Abstract
Cytoplasmic dynein-1 is an important microtubule-based motor in many eukaryotic cells. Dynein has critical roles both in interphase and during cell division. Here we focus on interphase cargoes of dynein, which include membrane-bound organelles, RNAs, protein complexes and viruses. A central challenge in the field is to understand how a single motor can transport such a diverse array of cargoes and how this process is regulated. The molecular basis by which each cargo is linked to dynein and its cofactor dynactin has started to emerge. Of particular importance for this process is a set of coiled coil proteins — ‘activating adaptors’ — which both recruit dynein–dynactin to their cargoes and activate dynein motility.
The microtubule cytoskeleton is responsible for long distance movements and spatial organization of intracellular vesicles, organelles, and large protein- and RNA-containing complexes in many eukaryotic cells. Microtubules are polarized structures with a minus and a plus end. In interphase cells microtubule plus ends are typically located near the periphery. Minus ends originate from microtubule organizing centres (MTOC), which are often located close to the nucleus, but are also found at other locations in the cell. In mitosis microtubules reorganize to form the spindle, with the microtubule minus ends focused at the two poles. The molecular motors, dyneins (minus-end-directed) and kinesins (primarily plus-end-directed), are responsible for many microtubule-based functions. Notably, plants and some algae lack dynein genes in their genome.
Dyneins were first discovered as the motors that drive flagellar beating in Tetrahymena pyriformis1. Subsequently, two dynein isoforms were found that are responsible for movement in the cytoplasm (cytoplasmic dynein-1) and cilia (cytoplasmic dynein-2)2–5. This review will focus on cytoplasmic dynein-1 (subsequently referred to as dynein). Dynein is an essential gene in a number of organisms, including Dropsophila melanogaster and mice6,7 and mutations in the dynein transport machinery have been linked to neurological diseases. These include neurodegenerative disorders such as the Parkinson’s-like Perry Syndrome, Spinal Muscle Atrophy with Lower Extremity Dominant (SMA-LED), Hereditary Motor Neuron disease, and Charcot-Marie-Tooth. They also include neurodevelopmental diseases such as lissencephaly [G], as well as other malformations of cortical development and intellectual disability8.
Remarkably, a single dynein functions in the cytoplasm in animal cells, in contrast to the ~40 kinesins that perform related functions. This suggests dynein uses a different strategy to interact with its cargoes compared to kinesin. Here we address how a single dynein transports such a diversity of cargoes. We first discuss the structure of the dynein-based transport machinery. We then describe the main cargoes of dynein, which we group under four categories: membranes, RNAs, proteins, and viruses. We have limited this Review to interphase functions of dynein, as its mitotic and meiotic roles have been covered elsewhere9.
The dynein transport machinery
After the discovery of dynein, it became clear that other factors were required for its activity. A large molecular weight complex, later named dynactin, was found to be necessary for vesicle movement along microtubules10,11. However, the two complexes only interacted weakly in vitro12. This interaction was shown to become stronger in the presence of an amino-terminal domain of BICD2 (BICD2-N), which is predominantly coiled coil12. BICD2 is the human homologue of bicaudal-D, which was originally identified as a polarity factor in D. melanogaster13. Recent studies demonstrated that BICD2-N dramatically activates the motility of isolated dynein–dynactin complexes to move long distances in vitro14,15. Additional coiled-coil-containing proteins have also been shown to activate dynein–dynactin motility (Table 1). We refer to these coiled-coil proteins as ‘activating adaptors’. These proteins have the dual property of both activating motility and linking dynein–dynactin to their cargoes. Collectively, these results suggest the ‘dynein transport machine’ consists of 1) the dynein complex, 2) the dynactin complex, and 3) a coiled coil-containing activating adaptor (such as BICD2) (Fig. 1 and Table 1). Below we provide an overview of these three central components, as well as two additional regulators, Lis1 (PAFA1H1) and Nudel (NDE1 and NDEL1; collectively referred to as “Nudel” here), which associate with the dynein complex and are required for many dynein functions. Beyond activating adaptors, other connections between dynein and its cargoes have been shown to be involved in dynein transport (Box 1).
Table 1 |.
Activating adaptors and candidate activating adaptors for dynein–dynactin.
| Activator or candidate activator | Evidence | Cargo |
|---|---|---|
| Confirmed activators (active in in vitro motility assays) | ||
| BICD2 | Reconstituted motility14,15, relocation assay 224 | COP1-independent Golgi-to-ER vesicles58, Golgi vesicles57, nuclear pore complexes133 |
| BICDL1 (BICDR1) | Reconstituted motility 16 | Rab6 vesicles61 |
| SPDL1 (Spindly) | Co-IP motility14 | Kinetochore227 |
| HOOK1 | Lysate motility and relocation assay226 | Rab5 early endosomes70
Clathrin-independent cargoes228 |
| HOOK3 | Reconstituted motility24, Co-IP motility14, lysate motility and relocation assay 226 | Rab5 early endosomes70, Golgi 229 |
| NIN (Ninein) | Reconstituted motility98 | Unknown |
| NINL (Ninein-like) | Reconstituted motility98 | MICAL3 and RAB8A containing vesicles63 |
| RAB11-FIP3 | Co-IP motility14 | Recycling endosomes28 |
| Candidate activators | ||
| BICD1 | Co-IP230 | COP1-independent Golgi-to-ER vesicles58, microtubule arrays230 |
| BICDL2 (BICDR2) | Homology to BICD proteins61 | Rab13 vesicles61 |
| HOOK2 | Homology to Hook proteins | Centrosomal proteins174, spermatid intramanchette trafficking231 |
| CCDC88A (Girdin) | Co-IP98 | Unknown |
| CCDC88B (Gipie) | Co-IP232 | Secretory lysosomes (lytic granules)232 |
| CCDC88C (Daple) | Co-IP98 | Unknown |
| NUMA | Co-IP233 | Minus ends of microtubules in the spindle131,234 |
| TRAK1 | Co-IP111 | Mitochondria111 |
| TRAK2 | Co-IP111 | Mitochondria111 |
| HAP1 | Co-IP75,76,207 | Many membrane cargos75 |
Each activator or candidate activator is listed along with its cargo(s). Cargos are defined by their reliance on dynein or dynactin for movement or localization. We have not included known activator interacting proteins where there is not yet evidence for dynein–dynactin involvement for their trafficking.
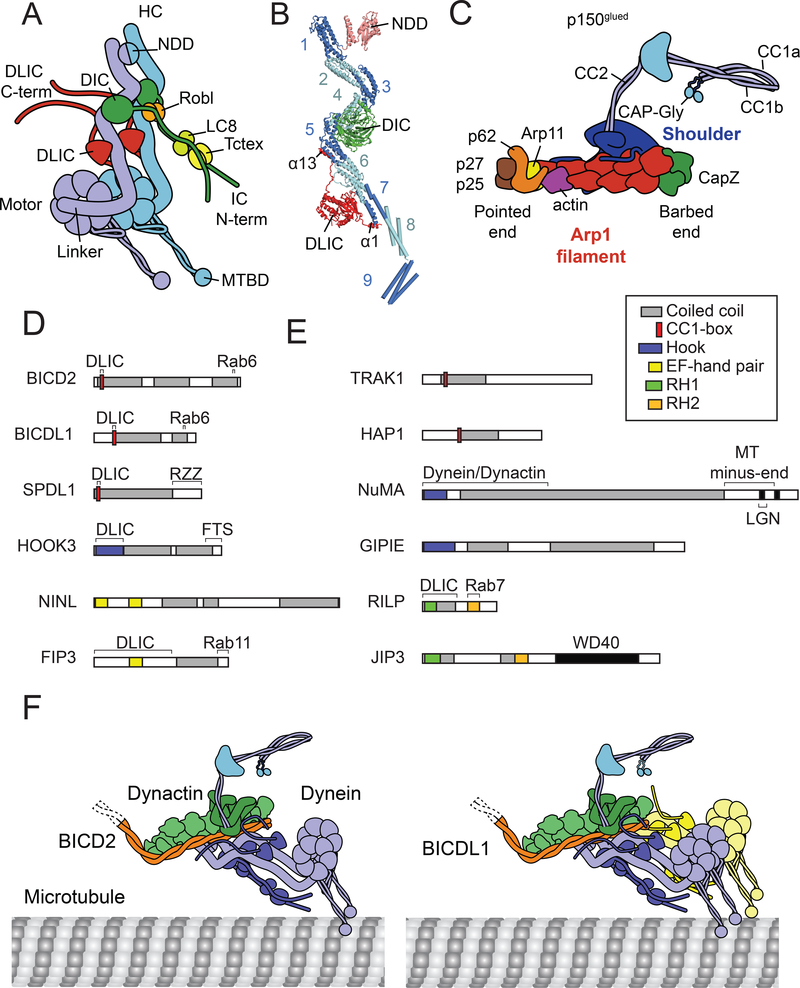
Figure 1 |. The dynein transport machinery.
a | Cartoon of cytoplasmic dynein-1. The two dynein heavy chains (DHCs) are linked together by an amino-terminal dimerization domain (NDD) and have a carboxy-terminal motor domain with a microtubule binding domain (MTDB) at the end of a long anti parallel coiled-coil stalk. The intermediate chains (DIC) have extended amino-termini that bind dimers of the light chains Roadblock (Robl), LC8 and Tctex. The light intermediate chain (DLIC) has an extended carboxy-terminus. b | Structure of the dynein tail16,18 (PDBs 6F1T and 5NVU) showing the amino-terminal dimerization domain (NDD) and nine helical bundles (1–9; neighbouring bundles are shown in different shades of blue to distinguish them) in the DHC and depicting interactions of DHC with DIC and DLIC. The DIC WD40 domain binds bundles 4 and 5, whereas the Ras-like domain of DLIC binds to bundles 5 and 7, using its amino- and carboxy-terminal helices (α1 and α13). The DIC and DLIC extended termini are not shown. The structure of helical bundles 8 and 9 is approximate. c | Dynactin is built around a filament of eight actin related proteins (Arp1). At the barbed end are capping proteins (CapZ). At the pointed end is an actin monomer, another actin related protein (Arp11) and a complex of three proteins (p62, p27 and p25). The shoulder domain binds the filament via extended amino-terminal peptides of p50/dynamitin. The p150 component extends from the shoulder, containing stretches of coiled-coil (CC2, CC1b and CC1a). At the amino-terminus of p150 are the basic and CAP-Gly domains that can interact with microtubules. d | Domain structure of known and candidate activating adaptors (see also Table 1). Reported sites of interactions are indicated above each cartoon. Key shows domains identified in the literature or by InterPro. RH1 and RH2, RILP homology 1 and 2. e | Dynein–dynactin–activator complexes on microtubules. The activating adaptors BICD2 or BICDL1 run along the dynactin filament and recruit the dynein heavy chains. BICD2 preferentially recruits one dynein dimer (left), whereas BICDL1 recruits two dimers (right).
Box 1 |. Other connections between dynein and its cargo.
In addition to the activating adaptors or candidate activating adaptors (Table 1) a number of other links between dynein–dynactin and its cargoes have been reported. Here we present some prominent examples.
RILP is required for dynein–dynactin recruitment to Rab7 lysosomes34. Because RILP interacts with the HOPS complex it may be involved in linking dynein–dynactin to Rab7 (Fig. 3c). Purified RILP co-precipitates with purified dynein DLIC1, suggesting a direct interaction with the complex23,35.
Huntingtin (Htt) and Huntingtin-associated-protein1 (HAP1) associate with membrane vesicles75 and are transported in neurons along microtubules206. Htt is linked to signalling endosomes74 and autophagosomes105. Htt and HAP1 knockdown decreases retrograde movement of autophagosomes105. Htt binds purified dynein75, suggesting a direct interaction that does not require dynactin. HAP1 co-immunoprecipitates with p150 from brain lysate76,207. HAP1 contains a coiledcoil and has sequence similarity to TRAK and BICDL160 (Fig. 1d, Table 1), raising the possibility that it is a BICD-like activating adaptor.
c-Jun N-terminal kinase (JNK) interacting proteins (JIPs) are implicated in the motility of dynein–dynactin cargoes. JIP1 depletion inhibits retrograde transport of amyloid precursor protein208 and autophagosomes106 in neurons. JIP3 co-immunoprecipitates with dynactin and colocalizes with dynein and dynactin on vesicles in neurons33. JIP1 lacks coiled-coils, whereas JIP3 contains two short stretches of coiled coil (Fig. 1d). JIP4 is closely related to JIP3 and shares the same domain architecture.
Ankyrin B (AnkB) knockout mice show reduction in the speed of fast axonal transport of early endosomes, lysosomes, mitochondria and synaptic vesicles. AnkB binds the lipid PI(3)P on membrane cargoes and directly contacts the pointed-end complex of dynactin85.
Sorting nexins (SNX) are a large family of membrane-associated proteins that contain a lipid binding PX domain. Affinity tagged SNX5 and SNX6 co-precipitate with dynein and/or dynactin from human cell lysates209,210.
Spectrin is a peripheral membrane protein that co-immunoprecipitates with dynactin87. While a two-hybrid screen reported an interaction between spectrin and Arp1211, the structure of the dynein– dynactin–BICD2 complex shows little fully exposed Arp1, which would be available for binding17.
Snapin, a SNARE [G] interacting protein that is part of the BLOC-1 complex [G]212, has been implicated in the retrograde movement of late endosomes and suggested to be a dynein adaptor86. Snapin is required for dynein to localize to late endosomes and GST-snapin co-precipitates with dynein and dynactin components from brain lysate86. However, because these in vitro experiments were done in the absence of other BLOC-1 complex members, they should be revisited.
Direct cargo binding to dynein. In addition to its interaction with activating adaptors, the DLIC can form direct interactions with cargoes, such as the adenoviral hexon protein35, pericentrin213 and Par3214. The dynein light chains LC8 and Tctex bind motifs found in many proteins, leading to the idea that DLCs directly recruit proteins to dynein. However, because these motif binding sites on the DLCs bind to the DIC amino-terminus, the current consensus is that this is not the case215. Many of the DLC binding motifs likely recruit LC8 or Tctex for dynein-independent functions, such as dimerization. However, there is some evidence for cargoes connecting to dynein via DLCs, including some viruses181, rhodopsin216,217, and the Rho GEF Lfc (ARHGEF2)218. Additional structural studies will be required to determine if peptides from these proteins can bind to DLCs that are already in complex with the DIC amino-terminal peptides; NMR mapping experiments suggest that this may be possible218,219.
Structure of dynein
Human dynein is a 1.4 MDa complex composed of six different polypeptides all of which are present in two copies (Fig. 1a). There is a single dynein heavy chain gene (DHC; DYNC1H1) and two isoforms of each of the other components: the intermediate chains (DIC; DYNC1IC1 and 2), the light intermediate chains (DLIC; DYNC1LI1 and 2) and the three light chain families (DLCs): Robl (DYNLRB1 and 2), LC8 (DYNLL1 and 2), and Tctex (DYNLT1 and 3).
The DHC is 4634 amino acids long, containing a carboxy-terminal motor domain and an amino-terminal tail domain. Starting from the amino-terminus of the DHC, the ‘tail’ contains a dimerization domain (residues 1–200), followed by an extended region made up of nine helical bundles (residues 201–1420)16–18 (Fig. 1b). The last helical bundle of the tail joins into the motor domain, which consists of the linker (itself also made of helical bundles), a ring of six AAA+ domains [G], and a carboxy-terminal domain. The motor binds to its track via a microtubule binding domain (MTBD) at the end of a coiled-coil stalk that emerges from the AAA+ ring19,20. Dynein moves along microtubules by coupling ATP induced conformational changes in the AAA+ ring with bending and straightening of the linker21.
The DIC contains a WD40 domain [G] that binds the DHC helical bundles 4 and 5, whereas the DLIC has a Ras-like domain [G] that contacts DHC helical bundles 6 and 716,18. The DLIC is further anchored onto the DHC by amino- and carboxy-terminal helices (α1 and α13, respectively) that span out from the Ras-like domain and contact DHC helical bundles 8 and 5, respectively (Fig. 1b). The DICs have extended ~230 residue amino-termini that contact dimers of DLCs. One of these DLCs, Robl, binds both this amino-terminus and docks onto the WD40 domain of one of the DICs. The two DLICs have extended ~130 residue carboxy-termini (Fig. 1a), which contains two alpha helices (α14 and α15)22. The DLIC carboxy-termini contact activating adaptors and in some cases may also provide a direct link to cargo23,24.
Structure of dynactin
The 1.1 MDa dynactin complex is composed of 23 subunits (11 different polypeptides; Fig. 1c)25. Its central feature is a short actin-like filament, which contains 8 copies of the actin related protein (Arp) 1 (ACTR1A and B), and 1 copy of ß actin (ACTB). The filament is capped at the barbed end [G] by the actin capping protein CapZ (CAPZA1 or CAPZA2, and CAPZB) and at the pointed end [G] by another actin related protein, Arp11 (ACTR10). Three other proteins p62 (DCTN4), p27 (DCTN6) and p25 (DCTN5) bind Arp11 to form a pointed end complex. A ‘shoulder’ domain sits on the filament near the barbed end. It is composed of 2 copies of p150glued (“p150”; DCTN1), four copies of p50/dynamitin (“p50”; DCTN2), and 2 copies of p24 (DCTN3). Extended peptides corresponding to the amino-terminus of p50 emerge from the shoulder and wrap around one side of the filament. The carboxy-terminus of p150 is buried in the shoulder and the amino-terminus forms a flexible extension with two stretches of coiled-coil (CC1 and CC2) interrupted by a globular domain. CC1 contains two halves, CC1a and CC1b, which form a hairpin structure. At the extreme amino-terminus of p150 are the CAP-Gly and basic domains, which have been implicated in microtubule binding26.
Activating adaptors
At the time of writing, eight activating adaptors have been shown to promote long distance movement of dynein–dynactin complexes in vitro (Table 1, Fig. 1d). There are no sequence motifs that are common to all of these activating adaptors. Instead the common features are 1) the presence of a long (> 200 residues) coiled coil, 2) a binding site for the DLIC carboxyterminus23,24,27, and 3) a binding site for proteins (e.g. Rab6, Rab11, RZZ, FTS) that link the adaptors to their cargoes (Fig. 1d).
Activating adaptors contain at least three different types of binding sites for the DLIC carboxy terminus. BICD2, the BICD-related protein BICDL1 (also called BICDR1), and the kinetochore binding adaptor SPDL1 (Spindly) likely bind the DLIC via a motif referred to a “CC1-box” in their coiled coil27. The CC1-box contains an AAxxG sequence (where x denotes any amino acid). In contrast, HOOK3 and HOOK1 use a small “Hook domain”, which is located amino-terminal to their coiled-coils24. The penultimate helix (α14) in the DLIC carboxy-terminus is the contact site for BICD2, SPDL1 and HOOK322. The activating adaptor RAB11FIP3 (Rab11 Family-interacting protein 3) also binds the DLIC carboxy-terminus. The exact site of interaction is not yet known, but the region of RAB11FIP3 (residues 2–43528) that interacts with DLIC contains a pair of EF-hands [G]. Interestingly the same type of domain is also found in the activating adaptors Ninein (NIN) and Ninein-like (NINL), although these proteins have not yet been shown to bind the DLIC. All of the activating adaptors are known or predicted to be dimers.
Cryo-EM structures have been solved for dynein and dynactin in complex with three different activating adaptors (BICD2, BICDL1 and HOOK3)16,17. These cryo-EM maps are at medium (BICDL1) to low (BICD2, HOOK3) resolution in the regions around the activating adaptors, but are sufficient to reveal the main ways in which the activating adaptors bring dynein and dynactin together. In all cases an ~250 residue coiled-coil of the activating adaptor runs along the length of the dynactin filament (Fig. 1e). Their amino termini lie close to the barbed end of the dynactin filament and their carboxy-termini contact the pointed end complex. The coiled-coils of the activating adaptors interact with dynactin slightly differently, especially toward the pointed end. This is consistent with the lack of conserved motifs in the coiled-coils of different activating adaptors. Interestingly, BICD2, HOOK3 and BICDL1 can all recruit two dynein dimers at a time16,29, although for BICD2 the recruitment of a second dynein dimer appears to be less efficient16. The second dynein lies next to the first along the dynactin filament (Fig. 1e). These interactions mediated by activating adaptors recruit dynein to dynactin so that the individual DHCs bind in grooves between dynactin filament subunits16,17. Importantly, these interactions induce large conformational changes in dynein that align its motor domains so that both microtubule binding domains can bind microtubules18. This likely underlies how activating adaptors increase the ability of dynein to move over long distances. The ability to recruit two dyneins to one dynactin further enhances this effect16,29.
Dynein and dynactin also interact via the amino-terminus of the DIC contacting CC1 of p15030. This interaction may reinforce the interactions described above. Alternatively, a recent report suggested that this DIC–p150 interaction inhibits dynein31, raising the possibility that binding of an activating adaptor could disrupt this interaction.
The activation of dynein includes a number of steps in addition to binding activating adaptors. Dynein in isolation can exist in an inhibited form, referred to as the “phi particle”32, which not only binds weakly to microtubules, but is also unable to bind dynactin and activating adaptors18. The mechanism by which the phi-particle opens up is not yet known.
In addition to the known activating adaptors there are a number of possible candidate activating adaptors (Table 1, Fig. 1d). These proteins contain long coiled-coils and co-immunoprecipiate with both dynein and dynactin. Sequence analysis suggests many of these putative activating adaptors have domains that bind the DLIC, although as of yet this has not been directly tested. TRAK1 (trafficking kinesin binding protein 1), TRAK2 and HAP1 (Huntington interacting protein 1) contain the CC1-box motif27, whereas NuMA (Nuclear mitotic apparatus protein), CCDC88A/Girdin, CCDC88B/Gipie, and CCDC88C/Daple all contain Hook domains (Fig. 1d).
An interesting question is whether RILP (Rab interacting lysosomal protein) and JIP3 (CJun-amino-terminal kinase-interacting protein3) are activating adaptors. Like known activating adaptors, both proteins can interact with dynein and dynactin33,34, with RILP directly binding to the DLIC23,35. While RILP and JIP3 both have regions of coiled-coil, they are not long enough to bind to dynein and dynactin in the same way as other activating adaptors (Fig. 1d). They may therefore fall into a category of non-activating adaptors (Box 1) that serve as links between the dynein complex and cargo without the activation function.
Dynein regulation by Lis1 and Nudel
Two other central regulators of dynein are Lis1 and Nudel. They have been linked to dynein function genetically in many organisms (reviewed recently in36). Lis1, a dimer of two β-propellers [G] binds directly to dynein’s motor domain at two distinct sites37,38. Nudel proteins are coiled coil-containing proteins that interact with the DIC and LC8, as well as Lis139–41. There is evidence that Nudel may tether Lis1 to dynein38,40. The molecular mechanism of Lis1mediated regulation appears to be complex as in vitro experiments have revealed a range of Lis1 functions including decreasing38,42,43 and increasing velocity of the dynein motor37,44,45. Based on experiments using Saccharomyces cerevisiae dynein, the effects Lis1 exerts on dynein depend on the nucleotide state at dynein’s 3rd AAA+ domain37. The function of Nudel also appears to be complex as it can both enhance38,46 and oppose Lis1 function in vitro42,43. Lis1 — likely in complex with Nudel proteins — has been implicated in multiple cellular processes, including localizing dynein to microtubule plus ends, initiating cargo transport, and supporting the ability of dynein to transport high-load cargoes (reviewed recently in36).
Dynein cargoes
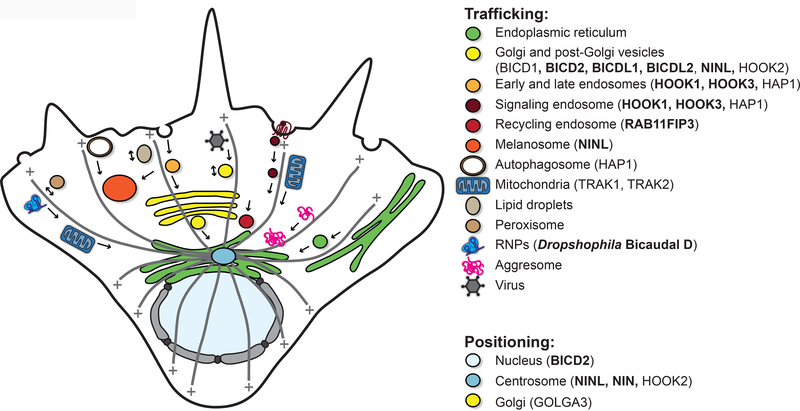
The remainder of this Review focuses on the wide range of cargoes that dynein transports (Fig. 2). Studies with dextrans suggested that, while 500kDa molecules can diffuse freely across a cell, complexes larger than 2MDa are confined and effectively immotile47. Thus, cargoes for dynein are typically large objects, such as organelles and ribonucleoprotein (RNP) or protein complexes. Dynein can also act while it is anchored at the cell cortex [G], where it can act as a tether and/or generate pulling forces. Although many of these cargoes move bi-directionally owing to the interplay between dynein and kinesin, here we focus on dynein-based motility. For each cargo we describe its physiological role and discuss the evidence for the involvement of dynein in its dynamics. We also describe the current state of knowledge for how each cargo is linked to dynein–dynactin, highlighting the role of known or candidate activating adaptors (Table 1, Fig. 1d and Fig. 2). Our focus will be on vertebrates, although results from other organisms, such as flies and filamentous fungi, are also discussed.
Figure 2 |. Many cargoes of dynein and their activating adaptors.
Many of the dynein cargoes discussed in this Review. Some cargoes are trafficked along microtubules, while in other cases the role of dynein is to position them. Known activating adaptors are marked with a star. Candidate activating adaptors are also listed.
Membrane cargoes
There are many discrete membrane-bound compartments in eukaryotic cells. Some of these are marked by small GTPases of the Rab family. Rabs bind effector proteins to direct trafficking processes including membrane tethering, fusion, and movement mediated by molecular motors48. Disrupting dynein function (Box 2) alters the cellular localization or motile properties of many of these membrane compartments.
Box 2 |. Methods to study dynein function and activation.
Components of the dynein machinery have been depleted using RNA interference and CRISPR methods and genetic studies have revealed the functions of many components. Several other widely used methods have been important for determining if dynein or dynactin are involved in a process of interest. Over-expression of dynactin components, such a dynamitin/p50220 (Fig. 1c) or one of the coiled coil segments (CC1) of p150221 (Fig. 1c) result in the disruption of many dynein–dynactin-dependent processes. The exact mechanism of disruption is not completely clear, but probably involves disruption of the dynein–dynactin interaction222. Antibodies raised against the DIC (Fig. 1a) have also been extensively used to block dynein function in cell-free systems or systems amenable to antibody injection223.
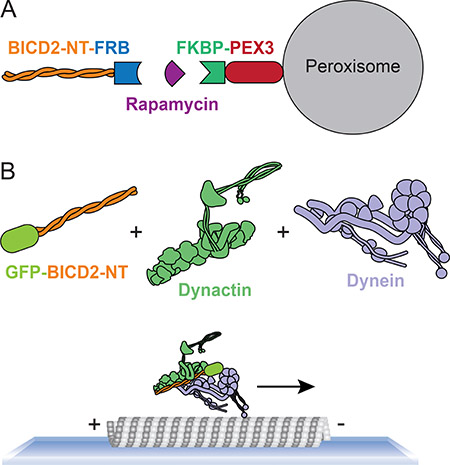
A number of methods have been used to provide evidence that a dynein adaptor is an activating adaptor. All activating adaptors co-immuoprecipitate with dynein and dynactin, providing an initial suggestion that a candidate adaptor could be an activating adaptor98. Cell-based “relocation assays” have also been used to provide evidence for dynein–dynactin activation224,225. In these experiments, a candidate activator is targeted to a largely non-motile organelle, such as the peroxisome. Any increase in minus-end-directed peroxisome motility provides indirect evidence that a candidate adaptor could be an activating adaptor. For example, the amino-terminus of BICD2 can be fused to the rapamycin-binding domain FRB. FKBP, which also binds rapamycin, is fused to an organelle targeting protein, such as the peroxisomal protein PEX3. In the presence of rapamycin BICD2 enhances peroxisome motiltiy225 (see figure part a).
In vitro motility experiments can also provide indirect evidence for activating adaptors. For example, candidate activators can be used to immunoprecipitate dynein and dynactin from cell lysates or tissue extracts and then the motile properties of the immunoprecipitated complex can be assessed using single-molecule motility assays14,98. A variation of this method is to visualize dynein or dynactin directly in lysates without prior immunoprecipitation226.
The gold standard for determining if a candidate adaptor is a dynein–dynactin activator is to reconstitute motility from purified components, as has been done for BICD2, BICDL1, HOOK3, NIN and NINL14–16,24,98. In this approach each protein or protein complex is purified separately and at least one component is tagged with a fluorescent marker. Motility is then monitored using total internal reflection microscopy [G]. Bond fide activators lead to processive dynein–dynactin motility (see figure part b).
Endoplasmic reticulum.
The endoplasmic reticulum (ER) is a meshwork of membranes consisting of tubules and sheets. Tubules are particularly dynamic and their movements require dynein as they are inhibited by overexpression of the dynactin component p50, which is a classic method for disrupting dynein–dynactin function (Box 2)49. Similarly, overexpression of p50 showed that dynein–dynactin is responsible for transporting ER membranes in neuronal dendrites50.
Dynein also moves vesicles originating from the ER, which is spread throughout the cell, to the cell centre. These vesicles coalesce to form the ERGIC (Endoplasmic Reticulum Golgi Intermediate Compartment), a precursor compartment of the Golgi. Evidence for this comes from visualization of a secreted viral protein (VSVG) that exits the ER and moves rapidly to the centre of the cell. This movement is disrupted by overexpression of p5051 and dynein colocalizes with markers of the ERGIC compartment52.
It is not yet clear how dynein associates with either the ER or the ERGIC. These interactions could either be direct, or mediated by attaching to another vesicle that then interacts with dynein. The latter process, called “hitchhiking”, has been observed in filamentous fungi53. In Ustilago maydis ER vesicles comigrate with early endosomes54, a cargo that requires dynein for movement in this organism55. It remains to be seen if ER hitchhiking is conserved in mammalian cells.
Golgi.
The Golgi apparatus is made up of stacks of membranes typically positioned near the nucleus. However in some organisms and cell types, the Golgi stacks are dispersed throughout the cytoplasm. In Golgi stacks, secreted proteins move from the nuclear proximal cis-Golgi compartment through the stack to the trans-Golgi network (TGN). From the TGN components are transported in Golgi-derived vesicles to the plasma membrane or to other organelles such as endosomes and lysosomes.
Dynein plays a key role in Golgi positioning. The Golgi is dispersed by deletion of dynein in mouse cells7, injection of anti DIC antibodies, or over-expression of p5056 (Box 2). The BICD adaptors (BICD1 and BICD2), which are involved in Golgi vesicle movement (see below), are not involved in this process, as over-expression of a carboxy-terminal fragment of BICD2 displaces endogenous BICD2 but has no effect on Golgi morphology57,58. A candidate adaptor for dynein-based Golgi positioning is GOLGA3 (Golgin 160), as its knockdown leads to Golgi dispersal and it co-immunoprecipitates with both dynein and dynactin. When GOLGA3 is ectopically recruited to non-Golgi membranes, it drives their movement towards the centre of the cell (“relocation assay”, Box 2)59, which would be consistent with it acting as an activating adaptor for dynein–dynactin. While GOLGA3 is rich in coiled-coils, it lacks any obvious DLIC interacting motif and the region of the protein used in the relocation assay is too short to activate in a BICD2-like manner. Therefore, it is an open question whether GOLGA3 is a true activating adaptor.
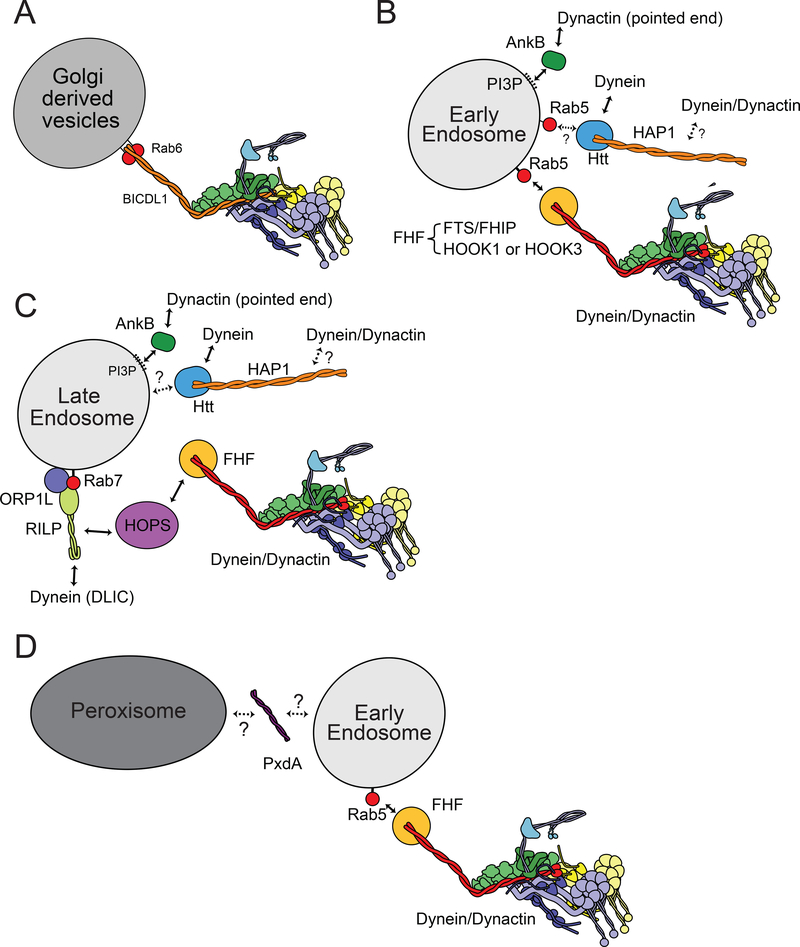
Dynein has been directly linked to the movement of Golgi-derived vesicles via activating adaptors of the BICD family. In addition to bridging the dynein–dynactin interaction, BICD2 binds the Rab6 GTPase and localizes to cytoplasmic vesicles and the TGN57,58 (Fig. 3a). Rab6-marked vesicles have been implicated in intra-Golgi, endosome to Golgi, Golgi to ER and Golgiderived exocytic vesicle trafficking48. Over-expression of a carboxy-terminal fragment of BICD2 displaces endogenous BICD2 from Rab6 vesicles and causes them to accumulate at the periphery of the cell57,58. This is presumably due to kinesin-driven transport dominating when dynein– dynactin is removed from the Rab6 vesicles60.
Figure 3 |. Mechanisms linking dynein and dynactin to membrane cargoes.
a | Dynein– dynactin associates with Golgi-derived vesicles using the activating adaptors BICD2 or BICDL1 depending on the cell type. Both BICD2 and BICDL1 bind, via their carboxy-terminal coiled-coils, to a dimer of the small GTPase Rab6. Rab6 binds to Golgi-derived membranes via its prenyl tails, as do other Rabs48. b | Early endosomes recruit dynein–dynactin via the activating adaptors HOOK1 or HOOK3. These adaptors bind the FTS and FHIP proteins to form the FHF complex. FHIP is reported to bind directly to the early endosome marker Rab5. Rab5 also binds Htt, which is linked to HAP1, another potential activating adaptor. The PI3P binding protein ANK-B binds the pointed end of dynactin and is also important for early endosome transport. How these and other dynein–dynactin adaptors work together is unknown. c | Late endosomes are marked with Rab7, which binds RILP and the cholesterol sensor ORP1L. RILP binds the DLIC. RILP also binds the HOPS complex, which interacts with the FHF complex, raising the possibility that HOOK proteins also link dynein–dynactin to late endosomes. As with early endosomes, other potential dynein–dynactin links have been reported for the movement of late endosomes. d | In filamentous fungi some cargos (peroxisomes, ER, lipid droplets, and RNPs) associate with the dynein transport machinery indirectly by hitchhiking on early endosomes, which can directly recruit the transport machinery via the Hook-containing FHF complex (see part b). PxdA is a putative tether that links peroxisomes to early endosomes although how it interacts with both peroxisomes and early endosomes is unknown.
BICDL1 is another Golgi-associated activating adaptor, which also binds Rab6 and is predominantly associated with exocytic Golgi-derived vesicles61. Unlike BICD1 and BICD2, BICDL1 is expressed predominantly early in embryonic development in neural and kidney tissue and regulates neurite outgrowth61. A related isoform, BICDL2, binds to Rab1361 and is also associated with post-Golgi trafficking62. The activating adaptor NINL and the candidate activating adaptor HOOK2 (Table 1), have both been implicated in trafficking Rab8-marked Golgi derived vesicles that are destined for the base of the primary cilium63,64.
Endolysosomal system.
Endocytosis is the process of internalizing portions of the plasma membrane, which can contain receptors and their ligands. Internalized membranes are sorted to various destinations including recycling endosomes [G], late endosomes [G], multivesicular bodies [G] (MVBs), the TGN, and the lysosome. Each compartment is marked by one or more Rab GTPases.
Dynein is required for trafficking throughout the endolysosomal system. For example, knockout of dynein in mouse cells disperses lysosomes and endosomes7. Overexpression of p50 disrupts early and late endosome and lysosome distribution56, inhibits trafficking of signalling receptors from the cell surface towards the centre of the cell65, and blocks transport of endosomes along nerve axons66.
Early endosomes are marked with the Rab5 GTPase. Activating adaptors of the Hook family have been implicated in linking these vesicles to dynein. Hook was first shown to have a role in endocytic trafficking in D. melanogaster67. Experiments in filamentous fungi subsequently linked Hook to dynein, showing that a Hook homolog was required to link Rab5marked early endosomes to the dynein machinery68,69. Mammals have three Hook-related proteins (HOOK1, HOOK2 and HOOK3). HOOK1 and HOOK3 have been directly linked to dynein-driven movement of early endosomes in axons70.
Hook proteins are part of a complex called FHF, named after its components FTS (AKTIP), Hook, and FHIP (FAM160A2)68,71,72. The GTP-bound form of Rab5 interacts with the FHF complex in both D. melagoaster73 and human cell70 extracts. In Aspergillus nidulans FHIP can bind to early endosomes in the absence of the other two components72 and two-hybrid interaction studies with human proteins suggest that FHIP can bind directly to the GTP-bound form of Rab570. This suggests the FHF complex binds directly to Rab5 on early endosomes via the FHIP component, and the Hook protein recruits the dynein–dynactin complex (Fig. 3b).
Another potential connection between dynein and early endosomes is via Huntingtin (Htt) and HAP1 (Box 1, Fig. 1d). Htt binds to the GTP-form of Rab573 and has been linked to the movement of signalling endosomes (see also below)74, as well as other membranous cargoes. In addition, Htt binds dynein75 and HAP1 binds dynactin76. HAP1 is a candidate activating adaptor based on its homology to other adaptors (Fig. 1d, Table 1). An intriguing question is whether both Htt/HAP1 and the FHF complex are found on the same early endosomes, or whether they provide alternate routes to recruit dynein–dynactin (Fig. 3b).
Late endosomes are marked with the GTPase Rab7. Rab7 forms a complex with the cholesterol sensor ORP1L (oxysterol-binding protein-related protein 1L) and RILP (Box 1)77. RILP binds to GTP-bound Rab778, directly binds the DLIC23,35, and is required to recruit dynein and dynactin to late endosomes and lysosomes34 (Fig. 3c). Interestingly the ORP1L–Rab7–RILP complex also interacts with the HOPS complex [G]71, which is implicated in late endosome trafficking79. The HOPS complex binds to the Hook-containing FHF complex80,81, raising the possibility that late endosomes also engage Hook proteins to link them to dynein82 (Fig. 3c). If this is confirmed, it will suggest that, as with early endosomes, there are multiple ways in which dynein can be recruited to late endosomes. Late endosomes mature into lysosomes, which are found in a perinculear region in some cell types. This positioning has also been linked to dynein– dynactin56 and there is evidence that lysosome motility and positioning is mediated by the same factors that are used by late endosomes83.
In addition to the adaptors described above, a number of other proteins have been implicated in dynein’s association with the endolysosomal system, including JIP333,84, AnkyrinB85, Snapin86 and Spectrin87 (Box 1, Figs. 3b and 3c). These proteins all lack long coiled-coils, suggesting they are not activating adaptors. This raises the question of whether these proteins act together with an activating adaptor or can provide an alternate means of activating dynein–dynactin.
A subset of endosomes contains receptors that signal in the cytoplasm after internalization. These endosomes, called signalling endosomes, have been well studied in neurons, where growth factors bind to receptors at the nerve synapse, undergo endocytosis, and are transported back along the axon to the cell body by dynein–dynactin. For example, nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) bind to the receptor tyrosine kinases TrkA and TrkB, respectively88. These NGF–TrkA and BDNF–TrkB complexes colocalize with both Rab5 early endosomes89 and Rab7 late endosomes in axons90, suggesting that signalling endosomes may use the same adaptors as early and late endosomes.
Many cells also have an endosomal compartment near the MTOC that is a nexus for receptor recycling to the plasma membrane and is distinct from early endosomes. These recycling endosomes are marked by Rab1191. RAB11FIP3 is a Rab11 interacting protein that is required to maintain the structure of recycling endosomes92 and is a dynein–dynactin activator in vitro14 (Table 1 and Fig. 1d). RAB11FIP3 is also required to deliver membranes to cilia93 and the cytokinetic furrow94. As with endosomes, other adaptors may also recruit dynein–dynactin to recycling endosomes. For example, JIP3 has been shown to have a role in the movement of recycling endosomes during cytokinesis95.
Melanosomes.
Melanosomes are pigment-containing organelles related to lysosomes. In many vertebrates, their movements are regulated to control skin colour changes. In pigmented cells of Xenopus laevis, antibodies against the DLIC block melanosome movement towards the cell centre96. In zebrafish lacking the protein ninein-like (NINL), melanosome transport is severely impaired. NINL interacts with components of the dynein and dynactin complex as shown via mass spectrometry experiments97,98, and human NINL is an activating adaptor98, suggesting that NINL is the activating adaptor for melanosome motility.
Autophagosomes.
Autophagosomes are double membrane vesicles formed after engulfment of organelles and proteins destined for destruction by autophagy. They are marked with the ubiquitin-related protein LC3. In HeLa cells autophagosomes move to the cell centre and cluster with lysosomes99. In neurons, autophagosomes can form at the axon tip100 and then fuse with LAMP1- or Rab7-marked late endosomes to initiate transport towards the cell body100–102. Autophagosomes can also form in the soma103.
A role for dynein in autophagy came from the observation that overexpression of p50 or p150 CC1 (Box 2) impairs autophagic clearance104. Furthermore, injection of an anti-DIC antibody inhibits clustering of LC3 autophagosomes in HeLa cells99 and p150 CC1 overexpression inhibits their movement in neurons100. Because moving autophagosomes can fuse with late endosomes, a Hook protein may be the activating adaptor for dynein-mediated autophagosome transport. There is also evidence that Htt and the candidate activating adaptor HAP1 are important for autophagosome motility105. The protein JIP1 (c-Jun N-terminal kinase interacting protein 1) is also necessary for autophagosome motility105,106. JIP1 is structurally unrelated to JIP3 and lacks any predicted coiled-coil, suggesting it is not an activating adaptor. Intriguingly, autophagosomes have different motile properties than endosomes and lysosomes107. It is not yet clear what accounts for these differences.
Mitochondria.
Mitochondria move bidirectionally along microtubules and pause frequently. Their motility can respond to changes in cell signalling or axon growth, for example, which can lead to their accumulation in areas with high metabolic requirements108. Mutations in D. melanogaster dynein impair movement of mitochondria in axons, suggesting a direct role of dynein in retrograde movement (that is from the periphery to the cell body) of mitochondria109. Both dynein and kinesin associate with mitochondria via Milton (TRAK 1 and 2 in humans), which binds to the mitochondrial outer membrane protein Miro (RHOT1 and RHOT2 in humans)110. TRAK proteins co-precipitate dynein and dynactin from brain extracts111, suggesting they interact with the dynein motor. TRAK1 and TRAK2 contain a region with similarity to BICDL1 and HAP160, raising the possibility that the TRAK proteins are activating adaptors of dynein–dynactin for mitochondrial motility (Box 1, Fig. 1d).
Peroxisomes.
Peroxisomes perform a variety of metabolic functions, including the breakdown of fatty acids and metabolism of hydrogen peroxide. Peroxisomes are relatively evenly distributed in cells and only 10–15% of them are mobile in many cell types112. Overexpression of p50 disrupts peroxisome motility in mammalian cells113. However, to date no peroxisome-specific dynein–dynactin adaptor has been identified.
In filamentous fungi, where peroxisomes move in a kinesin- and dynein-dependent manner114, they do so by hitchhiking on moving early endosomes54,115. Peroxisomes co-migrate with Rab5-marked early endosomes and require endosome motility for movement. In Aspergillus nidulans, PxdA, a long coiled-coil-containing protein is required for hitchhiking, and may act to tether the two organelles115 (Fig. 3d). It is not clear if the hitchhiking mechanism is used for peroxisome motility outside of filamentous fungi.
Lipid droplets.
Lipid droplets serve as lipid storage compartments and also provide a source of membrane lipid precursors116. Like peroxisomes they show both diffusive and microtubule-dependent movements117. Genetic studies in D. melanogaster and filamentous fungi have implicated kinesin and dynein in the bidirectional motility of lipid droplets54,118,119. In D. melanogaster BicD, which is related to the mammalian BICD proteins, has been shown to have a role in lipid droplet movement120. However, it is unclear if this role is direct and no other adaptor proteins have been identified for lipid droplets to date. In the filamentous fungus, U. maydis lipid droplets hitchhike on early endosomes, like ER vesicles and peroxisomes54.
Nuclei.
Dynein-dependent movement and positioning of nuclei occurs in many organisms ranging from yeast to humans. In mitosis dynein positions the spindle, which ultimately leads to nuclear positioning121,122. In interphase cells dynein also has direct roles in nuclear movement or positioning. Early demonstrations of this include defects in nuclear positioning in filamentous fungi with mutations in dynein or dynactin subunits123,124 and defects in the movement of pronuclei [G] in Caenorhabditis elegans embryos depleted of dynein or dynactin125. In vertebrate cells dynein is responsible for the movement of nuclei during cell migration126,127, interkinetic nuclear migration [G] in neural progenitor cells128, and distributing and positioning nuclei in developing muscle cells (myoblasts)129,130.
These different nuclear movements can be driven by dynein that is directly connected to the nucleus or by cortically anchored dynein pulling on microtubules emanating from MTOCs. A number of proteins have been identified as being important for linking dynein to the cell cortex or nuclear membrane. In mitosis NuMA and the LGN complex anchor dynein to the cortex131. In interphase, dynein is anchored to the nucleus by BICD2 via the nucleoporin RANBP2132,133. The nucleoporin Nup133 and the dynein regulator Nudel also have a role in localizing dynein to the nucleus132,134. In some cell types dynein may be recruited by different mechanisms. For example, Par6 is required for dynactin localization to the nuclear envelope in myotubes, and its depletion reduces the movement of recently fused myoblast nuclei within myotubes133. In C. elegans a protein related to the Hook family of activating adaptors, Zyg-12, is important for attaching dynein to the nuclear membrane135.
RNA cargoes
Subcellular localization of mRNAs is a mechanism to locally control gene expression in many organisms136. The role of dynein in RNA localization was discovered in D. melanogaster137, where mutations in the DHC or injection of antibodies against the DHC or p50 all lead to defects in mRNA localization in fly syncytial blastoderm [G]138. In mammalian cells there is no direct evidence for the role of dynein in RNA localization, although the DIC co-immunoprecipitates with the RNA binding proteins Staufen139 and La140.
mRNAs are transported in complex with proteins, as ribonucleoprotein particles (RNPs). In D. melanogaster, the localization of many RNAs requires BicD and the formation of transport competent mRNP complexes requires the RNA-binding protein Egalitarian (Egl). Egl binds directly to RNA and the carboxy-terminal cargo-binding domain of BicD, thus linking RNA to the dynein transport machinery141. As an example, the dynein–dynactin–BicD–Egl complex has an essential role during oogenesis in flies by controlling the localization of oskar and bicoid mRNAs, which are involved in anterior–posterior axis determination of the embryo142. These mRNAs, packaged as mRNPs, are transported from nurse cells [G] into the oocyte along polarized microtubule arrays, requiring dynein, dynactin, BicD, and Egl for movement143,144. Once inside the oocyte, bicoid continues to require dynein to maintain its localization145. BicD also contributes to RNA localization in fly neurons, where it interacts with the RNA-binding factor, Fragile X mental retardation protein (FMRP)146. BicD and FMRP interact and move together bidirectionally in fly neurons, and both proteins are required for normal levels of dendritic branching [G]146.
In filamentous fungi, RNAs are distributed by hitchhiking on early endosomes147. For example, in U. maydis mRNA-containing polysomes [G] are distributed throughout the cytoplasm by their association with dynein and kinesin-driven early endosomes148,149. It remains to be explored if hitchhiking is used by higher eukaryotes as a means to distribute or localize mRNA transcripts.
Protein cargoes
Aggresomes and misfolded proteins.
Misfolded proteins are processed by either chaperones, which refold them, or the ubiquitin-proteasome system, which removes them by proteolysis. When these pathways are overwhelmed, cells temporarily sequester these toxic protein species. The misfolded proteins are transported to juxtanuclear structures known as aggresomes that can be eventually processed by autophagy150. Overexpression of p50 (Box 1) prevents the formation of aggresomes, implicating dynein–dynactin in their formation151,152. Three pathways have been reported to recognize misfolded proteins and connect them to dynein– dynactin. The first depends on HDAC6, which links polyubiquitylated proteins to dynein– dynactin153. The precise protein–protein interactions underlying the connection of HDAC6 to dynein–dynactin are not known. A second pathway uses SQSTM1 to link polyubiquitylated proteins to dynein through an interaction with the DIC153. HDAC6 and SQSTM1 interact with each other154, although depletion of HDAC6 increases the amount of SQSTM1 that interacts with dynein, suggesting that HDAC6- and SQTSM1-dependent pathways might act competitively155. A third pathway uses the Hsp70 co-chaperone BAG3 to couple misfolded proteins to the dynein–dynactin complex. This pathway does not require ubiquitylation to target proteins to the aggresome156. The BAG3–dynein interaction is bridged by the 14–3-3 protein [G], which interacts with BAG3 and the DIC157. No known activating adaptor has been so far implicated in aggresome formation.
Transcription factors.
Although small proteins can move quickly by diffusion, there is evidence that some are transported by dynein. For example, transcription factors are translated locally in axons in response to nerve injury and then transported in a retrograde direction along microtubules. This relocalizes them to the nucleus where they activate a transcriptional response158. The transcription factor STAT3 immunoprecipitates with dynein and disruption of microtubules prevents its accumulation in the nucleus in response to axon damage159. The connection between dynein and STAT3 may involve importins [G], as a peptide that blocks the STAT3 interaction with importin reduces the ability of STAT3 to immunoprecipitate with dynein159,160.
Intermediate filaments and microtubules.
Intermediate filaments are cytoskeletal components that contribute to cell shape, motility and organelle positioning161. Dynein participates in the subcellular distribution of vimentin intermediate filaments. Vimentin moves along microtubules162, and overexpression of p50 (Box 1) redistributes vimentin from the perinuclear region to the cell periphery163. In addition, vimentin co-immunoprecipitates with dynactin164. Depletion of the DHC also impairs the retrograde motility of intermediate filaments present in neurons, known as neurofilaments165. This might reflect a direct interaction of dynein with some neurofilaments, as neurofilament-M interacts with the DIC166.
It has also been shown that dynein can contribute to the movement of microtubules. Cell body-originating microtubules are transported into developing axons in rat neuron cultures via a process that is blocked by expression of p50 (Box 1)167. Because these dynein-dependent microtubule movements are anterograde (in the opposite direction from normal dynein transport in the axon) it suggests that the microtubules move by sliding over dynein anchored to the cortex165. Cortically anchored dynein also drives the formation of a uniformly polarized microtubule network in D. melanogaster neurons168.
Centrosomal components.
Centrosomes are the sites of microtubule nucleation and typically anchor microtubule minus ends169. Dynein has a role in transporting proteins to the centrosome. For example, expression of the CC1 domain of the dynactin subunit p150 (Box 2) disrupts the localization of CDK5RAP2 (also known as CEP215) to the centrosome170. CDK5RAP2 is required for γ-tubulin [G] localization and microtubule polymerization171. CDK5RAP2 contains a long region of predicted coiled-coil and co-immunoprecipitates with the DIC, although further work will be required to determine if it is an activating adaptor for dynein cargos destined for the centrosome. There is evidence that the known or candidate activating adaptors, NIN, NINL and HOOK2 are enriched at the centrosome172–174. These proteins have a role in nucleating microtubules, suggesting that they may transport factors required for this process to centrosomes.
Centriolar satellites are smaller motile structures that contribute material to centrosomes and are important for ciliogenesis175. Dynein interacts with components of satellites176, and overexpression p50 (Box 1) disperses them177. In neuroblastoma cells, the activating adaptor HOOK3 localizes to and is important for the function of centriolar satellites, raising the possibility it activates dynein–dynactin to drive the movement of satellite components towards the MTOC178.
Finally, dynein also has a role in tethering centrosomes to the nucleus. Depletion of BICD2 or RANBP2, which are located on the nucleus, severs the tight association of centrosomes and the nucleus133.
Viruses
There are seven classes of viruses and members from every class use dynein for some aspect of their life cycle (Table 2). The most common roles are related to viral replication. For example, DNA and RNA viruses that replicate in the nucleus use dynein motility to reach it179–182. Some retroviruses, which reverse transcribe their RNA genomes into DNA in the cytoplasm, use dynein to deliver the DNA to the nucleus for integration into the nuclear genome183. In addition some DNA and RNA viruses that replicate in the cytoplasm in “perinuclear factories” also require dynein to accumulate at these sites184–188.
Table 2 |.
Viruses interacting with dynein during their life cycle.
| Virus | Presence of the viral envelope | Replication site | Role of dynein in viral life cycle | Evidence for dynein involvement |
|---|---|---|---|---|
| Class 1 (dsDNA) | ||||
| Herpes Simplex Virus (HSV) | Env | Nuc | Transport (direct) | HSV movement requires microtubules235, p50-blocks movement235 |
| Pseudorabies virus | Env | Nuc | Transport (direct) | GFP-capsids move retrograde in axons 236, pUL36 on mitochondria relocates them to perinuclear regions195 |
| Adenovirus | Non-Env | Nuc | Transport (vesicles) | Virus does not co-localize with endosomes by electron microscopy, p50 blocks perinuclear accumulation196 |
| Polyomavirus | Non-Env | Nuc | Transport (vesicles?) | p50-blocks virus at cell periphery237 |
| Bovine papliomaviruses | Non-Env | Nuc | Transport (vesicles) | Transport in endosomes. Viruses remain at the periphery with Nocodazole treatment238 |
| Vaccinia virus (Pox virus) | Env | Cyt | Assembly (direct) | p50-blocks perinuclear accumulation of newly assembled virus particles186 |
| African swine fever virus | Env | Cyt | Assembly (?) | p50-blocks viral replication at perinuclear regions184 |
| Class II (ssDNA) | ||||
| Adeno-associated virus (parvovirus) | Non-Env (replication defective) | Nuc | Transport (vesicles) | Axonal transport of labelled virus in Rab7 vesicles180 |
| Circovirus | Non-Env | Nuc | Transport (vesicles) | Nocodozaole disrupts perinuclear accumulation179 |
| Canine parvovirus | Non-Env | Nuc | Transport (direct?) | Anti-dynein antibody reduces perinuclear accumulation198 |
| Class III (dsRNA) | ||||
| Reoviruses (e.g. rotavirus) | Non-Env | Cyt | Transport (vesicles) | Viral particles in endosomes accumulate in perinuclear regions185 |
| Class IV (+ssRNA) | ||||
| Dengue virus (Flavivirus) | Env | Cyt | Transport (vesicles) & Assembly (vesicles) | Virus colocalizes with endosomes and accumulates in perinuclear regions. Subsequently newly synthesized E-protein accumulates in perinuclear regions (p50 blocks it)188 |
| Hepatitis C (Flavivirus) | Env | Cyt | Cell organization | HCV clustering of lipid droplets in perinuclear regions190 requires viral protein NS5A and dynein239 |
| Class V (-ssRNA) | ||||
| Influenza virus | Env | Cyt | Transport (vesicles) | Anti-dynein antibody inhibits rapid endosome transport to perinuclear regions182 |
| Rabies virus | Env | Cyt | Transport (vesicles) | Long distance tracking of labelled virus189, Rabies envelope-G protein can confer retrograde transport199 |
| Hantaan virus | Env | Cyt | Transport (direct) | p50 blocks perinuclear accumulation of nucleocapsid (N) protein from virus or recombinantly expressed187 |
| Class VI (ssRNA-RT) | ||||
| Human foamy virus | Env | Cyt&Nuc | Transport (vesicles) | p50 blocks perinuclear accumulation of virus or Gag protein alone183 |
| Mason-Pfizer monkey virus | Env | Cyt&Nuc | Transport (direct) | p50 blocks perinuclear accumulation of Gag protein240 |
| Class VII (dsDNA-RT) | ||||
| Hepatitis B | Env | ? | Cell organization | Triggers dynein clustering of mitochondria in perinuclear regions191 |
Viruses are listed by class: classification based on nucleic acid (DNA or RNA), strandedness (ss–single stranded, ds-double stranded), sense (+ strand or − strand), and whether they use a reverse transcriptase (RT). Dynein is involved in different aspects of the viral life cycle, irrespective of whether the viruses are enveloped (Env) or non-enveloped (Non-Env), or whether they replicate in the nucleus (Nuc) or cytoplasm (Cyt). Roles of dynein can include transport of viruses to the perinuclear region during infection (via a direct connection or by transporting them in endosomes). Dynein can also transport components during assembly of new viruses, or can have a role reorganizing the cell in response to viral infection. Evidence for the role of dynein in viral infection is accumulation near the nucleus (perinuclear). Further support comes from showing that the viral components are dispersed, or fail to accumulate, when nocodazole is used to depolymerize microtubules, p50 is over-expressed (p50-block), or an anti-dynein antibody is injected. Direct visualization of viruses moving in the retrograde direction in axons also indicates a role for dynein.
In addition to using dynein to reach their site of replication, some viruses use dynein at other stages of their life cycles. Rabies virus uses dynein for motility along neural axons, which allows it to spread from the site of infection to other parts of the nervous system189. Dengue virus uses dynein during the assembly of new viral particles188. Hepatitis C virus triggers dynein-dependent clustering of lipid droplets at the MTOC, which are then incorporated into newly formed viral particles190. Hepatitis B virus uses dynein to cluster mitochondria close to the nucleus, perhaps to provide energy for its replication191. Finally, there is evidence that Influenza virus requires dynein to disassemble the viral capsid to release its RNA to the cytoplasm. HDAC6 is also required for this release step, suggesting that HDAC6 serves as a dynein adaptor in this context192.
The connections between viruses and dynein during viral particle transport can either be direct or indirect. Examples of viruses that use direct interactions include Herpes Simplex Virus and Pseudorabies Virus193. In these cases, the viral inner tegument proteins (which are attached to the capsid) are required for dynein binding194. The prime candidate for interaction with dynein is pUL36, which is a potential activating adaptor of dynein, as it is active in a relocation assay (Box 2)195. Adenoviral particles may also travel by direct interaction with dynein after infection and may use this transport to reach the nucleus196. The adenovirus capsid protein, hexon, immunoprecipitates with dynein and interacts with both DLIC and DIC197. The interaction with dynein is stimulated by phosphorylation of the DLIC Ras-domain35, suggesting a direct interaction with dynein. However, the role of dynactin in adenoviral intracellular transport is less clear, as dynactin did not immunoprecipitate with hexon197, but p50 overexpression (Box 1) blocked nuclear accumulation of the adenovirus196. There is also evidence that other viruses interact directly with dynein–dynactin183,187,198.
Examples of indirect dynein-based transport involve viruses that move intracellularly within endosomes. Adeno-associated viral particles, for example, which move along neural axons, colocalize with Rab7 vesicles180. Rabies viral particles also move within vesicles and in this form are transported along neuronal axons189. Their dynein-directed movement depends on the envelope glycoprotein, as it was shown that incorporating this protein into retroviral capsids allows retrograde transport of retroviral particles along axons199. How the viral glycoprotein regulates dynein is a mystery, as it is localized within the moving endosome rather than on its cytoplasmic face.
Conclusions and perspectives
While a vast number of dynein cargoes have been described, there are likely to be many more. For example, recent proteomic experiments have identified a number of putative new cargoes linked to dynein by the BICD1, BICD2, HOOK1, HOOK3, NIN or NINL activating adaptors98. Here we have focused largely on cargoes that dynein translocates along microtubules, but dynein can also function anchored at the cell cortex, where it can capture the plus ends of dynamic microtubules200. New dynein “cargoes” may include specific cortical sites that anchor dynein, as suggested by studies that find dynein on the cortex at adherens junctions201 and focal adhesions202, and the numerous cortically localized proteins found in the dynein interactome98.
This large number of dynein cargoes raises many questions related to how dynein achieves cargo specificity. What molecular interactions will mediate binding of dynein to new cargoes? Do some cargoes hitchhike rather than recruit dynein directly53?
In this Review we have emphasized the role of activating adaptors in dynein–dynactin motility. A question for the future will be to determine if all dynein cargoes, including viruses, require an activating adaptor. Reconstitution experiments will be required to verify if all current candidate activating adaptors (i.e. BICD1, HOOK2, CCDC88A, CCDC88B, CCDC88C, TRAK1, TRAK2, NUMA, and HAP1) are indeed able to activate dynein–dynactin motility. If there are fewer activating adaptors than cargoes, how does dynein achieve cargo specificity? Additional proteins that are not activating adaptors are likely required to regulate dynein. For example, as discussed above, the dynein–dynactin interacting proteins, RILP, JIP3 and Htt may be involved in adding cargo specificity to an already activated dynein–dynactin complex.
There may also be mechanisms to activate dynein that do not require activating adaptors. Presumably these factors will also release dynein from its autoinhibited “phi” conformation18. It is also possible that large clusters of dynein motors could overcome the need for activating adaptors or even for the interaction with dynactin, as dynein groups can processively move beads in vitro in the absence of both dynactin and any activator203.
Finally, while we have focused on dynein-based movements in this Review, many cargoes move bi-directionally204. Some cargoes that move bidirectionally can switch directions rapidly implying that there is coordination between opposite polarity motors. What regulates this coordination? Do activating adaptors, some of which can also bind kinesins61,98,205, have a role in this process?
Acknowledgements
We thank Morgan DeSantis, John Salogiannis, and John Srouji for critical comments on the manuscript. SRP is a Howard Hughes Medical Institute-Simons Faculty Scholar and is funded by NIH grants R01GM107214 and R01GM121772. APC is funded by the Wellcome Trust (WT100387) and the Medical Research Council, UK (MC_UP_A025_1011). RDV is Howard Hughes Medical Institute investigator and funded by NIH grant R01 GM097312. We apologize to our colleagues whose work we did not have space to cite.
Glossary terms
- Lissencephaly
Derived from Greek for “smooth brain”, lissencephaly is a spectrum of developmental disorders characterized by defective neuronal migration and the resulting lack of brain folds and grooves.
- Cell cortex
The cytoplasmic face of the plasma membrane.
- AAA+ domain
An “ATPase associated with diverse cellular activities” domain is a highly conserved ATPase fold.
- WD 40 domain
A structural domain formed from WD40 repeats, themselves composed of approximately 40 amino acids and often ending in tryptophan (W), followed by aspartic acid (D).
- Ras-like domain
A protein domain with sequence similarity to the GTPase domain of Ras.
- Barbed end
When actin filaments are decorated with myosin motor domains and visualized by electron microscopy the barbed end is the end where myosins can be seen protruding; similar nomenclature is used to refer to the equivalent end of the Arp1 minifilament in dynactin.
- Pointed end
When actin filaments are decorated with myosin motor domains and visualized by electron microscopy the pointed end is the end where myosins cannot be seen protruding; similar nomenclature is used to refer to the equivalent end of the Arp1 minifilament in dynactin.
- EF-hands
A helix-loop-helix protein structural domain that often confers a protein with calcium-binding ability.
- β-propellers
A protein structural domain characterized by 4–8 wedge-shaped beta sheets arranged similarly to the blades on a propeller.
- Recycling endosomes
Endocytic vesicles characterized by the presence of the protein Rab11, which direct the anterograde trafficking of materials to the cell surface.
- Late endosomes
Pre-lysosomal endocytic vesicles with lower internal pH relative to early endosomes, and characterized by the presence of the protein Rab7.
- Multivesicular bodies
Late endosomes that contain multiple internalized vesicles.
- HOPS complex
The “homotypic fusion and vacuole protein sorting complex” is a multisubunit membrane tethering complex that participates in organelle fusion events within the endolysosomal system in concert with Rab proteins.
- Pronuclei
Refers to the distinct egg and sperm nuclei that are present within a single cell at the onset of fertilization before the fusion of their genetic material.
- Interkinetic nuclear migration
The cell cycle-dependent movement of nuclei observed in neural progenitor cells.
- Syncytial blastoderm
Drosophila melanogaster early embryos comprise a syncytial blastoderm, which is characterized by multiple nuclei residing in a shared cytoplasm, and is the result of multiple nuclear divisions in the absence of cytokinesis.
- Nurse cells
A group of fifteen polyploid Drosophila melanogaster ovarian cells that share a cytoplasm with each other and the developing oocyte and function to support the development of the oocyte by providing nutrients and biomolecules (mRNAs and proteins) through intercellular connections called ring canals
- Dendritic branching
The process by which a dendrite, the portion of neuron that receives signals from other cells, forms the cellular projections it contributes to synapses.
- Polysomes
The complex formed by two or more ribosomes simultaneously engaged in translation along the length of a single messenger RNA.
- 14–3-3 protein
A conserved family of adaptor proteins that interact with diverse proteins and regulate their function through, for example, altered localization, activity, or stability
- Importins
Proteins that recognize and deliver proteins with nuclear localization signals into the nucleus through nuclear pores.
- γ-tubulin
Tubulin family member that, as a component of γ-tubulin ring complexes, templates nascent microtubules.
- SNARE
Proteins that are anchored to either donor or acceptor membranes, mediating fusion between distinct membranes.
- BLOC-1 complex
(biogenesis of lysosome-related organelles complex-1) A multi-subunit protein complex that contributes to membrane tubulation, which is important for sorting and organelle biogenesis in the endolysosmal system.
- Total internal reflection microscopy
This microscopy techniques results in illumination of only a region approximately 100 nm from the coverslip surface, allowing high signal-to-noise ratios to be achieved, which makes it feasible to image and track single molecules.
Biography
Samara L Reck-Peterson is a Professor at the University of California, San Diego, USA and a Howard Hughes Medical Institute-Simons Faculty Scholar. She was previously on the Faculty at Harvard Medical School, Boston, USA. She was a postdoctoral fellow with Ron Vale at the University of California, San Francisco, USA and a graduate student at Yale University, New Haven, USA with Mark Mooseker and Peter Novick. Her laboratory studies the mechanisms and regulation of microtubule-based intracellular transport.
William Bret Redwine is a postdoctoral fellow in the laboratory of Samara Reck-Peterson. He was a graduate student at Harvard Medical School, Boston, USA with Andres Leschziner and Samara Reck-Peterson.
Ronald D. Vale is a Professor at the University of California, San Francisco, USA and an Investigator in the Howard Hughes Medical Institute. He received his Ph.D. in Neuroscience from Stanford University, Palo Alto, USA in 1985 where he trained with Dr. Eric Shooter, and was a Staff Fellow with the NIH at the Marine Biological Laboratory with Tom Reese in 19851986. His laboratory integrates biochemical, structural, and microscopy-based approaches to study spatial organization, movement, and signaling within cells.
Andrew Carter is a programme leader at the Medical Research Council Lab of Molecular Biology (MRC LMB), Cambridge, UK. He was a postdoctoral fellow with Ron Vale at the University of California, San Francisco, USA and a graduate student at the MRC LMB, Cambridge, UK with Venki Ramakrishnan. His lab studies the structure and mechanism of dynein based transport.
Footnotes
Competing interest
The authors have no competing interests to declare.
References
- 1.Gibbons IR & Rowe AJ Dynein: A Protein with Adenosine Triphosphatase Activity from Cilia. Science 149, 424–6 (1965).Describes the original discovey of dynein.
- 2.Paschal BM, Shpetner HS & Vallee RB MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol 105, 1273–82 (1987).Describes the discovery of cytoplasmic dynein-1.
- 3.Paschal BM & Vallee RB Retrograde transport by the microtubule-associated protein MAP 1C. Nature 330, 181–3 (1987).Shows that cytoplasmic dynein-1 is a minus-end-directed motor.
- 4.Pazour GJ, Dickert BL & Witman GB The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol 144, 473–81 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter ME, Bower R, Knott JA, Byrd P & Dentler W Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell 10, 693–712 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gepner J et al. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics 142, 865–78 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harada A et al. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol 141, 51–9 (1998).Characterization of dynein knockout mouse, which showed that dynein is essential during development.
- 8.Lipka J, Kuijpers M, Jaworski J & Hoogenraad CC Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem Soc Trans 41, 160512 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Raaijmakers JA & Medema RH Function and regulation of dynein in mitotic chromosome segregation. Chromosoma 123, 407–22 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Gill SR et al. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol 115, 1639–50 (1991).Along with the ref 11 describes the discovery of dynactin.
- 11.Schroer TA & Sheetz MP Two activators of microtubule-based vesicle transport. J Cell Biol 115, 1309–18 (1991).Along with the ref 10 describes the discovery of dynactin.
- 12.Splinter D et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell 23, 4226–41 (2012).Shows that the N-terminal domain of BICD2 strengthens the interaction between dynein and dyanctin and activates motility in cells.
- 13.Mohler J & Wieschaus EF Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics 112, 803–22 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G & Vale RD Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–41 (2014).Along with the ref 15, showed that coiled-coil containing activators like BICD2, along with dynactin activate mammalian dynein to move processively.
- 15.Schlager MA, Hoang HT, Urnavicius L, Bullock SL & Carter AP In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33, 1855–68 (2014).Along with the ref 15, showed that coiled-coil containing activators like BICD2, along with dynactin activate mammalian dynein to move processively.
- 16.Urnavicius L et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. bioRxiv 10.1101/183160 (2017).High resolution structure of the dynein tail and shows that some activating adaptors preferentially recruit two dynein dimers.
- 17.Urnavicius L et al. The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–6 (2015).Revealed the structural basis for dynein activation by dynactin and the activating adaptor, BICD2.
- 18.Zhang K et al. Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell 169, 1303–1314 e18 (2017).Demonstrates how dynactin activates dynein’s ability to move long distances.
- 19.Kon T et al. The 2.8 A crystal structure of the dynein motor domain. Nature 484, 345–50 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Schmidt H, Gleave ES & Carter AP Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol 19, 492–7, S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt H, Zalyte R, Urnavicius L & Carter AP Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518, 435–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee IG et al. A Conserved Interaction of the Dynein Light Intermediate Chain with Dynein-Dynactin Effectors Necessary for Processivity. Nature Comm in press(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder CM, Ostrem JM, Hertz NT & Vale RD A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. Elife 3, e03351 (2014).Identified the carboxy terminus of DLIC as a binding site for activating adaptors.
- 24.Schroeder CM & Vale RD Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol 214, 309–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter AP, Diamant AG & Urnavicius L How dynein and dynactin transport cargos: a structural perspective. Curr Opin Struct Biol 37, 62–70 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Waterman-Storer CM, Karki S & Holzbaur EL The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1). Proc Natl Acad Sci U S A 92, 1634–8 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gama JB et al. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J Cell Biol 216, 943–960 (2017).Identified the CC1 box for DLIC binding in activating adaptors.
- 28.Horgan CP, Hanscom SR, Jolly RS, Futter CE & McCaffrey MW Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J Cell Sci 123, 181–91 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Grotjahn DA et al. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. bioRxiv 10.1101/182576 (2017).Together with ref 19 shows that dynactin can recruit two dynein dimers.
- 30.Siglin AE et al. Dynein and dynactin leverage their bivalent character to form a high-affinity interaction. PLoS One 8, e59453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi T, Miyashita T, Murayama T & Toyoshima YY Dynactin has two antagonistic regulatory domains and exerts opposing effects on dynein motility. PLoS One 12, e0183672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torisawa T et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol 16, 1118–24 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Cavalli V, Kujala P, Klumperman J & Goldstein LS Sunday Driver links axonal transport to damage signaling. J Cell Biol 168, 775–87 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordens I et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 11, 1680–5 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Scherer J, Yi J & Vallee RB PKA-dependent dynein switching from lysosomes to adenovirus: a novel form of host-virus competition. J Cell Biol 205, 163–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cianfrocco MA, DeSantis ME, Leschziner AE & Reck-Peterson SL Mechanism and regulation of cytoplasmic dynein. Annu Rev Cell Dev Biol 31, 83–108 (2015).Showed that Lis1 has two functionally distinct binding sites on the dynein motor domain.
- 37.DeSantis ME et al. Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein. Cell 170, 11971208 e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Roberts AJ, Leschziner AE & Reck-Peterson SL Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell 150, 975–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenney RJ, Weil SJ, Scherer J & Vallee RB Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem 286, 39615–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S & Zheng Y Identification of a novel dynein binding domain in nudel essential for spindle pole organization in Xenopus egg extract. J Biol Chem 286, 587–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zylkiewicz E et al. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J Cell Biol 192, 433–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torisawa T et al. Functional dissection of LIS1 and NDEL1 towards understanding the molecular mechanisms of cytoplasmic dynein regulation. J Biol Chem 286, 1959–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada M et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J 27, 2471–83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumbach J et al. Lissencephaly-1 is a context-dependent regulator of the human dynein complex. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez PA, Ackermann BE, Vershinin M & McKenney RJ Differential effects of the dyneinregulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J Biol Chem 292, 12245–12255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S et al. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell 24, 3522–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seksek O, Biwersi J & Verkman AS Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J Cell Biol 138, 131–42 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutagalung AH & Novick PJ Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91, 119–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wozniak MJ et al. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci 122, 1979–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valenzuela JI et al. Transport along the dendritic endoplasmic reticulum mediates the trafficking of GABAB receptors. J Cell Sci 127, 3382–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Presley JF et al. ER-to-Golgi transport visualized in living cells. Nature 389, 81–5 (1997).Elegant demonstration of the role of dynein in transporting proteins from the ER to Golgi.
- 52.Roghi C & Allan VJ Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J Cell Sci 112 (Pt 24), 4673–85 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Salogiannis J & Reck-Peterson SL Hitchhiking: A Non-Canonical Mode of Microtubule-Based Transport. Trends Cell Biol 27, 141–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guimaraes SC et al. Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J Cell Biol 211, 945–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wedlich-Soldner R, Straube A, Friedrich MW & Steinberg G A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J 21, 2946–57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burkhardt JK, Echeverri CJ, Nilsson T & Vallee RB Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol 139, 469–84 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoogenraad CC et al. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J 20, 4041–54 (2001).Demonstrated a link between BICD2 and dynein-dynactin.
- 58.Matanis T et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dyneindynactin motor complex. Nat Cell Biol 4, 986–92 (2002).Showed that BICD proteins receruit dynein to Rab6-labelled Golgi vesicles.
- 59.Yadav S, Puthenveedu MA & Linstedt AD Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev Cell 23, 153–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlager MA et al. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell Rep 8, 1248–56 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Schlager MA et al. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J 29, 1637–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nokes RL, Fields IC, Collins RN & Folsch H Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J Cell Biol 182, 845–53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bachmann-Gagescu R et al. The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking. PLoS Genet 11, e1005575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baron Gaillard CL et al. Hook2 is involved in the morphogenesis of the primary cilium. Mol Biol Cell 22, 4549–62 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Driskell OJ, Mironov A, Allan VJ & Woodman PG Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat Cell Biol 9, 113–20 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Heerssen HM, Pazyra MF & Segal RA Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nat Neurosci 7, 596–604 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Kramer H & Phistry M Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J Cell Biol 133, 1205–15 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bielska E et al. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol 204, 989–1007 (2014).Along with ref 69 identified a role for Hook proteins in dynein-based endosome motility.
- 69.Zhang J, Qiu R, Arst HN Jr., Penalva MA & Xiang X HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J Cell Biol 204, 1009–26 (2014).Along with ref 68 identified a role for Hook proteins in dynein-based endosome motility.
- 70.Guo X, Farias GG, Mattera R & Bonifacino JS Rab5 and its effector FHF contribute to neuronal polarity through dynein-dependent retrieval of somatodendritic proteins from the axon. Proc Natl Acad Sci U S A 113, E5318–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu L et al. An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol Biol Cell 19, 5059–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao X, Wang X & Xiang X FHIP and FTS proteins are critical for dynein-mediated transport of early endosomes in Aspergillus. Mol Biol Cell 25, 2181–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gillingham AK, Sinka R, Torres IL, Lilley KS & Munro S Toward a comprehensive map of the effectors of rab GTPases. Dev Cell 31, 358–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gauthier LR et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118, 127–38 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Caviston JP, Ross JL, Antony SM, Tokito M & Holzbaur EL Huntingtin facilitates dynein/dynactinmediated vesicle transport. Proc Natl Acad Sci U S A 104, 10045–50 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engelender S et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet 6, 2205–12 (1997). [DOI] [PubMed] [Google Scholar]
- 77.Johansson M et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILPp150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol 176, 459–71 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu M, Wang T, Loh E, Hong W & Song H Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J 24, 1491–501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spang A Membrane Tethering Complexes in the Endosomal System. Front Cell Dev Biol 4, 35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin X et al. RILP interacts with HOPS complex via VPS41 subunit to regulate endocytic trafficking. Sci Rep 4, 7282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van der Kant R et al. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci 126, 3462–74 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Szatmari Z et al. Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Mol Biol Cell 25, 522–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pu J, Guardia CM, Keren-Kaplan T & Bonifacino JS Mechanisms and functions of lysosome positioning. J Cell Sci 129, 4329–4339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Drerup CM & Nechiporuk AV JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet 9, e1003303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lorenzo DN et al. A PIK3C3-ankyrin-B-dynactin pathway promotes axonal growth and multiorganelle transport. J Cell Biol 207, 735–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cai Q et al. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 68, 73–86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holleran EA, Tokito MK, Karki S & Holzbaur EL Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J Cell Biol 135, 1815–29 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmieg N, Menendez G, Schiavo G & Terenzio M Signalling endosomes in axonal transport: travel updates on the molecular highway. Semin Cell Dev Biol 27, 32–43 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Delcroix JD et al. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron 39, 69–84 (2003). [DOI] [PubMed] [Google Scholar]
- 90.Saxena S, Bucci C, Weis J & Kruttgen A The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J Neurosci 25, 10930–40 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Welz T, Wellbourne-Wood J & Kerkhoff E Orchestration of cell surface proteins by Rab11. Trends Cell Biol 24, 407–15 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Horgan CP et al. Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic 8, 414–30 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Wang J & Deretic D The Arf and Rab11 effector FIP3 acts synergistically with ASAP1 to direct Rabin8 in ciliary receptor targeting. J Cell Sci 128, 1375–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilson GM et al. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell 16, 849–60 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montagnac G et al. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr Biol 19, 184–95 (2009). [DOI] [PubMed] [Google Scholar]
- 96.Reilein AR et al. Differential regulation of dynein-driven melanosome movement. Biochem Biophys Res Commun 309, 652–8 (2003). [DOI] [PubMed] [Google Scholar]
- 97.Dona M et al. NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish. PLoS Genet 11, e1005574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Redwine WB et al. The human cytoplasmic dynein interactome reveals novel activators of motility. eLife 6:e28257(2017).A proteomics resource of the cytoplasmic dynein interactome.
- 99.Kimura S, Noda T & Yoshimori T Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 33, 109–22 (2008). [DOI] [PubMed] [Google Scholar]
- 100.Maday S, Wallace KE & Holzbaur EL Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 196, 407–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng XT, Zhou B, Lin MY, Cai Q & Sheng ZH Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol 209, 377–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S, Sato Y & Nixon RA Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci 31, 7817–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maday S & Holzbaur EL Compartment-Specific Regulation of Autophagy in Primary Neurons. J Neurosci 36, 5933–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ravikumar B et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 37, 771–6 (2005). [DOI] [PubMed] [Google Scholar]
- 105.Wong YC & Holzbaur EL The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci 34, 1293305 (2014).Demonstrates the roles of Htt and HAP1 in autophagy.
- 106.Fu MM, Nirschl JJ & Holzbaur EL LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell 29, 577–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klinman E & Holzbaur EL Comparative analysis of axonal transport markers in primary mammalian neurons. Methods Cell Biol 131, 409–24 (2016).Paper showing different types of retrograde transport of many key cargoes in axons.
- 108.Hollenbeck PJ & Saxton WM The axonal transport of mitochondria. J Cell Sci 118, 5411–9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pilling AD, Horiuchi D, Lively CM & Saxton WM Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell 17, 2057–68 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwarz TL Mitochondrial trafficking in neurons. Cold Spring Harb Perspect Biol 5(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Spronsen M et al. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron 77, 485–502 (2013). [DOI] [PubMed] [Google Scholar]
- 112.Neuhaus A, Eggeling C, Erdmann R & Schliebs W Why do peroxisomes associate with the cytoskeleton? Biochim Biophys Acta 1863, 1019–26 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Schrader M, King SJ, Stroh TA & Schroer TA Real time imaging reveals a peroxisomal reticulum in living cells. J Cell Sci 113 (Pt 20), 3663–71 (2000). [DOI] [PubMed] [Google Scholar]
- 114.Egan MJ, Tan K & Reck-Peterson SL Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol 197, 971–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salogiannis J, Egan MJ & Reck-Peterson SL Peroxisomes move by hitchhiking on early endosomes using the novel linker protein PxdA. J Cell Biol 212, 289–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thiam AR, Farese RV Jr. & Walther TC The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14, 775–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Targett-Adams P et al. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J Biol Chem 278, 15998–6007 (2003). [DOI] [PubMed] [Google Scholar]
- 118.Gross SP, Welte MA, Block SM & Wieschaus EF Dynein-mediated cargo transport in vivo. A switch controls travel distance. J Cell Biol 148, 945–56 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shubeita GT et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell 135, 1098–107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Larsen KS, Xu J, Cermelli S, Shu Z & Gross SP BicaudalD actively regulates microtubule motor activity in lipid droplet transport. PLoS One 3, e3763 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McNally FJ Mechanisms of spindle positioning. J Cell Biol 200, 131–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moore JK, Stuchell-Brereton MD & Cooper JA Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton 66, 546–55 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Plamann M, Minke PF, Tinsley JH & Bruno KS Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol 127, 139–149 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xiang X, Beckwith SM & Morris NR Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci U S A 91, 2100–4 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gonczy P, Pichler S, Kirkham M & Hyman AA Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol 147, 135–50 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dujardin DL et al. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol 163, 1205–11 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsai JW, Bremner KH & Vallee RB Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci 10, 970–9 (2007). [DOI] [PubMed] [Google Scholar]
- 128.Tsai JW, Lian WN, Kemal S, Kriegstein AR & Vallee RB Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci 13, 1463–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cadot B, Gache V & Gomes ER Moving and positioning the nucleus in skeletal muscle - one step at a time. Nucleus 6, 373–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Folker ES, Schulman VK & Baylies MK Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development 141, 355–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Radulescu AE & Cleveland DW NuMA after 30 years: the matrix revisited. Trends Cell Biol 20, 214–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hu DJ et al. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell 154, 1300–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Splinter D et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8, e1000350 (2010).Demonstrated a role for BICD2 in dynein recruitment to nuclear pores.
- 134.Bolhy S et al. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 192, 855–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malone CJ et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–36 (2003). [DOI] [PubMed] [Google Scholar]
- 136.Bullock SL Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem Soc Trans 39, 1161–5 (2011). [DOI] [PubMed] [Google Scholar]
- 137.Bullock SL & Ish-Horowicz D Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414, 611–6 (2001).Demonstrated a role for BicD in RNA localization.
- 138.Wilkie GS & Davis I Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 105, 209–19 (2001). [DOI] [PubMed] [Google Scholar]
- 139.Gershoni-Emek N et al. Proteomic Analysis of Dynein-Interacting Proteins in Amyotrophic Lateral Sclerosis Synaptosomes Reveals Alterations in the RNA-Binding Protein Staufen1. Mol Cell Proteomics 15, 506–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van Niekerk EA et al. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci U S A 104, 12913–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dienstbier M, Boehl F, Li X & Bullock SL Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 23, 1546–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weil TT mRNA localization in the Drosophila germline. RNA Biol 11, 1010–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Clark A, Meignin C & Davis I A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134, 1955–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mische S, Li M, Serr M & Hays TS Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell 18, 2254–63 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weil TT, Forrest KM & Gavis ER Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell 11, 251–62 (2006). [DOI] [PubMed] [Google Scholar]
- 146.Bianco A, Dienstbier M, Salter HK, Gatto G & Bullock SL Bicaudal-D regulates fragile X mental retardation protein levels, motility, and function during neuronal morphogenesis. Curr Biol 20, 1487–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Haag C, Steuten B & Feldbrugge M Membrane-Coupled mRNA Trafficking in Fungi. Annu Rev Microbiol 69, 265–81 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Baumann S, Konig J, Koepke J & Feldbrugge M Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep 15, 94–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Higuchi Y, Ashwin P, Roger Y & Steinberg G Early endosome motility spatially organizes polysome distribution. J Cell Biol 204, 343–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Garcia-Mata R, Gao YS & Sztul E Hassles with taking out the garbage: aggravating aggresomes. Traffic 3, 388–96 (2002). [DOI] [PubMed] [Google Scholar]
- 151.Garcia-Mata R, Bebok Z, Sorscher EJ & Sztul ES Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol 146, 1239–54 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Johnston JA, Illing ME & Kopito RR Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton 53, 26–38 (2002). [DOI] [PubMed] [Google Scholar]
- 153.Kawaguchi Y et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–38 (2003). [DOI] [PubMed] [Google Scholar]
- 154.Yan J et al. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS One 8, e76016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Calderilla-Barbosa L et al. Interaction of SQSTM1 with the motor protein dynein--SQSTM1 is required for normal dynein function and trafficking. J Cell Sci 127, 4052–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gamerdinger M, Kaya AM, Wolfrum U, Clement AM & Behl C BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep 12, 149–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xu Z et al. 14–3-3 protein targets misfolded chaperone-associated proteins to aggresomes. J Cell Sci 126, 4173–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Perry RB & Fainzilber M Local translation in neuronal processes--in vivo tests of a “heretical hypothesis”. Dev Neurobiol 74, 210–7 (2014). [DOI] [PubMed] [Google Scholar]
- 159.Ben-Yaakov K et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J 31, 1350–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hanz S et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40, 1095104 (2003). [DOI] [PubMed] [Google Scholar]
- 161.Lowery J, Kuczmarski ER, Herrmann H & Goldman RD Intermediate Filaments Play a Pivotal Role in Regulating Cell Architecture and Function. J Biol Chem 290, 17145–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hookway C et al. Microtubule-dependent transport and dynamics of vimentin intermediate filaments. Mol Biol Cell 26, 1675–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Helfand BT, Mikami A, Vallee RB & Goldman RD A requirement for cytoplasmic dynein and dynactin in intermediate filament network assembly and organization. J Cell Biol 157, 795–806 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Payne C, Rawe V, Ramalho-Santos J, Simerly C & Schatten G Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J Cell Sci 116, 4727–38 (2003). [DOI] [PubMed] [Google Scholar]
- 165.He Y et al. Role of cytoplasmic dynein in the axonal transport of microtubules and neurofilaments. J Cell Biol 168, 697–703 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wagner OI et al. The interaction of neurofilaments with the microtubule motor cytoplasmic dynein. Mol Biol Cell 15, 5092–100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ahmad FJ, Echeverri CJ, Vallee RB & Baas PW Cytoplasmic dynein and dynactin are required for the transport of microtubules into the axon. J Cell Biol 140, 391–401 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.del Castillo U, Winding M, Lu W & Gelfand VI Interplay between kinesin-1 and cortical dynein during axonal outgrowth and microtubule organization in Drosophila neurons. Elife 4, e10140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tang N & Marshall WF Centrosome positioning in vertebrate development. J Cell Sci 125, 4951–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jia Y, Fong KW, Choi YK, See SS & Qi RZ Dynamic recruitment of CDK5RAP2 to centrosomes requires its association with dynein. PLoS One 8, e68523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fong KW, Choi YK, Rattner JB & Qi RZ CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell 19, 115–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Casenghi M et al. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell 5, 113–25 (2003). [DOI] [PubMed] [Google Scholar]
- 173.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V & Bornens M Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci 113 (Pt 17), 3013–23 (2000). [DOI] [PubMed] [Google Scholar]
- 174.Szebenyi G, Hall B, Yu R, Hashim AI & Kramer H Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic 8, 32–46 (2007). [DOI] [PubMed] [Google Scholar]
- 175.Hori A & Toda T Regulation of centriolar satellite integrity and its physiology. Cell Mol Life Sci 74, 213229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim JC et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet 36, 462–70 (2004). [DOI] [PubMed] [Google Scholar]
- 177.Dammermann A & Merdes A Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol 159, 255–66 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ge X, Frank CL, Calderon de Anda F & Tsai LH Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron 65, 191–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cao J et al. Circovirus transport proceeds via direct interaction of the cytoplasmic dynein IC1 subunit with the viral capsid protein. J Virol 89, 2777–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Castle MJ, Perlson E, Holzbaur EL & Wolfe JH Long-distance axonal transport of AAV9 is driven by dynein and kinesin-2 and is trafficked in a highly motile Rab7-positive compartment. Mol Ther 22, 554–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dodding MP & Way M Coupling viruses to dynein and kinesin-1. EMBO J 30, 3527–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lakadamyali M, Rust MJ, Babcock HP & Zhuang X Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A 100, 9280–5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Petit C et al. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J Cell Sci 116, 3433–42 (2003). [DOI] [PubMed] [Google Scholar]
- 184.Alonso C et al. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J Virol 75, 9819–27 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mainou BA et al. Reovirus cell entry requires functional microtubules. MBio 4(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ploubidou A et al. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J 19, 3932–44 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ramanathan HN et al. Dynein-dependent transport of the hantaan virus nucleocapsid protein to the endoplasmic reticulum-Golgi intermediate compartment. J Virol 81, 8634–47 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shrivastava N, Sripada S, Kaur J, Shah PS & Cecilia D Insights into the internalization and retrograde trafficking of Dengue 2 virus in BHK-21 cells. PLoS One 6, e25229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klingen Y, Conzelmann KK & Finke S Double-labeled rabies virus: live tracking of enveloped virus transport. J Virol 82, 237–45 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Boulant S et al. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9, 1268–82 (2008). [DOI] [PubMed] [Google Scholar]
- 191.Kim S et al. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol 81, 1714–26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Banerjee I et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346, 473–7 (2014). [DOI] [PubMed] [Google Scholar]
- 193.Lycke E et al. Herpes simplex virus infection of the human sensory neuron. An electron microscopy study. Arch Virol 101, 87–104 (1988). [DOI] [PubMed] [Google Scholar]
- 194.Radtke K et al. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6, e1000991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zaichick SV et al. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13, 193–203 (2013).Evidence that a herpes virus contains a protein that can activate dynein–dynactin transport.
- 196.Suomalainen M et al. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol 144, 657–72 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bremner KH et al. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6, 523–35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Suikkanen S et al. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J Virol 77, 10270–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mazarakis ND et al. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum Mol Genet 10, 2109–21 (2001). [DOI] [PubMed] [Google Scholar]
- 200.Laan L et al. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 148, 502–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ligon LA, Karki S, Tokito M & Holzbaur EL Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat Cell Biol 3, 913–7 (2001). [DOI] [PubMed] [Google Scholar]
- 202.Rosse C et al. Binding of dynein intermediate chain 2 to paxillin controls focal adhesion dynamics and migration. J Cell Sci 125, 3733–8 (2012). [DOI] [PubMed] [Google Scholar]
- 203.Mallik R, Rai AK, Barak P, Rai A & Kunwar A Teamwork in microtubule motors. Trends Cell Biol 23, 575–82 (2013). [DOI] [PubMed] [Google Scholar]
- 204.Hancock WO Bidirectional cargo transport: moving beyond tug of war. Nat Rev Mol Cell Biol 15, 615–28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Grigoriev I et al. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell 13, 305–14 (2007). [DOI] [PubMed] [Google Scholar]
- 206.Block-Galarza J et al. Fast transport and retrograde movement of huntingtin and HAP 1 in axons. Neuroreport 8, 2247–51 (1997). [DOI] [PubMed] [Google Scholar]
- 207.Li SH, Gutekunst CA, Hersch SM & Li XJ Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci 18, 1261–9 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fu MM & Holzbaur EL JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J Cell Biol 202, 495–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hong Z et al. The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res 19, 1334–49 (2009). [DOI] [PubMed] [Google Scholar]
- 210.Wassmer T et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell 17, 110–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Holleran EA et al. beta III spectrin binds to the Arp1 subunit of dynactin. J Biol Chem 276, 36598–605 (2001). [DOI] [PubMed] [Google Scholar]
- 212.Starcevic M & Dell’Angelica EC Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC1). J Biol Chem 279, 28393–401 (2004). [DOI] [PubMed] [Google Scholar]
- 213.Tynan SH, Purohit A, Doxsey SJ & Vallee RB Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J Biol Chem 275, 32763–8 (2000). [DOI] [PubMed] [Google Scholar]
- 214.Schmoranzer J et al. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol 19, 1065–74 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Williams JC et al. Structural and thermodynamic characterization of a cytoplasmic dynein light chainintermediate chain complex. Proc Natl Acad Sci U S A 104, 10028–33 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tai AW, Chuang JZ, Bode C, Wolfrum U & Sung CH Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell 97, 877–87 (1999). [DOI] [PubMed] [Google Scholar]
- 217.Tai AW, Chuang JZ & Sung CH Cytoplasmic dynein regulation by subunit heterogeneity and its role in apical transport. J Cell Biol 153, 1499–509 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Meiri D et al. Mechanistic insight into the microtubule and actin cytoskeleton coupling through dyneindependent RhoGEF inhibition. Mol Cell 45, 642–55 (2012). [DOI] [PubMed] [Google Scholar]
- 219.Wu H, Maciejewski MW, Takebe S & King SM Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure 13, 213–23 (2005). [DOI] [PubMed] [Google Scholar]
- 220.Echeverri CJ, Paschal BM, Vaughan KT & Vallee RB Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol 132, 617–33 (1996).Discovery that p50 overexpression inhibits dynein function.
- 221.Quintyne NJ et al. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol 147, 321–34 (1999).Discovery that overexpression of the CC1 fragment of the p150 dynactin subunit inhibits dynein function.
- 222.Schroer TA Dynactin. Annu Rev Cell Dev Biol 20, 759–79 (2004). [DOI] [PubMed] [Google Scholar]
- 223.Steffen W et al. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol Biol Cell 8, 2077–88 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hoogenraad CC et al. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J 22, 6004–15 (2003).The first use of a “relocation assay” to monitor dynein recruitment and activation.
- 225.Kapitein LC et al. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys J 99, 2143–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Olenick MA, Tokito M, Boczkowska M, Dominguez R & Holzbaur EL Hook Adaptors Induce Unidirectional Processive Motility by Enhancing the Dynein-Dynactin Interaction. J Biol Chem 291, 1823951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Griffis ER, Stuurman N & Vale RD Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177, 1005–15 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maldonado-Baez L, Cole NB, Kramer H & Donaldson JG Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J Cell Biol 201, 233–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Walenta JH, Didier AJ, Liu X & Kramer H The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol 152, 923–34 (2001).Describes the discovery of the Hook family of proteins.
- 230.Fumoto K, Hoogenraad CC & Kikuchi A GSK-3beta-regulated interaction of BICD with dynein is involved in microtubule anchorage at centrosome. EMBO J 25, 5670–82 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Okuda H et al. LRGUK1 is part of a multiprotein complex required for manchette function and male fertility. FASEB J 31, 1141–1152 (2017). [DOI] [PubMed] [Google Scholar]
- 232.Ham H, Huynh W, Schoon RA, Vale RD & Billadeau DD HkRP3 is a microtubule-binding protein regulating lytic granule clustering and NK cell killing. J Immunol 194, 3984–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Merdes A, Heald R, Samejima K, Earnshaw WC & Cleveland DW Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol 149, 851–62 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hueschen C, Kenny SJ, Xu K & Dumont S NuMA Targets Dynein to Microtubule Minus-Ends at Mitosis. bioRxiv doi: 10.1101/148692(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Dohner K et al. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell 13, 2795–809 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Smith GA, Pomeranz L, Gross SP & Enquist LW Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc Natl Acad Sci U S A 101, 16034–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zila V, Difato F, Klimova L, Huerfano S & Forstova J Involvement of microtubular network and its motors in productive endocytic trafficking of mouse polyomavirus. PLoS One 9, e96922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liu WJ et al. Association of bovine papillomavirus type 1 with microtubules. Virology 282, 237–44 (2001). [DOI] [PubMed] [Google Scholar]
- 239.Eyre NS et al. Dynamic imaging of the hepatitis C virus NS5A protein during a productive infection. J Virol 88, 3636–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sfakianos JN, LaCasse RA & Hunter E The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4, 660–70 (2003). [DOI] [PubMed] [Google Scholar]