Abstract
Purpose of review:
To summarize recent advances in our understanding of HIV adaptation to Human Leukocyte Antigen (HLA)-associated immune pressures, and its relevance to HIV prevention and cure research.
Recent findings:
Recent research has confirmed that HLA is a major driver of individual- and population-level HIV evolution, that HIV strains are adapting to the immunogenetic profiles of the different human ethnic groups in which they circulate, and that HIV adaptation has substantial clinical and immunologic consequences. As such, adaptation represents a major challenge to HIV prevention and cure. At the same time, there are opportunities: Studies of HIV adaptation are revealing why certain HLA alleles are protective in some populations and not others; they are identifying immunogenic viral epitopes that harbor high mutational barriers to escape, and they may help illuminate novel, vaccine-relevant HIV epitopes in regions where circulating adaptation is extensive. Elucidation of HLA-driven adapted and nonadapted viral forms in different human populations and HIV subtypes also renders “personalized” immunogen selection, as a component of HIV cure strategies, conceptually feasible.
Summary:
Though adaptation represents a major challenge to HIV prevention and cure, achieving an in-depth understanding of this phenomenon can help move the design of such strategies forward.
Keywords: HIV, HLA, immune escape, adaptation, evolution, vaccine
Introduction
Human Leukocyte Antigen (HLA) class I-restricted CD8+ T-lymphocytes (CTL) play a critical role in HIV control: the initial T cell responses to the transmitted/founder virus help control acute-phase viremia to setpoint levels [1–3], and the expression of specific protective HLA alleles is firmly linked to slower HIV progression [4]. In the early 1990s however, the first reports of mutational HIV escape from HLA-restricted CTL pressures in individuals began to emerge [5]; subsequent studies confirmed that the earliest HIV escape variants emerge rapidly following infection [6–11] and continue to be selected thereafter [12,13] (Figure 1). Mechanisms were also elucidated: escape mutations allow HIV-infected cells to avoid CTL detection by disrupting intracellular epitope processing [14], abrogating epitope-HLA binding [12] and/or altering epitope-HLA interactions with the T-cell receptor [15,16]. It is now appreciated that CTL escape represents a major challenge for immune- and vaccine-mediated HIV control [17,18].
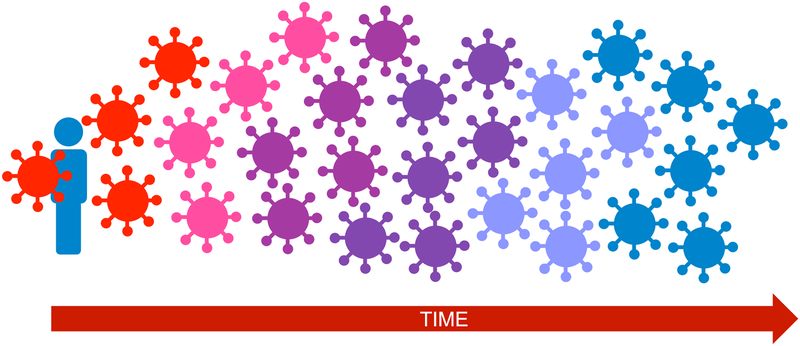
Figure 1. HIV adaptation to HLA within-host.
After infection is established by (usually) a single transmitted/founder virus (red), descendant within-host viral populations display progressively increasing adaptation to host HLA. Immune escape mutations can, at least in theory, accumulate until HIV has fully adapted to host HLA (shown here by the virus population that gradually shifts to the same color as the host).
Identification of HLA-associated polymorphisms in HIV using statistical approaches
A significant advance in our understanding of the extent and locations of HLA-driven escape in HIV occurred in 2002, when Moore et al developed a statistical approach to identify these [19]. If escape is HLA-specific, they reasoned, then these pathways should be identifiable by analyzing large HIV sequence datasets annotated with HLA information: specifically, HIV amino acids that were overrepresented in individuals carrying a specific HLA allele (representing the HLA-adapted or inferred escaped form) and, conversely, HIV amino acids that were underrepresented in individuals carrying this allele (representing the HLA-nonadapted or inferred susceptible form) could be identified. This approach was unbiased to the location of CTL epitopes (in fact, it was later used to discover new ones [20]); in addition it corrected for multiple comparisons and addressed possible confounders including co-expressed HLA alleles and covarying HIV amino acids. Over 100 HLA-associated polymorphisms in HIV reverse transcriptase were identified, confirming that escape was reproducible and HLA-specific, and establishing that HLA extensively impacted HIV sequence diversity [19]. Moore et al also recognized that immunogenicity sometimes equated with benefit to the virus rather than the host and coined the broader term “HIV adaptation” (as opposed to escape mutation) to remain agnostic to mechanism and include the paradoxical situation in which the virus adapts to increase, rather than decrease, HLA-restricted recognition [19]. This phenomenon has since been described in more detail [21] and a potential mechanism described experimentally [22]. Later studies further refined the methodology. Critically, it was demonstrated that formal phylogenetic correction for the underlying evolutionary relationships between HIV sequences substantially reduced the number of spurious associations (both false-positive and false-negative) by controlling for viral founder effects [23,24], and more sophisticated approaches to address HLA co-expression and HIV codon covariation were also later developed [25–27]. Nevertheless, the original observation that HLA extensively impacted HIV sequence diversity [19] still held [25].
Phylogenetically-informed approaches have since been used to elucidate HLA-driven escape pathways in all major HIV subtypes including A [28], B [29–31] C [32,33], D [28] and Circulating Recombinant form (CRF)_01 (AE) [34,35], confirming that adaptation is widespread (e.g. >2100 HLA-associated polymorphisms have been mapped across HIV subtype B [29]) and that its specific mutational pathways are reproducible and HLA-specific. Over eighty percent of subtype B-infected, HLA-A*24:02-expressing persons, for example, will select Nef-Y135F [29], which confers partial escape from CTL responses to the overlapping Nef-RW8 and Nef-RF10 epitopes [36,37]. HLA-class II-associated HIV polymorphisms have also been mapped [38], indicating that selective pressure exerted by CD4+ T cells drive HIV evolution as well.
Quantifying HIV adaptation to HLA, and demonstrating clinical relevance
While these studies yielded comprehensive knowledge of HLA-driven escape pathways [25,26,29,33,39–42], most did not address their clinical relevance. While carriage of HLA alleles such as HLA-B*57:01 (e.g. [4,43–46]), CTL targeting of HIV proteins such as Gag [47–50] and other CD8+ T-cell properties such as variant cross-reactivity [51] were clearly linked to viral control, evidence that CTL escape led to loss of this control was limited. Well into the 2000s, evidence was still largely restricted to case-reports of viremia breakthrough following isolated escape events [12,52–54], though some exceptions exist (e.g. [55,56]). Clinical relevance was challenging to establish because a single escape event was unlikely to precipitate loss of viral control in most cases (an average of 3 HIV epitopes are targeted during acute infection [57], broadening to an average of 20 by the chronic phase [58]), and because immune responses are highly dynamic (variant-specific and/or cross-reactive CD8+ T-cells responses often emerge following escape [59,60], and escape within one epitope may create another nearby [61]).
Population-level analyses were performed to link overall escape burden with clinical prognosis, but early results were unconvincing (e.g. [62]), largely for two reasons. First, early algorithms treated all HLA-associated polymorphisms equally, even though their strengths of selection varied widely [29]. A more nuanced metric was thus needed. Secondly, inference of clinical impact from HIV/HLA genotypes alone had its limitations. Specifically, while the presence of escaped HIV in an individual harboring the restricting HLA supported a prior CTL response from which HIV had managed to escape (though transmitted escape cannot be ruled out), the presence of HLA-susceptible HIV could indicate two opposing scenarios. It could indicate a sustained CTL response from which HIV had not yet managed to escape (a protective scenario with respect to viremia control), or it could indicate an inability to target the epitope in the first place (a detrimental scenario). Analyses that integrated HIV/HLA genotypes with CTL response data were thus needed.
These limitations were only recently overcome as part of an international collaborative effort, which included the development of a nuanced metric to quantify the level of adaptation of a given HIV sequence to a given HLA allele, termed the “adaptation score” [63]. The metric employs a probabilistic model to compare what an HIV sequence would ‘look like’ if it were to evolve indefinitely in a host whose CTL response solely targeted HIV epitopes restricted by the HLA allele(s) in question, versus what it would be if the virus were to evolve indefinitely under no immune pressure. Researchers first validated the metric by demonstrating that transmitted HIV sequences initially harbored high adaptation to the donor’s HLA profile but thereafter displayed increasing adaptation to the recipient [63]. Elite controllers [64] also displayed significantly lower HIV adaptation than non-controllers, independent of protective HLA allele carriage, confirming that low autologous HIV adaptation is a correlate of control [63]. Importantly, researchers demonstrated that individuals carrying protective HLA alleles exhibited viral load benefits only if they harbored HLA-susceptible HIV, indicating that HLA-specific protective effects disappear with adaptation [63]. Furthermore, by incorporating CTL response data, researchers confirmed that the best viremia control occurred in individuals with broad anti-HIV CTL responses but low HIV adaptation [63], confirming that sustained CTL targeting of multiple epitopes is critical to HIV control, and providing strong evidence that escape from these responses has negative clinical consequences.
Clinical consequences of transmitted escape
Transmission of HLA-adapted HIV sequences should accelerate clinical progression, but this too had been challenging to demonstrate (Figure 2). It was known that escape mutations could be stably transmitted [65]; that recipients who shared HLA alleles with their donors tended to experience higher setpoint viremia [66]; that rapid progressors tended to harbor a higher mutational burden within optimal CTL epitopes [56]; it had even been specifically demonstrated that transmission of B*57:03-associated escape mutations to B*57:03-expressing recipients compromised HIV control [67]. But, it was not until the adaptation metric was developed that it was possible to broadly demonstrate that transmission of “pre-adapted” HIV had negative clinical consequences [63]. Furthermore, HLA adaptation provided an alternative mechanism to explain the longstanding observation that the pVL setpoint was to a certain extent “heritable” [68]: donor and recipient pVL correlated significantly only in transmission pairs sharing high HLA-B adaptation-similarity, suggesting that the “heritable” viral factor is, in part, HLA adaptation [63]. The reason why pre-adapted HIV transmission led to poorer prognosis was also intuitive: acute-phase immune responses to founder virus epitopes transmitted in their HLA-adapted form were limited, and those that did exist were largely dysfunctional, confirming that pre-adapted HIV sequences were less immunogenic than their nonadapted counterparts [56,63,69].
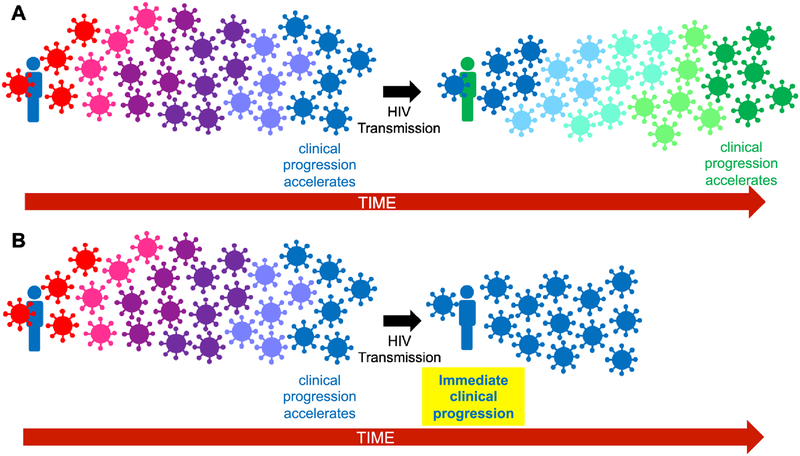
Figure 2. Clinical consequences of adapted HIV transmission.
Panel A. HIV transmission between HLA-unmatched individuals: normal clinical progression. As illustrated in Figure 1, it takes time for descendant within-host HIV populations to fully adapt to host HLA (shown by virus color change to match that of the host). Upon transmission of this strain to an HLA-unmatched host (black arrow), descendant within-host viral populations will need to adapt anew. In both the donor and recipient, clinical progression will accelerate as HIV increasingly adapts to host HLA, but this takes time due to the required viral evolutionary process. Panel B. HIV transmission to an HLA-matched host: rapid clinical progression in the recipient. Transmission of an HIV strain that matches the recipient’s HLA profile requires no further viral adaptation to circumvent immune responses in the new host, leading to rapid clinical progression.
The full immunologic consequences of HIV preadaptation to HLA are best appreciated at the population level. Given that HIV sequences reproducibly adapt to the HLA alleles expressed by their hosts, HIV sequences circulating in a human population will harbor adaptations specific to the HLA alleles expressed in that population, at frequencies that roughly correlate with that of the restricting HLA (and will exceed the HLA frequency if reversion is sufficiently low) (Figure 3). This was first established by Kawashima et al, who showed, using linked HIV/HLA genotypes from nearly 3000 participants of 9 cohorts spanning 5 continents, that the prevalence of select escape mutations correlated with the frequency of the restricting HLA in the population [70]. The most striking example was the B*51-restricted I135X escape mutation in Reverse Transcriptase, which occurs at the C-terminus of the TI8 epitope, whose circulating prevalence correlated significantly with population B*51 prevalence. Indeed, Japan differs from nearly all other subtype B epidemics in that the consensus residue at RT-135 is not Isoleucine (I), but rather Threonine (T), presumably due to its selection by B*51 and the related allele B*52, which are expressed in more than 40% of Japanese individuals [71]. There is only one other subtype B epidemic where RT-I135T is consensus: Saskatchewan, Canada [72], where 80% of infected persons have Indigenous ancestry, and where B*51 represents the most common HLA-B allele in this population [73]. The A*24:02-associated Nef-Y135F escape mutation provides another example: whereas the global HIV subtype B consensus is tyrosine (Y), in Japan, where >60% of persons express A*24:02, the consensus is Phenylalanine (F), the A*24:02-associated escape variant [74].
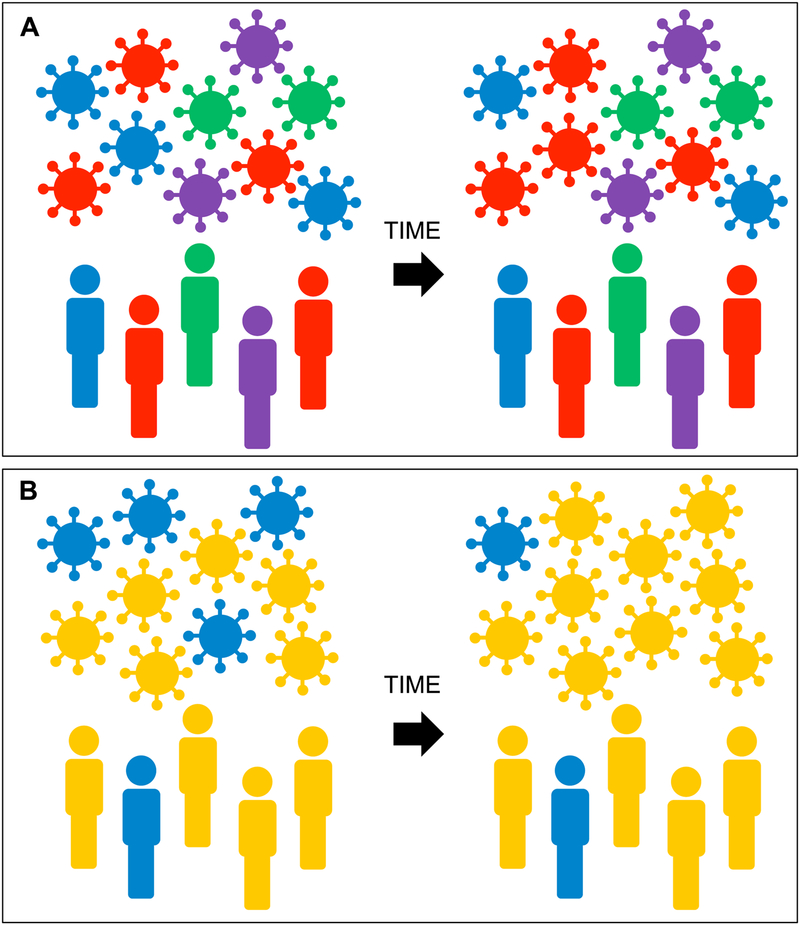
Figure 3. HIV adaptation to HLA at the population level.
HIV sequences circulating in a human population will harbor adaptations that reflect the HLA alleles expressed in that population. Panel A. In a population with high HLA diversity (shown by different host colors), the risk of acquiring an HIV strain pre-adapted to host HLA (i.e. the risk of acquiring a virus whose color matches that of the host) is relatively low. In such a setting, population-level HIV adaptation to HLA will occur relatively slowly (indicated by minimal shifts in virus color composition over time). Panel B. In a population with low HLA diversity, the risk of being infected with HIV pre-adapted to host HLA is far higher. Furthermore, over time, HIV strains harboring adaptations specific for common HLA alleles in the population (shown here in yellow) will increase in frequency more rapidly.
This led Kawashima et al to postulate that “viral adaptation may dismantle the well-established HLA associations with control of HIV infection” [70], a prediction that has been borne out in some populations, in particular those where HLA diversity is limited and/or HIV seroprevalence is high. In Japan, HIV adaptation has occurred due to low population HLA diversity. More than 60% of Japanese persons express A*24:02 and >40% express B*51 or B*52, and escape mutations selected by these alleles (Nef-Y135F and RT-I135X respectively) represent the population consensus [70,74]. As a result, B*51’s protective allele status in Japan disappeared by 2001 [75], and accelerated clinical progression has been documented in A*24:02-expressing individuals inferred to have been transmitted Nef-Y135F-containing HIV [37]. In Botswana by contrast, HIV adaptation has occurred due to high HIV seroprevalence maintained over decades: here, HIV adaptation to multiple HLA alleles, including B*57/58:01, is significantly elevated compared to neighboring regions, and as a result B*57 and 58:01 are no longer protective [76]. Saskatchewan, Canada, provides an example where adaptation has occurred in a population with limited HLA diversity and high HIV seroprevalence: extensive HIV adaptation to HLA, and B*51 in particular [72] likely explain, at least in part, regional reports of rapid progression [77], in particular among HLA-B*51-expressing persons [78].
Elsewhere, adaptation is occurring more slowly. Surveys in North America have indicated that, while CTL escape mutation frequencies have approximately doubled during the past 30 years, their absolute magnitudes generally remain low, indicating that viral adaptation will not erode HLA-associated protective effects anytime soon [79,80]. The recent demonstration however that, regardless of location, “protective” HLA alleles are those to which circulating HIV sequences are not (yet) well adapted, confirms that the immunological landscape presented by HIV is dynamic [63]. For example, while B*57:03 and B*58:01 retain their protective status in South Africa, where circulating HIV adaptation to these alleles remains low, they have lost much of their protective status in Botswana and Zambia, where adaptation is higher [63].
Will HIV adaptation to HLA undermine efforts to develop a prophylactic vaccine?
Given HIV’s ongoing adaptation to HLA, are efforts to develop CTL-based vaccines becoming increasingly futile? Indeed, recent HIV epidemic models that simulated large-scale vaccination programmes indicated that population-level HIV adaptation would substantially undermine HIV vaccine efficacy, in terms of the number of new infections averted by vaccination [81]. Recent evidence from Japan however, where population-level HIV adaptation to HLA is extensive, suggests that hope nevertheless remains. Immunogenicity studies of the second-generation tHIVconsvX “conserved mosaic” T-cell vaccine, which comprises functionally conserved HIV regions that include CTL epitopes associated with viremia control, delivered as bivalent complementary mosaic immunogens (to maximize HIV diversity coverage), revealed significant correlations between response breadth/magnitude and favorable clinical profiles in chronically HIV subtype B-infected treatment-naive Japanese persons, in a cross-sectional analysis [82]. Furthermore, response fine-mapping uncovered more than 15 novel subdominant CD8+ T-cell epitopes within the immunogen in this population [50,83], where CD8+ T-cells specific to five of these epitopes cross-recognized diverse viral variants and suppressed HIV replication in vivo [50]. This, taken together with the recent results of the phase 1/2a APPROACH trial, which reported an 83% T-cell response rate to the mosaic HIV envelope/Gag/Pol immunogens delivered via an adenovirus serotype 26 vector [84], as well as the striking vaccine-mediated protection from Simian Immunodeficiency Virus infection in a rhesus monkey cytomegalovirus-vectored vaccine [85–87] mediated by non-canonical CD8+ T-cell responses [88,89], gives continued hope that vaccine-mediated stimulation of effective T-cell responses will be achievable despite population-level HIV adaptation to HLA. However, an important caveat may be to exclude epitopes from the immunogen that are known to induce T-cell responses associated with increased (rather than decreased or equivalent) viral loads in vivo [21,22].
Is HIV adaptation to HLA changing viral virulence?
Although escape mutations clearly advantage HIV in terms of immune evasion, some weaken the virus by compromising viral protein function and/or replication [8,9,13,90–92]. And, when these mutations are transmitted to individuals lacking the restricting HLA, the clinical effects are measurable. Individuals harboring HIV with low replication capacity display favorable clinical profiles [32,56,93–95]; transmission of HIV sequences containing fitness-costly escape mutations are linked to lower viral loads in the recipient [33,96,97]; and the presence of these mutations offset the negative clinical consequences of transmitted escape [69]. These observations have led some to hypothesize that adaptation may have a “silver lining”: specifically, that HIV adaptation to the most protective HLA alleles (which tend to drive the most fitness-costly escape mutations [98,99]) will gradually lower viral replication capacity at the population level, thereby driving down viral loads and reducing HIV virulence over time [76,100]. Comparative studies in Southern Africa [76], as well as Mexico and the Caribbean [100] support this notion, and a recent report from Uganda indicated that setpoint viral loads among newly-infected individuals declined by 0.4 log10 copies/mL from 1995 to 2012 (though this study did not investigate HLA adaptation as a possible cause) [101]. Studies in other regions however suggest that HIV virulence may be increasing [102,103]. Future analyses of virulence dynamics that explicitly incorporate HIV adaptation to HLA may therefore aid in resolving these conflicting - or perhaps region-specific - differences.
Can the study of HIV adaptation to HLA help inform the design of a preventive vaccine?
Ending the HIV pandemic will likely require an effective preventive HIV vaccine, and the best strategies will likely induce broadly-neutralizing antibodies as well as effective cellular responses across the range of HLA alleles expressed in the population. Three approaches are being pursued to achieve the latter. The first two are guided by HIV sequence information: these include “conserved element” strategies which aim to focus CTL responses against HIV regions with a high mutational barrier to escape [82,104–106], and “mosaic” (polyvalent) approaches which seek to maximize coverage of global HIV diversity, including common escape variants, with the goal of priming immune responses against the most diverse possible array of infecting strains [107,108]. A third strategy is guided by human immune response data, by identifying epitopes and/or viral regions associated with viral control for immunogen design [109,110]. These strategies are not mutually exclusive; the tHIVconsvX vaccine for example incorporates elements of all three [82].
Although the above strategies show promise, substantial gaps remain in our knowledge of which HIV epitopes/regions are immunogenic and protective (versus immunogenic and neutral or harmful), which harbor the highest mutational barriers to escape, and which HLA alleles mediate responses to these key regions, across different global populations and HIV subtypes. This information is essential if we wish to achieve effective vaccine-induced antiviral responses across a broad range of HIV epitopes in immunogenetically diverse human populations. Studies of HIV adaptation to HLA can narrow these knowledge gaps. In particular, integrated bioinformatics and mechanistic analyses of HLA-driven adaptation pathways across different populations and HIV subtypes can inform CTL-based immunogen selection for region-specific or universal HIV vaccines [28,30,31]. This is because HLA-associated polymorphisms mark viral sites under intense and reproducible in vivo immune pressure [29]; as such, their identification can reveal novel immunogenic viral regions, including epitopes targeted by understudied and/or population-specific HLA alleles [20,111–113]. Comparative analyses across distinct host populations harboring the same HIV subtype can illuminate the extent to which HIV immunogenic regions and escape pathways are universal versus population-specific [30,31,41], while analyses of HIV epidemics where multiple viral subtypes co-circulate can illuminate the extent to which HIV immunogenic regions are universal versus HIV subtype-specific [28]. Recently, both types of studies have been undertaken, yielding novel insights.
It is now clear, for example, that HIV adaptation pathways differ markedly around the globe. Even though the Japanese and North American HIV epidemics are predominantly subtype B, two-thirds of HLA-driven HIV adaptations in Japan are not observed in North America because the HLA distributions of these populations differ so markedly [30]. Effects can be highly region-specific: over 60% of HLA-associated polymorphisms identified in the unique and highly genetically admixed Mexican population are not observed in Canada/USA, despite HIV subtype B predominating throughout North America [31]. Effects are also HLA-specific: closely related HLA alleles, targeting the same CTL epitope, often drive distinct escape pathways [114]. HIV adaptation can also yield unanticipated insight into population immunity: for example, the overall strength of HLA-driven selection on HIV in Mexico was found to be significantly lower than that in Canada/USA, suggesting that antiviral immunity in Mexico may be weaker, and/or HIV escape pathways less consistent, than elsewhere [31]. If this phenomenon were to be observed in other epidemics, it could have implications for natural and vaccine-induced anti-HIV immunity in these regions.
HIV genetic context also matters [28,115]. This was originally described for a single HLA-restricted CTL epitope: B*57:03-mediated escape within KF11 (Gag codon 162–172) differed in subtypes B and C due to viral backbone-specific functional constraints [115]. Recently however the first comprehensive characterization of differential HLA-driven escape across HIV subtypes (in this case, A1 and D, which cocirculate in East Africa) was undertaken, revealing that one-third of HLA-associated polymorphisms were differentially selected between them, confirming that viral genetic context markedly influences adaptation [28]. Notably, researchers identified epitopes that were universally immunogenic but where the mutational barrier to escape was particularly high in one HIV subtype, thereby illuminating potentially useful vaccine immunogen(s) for that HIV subtype [28].
How else may HIV adaptation to HLA influence HIV prevention strategies?
Escape mutations rarely overlap with drug resistance sites, but some exceptions exist. RT-I135T/V/L (selected by B*51 and B*52 [39]) and RT-283I/L (selected by B*15 [39]) confer up to 3-fold reduced in vitro susceptibility to first-generation Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs) [116], while RT-E138G/A/K, selected by B*18 [29,117] can mediate up to 7-fold decreased susceptibility to the second-generation NNRTI rilpivirine [117]. The latter was recently highlighted as an example of a natural immune-driven viral polymorphism that could compromise antiretroviral-based HIV prevention [118], otherwise known as Pre-exposure prophylaxis (PrEP). Rilpivirine was initially favored as a potential PrEP agent as it was available in a long-acting form [119]. However, analysis of linked viral/HLA genotypes from nearly 8000 antiretroviral-naive individuals confirmed that RT-E138G/A/K prevalence varied markedly across the globe, with frequencies exceeding 10% in key epidemic regions (including Sub-Saharan Africa and Eastern Europe) where HLA-B*18 carriage was common [118]. These results underscored the potential for HIV adaptation to HLA to compromise PrEP efficacy in the very regions that need it most, leading researchers to recommend regional HIV polymorphism surveillance prior to PrEP rollout and call for enhanced collaboration across the immune and drug resistance fields.
Can the study of HIV adaptation to HLA inform cure strategies?
HIV cure will require the elimination of the latent HIV reservoir, a pool of long-lived, predominantly CD4+ T-cells that persistently harbor integrated proviral DNA despite suppressive antiretroviral therapy [120,121]. Seeding of HIV sequences into the reservoir begins shortly after infection and continues as long as viral replication remains uncontrolled [122–124]. And, these proviruses can persist for years thereafter [125,126], either in the original latent cell or clonal descendants thereof [127]. As such, the within-host latent HIV reservoir is genetically diverse [128–136] and contains immune escape mutations [137,138], features that represent barriers to cure. While vaccines being pursued for HIV prophylaxis may also be appropriate as therapeutic vaccines to achieve HIV reservoir reduction or elimination (and some are being evaluated in this context [139]), the design of “personalized” immunogens, for example by identifying epitopes within the reservoir that remain immunologically susceptible to autologous HLA-restricted CTL, are being considered [140,141]. The comprehensive elucidation of HLA-susceptible and adapted forms across human populations and HIV subtypes makes such “personalized” approaches increasingly feasible.
Conclusion
Recent international collaborative efforts have advanced our understanding of the clinical and immunologic consequences of HLA-driven HIV adaptation at the individual and population levels. And, because HIV adaptation continues to represent a major challenge to HIV prevention and eradication, its study continues to be relevant. Studies of HIV adaptation to HLA are revealing why certain HLA alleles are protective in some human populations and not others; they are identifying regions which are immunogenic, yet harbor high mutational barriers to escape in certain HIV subtypes, and they can help illuminate novel subdominant HIV epitopes in regions where circulating adaptation is extensive. Furthermore, the comprehensive elucidation of HLA-specific adapted and nonadapted HIV forms across a growing number of host populations and HIV subtypes renders the design of personalized vaccine immunogens conceptually feasible, and we predict that such strategies will soon be pursued as a component of HIV cure strategies.
Key Points.
Mutational pathways of HIV adaptation to HLA are increasingly being elucidated in different human populations and HIV subtypes, yielding information that can inform HIV prevention and cure strategies
Population-level HIV adaptation to HLA is indeed occurring, but at rates that differ markedly between epidemics and regions globally
Transmission of HIV “pre-adapted” to host HLA has significant clinical and immunologic consequences, but not to the point where these will fully undermine HIV vaccine development efforts
Enhanced and comprehensive understanding of HLA-associated escape pathways render “personalized” immunogen selection approaches to achieve HIV reservoir elimination conceptually feasible.
Acknowledgements
We thank the participants of HIV research studies worldwide.
Financial support and sponsorship
This work was supported in part by the Canadian Institutes of Health Research (through project grant PJT-148621 to SAR, MJ, SM and ZLB and project grant PJT-159625) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number UM1AI126617, with co-funding support from the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke (to SAR and ZLB) and by award P30 AI110527 (to SAM). ZLB is supported by a Scholar Award from the Michael Smith Foundation for Health Research (MSFHR).
This work was supported in part by funds from the National Institutes of Health (NIH) and other funding organizations, as detailed in the “Financial support and sponsorship” section at the end of the manuscript.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.Koup RA, Safrit JT, Cao Y, et al. : Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol 1994, 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow P, Lewicki H, Hahn BH, et al. : Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol 1994, 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, et al. : The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 2009, 206:1253–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington M, O’Brien SJ: The influence of HLA genotype on AIDS. Annu Rev Med 2003, 54:535–551. [DOI] [PubMed] [Google Scholar]
- 5.Phillips RE, Rowland-Jones S, Nixon DF, et al. : Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 1991, 354:453–459. [DOI] [PubMed] [Google Scholar]
- 6.Borrow P, Lewicki H, Wei X, et al. : Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med 1997, 3:205–211. [DOI] [PubMed] [Google Scholar]
- 7.Price DA, Goulder PJ, Klenerman P, et al. : Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci U S A 1997, 94:1890–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leslie AJ, Pfafferott KJ, Chetty P, et al. : HIV evolution: CTL escape mutation and reversion after transmission. Nat Med 2004, 10:282–289. [DOI] [PubMed] [Google Scholar]
- 9.Henn MR, Boutwell CL, Charlebois P, et al. : Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 2012, 8:e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu MK, Hawkins N, Ritchie AJ, et al. : Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest 2013, 123:380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbeck JT, Rolland M, Liu Y, et al. : Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol 2011, 85:7523–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulder PJ, Phillips RE, Colbert RA, et al. : Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med 1997, 3:212–217. [DOI] [PubMed] [Google Scholar]
- 13.Brumme ZL, Brumme CJ, Carlson J, et al. : Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol 2008, 82:9216–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Draenert R, Le Gall S, Pfafferott KJ, et al. : Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med 2004, 199:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iglesias MC, Almeida JR, Fastenackels S, et al. : Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood 2011, 118:2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogunshola F, Anmole G, Miller RL, et al. : Dual HLA B*42 and B*81-reactive T cell receptors recognize more diverse HIV-1 Gag escape variants. Nat Commun 2018, 9:5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloverpris HN, Leslie A, Goulder P: Role of HLA Adaptation in HIV Evolution. Front Immunol 2015, 6:665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson JM, Le AQ, Shahid A, Brumme ZL: HIV-1 adaptation to HLA: a window into virus-host immune interactions. Trends Microbiol 2015. [DOI] [PubMed] [Google Scholar]
- 19.Moore CB, John M, James IR, et al. : Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2002, 296:1439–1443. [DOI] [PubMed] [Google Scholar]
- 20.Almeida CA, Roberts SG, Laird R, et al. : Exploiting knowledge of immune selection in HIV-1 to detect HIV-specific CD8 T-cell responses. Vaccine 2010, 28:6052–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keane NM, Roberts SG, Almeida CA, et al. : High-avidity, high-IFNgamma-producing CD8 T-cell responses following immune selection during HIV-1 infection. Immunol Cell Biol 2012, 90:224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mailliard RB, Smith KN, Fecek RJ, et al. : Selective induction of CTL helper rather than killer activity by natural epitope variants promotes dendritic cell-mediated HIV-1 dissemination. J Immunol 2013, 191:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya T, Daniels M, Heckerman D, et al. : Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 2007, 315:1583–1586. [DOI] [PubMed] [Google Scholar]
- 24.Carlson J, Kadie C, Mallal S, Heckerman D: Leveraging hierarchical population structure in discrete association studies. PLoS ONE 2007, 2:e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brumme ZL, Brumme CJ, Heckerman D, et al. : Evidence of Differential HLA Class I-Mediated Viral Evolution in Functional and Accessory/Regulatory Genes of HIV-1. PLoS Pathog 2007, 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson JM, Brumme ZL, Rousseau CM, et al. : Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol 2008, 4:e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poon AF, Swenson LC, Dong WW, et al. : Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol Biol Evol 2010, 27:819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.*.Kinloch NN, Lee GQ, Carlson JM, et al. : Genotypic and Mechanistic Characterization of Subtype-Specific HIV Adaptation to Host Cellular Immunity. J Virol 2019, 93.First comprehensive documentation of the marked effects of the HIV genetic backbone on modulating viral adaptation to HLA
- 29.Carlson JM, Brumme CJ, Martin E, et al. : Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol 2012, 86:13202–13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chikata T, Carlson JM, Tamura Y, et al. : Host-specific adaptation of HIV-1 subtype B in the Japanese population. J Virol 2014, 88:4764–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.*.Soto-Nava M, Avila-Rios S, Valenzuela-Ponce H, et al. : Weaker HLA Footprints on HIV in the Unique and Highly Genetically Admixed Host Population of Mexico. J Virol 2018, 92.Highlights region and host population-specific nature of HIV adaptation to HLA: over 60% of HLA-associated polymorphisms in Mexico differ from those in Canada/USA, despite HIV subtype B dominating in North America
- 32.Prince JL, Claiborne DT, Carlson JM, et al. : Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog 2012, 8:e1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson JM, Schaefer M, Monaco DC, et al. : Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 2014, 345:1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gesprasert G, Wichukchinda N, Mori M, et al. : HLA-associated immune pressure on Gag protein in CRF01_AE-infected individuals and its association with plasma viral load. PLoS ONE 2010, 5:e11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Tran G, Chikata T, Carlson JM, et al. : A strong association of human leukocyte antigen-associated Pol and Gag mutations with clinical parameters in HIV-1 subtype A/E infection. AIDS 2016, 30:681–689. [DOI] [PubMed] [Google Scholar]
- 36.Sun X, Shi Y, Akahoshi T, et al. : Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation. Cell Rep 2016, 15:2279–2291. [DOI] [PubMed] [Google Scholar]
- 37.Katoh J, Kawana-Tachikawa A, Shimizu A, et al. : Rapid HIV-1 Disease Progression in Individuals Infected with a Virus Adapted to Its Host Population. PLoS One 2016, 11:e0150397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdmann N, Du VY, Carlson J, et al. : HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses. PLoS Pathog 2015, 11:e1005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brumme ZL, John M, Carlson JM, et al. : HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS ONE 2009, 4:e6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseau CM, Daniels MG, Carlson JM, et al. : HLA Class-I Driven Evolution of Human Immunodeficiency Virus Type 1 Subtype C Proteome: Immune Escape and Viral Load. J Virol 2008, 82:6434–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avila-Rios S, Ormsby CE, Carlson JM, et al. : Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology 2009, 6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rolland M, Carlson JM, Manocheewa S, et al. : Amino-acid co-variation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS ONE 2010, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiepiela P, Leslie AJ, Honeyborne I, et al. : Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004, 432:769–775. [DOI] [PubMed] [Google Scholar]
- 44.Naruto T, Gatanaga H, Nelson G, et al. : HLA class I-mediated control of HIV-1 in the Japanese population, in which the protective HLA-B*57 and HLA-B*27 alleles are absent. J Virol 2012, 86:10870–10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chikata T, Tran GV, Murakoshi H, et al. : HLA Class I-Mediated HIV-1 Control in Vietnamese Infected with HIV-1 Subtype A/E. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valenzuela-Ponce H, Alva-Hernandez S, Garrido-Rodriguez D, et al. : Novel HLA class I associations with HIV-1 control in a unique genetically admixed population. Sci Rep 2018, 8:6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiepiela P, Ngumbela K, Thobakgale C, et al. : CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 2007, 13:46–53. [DOI] [PubMed] [Google Scholar]
- 48.Edwards BH, Bansal A, Sabbaj S, et al. : Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol 2002, 76:2298–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuniga R, Lucchetti A, Galvan P, et al. : Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol 2006, 80:3122–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.*.Murakoshi H, Zou C, Kuse N, et al. : CD8(+) T cells specific for conserved, cross-reactive Gag epitopes with strong ability to suppress HIV-1 replication. Retrovirology 2018, 15:46.Demonstrated that effective vaccine-elicited CTL responses can be elicited, even in populations where HIV adaptation to HLA is extensive
- 51.Mothe B, Llano A, Ibarrondo J, et al. : CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PLoS ONE 2012, 7:e29717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feeney ME, Tang Y, Roosevelt KA, et al. : Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J Virol 2004, 78:8927–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Streeck H, Li B, Poon AF, et al. : Immune-driven recombination and loss of control after HIV superinfection. J Exp Med 2008, 205:1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kemal KS, Beattie T, Dong T, et al. : Transition from long-term nonprogression to HIV-1 disease associated with escape from cellular immune control. J Acquir Immune Defic Syndr 2008, 48:119–126. [DOI] [PubMed] [Google Scholar]
- 55.Karlsson AC, Iversen AK, Chapman JM, et al. : Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS ONE 2007, 2:e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalmau J, Rotger M, Erkizia I, et al. : Highly pathogenic adapted HIV-1 strains limit host immunity and dictate rapid disease progression. AIDS 2014, 28:1261–1272. [DOI] [PubMed] [Google Scholar]
- 57.Radebe M, Gounder K, Mokgoro M, et al. : Broad and persistent Gag-specific CD8+ T-cell responses are associated with viral control but rarely drive viral escape during primary HIV-1 infection. AIDS 2015, 29:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frahm N, Korber BT, Adams CM, et al. : Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol 2004, 78:2187–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen TM, Yu XG, Kalife ET, et al. : De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J Virol 2005, 79:12952–12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ladell K, Hashimoto M, Iglesias MC, et al. : A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 2013, 38:425–436. [DOI] [PubMed] [Google Scholar]
- 61.Han C, Kawana-Tachikawa A, Shimizu A, et al. : Switching and emergence of CTL epitopes in HIV-1 infection. Retrovirology 2014, 11:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brumme ZL, Tao I, Szeto S, et al. : Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS 2008, 22:1277–1286. [DOI] [PubMed] [Google Scholar]
- 63.**.Carlson JM, Du VY, Pfeifer N, et al. : Impact of pre-adapted HIV transmission. Nat Med 2016, 22:606–613.International collaborative effort that led to the development of a nuanced metric to quantify HIV adaptation to HLA and to demonstrated that transmission of HIV “pre-adapted” to host HLA had significant negative clinical consequences.
- 64.Walker BD: HIV controllers: an untapped source of clues to overcoming HIV infection. Res Initiat Treat Action 2007, 12:21–22. [PubMed] [Google Scholar]
- 65.Schneidewind A, Brumme ZL, Brumme CJ, et al. : Transmission and long-term stability of compensated CD8 escape mutations. J Virol 2009, 83:3993–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yue L, Prentice HA, Farmer P, et al. : Cumulative impact of host and viral factors on HIV-1 viral-load control during early infection. J Virol 2013, 87:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crawford H, Lumm W, Leslie A, et al. : Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med 2009, 206:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alizon S, von Wyl V, Stadler T, et al. : Phylogenetic approach reveals that virus genotype largely determines HIV set-point viral load. PLoS Pathog 2010, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.*.Monaco DC, Dilernia DA, Fiore-Gartland A, et al. : Balance between transmitted HLA preadapted and nonassociated polymorphisms is a major determinant of HIV-1 disease progression. J Exp Med 2016, 213:2049–2063.demonstrated that the transmission of fitness-costly immune-driven mutations offsets the negative clinical consequences of transmitted escape
- 70.Kawashima Y, Pfafferott K, Frater J, et al. : Adaptation of HIV-1 to human leukocyte antigen class I. Nature 2009, 458:641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yagita Y, Kuse N, Kuroki K, et al. : Distinct HIV-1 escape patterns selected by cytotoxic T cells with identical epitope specificity. J Virol 2013, 87:2253–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.*.Brumme ZL, Kinloch NN, Sanche S, et al. : Extensive host immune adaptation in a concentrated North American HIV epidemic. AIDS 2018, 32:1927–1938.First description of a North American epidemic featuring elevated levels of HIV adaptation to HLA
- 73.Dyck R, Bohm C, Klomp H: Increased frequency of HLA A2/DR4 and A2/DR8 haplotypes in young saskatchewan aboriginal people with diabetic end-stage renal disease. Am J Nephrol 2003, 23:178–185. [DOI] [PubMed] [Google Scholar]
- 74.Furutsuki T, Hosoya N, Kawana-Tachikawa A, et al. : Frequent transmission of cytotoxic-T-lymphocyte escape mutants of human immunodeficiency virus type 1 in the highly HLA-A24-positive Japanese population. J Virol 2004, 78:8437–8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koga M, Kawana-Tachikawa A, Heckerman D, et al. : Changes in impact of HLA class I allele expression on HIV-1 plasma virus loads at a population level over time. Microbiol Immunol 2010, 54:196–205. [DOI] [PubMed] [Google Scholar]
- 76.Payne R, Muenchhoff M, Mann J, et al. : Impact of HLA-driven HIV adaptation on virulence in populations of high HIV seroprevalence. Proc Natl Acad Sci U S A 2014, 111:E5393–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hunt K, Mondal P, Konrad S, et al. : Identifying factors associated with changes in CD4(+) count in HIV-infected adults in Saskatoon, Saskatchewan. Can J Infect Dis Med Microbiol 2015, 26:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keynan Y, Rueda ZV, Bresler K, et al. : HLA B51 is associated with faster AIDS progression among newly diagnosed HIV-infected individuals in Manitoba, Canada. Int J Immunogenet 2015, 42:336–340. [DOI] [PubMed] [Google Scholar]
- 79.Cotton LA, Kuang XT, Le AQ, et al. : Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic. PLoS Genet 2014, 10:e1004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kinloch NN, MacMillan DR, Le AQ, et al. : Population-level Immune-mediated Adaptation in HIV-1 Polymerase during the North American Epidemic. J Virol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.*.Herbeck JT, Peebles K, Edlefsen PT, et al. : HIV population-level adaptation can rapidly diminish the impact of a partially effective vaccine. Vaccine 2018, 36:514–520.Epidemic modeling demonstrates that rapid population-level HIV adaptation to HLA can compromise vaccine efficacy
- 82.*.Ondondo B, Murakoshi H, Clutton G, et al. : Novel Conserved-region T-cell Mosaic Vaccine With High Global HIV-1 Coverage Is Recognized by Protective Responses in Untreated Infection. Mol Ther 2016, 24:832–842.Promising CTL-based HIV vaccine strategy that incorporates bivalent conserved mosaic antigens to maximize global epitope coverage and block common escape paths
- 83.Borthwick N, Lin Z, Akahoshi T, et al. : Novel, in-natural-infection subdominant HIV-1 CD8+ T-cell epitopes revealed in human recipients of conserved-region T-cell vaccines. PLoS One 2017, 12:e0176418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barouch DH, Tomaka FL, Wegmann F, et al. : Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13–19). Lancet 2018, 392:232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hansen SG, Ford JC, Lewis MS, et al. : Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011, 473:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hansen SG, Piatak M Jr., Ventura AB, et al. : Immune clearance of highly pathogenic SIV infection. Nature 2013, 502:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hansen SG, Wu HL, Burwitz BJ, et al. : Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science 2016, 351:714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McMichael AJ, Picker LJ: Unusual antigen presentation offers new insight into HIV vaccine design. Curr Opin Immunol 2017, 46:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen SG, Sacha JB, Hughes CM, et al. : Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013, 340:1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li B, Gladden AD, Altfeld M, et al. : Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J Virol 2007, 81:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duda A, Lee-Turner L, Fox J, et al. : HLA-associated clinical progression correlates with epitope reversion rates in early human immunodeficiency virus infection. J Virol 2009, 83:1228–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Treurnicht FK, Seoighe C, Martin DP, et al. : Adaptive changes in HIV-1 subtype C proteins during early infection are driven by changes in HLA-associated immune pressure. Virology 2010, 396:213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brockman MA, Brumme ZL, Brumme CJ, et al. : Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol 2010, 84:11937–11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wright J, Brumme Z, Carlson J, et al. : Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. Journal of Virology 2010, 84:10820–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wright JK, Novitsky V, Brockman MA, et al. : Influence of Gag-protease-mediated replication capacity on disease progression in individuals recently infected with HIV-1 subtype C. J Virol 2011, 85:3996–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chopera DR, Woodman Z, Mlisana K, et al. : Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog 2008, 4:e1000033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goepfert PA, Lumm W, Farmer P, et al. : Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med 2008, 205:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matthews PC, Prendergast A, Leslie A, et al. : Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol 2008, 82:8548–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boutwell CL, Carlson JM, Lin TH, et al. : Frequent and variable cytotoxic-T-lymphocyte escape-associated fitness costs in the human immunodeficiency virus type 1 subtype B Gag proteins. J Virol 2013, 87:3952–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Juarez-Molina CI, Payne R, Soto-Nava M, et al. : Impact of HLA selection pressure on HIV fitness at a population level in Mexico and Barbados. J Virol 2014, 88:10392–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blanquart F, Grabowski MK, Herbeck J, et al. : A transmission-virulence evolutionary trade-off explains attenuation of HIV-1 in Uganda. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pantazis N, Porter K, Costagliola D, et al. : Temporal trends in prognostic markers of HIV-1 virulence and transmissibility: an observational cohort study. Lancet HIV 2014, 1:e119–126. [DOI] [PubMed] [Google Scholar]
- 103.Herbeck JT, Muller V, Maust BS, et al. : Is the virulence of HIV changing? A meta-analysis of trends in prognostic markers of HIV disease progression and transmission. AIDS 2012, 26:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Borthwick N, Ahmed T, Ondondo B, et al. : Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rolland M, Nickle DC, Mullins JI: HIV-1 group M conserved elements vaccine. PLoS Pathog 2007, 3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Letourneau S, Im EJ, Mashishi T, et al. : Design and Pre-Clinical Evaluation of a Universal HIV-1 Vaccine. PLoS ONE 2007, 2:e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fischer W, Perkins S, Theiler J, et al. : Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 2007, 13:100–106. [DOI] [PubMed] [Google Scholar]
- 108.Barouch DH, O’Brien KL, Simmons NL, et al. : Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med 2010, 16:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mothe B, Hu X, Llano A, et al. : A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J Transl Med 2015, 13:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mothe B, Llano A, Ibarrondo J, et al. : Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med 2011, 9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berger CT, Carlson JM, Brumme CJ, et al. : Viral adaptation to immune selection pressure by HLA class I-restricted CTL responses targeting epitopes in HIV frameshift sequences. J Exp Med 2010, 207:61–75, S61–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brockman MA, Chopera DR, Olvera A, et al. : Uncommon pathways of immune escape attenuate HIV-1 integrase replication capacity. J Virol 2012, 86:6913–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shahid A, Olvera A, Anmole G, et al. : Consequences of HLA-B*13-Associated Escape Mutations on HIV-1 Replication and Nef Function. J Virol 2015, 89:11557–11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carlson JM, Listgarten J, Pfeifer N, et al. : Widespread Impact of HLA Restriction on Immune Control and Escape Pathways of HIV-1. J Virol 2012, 86:5230–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Payne RP, Branch S, Kloverpris H, et al. : Differential escape patterns within the dominant HLA-B*57:03-restricted HIV Gag epitope reflect distinct clade-specific functional constraints. J Virol 2014, 88:4668–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brown AJ, Precious HM, Whitcomb JM, et al. : Reduced susceptibility of human immunodeficiency virus type 1 (HIV-1) from patients with primary HIV infection to nonnucleoside reverse transcriptase inhibitors is associated with variation at novel amino acid sites. J Virol 2000, 74:10269–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.*.Gatanaga H, Murakoshi H, Hachiya A, et al. : Naturally selected rilpivirine-resistant HIV-1 variants by host cellular immunity. Clin Infect Dis 2013, 57:1051–1055.demonstrates that natural HLA-driven HIV polymorphisms could, in theory compromise PrEP if proper surveillance is not undertaken
- 118.Gatanaga H, Brumme ZL, Adland E, et al. : Potential for immune-driven viral polymorphisms to compromise antiretroviral-based preexposure prophylaxis for prevention of HIV-1 infection. AIDS 2017, 31:1935–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McGowan I, Dezzutti CS, Siegel A, et al. : Long-acting rilpivirine as potential pre-exposure prophylaxis for HIV-1 prevention (the MWRI-01 study): an open-label, phase 1, compartmental, pharmacokinetic and pharmacodynamic assessment. Lancet HIV 2016, 3:e569–e578. [DOI] [PubMed] [Google Scholar]
- 120.International ASSWGoHIVC, Deeks SG, Autran B, et al. : Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012, 12:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martin AR, Siliciano RF: Progress Toward HIV Eradication: Case Reports, Current Efforts, and the Challenges Associated with Cure. Annu Rev Med 2016, 67:215–228. [DOI] [PubMed] [Google Scholar]
- 122.Ruff CT, Ray SC, Kwon P, et al. : Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol 2002, 76:9481–9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Whitney JB, Hill AL, Sanisetty S, et al. : Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014, 512:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ledford H: HIV rebound dashes hope of ‘Mississippi baby’ cure. Edited by: Nature News; 2014. [Google Scholar]
- 125.Finzi D, Blankson J, Siliciano JD, et al. : Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999, 5:512–517. [DOI] [PubMed] [Google Scholar]
- 126.Siliciano JD, Kajdas J, Finzi D, et al. : Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003, 9:727–728. [DOI] [PubMed] [Google Scholar]
- 127.Mullins JI, Frenkel LM: Clonal Expansion of Human Immunodeficiency Virus-Infected Cells and Human Immunodeficiency Virus Persistence During Antiretroviral Therapy. J Infect Dis 2017, 215:S119–S127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bruner KM, Murray AJ, Pollack RA, et al. : Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016, 22:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee GQ, Orlova-Fink N, Einkauf K, et al. : Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 2017, 127:2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buzon MJ, Sun H, Li C, et al. : HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 2014, 20:139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hiener B, Horsburgh BA, Eden JS, et al. : Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+ T Cells from Effectively Treated Participants. Cell Rep 2017, 21:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Josefsson L, von Stockenstrom S, Faria NR, et al. : The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013, 110:E4987–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Evering TH, Mehandru S, Racz P, et al. : Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog 2012, 8:e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chomont N, El-Far M, Ancuta P, et al. : HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009, 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.von Stockenstrom S, Odevall L, Lee E, et al. : Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy. J Infect Dis 2015, 212:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rothenberger MK, Keele BF, Wietgrefe SW, et al. : Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 2015, 112:E1126–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brumme ZL, Sudderuddin H, Ziemniak C, et al. : Genetic complexity in the replication-competent latent HIV reservoir increases with untreated infection duration in infected youth. AIDS 2019, 33:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deng K, Pertea M, Rongvaux A, et al. : Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 2015, 517:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Leal L, Guardo AC, Moron-Lopez S, et al. : Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS 2018, 32:2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Theiler J, Yoon H, Yusim K, et al. : Epigraph: A Vaccine Design Tool Applied to an HIV Therapeutic Vaccine and a Pan-Filovirus Vaccine. Sci Rep 2016, 6:33987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tumiotto C, Riviere L, Bellecave P, et al. : Sanger and Next-Generation Sequencing data for characterization of CTL epitopes in archived HIV-1 proviral DNA. PLoS One 2017, 12:e0185211. [DOI] [PMC free article] [PubMed] [Google Scholar]