substrate-specificity is conferred by non-catalytic interactions between substrate and, most often, the C-domain of ADAMs (Janes et al., 2005; Levy et al., 2005; Reddy et al., 2000). As a logical alternative, antibodies raised against the ectodomain, or more specifically, the non-catalytic domains (D or C) of ADAM can serve as potential tools for therapeutic intervention. In cell-based studies with breast cancer cell-lines, polyclonal antibodies to ADAM15 and ADAM17 significantly reduce proliferation of the cell lines (Lendeckel et al., 2005).
In the context of Eph/ephrin signaling, another therapeutic antibody approach has made significant headway (Atapattu et al., 2012). Monoclonal antibodies (mAb’s) were raised against the substrate-binding domain of ADAM10 and one of the mAb’s, 8C7, was found to block ephrin cleavage and internalization (Figure 6). It also inhibits phosphorylation of the Eph receptor, which leads to the onset of biological responses. Consequently Eph/ephrin-mediated cell repulsion and cell segregation was abrogated. Additionally, in stripe assays it inhibited the repulsion of cells mediated by EphB2 and ephrin-A5 (Atapattu et al., 2012). The inhibitory effect of the mAb toward EphB2-mediated ephrin shedding is in conformity with the report that ADAM10 interacts with EphB (Solanas et al., 2011) and demonstrates the efficacy of the mAb 8C7 in blocking ADAM10-mediated biological response of both Eph subtypes (Atapattu et al., 2012).
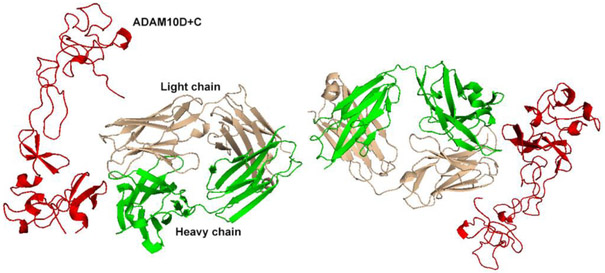
Figure 6.
Crystal structure of mAb/8C7-bound ADAM10. The heavy chain of the 8C7 mAb is in green and the light in wheat. The D+C domains of ADAM10 are represented in red.
Future prospects
The availability of large and ever-growing amount of sequencing data from healthy individuals and patients with various diseases provides a great opportunity to study the molecular mechanisms of Eph signaling. Particularly, critical analysis of disease-related mutations in the Eph receptors offers a way to evaluate the precise role and functional importance of the various (Eph-ephrin and Eph-Eph) protein-protein interactions and interfaces for receptor activation and initiation of downstream signaling. Importantly, the genomic data can reveal previously unrecognized or under-appreciated function-altering sites in the Eph receptors that can be studied at the mechanistic level. These studies are expected to advance our understanding of the basic molecular mechanisms underlying Eph/ephrin-mediated signaling and will illuminate how the genetic alterations destabilize normal cellular processes, leading to new therapeutic possibilities (Stewart et al., 2017). For example, previous structural studies have identified Eph receptor “hot spots”, and many mutations in various cancer types are found at these sites (http://www.cbioportal.org/; (Cerami et al., 2012; Gao et al., 2013). The use of genomic/proteomic databases will be in future the main theme of the ‘precision cancer medicine’ and other personalized medicine or ‘targeted therapeutics’ developments (Cheng et al., 2015; Sirintrapun et al., 2015). This new approach should enable us to identify individual’s susceptibility to disease, predict how a given patient will respond to a particular drug, and match patients with the right therapeutics (Gymrek and Erlich, 2011; Hood et al., 2012). This new science of personalized medicine has the potential to eliminate unnecessary treatments, reduce the incidence of adverse reactions to drugs, increase the efficacy of treatments and ultimately, improve health outcomes.
As mentioned above, Eph’s have two types of surface regions/protein-protein interfaces that control their function. The first type is directly responsible for Eph-ephrin ligand/receptor recognition and binding (Interface-1), while the second type includes two distinct surfaces (Interface-2 and -3) that mediate Eph-Eph clustering interactions (Nikolov et al., 2013; 2014). The genetic alleles identified in the latter (Eph-Eph interface) regions are particularly interesting for future drug development because they alter functions that have been previously unappreciated in Eph signaling and tumorigenesis. Structural studies have shown that Interface-2 is the Eph/Eph interface that mediates the formation of Eph/ephrin clusters upon ligand binding. Interface-3, on the other hand, is the Eph/Eph pre-clustering interface, which mediates Eph-Eph interactions (and receptor pre-clustering) on the cell surface prior to ligand binding. Once a mutation – expected to have a major impact for receptor function – is identified, its significance for e.g. cluster formation dynamics or cluster size can be evaluated using live imaging. Further down the line, a medication targeted specifically against the mutated residue or the surrounding surface interface can be developed. Interestingly, recent structural studies have shown how the un-liganded Eph receptor can, unexpectedly, form head-to-tail homotypic complexes (Nikolov et al., 2014; Xu et al., 2013b). Specifically, the ligand-binding domain of one Eph ectodomain is associated with the second fibronectin domain of the neighboring receptor. These interactions are likely a way for the cell to introduce subtle changes into the Eph/ephrin signaling process. Indeed, in some cases the pre-clustering interactions are inhibitory, while in others they appear to promote signaling. Future studies on the mutations in this and the other two interfaces should be designed based on the available genomic data. Such an approach will be extremely informative because the information within the genetic databases provides an ideal tool allowing functional and mechanistic characterization of the Eph clustering and pre-clustering events.
Table 1:
Summary of the proteases discussed in the context of Eph-ephrin signaling and their targets
| Protease | Target |
|---|---|
| ADAM10 | ephrin-A2, ephrin-A5, ephrin-B2 |
| ADAM12 | ephrin-A1 |
| ADAM13 | ephrin-B1, ephrin-B2 |
| MMP-2/MMP-9 | EphB2 |
| Tissue Factor/ serine protease factor VIIa | EphB2 |
Highlights.
Eph receptors have emerged as exciting targets for drug development
3-D structures of Eph/ephrin provide information for therapeutic intervention
Small molecules and antibodies can intervene with Eph/ephrin signaling
ADAM metalloproteinases can be inhibited to block Eph activation
Acknowledgements
Our studies are supported by the National Institutes of Health [R21CA185930 and R01 NS038486 to D.B.N and 5RO1NS096956-03 / RES510708 to JPH]; The Experimental Therapeutics Center of Memorial Sloan-Kettering, support from Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research [2013-2015 and 2018-2019 to D.B.N]; The Memorial Sloan-Kettering Functional Genomics Initiative [2015-2017 to D.B.N.]
Abbreviations
- RTK
Receptor Tyrosine Kinase
- LBD
Ligand-Binding Domain
- CRD
Cysteine-Rich Domain
- FN
Fibronectin
- SAM
Sterile Alpha Motif
- ADAM
A Disintegrin And Metalloproteinase
- Ab
Antibody
- mAb
monoclonal Antibody
- scFv
single-chain variable fragment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abengozar MA, de Frutos S, Ferreiro S, Soriano J, Perez-Martinez M, Olmeda D, Marenchino M, Canamero M, Ortega S, Megias D, Rodriguez A and Martinez-Torrecuadrada JL (2012) Blocking ephrinB2 with highly specific antibodies inhibits angiogenesis, lymphangiogenesis, and tumor growth. Blood 119:4565–4576. [DOI] [PubMed] [Google Scholar]
- Adams RH (2003) Molecular control of arterial-venous blood vessel identity. J Anat 202:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amero P, Esposito CL, Rienzo A, Moscato F, Catuogno S and de Franciscis V (2016) Identification of an Interfering Ligand Aptamer for EphB2/3 Receptors. Nucleic Acid Ther 26:102–110. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Kohn EC, LoRusso P, Houston ND, Coleman RL, Buzoianu M, Robbie G and Lechleider R (2013) Phase 1, open-label study of MEDI-547 in patients with relapsed or refractory solid tumors. Investigational new drugs 31:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous (2013) Nipah encephalitis, human - Bangladesh (05), in Pro-med, International Society for Infectious Diseases. [Google Scholar]
- Atapattu L, Lackmann M and Janes PW (2014) The role of proteases in regulating Eph/ephrin signaling. Cell adhesion & migration 8:294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atapattu L, Saha N, Llerena C, Vail ME, Scott AM, Nikolov DB, Lackmann M and Janes PW (2012) Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function. Journal of cell science 125:6084–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood BK, Bourgognon JM, Patel S, Mucha M, Schiavon E, Skrzypiec AE, Young KW, Shiosaka S, Korostynski M, Piechota M, Przewlocki R and Pawlak R (2011) Neuropsin cleaves EphB2 in the amygdala to control anxiety. Nature 473:372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile E, Wang S, Das SK, Noberini R, Dahl R, Stebbins JL, Pasquale EB, Fisher PB and Pellecchia M (2014) Design, synthesis and bioevaluation of an EphA2 receptor-based targeted delivery system. ChemMedChem 9:1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A and Pasquale EB (2015) Eph receptors and ephrins: therapeutic opportunities. Annual review of pharmacology and toxicology 55:465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton WA, Himanen JP, Antipenko A and Nikolov DB (2004) Structures of axon guidance molecules and their neuronal receptors. Advances in protein chemistry 68:65–106. [DOI] [PubMed] [Google Scholar]
- Biao-xue R, Xi-guang C, Shuan-ying Y, Wei L and Zong-juan M (2011) EphA2-dependent molecular targeting therapy for malignant tumors. Current cancer drug targets 11:1082–1097. [DOI] [PubMed] [Google Scholar]
- Bossart KN, Fusco DL and Broder CC (2013) Paramyxovirus Entry, in Viral Entry into Host Cells (Pöhlmann S and Simmons G eds) pp 95–127, Landes Bioscience and Springer Science+Business Media, Austin, TX [Google Scholar]
- Bossart KN, Geisbert TW, Feldmann H, Zhu Z, Feldmann F, Geisbert JB, Yan L, Feng YR, Brining D, Scott D, Wang Y, Dimitrov AS, Callison J, Chan YP, Hickey AC, Dimitrov DS, Broder CC and Rockx B (2011) A neutralizing human monoclonal antibody protects african green monkeys from hendra virus challenge. Science translational medicine 3:105ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Zhu Z, Middleton D, Klippel J, Crameri G, Bingham J, McEachern JA, Green D, Hancock TJ, Chan YP, Hickey AC, Dimitrov DS, Wang LF and Broder CC (2009) A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS pathogens 5:e1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY and Stuart DI (2008a) Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nature structural & molecular biology 15:567–572. [DOI] [PubMed] [Google Scholar]
- Bowden TA, Crispin M, Harvey DJ, Aricescu AR, Grimes JM, Jones EY and Stuart DI (2008b) Crystal Structure and Carbohydrate Analysis of Nipah Virus Attachment Glycoprotein: A Template for Antiviral and Vaccine Design. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd AW, Bartlett PF and Lackmann M (2014) Therapeutic targeting of EPH receptors and their ligands. Nature reviews Drug discovery 13:39–62. [DOI] [PubMed] [Google Scholar]
- Boyd AW and Lackmann M (2001) Signals from Eph and ephrin proteins: a developmental tool kit. Science's STKE : signal transduction knowledge environment 2001:re20. [DOI] [PubMed] [Google Scholar]
- Brantley-Sieders DM and Chen J (2004) Eph receptor tyrosine kinases in angiogenesis: from development to disease. Angiogenesis 7:17–28. [DOI] [PubMed] [Google Scholar]
- Brantley DM, Cheng N, Thompson EJ, Lin Q, Brekken RA, Thorpe PE, Muraoka RS, Cerretti DP, Pozzi A, Jackson D, Lin C and Chen J (2002) Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene 21:7011–7026. [DOI] [PubMed] [Google Scholar]
- Broder CC (2010) Therapeutics and Vaccines against Hendra and Nipah Viruses, in New Generation Vaccines (Levine MM, Dougan G, Good MF, Liu MA, Nabel GJ, Nataro JP and Rappuoli R eds) pp 885–894, Informa Healthcare; USA, New York. [Google Scholar]
- Broder CC (2013) Passive Immunization and Active Vaccination against Hendra and Nipah Viruses. Developments in biologicals 135:125–138. [DOI] [PubMed] [Google Scholar]
- Broder CC, Geisbert TW, Xu K, Nikolov DB, Wang LF, Middleton D, Pallister J and Bossart KN (2012) Immunization strategies against henipaviruses. Current topics in microbiology and immunology 359:197–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder CC, Xu K, Nikolov DB, Zhu Z, Dimitrov DS, Middleton D, Pallister J, Geisbert TW, Bossart KN and Wang LF (2013) A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral research 100:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli R, Tognolini M, Vacondio F, Incerti M, Pala D, Callegari D, Bertoni S, Giorgio C, Hassan-Mohamed I, Zanotti I, Bugatti A, Rusnati M, Festuccia C, Rivara S, Barocelli E, Mor M and Lodola A (2015) Delta(5)-Cholenoyl-amino acids as selective and orally available antagonists of the Eph-ephrin system. European journal of medicinal chemistry 103:312–324. [DOI] [PubMed] [Google Scholar]
- Cazzamalli S, Dal Corso A, Widmayer F and Neri D (2018) Chemically Defined Antibody- and Small Molecule-Drug Conjugates for in Vivo Tumor Targeting Applications: A Comparative Analysis. Journal of the American Chemical Society 140:1617–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C and Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmsaz S, Beckett K, Smith FM, Bruedigam C, Moore AS, Al-Ejeh F, Lane SW and Boyd AW (2015) EphA2 Is a Therapy Target in EphA2-Positive Leukemias but Is Not Essential for Normal Hematopoiesis or Leukemia. PloS one 10:e0130692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmsaz S and Boyd AW (2017) Eph receptors as oncotargets. Oncotarget 8:81727–81728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmsaz S, Scott AM and Boyd AW (2017) Targeted therapies in hematological malignancies using therapeutic monoclonal antibodies against Eph family receptors. Exp Hematol 54:31–39. [DOI] [PubMed] [Google Scholar]
- Chaudhari A, Mahfouz M, Fialho AM, Yamada T, Granja AT, Zhu Y, Hashimoto W, Schlarb-Ridley B, Cho W, Das Gupta TK and Chakrabarty AM (2007) Cupredoxin-cancer interrelationship: azurin binding with EphB2, interference in EphB2 tyrosine phosphorylation, and inhibition of cancer growth. Biochemistry 46:1799–1810. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang H and Zhang Y (2017) Targeting receptor tyrosine kinase EphB4 in cancer therapy. Semin Cancer Biol. [DOI] [PubMed] [Google Scholar]
- Cheng PF, Dummer R and Levesque MP (2015) Data mining The Cancer Genome Atlas in the era of precision cancer medicine. Swiss medical weekly 145:w14183. [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Brooun A, Recht MI, Kraus ML, Koolpe M, Kolatkar AR, Bruce RH, Martiny-Baron G, Widmer H, Pasquale EB and Kuhn P (2006) Structure and thermodynamic characterization of the EphB4/Ephrin-B2 antagonist peptide complex reveals the determinants for receptor specificity. Structure 14:321–330. [DOI] [PubMed] [Google Scholar]
- Colgrave ML, Snelling HJ, Shiell BJ, Feng YR, Chan YP, Bossart KN, Xu K, Nikolov DB, Broder CC and Michalski WP (2011) Site occupancy and glycan compositional analysis of two soluble recombinant forms of the attachment glycoprotein of Hendra virus. Glycobiology 22:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik DJ, Fairlie DP, Liras S and Price D (2013) The future of peptide-based drugs. Chemical biology & drug design 81:136–147. [DOI] [PubMed] [Google Scholar]
- Damelin M, Bankovich A, Park A, Aguilar J, Anderson W, Santaguida M, Aujay M, Fong S, Khandke K, Pulito V, Ernstoff E, Escarpe P, Bernstein J, Pysz M, Zhong W, Upeslacis E, Lucas J, Lucas J, Nichols T, Loving K, Foord O, Hampl J, Stull R, Barletta F, Falahatpisheh H, Sapra P, Gerber HP and Dylla SJ (2015) Anti-EFNA4 Calicheamicin Conjugates Effectively Target Triple-Negative Breast and Ovarian Tumor-Initiating Cells to Result in Sustained Tumor Regressions. Clin Cancer Res 21:4165–4173. [DOI] [PubMed] [Google Scholar]
- DasGupta S, Murumkar PR, Giridhar R and Yadav MR (2009) Current perspective of TACE inhibitors: a review. Bioorganic & medicinal chemistry 17:444–459. [DOI] [PubMed] [Google Scholar]
- DeVeale B, Bausch-Fluck D, Seaberg R, Runciman S, Akbarian V, Karpowicz P, Yoon C, Song H, Leeder R, Zandstra PW, Wollscheid B and van der Kooy D (2014) Surfaceome profiling reveals regulators of neural stem cell function. Stem cells 32:258–268. [DOI] [PubMed] [Google Scholar]
- Dines M and Lamprecht R (2014) EphrinA4 mimetic peptide targeted to EphA binding site impairs the formation of long-term fear memory in lateral amygdala. Translational psychiatry 4:e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzanski P, Hunter K, Jones-Bolin S, Chang H, Robinson C, Pritchard S, Zhao H and Ruggeri B (2004) Antiangiogenic and antitumor efficacy of EphA2 receptor antagonist. Cancer research 64:910–919. [DOI] [PubMed] [Google Scholar]
- Dong J, Zhao H, Zhou T, Spiliotopoulos D, Rajendran C, Li XD, Huang D and Caflisch A (2015) Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases. ACS medicinal chemistry letters 6:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M and Salvesen GS (2010) Emerging principles in protease-based drug discovery. Nature reviews Drug discovery 9:690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Wu A, Chen Z, Yang Y, Liu L and Shu Q (2017) MiR-204 Regulates Cell Proliferation and Invasion by Targeting EphB2 in Human Cervical Cancer. Oncol Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE (2010) Entry and fusion of emerging paramyxoviruses. PLoS Pathog 6:e1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta P, Siddiqui A, Botlani M and Varma S (2016) Stimulation of Nipah Fusion: Small Intradomain Changes Trigger Extensive Interdomain Rearrangements. Biophys J 111:1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BT, Broder CC, Middleton D and Wang LF (2006) Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol 4:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BT, Wright PJ, Wang LF, Sergeyev O, Michalski WP, Bossart KN and Broder CC (2004) Henipaviruses: recent observations on regulation of transcription and the nature of the cell receptor. Archives of virology Supplementum:122–131. [PubMed] [Google Scholar]
- Egea J and Klein R (2007) Bidirectional Eph-ephrin signaling during axon guidance. Trends in cell biology 17:230–238. [DOI] [PubMed] [Google Scholar]
- Eriksson O, Ramstrom M, Hornaeus K, Bergquist J, Mokhtari D and Siegbahn A (2014) The Eph tyrosine kinase receptors EphB2 and EphA2 are novel proteolytic substrates of tissue factor/coagulation factor VIIa. The Journal of biological chemistry 289:32379–32391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabes J, Anderson P, Brennan C and Bolsover S (2007) Regeneration-enhancing effects of EphA4 blocking peptide following corticospinal tract injury in adult rat spinal cord. The European journal of neuroscience 26:2496–2505. [DOI] [PubMed] [Google Scholar]
- Ferluga S, Tome CM, Herpai DM, D'Agostino R and Debinski W (2016) Simultaneous targeting of Eph receptors in glioblastoma. Oncotarget 7:59860–59876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia C, Gravina GL, Giorgio C, Mancini A, Pellegrini C, Colapietro A, Delle Monache S, Maturo MG, Sferra R, Chiodelli P, Rusnati M, Cantoni A, Castelli R, Vacondio F, Lodola A and Tognolini M (2018) UniPR1331, a small molecule targeting Eph/ephrin interaction, prolongs survival in glioblastoma and potentiates the effect of antiangiogenic therapy in mice. Oncotarget 9:24347–24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H, Crameri G, Kung NY and Wang LF (2012) Ecological aspects of hendra virus. Current topics in microbiology and immunology 359:11–23. [DOI] [PubMed] [Google Scholar]
- Field HE (2016) Hendra virus ecology and transmission. Current opinion in virology 16:120–125. [DOI] [PubMed] [Google Scholar]
- Fosgerau K and Hoffmann T (2015) Peptide therapeutics: current status and future directions. Drug discovery today 20:122–128. [DOI] [PubMed] [Google Scholar]
- Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyo D, Chait BT and Rout MP (2014) A robust pipeline for rapid production of versatile nanobody repertoires. Nature methods 11:1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu AK, Hung KW, Huang H, Gu S, Shen Y, Cheng EY, Ip FC, Huang X, Fu WY and Ip NY (2014) Blockade of EphA4 signaling ameliorates hippocampal synaptic dysfunctions in mouse models of Alzheimer's disease. Proc Natl Acad Sci U S A 111:9959–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimberti I, Bednarek E, Donato F and Caroni P (2010) EphA4 signaling in juveniles establishes topographic specificity of structural plasticity in the hippocampus. Neuron 65:627–642. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C and Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio C, Hassan Mohamed I, Flammini L, Barocelli E, Incerti M, Lodola A and Tognolini M (2011) Lithocholic acid is an Eph-ephrin ligand interfering with Eph-kinase activation. PloS one 6:e18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio C, Incerti M, Corrado M, Rusnati M, Chiodelli P, Russo S, Callegari D, Ferlenghi F, Ballabeni V, Barocelli E, Lodola A and Tognolini M (2018) Pharmacological evaluation of new bioavailable small molecules targeting Eph/ephrin interaction. Biochemical pharmacology 147:21–29. [DOI] [PubMed] [Google Scholar]
- Giorgio C, Russo S, Incerti M, Bugatti A, Vacondio F, Barocelli E, Mor M, Pala D, Hassan-Mohamed I, Gioiello A, Rusnati M, Lodola A and Tognolini M (2016) Biochemical characterization of EphA2 antagonists with improved physico-chemical properties by cell-based assays and surface plasmon resonance analysis. Biochemical pharmacology 99:18–30. [DOI] [PubMed] [Google Scholar]
- Gokmen-Polar Y, Toroni RA, Hocevar BA, Badve S, Zhao Q, Shen C, Bruckheimer E, Kinch MS and Miller KD (2011) Dual targeting of EphA2 and ER restores tamoxifen sensitivity in ER/EphA2-positive breast cancer. Breast cancer research and treatment 127:375–384. [DOI] [PubMed] [Google Scholar]
- Goldgur Y, Susi P, Karelehto E, Sanmark H, Lamminmaki U, Oricchio E, Wendel HG, Nikolov DB and Himanen JP (2014) Generation and characterization of a single-chain anti-EphA2 antibody. Growth factors 32:214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Korner R, Gaitanos L and Klein R (2016) Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance. The Journal of cell biology 214:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M and Erlich Y (2011) Using DNA sequencers as stethoscopes. Genome medicine 3:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KD, Bidlingmaier SM, Zhang Y, Su Y and Liu B (2014) High-content analysis of antibody phage-display library selection outputs identifies tumor selective macropinocytosis-dependent rapidly internalizing antibodies. Molecular & cellular proteomics : MCP 13:3320–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer CE, von Zur Muhlen C, von Elverfeldt D and Peter K (2009) Single-chain antibodies as diagnostic tools and therapeutic agents. Thrombosis and haemostasis 101:1012–1019. [PubMed] [Google Scholar]
- Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, Daszak P and The H (2011) Pteropid Bats are Confirmed as the Reservoir Hosts of Henipaviruses: A Comprehensive Experimental Study of Virus Transmission. Am J Trop Med Hyg 85:946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Lutterbuese R, Roff S, Lutterbuese P, Schlereth B, Bruckheimer E, Kinch MS, Coats S, Baeuerle PA, Kufer P and Kiener PA (2007) Selective targeting and potent control of tumor growth using an EphA2/CD3-Bispecific single-chain antibody construct. Cancer research 67:3927–3935. [DOI] [PubMed] [Google Scholar]
- Han X, Xu Y, Yang Y, Xi J, Tian W, Duggineni S, Huang Z and An J (2013) Discovery and characterization of a novel cyclic peptide that effectively inhibits ephrin binding to the EphA4 receptor and displays anti-angiogenesis activity. PloS one 8:e80183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Sue M, Yamato M, Ichikawa J, Ishida S, Shibutani T, Kitamura M, Wada T and Agatsuma T (2016) Novel anti-EPHA2 antibody, DS-8895a for cancer treatment. Cancer biology & therapy 17:1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Mohamed I, Giorgio C, Incerti M, Russo S, Pala D, Pasquale EB, Zanotti I, Vicini P, Barocelli E, Rivara S, Mor M, Lodola A and Tognolini M (2014) UniPR129 is a competitive small molecule Eph-ephrin antagonist blocking in vitro angiogenesis at low micromolar concentrations. British journal of pharmacology 171:5195–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Osterfield M and Flanagan JG (2000) Regulated cleavage of a contact-mediated axon repellent. Science 289:1360–1365. [DOI] [PubMed] [Google Scholar]
- Heinzlmeir S, Lohse J, Treiber T, Kudlinzki D, Linhard V, Gande SL, Sreeramulu S, Saxena K, Liu X, Wilhelm M, Schwalbe H, Kuster B and Medard G (2017) Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors. ChemMedChem 12:999–1011. [DOI] [PubMed] [Google Scholar]
- Himanen JP (2012) Ectodomain structures of Eph receptors. Seminars in cell & developmental biology 23:35–42. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Goldgur Y, Miao H, Myshkin E, Guo H, Buck M, Nguyen M, Rajashankar KR, Wang B and Nikolov DB (2009) Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep 10:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, Saha N and Nikolov DB (2007) Cell-cell signaling via Eph receptors and ephrins. Current opinion in cell biology 19:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homaira N, Rahman M, Hossain MJ, Epstein JH, Sultana R, Khan MS, Podder G, Nahar K, Ahmed B, Gurley ES, Daszak P, Lipkin WI, Rollin PE, Comer JA, Ksiazek TG and Luby SP (2010) Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiol Infect 138:1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Balling R and Auffray C (2012) Revolutionizing medicine in the 21st century through systems approaches. Biotechnology journal 7:992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husa AM, Magic Z, Larsson M, Fornander T and Perez-Tenorio G (2016) EPH/ephrin profile and EPHB2 expression predicts patient survival in breast cancer. Oncotarget 7:21362–21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieguchi K, Tomita T, Omori T, Komatsu A, Deguchi A, Masuda J, Duffy SL, Coulthard MG, Boyd A and Maru Y (2014) ADAM12-cleaved ephrin-A1 contributes to lung metastasis. Oncogene 33:2179–2190. [DOI] [PubMed] [Google Scholar]
- Janes PW, Nievergall E and Lackmann M (2012) Concepts and consequences of Eph receptor clustering. Seminars in cell & developmental biology 23:43–50. [DOI] [PubMed] [Google Scholar]
- Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M and Nikolov DB (2005) Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123:291–304. [DOI] [PubMed] [Google Scholar]
- Janes PW, Slape CI, Farnsworth RH, Atapattu L, Scott AM and Vail ME (2014) EphA3 biology and cancer. Growth factors 32:176–189. [DOI] [PubMed] [Google Scholar]
- Ji YJ, Hwang YS, Mood K, Cho HJ, Lee HS, Winterbottom E, Cousin H and Daar IO (2014) EphrinB2 affects apical constriction in Xenopus embryos and is regulated by ADAM10 and flotillin-1. Nature communications 5:3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Wang W and Fang G (2014) Targeting protein-protein interaction by small molecules. Annual review of pharmacology and toxicology 54:435–456. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Ventrella R, Peng H, Pal-Ghosh S, Arvanitis C, Rappoport JZ, Mitchell BJ, Stepp MA, Lavker RM and Getsios S (2018) EphA2/Ephrin-A1 Mediate Corneal Epithelial Cell Compartmentalization via ADAM10 Regulation of EGFR Signaling. Invest Ophthalmol Vis Sci 59:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolpe M, Dail M and Pasquale EB (2002) An ephrin mimetic peptide that selectively targets the EphA2 receptor. The Journal of biological chemistry 277:46974–46979. [DOI] [PubMed] [Google Scholar]
- Krasnoperov V, Kumar SR, Ley E, Li X, Scehnet J, Liu R, Zozulya S and Gill PS (2010) Novel EphB4 monoclonal antibodies modulate angiogenesis and inhibit tumor growth. The American journal of pathology 176:2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung A, Chen YC, Schimpl M, Ni F, Zhu J, Turner M, Molina H, Overman R and Zhang C (2016) Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3. Journal of the American Chemical Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagares D, Ghassemi-Kakroodi P, Tremblay C, Santos A, Probst CK, Franklin A, Santos DM, Grasberger P, Ahluwalia N, Montesi SB, Shea BS, Black KE, Knipe R, Blati M, Baron M, Wu B, Fahmi H, Gandhi R, Pardo A, Selman M, Wu J, Pelletier JP, Martel-Pelletier J, Tager AM and Kapoor M (2017) ADAM10-mediated ephrin-B2 shedding promotes myofibroblast activation and organ fibrosis. Nature medicine 23:1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberto I, Lechtenberg BC, Olson EJ, Mace PD, Dawson PE, Riedl SJ and Pasquale EB (2014) Development and structural analysis of a nanomolar cyclic peptide antagonist for the EphA4 receptor. ACS chemical biology 9:2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamminmaki U, Nikolov D and Himanen J (2015) Eph Receptors as Drug Targets: Single-Chain Antibodies and Beyond. Current drug targets 16:1021–1030. [DOI] [PubMed] [Google Scholar]
- Landen CN Jr., Lu C, Han LY, Coffman KT, Bruckheimer E, Halder J, Mangala LS, Merritt WM, Lin YG, Gao C, Schmandt R, Kamat AA, Li Y, Thaker P, Gershenson DM, Parikh NU, Gallick GE, Kinch MS and Sood AK (2006) Efficacy and antivascular effects of EphA2 reduction with an agonistic antibody in ovarian cancer. Journal of the National Cancer Institute 98:1558–1570. [DOI] [PubMed] [Google Scholar]
- Lee B, Pernet O, Ahmed AA, Zeltina A, Beaty SM and Bowden TA (2015) Molecular recognition of human ephrinB2 cell surface receptor by an emergent African henipavirus. Proc Natl Acad Sci U S A 112:E2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donat H and Rocken C (2005) Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. Journal of cancer research and clinical oncology 131:41–48. [DOI] [PubMed] [Google Scholar]
- Levy GG, Motto DG and Ginsburg D (2005) ADAMTS13 turns 3. Blood 106:11–17. [DOI] [PubMed] [Google Scholar]
- Li D, Liu S, Liu R, Park R, Hughes L, Krasnoperov V, Gill PS, Li Z, Shan H and Conti PS (2013) Targeting the EphB4 receptor for cancer diagnosis and therapy monitoring. Mol Pharm 10:329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KT, Sloniowski S, Ethell DW and Ethell IM (2008) Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. The Journal of biological chemistry 283:28969–28979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisabeth EM, Falivelli G and Pasquale EB (2013) Eph receptor signaling and ephrins. Cold Spring Harbor perspectives in biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tao Z, Zhang Q, Wan S, Zhang F, Zhang Y, Wu G and Wang J (2018) YSA-conjugated mesoporous silica nanoparticles effectively target EphA2-overexpressing breast cancer cells. Cancer chemotherapy and pharmacology. [DOI] [PubMed] [Google Scholar]
- Lodola A, Giorgio C, Incerti M, Zanotti I and Tognolini M (2017) Targeting Eph/ephrin system in cancer therapy. European journal of medicinal chemistry 142:152–162. [DOI] [PubMed] [Google Scholar]
- Mao W, Luis E, Ross S, Silva J, Tan C, Crowley C, Chui C, Franz G, Senter P, Koeppen H and Polakis P (2004) EphB2 as a therapeutic antibody drug target for the treatment of colorectal cancer. Cancer research 64:781–788. [DOI] [PubMed] [Google Scholar]
- Merchant AA, Jorapur A, McManus A, Liu R, Krasnoperov V, Chaudhry P, Singh M, Harton L, Agajanian M, Kim M, Triche TJ Jr., Druker BJ, Tyner JW and Gill PS (2017) EPHB4 is a therapeutic target in AML and promotes leukemia cell survival via AKT. Blood Adv 1:1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio FA and Leone M (2016) The Sam Domain of EphA2 Receptor and its Relevance to Cancer: A Novel Challenge for Drug Discovery? Curr Med Chem 23:4718–4734. [DOI] [PubMed] [Google Scholar]
- Mercurio FA, Marasco D, Di Natale C, Pirone L, Costantini S, Pedone EM and Leone M (2016) Targeting EphA2-Sam and Its Interactome: Design and Evaluation of Helical Peptides Enriched in Charged Residues. Chembiochem 17:2179–2188. [DOI] [PubMed] [Google Scholar]
- Micewicz ED, Jung CL, Schaue D, Luong H, McBride WH and Ruchala P (2011) Small Azurin Derived Peptide Targets Ephrin Receptors for Radiotherapy. Int J Pept Res Ther 17:247–257. [Google Scholar]
- Mohamed IH, Giorgio C, Bruni R, Flammini L, Barocelli E, Rossi D, Domenichini G, Poli F and Tognolini M (2011) Polyphenol rich botanicals used as food supplements interfere with EphA2-ephrinA1 system. Pharmacological research 64:464–470. [DOI] [PubMed] [Google Scholar]
- Moss ML and Lambert MH (2002) Shedding of membrane proteins by ADAM family proteases. Essays in biochemistry 38:141–153. [DOI] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y and Pasquale EB (2003) Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nature neuroscience 6:153–160. [DOI] [PubMed] [Google Scholar]
- Nero TL, Morton CJ, Holien JK, Wielens J and Parker MW (2014) Oncogenic protein interfaces: small molecules, big challenges. Nature reviews Cancer 14:248–262. [DOI] [PubMed] [Google Scholar]
- Nievergall E, Lackmann M and Janes PW (2012) Eph-dependent cell-cell adhesion and segregation in development and cancer. Cell Mol Life Sci 69:1813–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov DB, Xu K and Himanen JP (2013) Eph/ephrin recognition and the role of Eph/ephrin clusters in signaling initiation. Biochimica et biophysica acta 1834:2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov DB, Xu K and Himanen JP (2014) Homotypic receptor-receptor interactions regulating Eph signaling. Cell adhesion & migration 8:360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, De SK, Zhang Z, Wu B, Raveendra-Panickar D, Chen V, Vazquez J, Qin H, Song J, Cosford ND, Pellecchia M and Pasquale EB (2011) A disalicylic acid-furanyl derivative inhibits ephrin binding to a subset of Eph receptors. Chemical biology & drug design 78:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, Koolpe M, Lamberto I and Pasquale EB (2012a) Inhibition of Eph receptor-ephrin ligand interaction by tea polyphenols. Pharmacological research 66:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, Koolpe M, Peddibhotla S, Dahl R, Su Y, Cosford ND, Roth GP and Pasquale EB (2008) Small molecules can selectively inhibit ephrin binding to the EphA4 and EphA2 receptors. The Journal of biological chemistry 283:29461–29472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, Lamberto I and Pasquale EB (2012b) Targeting Eph receptors with peptides and small molecules: progress and challenges. Seminars in cell & developmental biology 23:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EJ, Lechtenberg BC, Zhao C, Rubio de la Torre E, Lamberto I, Riedl SJ, Dawson PE and Pasquale EB (2016) Modifications of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor To Achieve High Plasma Stability. ACS medicinal chemistry letters 7:841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KC and Wong KT (2015) Henipavirus Encephalitis: Recent Developments and Advances. Brain pathology 25:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, Liu X, Bruno J, Heguy A, Olshen AB, Socci ND, Teruya-Feldstein J, Weis-Garcia F, Tam W, Shaknovich R, Melnick A, Himanen JP, Chaganti RS and Wendel HG (2011) The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell 147:554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pala D, Castelli R, Incerti M, Russo S, Tognolini M, Giorgio C, Hassan-Mohamed I, Zanotti I, Vacondio F, Rivara S, Mor M and Lodola A (2014) Combining ligand- and structure-based approaches for the discovery of new inhibitors of the EPHA2-ephrin-A1 interaction. Journal of chemical Information and modeling 54:2621–2626. [DOI] [PubMed] [Google Scholar]
- Pasquale EB (2005) Eph receptor signalling casts a wide net on cell behaviour. Nature reviews Molecular cell biology 6:462–475. [DOI] [PubMed] [Google Scholar]
- Patel K, Doddapaneni R, Sekar V, Chowdhury N and Singh M (2016) Combination Approach of YSA Peptide Anchored Docetaxel Stealth Liposomes with Oral Antifibrotic Agent for the Treatment of Lung Cancer. Mol Pharm 13:2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg CL, Cooper LT, Zhao J, Gerometta M, Smith FM, Yeh M, Bartlett PF, Gorman JJ and Boyd AW (2017) Glycoengineering of EphA4 Fc leads to a unique, long-acting and broad spectrum, Eph receptor therapeutic antagonist. Sci Rep 7:6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty A, Idippily N, Bobba V, Geldenhuys WJ, Zhong B, Su B and Wang B (2018) Design and synthesis of small molecule agonists of EphA2 receptor. European journal of medicinal chemistry 143:1261–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty A, Myshkin E, Qin H, Guo H, Miao H, Tochtrop GP, Hsieh JT, Page P, Liu L, Lindner DJ, Acharya C, MacKerell AD Jr., Ficker E, Song J and Wang B (2012) A small molecule agonist of EphA2 receptor tyrosine kinase inhibits tumor cell migration in vitro and prostate cancer metastasis in vivo. PloS one 7:e42120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, Cotrina M and Wilkinson DG (2004) Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Developmental cell 7:465–480. [DOI] [PubMed] [Google Scholar]
- Porotto M, Carta P, Deng Y, Kellogg GE, Whitt M, Lu M, Mungall BA and Moscona A (2007) Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. Journal of virology 81:10567–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Hossain MJ, Sultana S, Homaira N, Khan SU, Rahman M, Gurley ES, Rollin PE, Lo MK, Comer JA, Lowe L, Rota PA, Ksiazek TG, Kenah E, Sharker Y and Luby SP (2011) Date Palm Sap Linked to Nipah Virus Outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis. [DOI] [PubMed] [Google Scholar]
- Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ and Black RA (2000) Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. The Journal of biological chemistry 275:14608–14614. [DOI] [PubMed] [Google Scholar]
- Riedl SJ and Pasquale EB (2015) Targeting the Eph System with Peptides and Peptide Conjugates. Current drug targets 16:1031–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht W and Philips T (2013) The changing scene of amyotrophic lateral sclerosis. Nature reviews Neuroscience 14:248–264. [DOI] [PubMed] [Google Scholar]
- Saftig P and Reiss K (2011) The "A Disintegrin And Metalloproteases" ADAM10 and ADAM17: novel drug targets with therapeutic potential? European journal of cell biology 90:527–535. [DOI] [PubMed] [Google Scholar]
- Salem AF, Wang S, Billet S, Chen JF, Udompholkul P, Gambini L, Baggio C, Tseng HR, Posadas EM, Bhowmick NA and Pellecchia M (2018a) A potent EphA2 agonistic peptide-drug conjugate reduces circulating cancer cells and metastases in breast cancer models. J Med Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AF, Wang S, Billet S, Chen JF, Udompholkul P, Gambini L, Baggio C, Tseng HR, Posadas EM, Bhowmick NA and Pellecchia M (2018b) Reduction of Circulating Cancer Cells and Metastases in Breast- Cancer Models by a Potent EphA2-Agonistic Peptide-Drug Conjugate. J Med Chem 61:2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgia R, Kulkarni P and Gill PS (2018) EphB4: A promising target for upper aerodigestive malignancies. Biochimica et biophysica acta 1869:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Liu J, Joshi SB, Krasnoperov V, Gill P, Middaugh CR and Volkin DB (2012) Biophysical characterization and stabilization of the recombinant albumin fusion protein sEphB4-HSA. J Pharm Sci 101:1969–1984. [DOI] [PubMed] [Google Scholar]
- Shi X, Hapiak V, Zheng J, Muller-Greven J, Bowman D, Lingerak R, Buck M, Wang BC and Smith AW (2017) A role of the SAM domain in EphA2 receptor activation. Sci Rep 7:45084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirintrapun SJ, Zehir A, Syed A, Gao J, Schultz N and Cheng DT (2015) Translational Bioinformatics and Clinical Research (Biomedical) Informatics. Surgical pathology clinics 8:269–288. [DOI] [PubMed] [Google Scholar]
- Skotland T, Iversen TG, Torgersen ML and Sandvig K (2015) Cell-penetrating peptides: possibilities and challenges for drug delivery in vitro and in vivo. Molecules 20:13313–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G, Cortina C, Sevillano M and Batlle E (2011) Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nature cell biology 13:1100–1107. [DOI] [PubMed] [Google Scholar]
- Spanevello MD, Tajouri SI, Mirciov C, Kurniawan N, Pearse MJ, Fabri LJ, Owczarek CM, Hardy MP, Bradford RA, Ramunno ML, Turnley AM, Ruitenberg MJ, Boyd AW and Bartlett PF (2013) Acute delivery of EphA4-Fc improves functional recovery after contusive spinal cord injury in rats. J Neurotrauma 30:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen DL, Xu K, Nikolov DB and Broder CC (2012) Henipavirus mediated membrane fusion, virus entry and targeted therapeutics. Viruses 4:280–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Deffrasnes C, Foo CH, Bean AGD and Wang LF (2017) A Functional Genomics Approach to Henipavirus Research: The Role of Nuclear Proteins, MicroRNAs and Immune Regulators in Infection and Disease. Current topics in microbiology and immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords RT, Greenberg PL, Wei AH, Durrant S, Advani AS, Hertzberg MS, Lewis ID, Rivera G, Gratzinger D, Fan AC, Felsher DW, Cortes JE, Watts JM, Yarranton GT, Walling JM and Lancet JE (2016) KB004, a first in class monoclonal antibody targeting the receptor tyrosine kinase EphA3, in patients with advanced hematologic malignancies: Results from a phase 1 study. Leuk Res 50:123–131. [DOI] [PubMed] [Google Scholar]
- Takano H, Nakamura T, Tsuchikawa T, Kushibiki T, Hontani K, Inoko K, Takahashi M, Sato S, Abe H, Takeuchi S, Sato N, Hiraoka K, Nishihara H, Shichinohe T and Hirano S (2015) Inhibition of Eph receptor A4 by 2,5-dimethylpyrrolyl benzoic acid suppresses human pancreatic cancer growing orthotopically in nude mice. Oncotarget 6:41063–41076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki S, Kamada H, Inoue M, Nagano K, Mukai Y, Higashisaka K, Yoshioka Y, Tsutsumi Y and Tsunoda S (2015) A Novel Bispecific Antibody against Human CD3 and Ephrin Receptor A10 for Breast Cancer Therapy. PloS one 10:e0144712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang FY, Chiang EP and Shih CJ (2007) Green tea catechin inhibits ephrin-A1-mediated cell migration and angiogenesis of human umbilical vein endothelial cells. The Journal of nutritional biochemistry 18:391–399. [DOI] [PubMed] [Google Scholar]
- Taylor H, Campbell J and Nobes CD (2017) Ephs and ephrins. Curr Biol 27:R90–R95. [DOI] [PubMed] [Google Scholar]
- Tognolini M, Giorgio C, Hassan Mohamed I, Barocelli E, Calani L, Reynaud E, Dangles O, Borges G, Crozier A, Brighenti F and Del Rio D (2012) Perturbation of the EphA2-EphrinA1 system in human prostate cancer cells by colonic (poly)phenol catabolites. Journal of agricultural and food chemistry 60:8877–8884. [DOI] [PubMed] [Google Scholar]
- Tognolini M, Incerti M and Lodola A (2014) Are we using the right pharmacological tools to target EphA4? ACS chemical neuroscience 5:1146–1147. [DOI] [PubMed] [Google Scholar]
- Tognolini M and Lodola A (2015) Targeting the Eph-ephrin System with Protein-Protein Interaction (PPI) Inhibitors. Current drug targets 16:1048–1056. [DOI] [PubMed] [Google Scholar]
- Truitt L and Freywald A (2011) Dancing with the dead: Eph receptors and their kinase-null partners. Biochemistry and cell biology = Biochimie et biologie cellulaire 89:115–129. [DOI] [PubMed] [Google Scholar]
- Turk B (2006) Targeting proteases: successes, failures and future prospects. Nature reviews Drug discovery 5:785–799. [DOI] [PubMed] [Google Scholar]
- Unzue A, Lafleur K, Zhao H, Zhou T, Dong J, Kolb P, Liebl J, Zahler S, Caflisch A and Nevado C (2016) Three stories on Eph kinase inhibitors: From in silico discovery to in vivo validation. European journal of medicinal chemistry 112:347–366. [DOI] [PubMed] [Google Scholar]
- Vail ME, Murone C, Tan A, Hii L, Abebe D, Janes PW, Lee FT, Baer M, Palath V, Bebbington C, Yarranton G, Llerena C, Garic S, Abramson D, Cartwright G, Scott AM and Lackmann M (2014) Targeting EphA3 inhibits cancer growth by disrupting the tumor stromal microenvironment. Cancer research 74:4470–4481. [DOI] [PubMed] [Google Scholar]
- Vargas LM, Leal N, Estrada LD, Gonzalez A, Serrano F, Araya K, Gysling K, Inestrosa NC, Pasquale EB and Alvarez AR (2014) EphA4 activation of c-Abl mediates synaptic loss and LTP blockade caused by amyloid-beta oligomers. PloS one 9:e92309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vearing C, Lee FT, Wimmer-Kleikamp S, Spirkoska V, To C, Stylianou C, Spanevello M, Brechbiel M, Boyd AW, Scott AM and Lackmann M (2005) Concurrent binding of anti-EphA3 antibody and ephrin-A5 amplifies EphA3 signaling and downstream responses: potential as EphA3-specific tumor-targeting reagents. Cancer research 65:6745–6754. [DOI] [PubMed] [Google Scholar]
- Wang L-F, Mackenzie JS and Broder CC (2013) Henipaviruses, in Fields Virology (Knipe DM and Howley PM eds) pp 1070–1085, Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- Wang Y, Li Q, Zheng Y, Li G and Liu W (2016) Systematic biochemical characterization of the SAM domains in Eph receptor family from Mus Musculus. Biochemical and biophysical research communications 473:1281–1287. [DOI] [PubMed] [Google Scholar]
- Weatherman S, Feldmann H and de Wit E (2017) Transmission of henipaviruses. Current opinion in virology 28:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Xu G, Bridges LC, Williams P, White JM and DeSimone DW (2010) ADAM13 induces cranial neural crest by cleaving class B Ephrins and regulating Wnt signaling. Developmental cell 19:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer-Kleikamp SH, Janes PW, Squire A, Bastiaens PI and Lackmann M (2004) Recruitment of Eph receptors into signaling clusters does not require ephrin contact. The Journal of cell biology 164:661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer-Kleikamp SH and Lackmann M (2005) Eph-modulated cell morphology, adhesion and motility in carcinogenesis. IUBMB life 57:421–431. [DOI] [PubMed] [Google Scholar]
- Wu B, Barile E, De SK, Wei J, Purves A and Pellecchia M (2015a) High-Throughput Screening by Nuclear Magnetic Resonance (HTS by NMR) for the Identification of PPIs Antagonists. Curr Top Med Chem 15:2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, De SK, Kulinich A, Salem AF, Koeppen J, Wang R, Barile E, Wang S, Zhang D, Ethell I and Pellecchia M (2017) Potent and Selective EphA4 Agonists for the Treatment of ALS. Cell Chem Biol 24:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Wang S, De SK, Barile E, Quinn BA, Zharkikh I, Purves A, Stebbins JL, Oshima RG, Fisher PB and Pellecchia M (2015b) Design and Characterization of Novel EphA2 Agonists for Targeted Delivery of Chemotherapy to Cancer Cells. Chem Biol 22:876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, Huang M, Zhang R, Song S, Lu W, Flores L 2nd, Gelovani J and Li C (2011) In vivo small-animal PET/CT of EphB4 receptors using 64Cu-labeled peptide. J Nucl Med 52:241–248. [DOI] [PubMed] [Google Scholar]
- Xu D, Bum-Erdene K, Si Y, Zhou D, Ghozayel MK and Meroueh SO (2017) Mimicking Intermolecular Interactions of Tight Protein-Protein Complexes for Small-Molecule Antagonists. ChemMedChem 12:1794–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jalal SI, Sledge GW and Meroueh SO (2016) Small-molecule binding sites to explore protein-protein interactions in the cancer proteome. Mol Biosyst 12:3067–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Broder CC and Nikolov DB (2012a) Ephrin-B2 and ephrin-B3 as functional henipavirus receptors. Seminars in cell & developmental biology 23:116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Chan YP, Rajashankar KR, Khetawat D, Yan L, Kolev MV, Broder CC and Nikolov DB (2012b) New insights into the Hendra virus attachment and entry process from structures of the virus G glycoprotein and its complex with Ephrin-B2. PloS one 7:e48742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC and Nikolov DB (2008) Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A 105:9953–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Rockx B, Xie Y, DeBuysscher BL, Fusco DL, Zhu Z, Chan YP, Luu T, Cer R, Feldmann H, Mokashi V, Dimitrov DS, Bishop-Lilly KA, Broder CC and Nikolov DB (2013a) Crystal structure of the Hendra virus attachment G glycoprotein bound to a potent cross-reactive neutralizing human monoclonal antibody. PLoS Pathog in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Tzvetkova-Robev D, Xu Y, Goldgur Y, Chan YP, Himanen JP and Nikolov DB (2013b) Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5. Proc Natl Acad Sci U S A 110:14634–14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zhang R, Xiong C, Zhong M, Melancon M, Gupta S, Nick AM, Sood AK and Li C (2012) Effective photothermal chemotherapy using doxorubicin-loaded gold nanospheres that target EphB4 receptors in tumors. Cancer research 72:4777–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X, Ding H, Zhao X, Li X, Du Z, Hu H, Qiao M, Chen D, Deng Y and Zhao X (2016) Anti-EphA10 antibody-conjugated pH-sensitive liposomes for specific intracellular delivery of siRNA. Int J Nanomedicine 11:3951–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Bossart KN, Bishop KA, Crameri G, Dimitrov AS, McEachern JA, Feng Y, Middleton D, Wang LF, Broder CC and Dimitrov DS (2008) Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. The Journal of infectious diseases 197:846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang G, Brantley-Sieders DM, Vaught D, Yu J, Xie L, Wells S, Jackson D, Muraoka-Cook R, Arteaga C and Chen J (2010) Elevation of receptor tyrosine kinase EphA2 mediates resistance to trastuzumab therapy. Cancer research 70:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]