Abstract
While recent clinical studies demonstrate the promise of cancer immunotherapy, a barrier for broadening the clinical benefit is identifying how tumors locally suppress cytotoxic immunity. As an emerging mode of intercellular communication, exosomes secreted by malignant cells can deliver a complex payload of coding and non-coding RNA to cells within the tumor microenvironment. Here, we quantified the RNA pay-load within tumor-derived exosomes and the resulting dynamic transcriptomic response to cytotoxic T cells upon exosome delivery to better understand how tumor-derived exosomes can alter immune cell function. Exosomes derived from B16F0 melanoma cells were enriched for a subset of coding and non-coding RNAs that did not reflect the abundance in the parental cell. Upon exosome delivery, RNAseq revealed the dynamic changes in the transcriptome of CTLL2 cytotoxic T cells. In analyzing transiently co-expressed gene clusters, pathway enrichment suggested that the B16F0 exosomal payload altered mitochondrial respiration, which was confirmed independently, and upregulated genes associated with the Notch signaling pathway. Interestingly, exosomal miRNA appeared to have no systematic effect on downregulating target mRNA levels.
Keywords: tumor microenvironment, cell-mediated immunity, exosomes, RNA sequencing, heterotypic cell communication
Introduction
Cancer immunosurveillance is the process by which the host’s immune system recognizes and eradicates nascent neoplastic cells, limiting cancer development. However, in some cases, transformed cells are not completely eliminated and tumor escape can occur after a dynamic equilibrium with immune cells. In this process, new tumor cell variants are selected due to their loss of immunogenicity and their increased ability to suppress anti-tumor immune effectors [1]. Cancer cells can actively induce T cell dysfunction through several mechanisms that involve conditioning the tumor microenvironment via soluble cues. Myeloid-derived suppressor cells (MDSC), tumor-associated macrophages (TAM), tumor-associated neutrophils (TAN) and regulatory T cells (Tregs) are expanded by tumor-derived soluble cues and inhibit anti-tumor T cell responses [2–7]. MDSC, TAM and TAN contribute to T cell dysfunction by expressing arginase 1 that can deplete L-arginine and MDSC limit L-cysteine availability, which are both essential amino acids for the proliferation of T cells [2, 3, 8]. MDSC-mediated production of reactive oxygen and nitrogen species, through NADPH oxidase and inducible nitric oxide synthase (iNOS), impairs antigen recognition, activation and function of T cells [9, 10]. Additionally, immunosuppressive molecules like IL-10, TGF-β, extracellular adenosine and prostaglandin E2 are preferentially released by Tregs and TAM, whereas IL-12 and TNF production by TAM and TAN, respectively, are significantly reduced [11, 3, 7, 5]. Other studies demonstrate that hypoxic conditions and the predominant metabolic pathways in cancer cells generate an acidic tumor microenvironment and deprive T cells of essential metabolites like glucose, both mechanisms associated with the impairment of T cell survival and effector functions [12–14].
Cell-to-cell communication is important in understanding the dynamic interaction between tumor and immune cells in the tumor microenvironment. In contrast to the soluble cues that condition the tumor microenvironment, exosomes are emerging as an important mode of intercellular communication. These nanoscaled extracellular vesicles are actively released by cells to transfer proteins, mRNA, and non-coding RNAs between cells. Interestingly, tumor cells release exosomes at a greater rate than normal cells, which suggests that they may play an important role in cancer [15]. Some of the earliest exosome studies suggest that exosomes mediate B cell transformations [16, 17]. Subsequent studies reinforce the notion of exosomes as immunoregulatory modulators and illustrate the ways in which tumor exosomes are capable of altering their microenvironment. Proteomic analysis suggests that exosomes from human breast cancer cell lines and not from normal human mammary cells are enriched in proteins involved in glycolytic metabolism [18]. Melanoma exosomes promote metastasis by delivering a payload that upregulates oncoproteins [19]. In mouse melanoma cell models, exosomes from B16F0 but not Cloudman S-91 or Melan-A cells inhibit cytotoxic T cell proliferation and are enriched for mRNA transcripts that target immune pathways, such as Ptpn11 and Dnmt3a [20]. While many prior studies focus on characterizing exosomal payloads or systemic response at a single point in time to exosomal delivery, a systematic analysis of how gene expression changes with time in response to delivery of an exosomal payload to a target cell remains unclear. To better understand how exosomes can regulate anti-tumor immunity, the goal of this study was to evaluate how the transcriptional landscape in cytotoxic T cells is changed with time following the delivery of a exosomal payload from tumor cells using a mouse melanoma model. In short, the transcriptional landscape reflected the delivery of an exosomal mRNA payload while exosomal miRNA appeared to have no effect on transcript abundance. In addition, exosomal payload elicited a dynamic non-linear response associated with a number of pathways, which was supported by results from a mitochondrial function assay.
Results and Discussion
B16F0 exosomes contain mRNA and miRNA
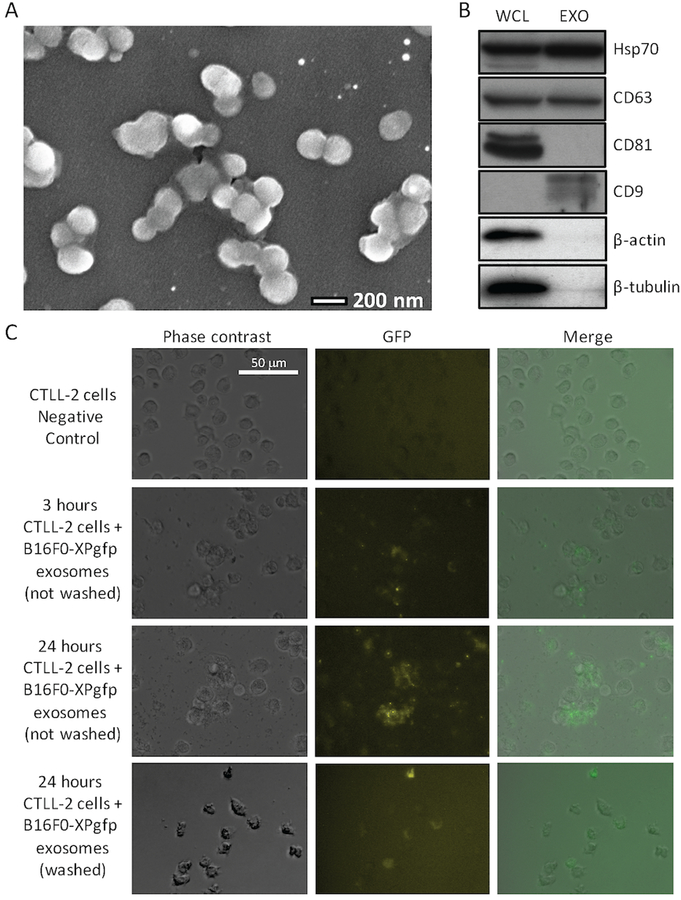
Extracellular vesicles were isolated using a staged centrifugation protocol from serum-free media conditioned by B16F0 cells. These extracellular vesicles were then characterized in terms of morphology using scanning electron microscopy and protein biomarker context (Fig. 1). Fresh extracellular vesicles exhibited a round morphology with a Feret diameter of 166 ± 15 nm (n = 26), which is consistent with prior studies of fresh exosomes [21, 20]. Probing for protein biomarkers associated with exosomes, we found that samples containing extracellular vesicles were positive for Hsp70, CD63, and CD9 and negative for CD81, β-actin and β-tubulin. Compared to B16F0 whole cell lysates, Hsp70 and CD63 were present in both samples while the remainder were not. The absence of CD81 and enrichment of CD9 in the extracellular vesicle samples has been previously reported for B16F0 exosomes [20]. The absence of β-actin and β-tubulin suggested that the extracellular vesicle sample does not contain cell fragments, such as vesicles released during cell death. These exosomes were also able to deliver GFP synthesized by B16F0 cells to CTLL2 cells (Fig. 1C). GFP was initially observed as bright punctae both in the media and within CTLL2 cells that became more dispersed with time within the CTLL2 cells. Functionally, 48 hour treatment with 0.2 mg/mL of B16F0 exosomes had no impact on the proliferation or viability of CTLL2 cells (data not shown). Collectively, the results suggest that the extracellular vesicles isolated from the B16F0 cell-conditioned media are exosomes and that these exosomes can deliver a biological payload to CTLL2 cells.
Figure 1: B16F0 release extracellular vesicles with uniform morphology that contain proteins commonly associated with exosomes and that can deliver a biological payload to CTLL2 cells.
(A) A SEM picture of extracellular vesicles isolated by ultracentrifugation of media conditioned for 24 hours by B16F0 cells (bar indicates 200 nm). (B) Western blot analysis of B16F0 whole cell lysates (WCL) and extracellular vesicles (EXO) for proteins commonly associated with exosomes, where 20 µg of protein was loaded in each lane. (C) B16F0 cells were transfected with a lentivirus-based XPack-GFP plasmid that targets GFP to exosomes. Untreated CTLL2 cells and CTLL2 co-cultured with exosomes isolated from B16F0-XPgfp cells for either 3 or 24 hours and imaged by fluorescence microscopy. After 24 hours, CTLL2 cells treated with B16F0-XPgfp exosomes were washed three times with DPBS and also imaged by fluorescence microscopy. Phase contrast and merged images are shown for comparison. The fluorescence intensity in GFP images has been colored using a black-yellow-blue color scale. Scale bar indicates 50 µm.
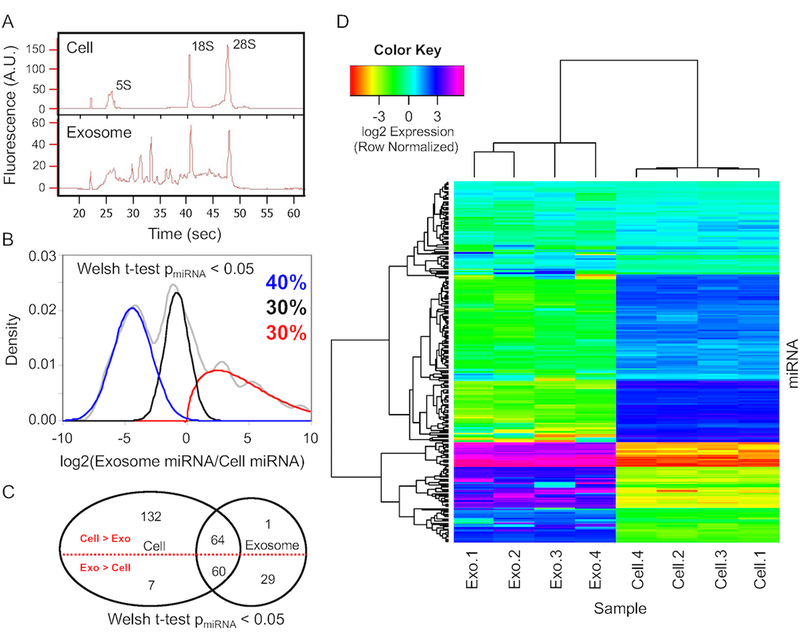
As exosomes can carry a complex payload of coding and non-coding RNA, we next characterized the RNA payload within B16F0 exosomes compared to the parental cells. A Bioanalyzer was first used to characterize the overall RNA quality (Fig. 2A). Bioanalyzer results comparing RNA in the parental cells versus exosome samples suggested that exosomes appear to contain full-length mRNA transcripts in addition to smaller miRNA fragments while ribosomal RNAs dominate the signature derived from the parental cells. The broad distribution of RNAs between the characteristic 5S and 18S ribosomal RNA peaks indicated that much of the content of the RNAs are between 120 to 1869 nucleotides long. The presence of characteristic 5S, 18S, and 28S rRNA peaks within the exosome samples was likely residual cellular debris attached to the outside of the exosomes as washing the exosome samples in PBS prior to analysis reduced these ribosomal RNA peaks (see Fig. 3A in [20]). Affymetrix microarrays were then used to quantify mRNA and miRNA within the exosome samples compared to parental B16F0 cells. Following isolation of mRNA from exosome and whole cell samples, exon-level mRNA microarrays were used to quantify the relative abundance of transcripts (see Fig. 3 in [20]). Similarly, the exosomal miRNA payload was characterized using an Affymetrix GeneChip miRNA 2.0 microarray (see Fig. 2). A Welch t-test was used to identify miRNAs that were present above background, where a cut-off p-value of 0.05 was used.
Figure 2: B16F0 exosomes contain miRNAs that are differentially packaged into exosomes relative to parental cells.
(A) The distribution of RNA isolated from parental B16F0 cells (top panel) and B16F0 exosomes (bottom panel) was quantified using microfluidic electrophoresis. (B) The overall distribution in exosome versus cellular abundance of miRNAs (gray curve) was deconvoluted into three normally distributed populations: miR-NAs enriched in cells (blue curve - 40% of total), miRNAs equally distributed between cell and exosomes (black curve - 30% of total), and miRNAs enriched in exosomes (red curve - 30% of total). (C) A Venn diagram summarizes the number of miRNAs that are present above background and are differentially expressed between cell and exosome samples. (D) Hierarchical clustering of miRNA detected above background (Welch t-test p-value < 0.05) from four B16F0 exosomes and four B16F0 cell samples using Affymetrix miRNA microarrays.
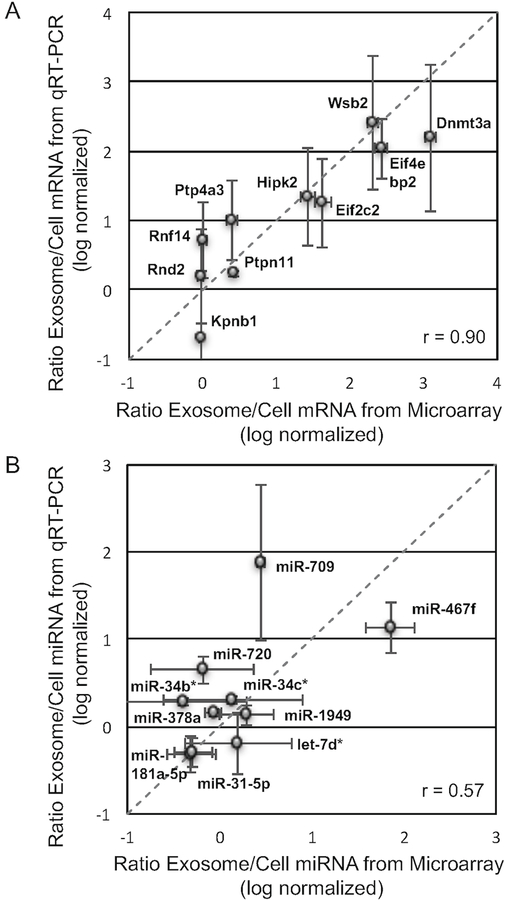
Figure 3: Relative abundance of mRNA and miRNA between B16F0 exosomes and cells were consistent between qRT-PCR and microarray analyses.
(A) As reported in [20], the abundance of ten transcripts (Kpnb1, Rnf14, Rnd2, Ptp4a3, Ptpn11, Eif2c2, Hipk2, Eif4ebp2, Dnmt3a, and Wsb2) in B16F0 exosomes versus B16F0 cells were quantified by quantitative RT-PCR (mean ± SD, n = 3) and compared against the relative abundance of mRNAs assayed by cDNA microarray. The linear relationship was quantified by a correlation coefficient of 0.90. (B) The abundance of ten miRNAs (miR-181a-5p, miR-31–5p, miR-378a, miR-34b*, miR-720, miR-34c*, miR-1949, let-7d*, miR-709, and miR-467f) in B16F0 exosomes versus B16F0 cells were quantified by quantitative RT-PCR (mean ± SD, n = 3) and compared against the relative abundance of miRNAs assayed by miRNA microarray. The linear relationship was quantified by a correlation coefficient of 0.57.
Similar to the distribution of mRNA between exosomes and parental cells, the distribution of miRNAs between exosomes and parental cells exhibited one of three different phenotypes: selectively enriched in exosomes, equally distributed between exosomes and parental cells, and more abundant in parental cells. To identify the fraction of the total number of miRNAs that are associated with each of these three phenotypes, we deconvoluted the cumulative distribution into three different subsets (Fig. 2B). Of the miRNAs present above background in either the exosome or parental cell samples, 30% of the miRNAs were more abundant in the exosomes, whereas 40% were more abundant in the samples obtained from the parental B16F0 cells. The remaining percentage of miRNAs were equally distributed between parental cell and exosomes, accounting for the last 30% of total recorded miRNA transcripts. A Venn diagram also summarizes the number of miRNAs detected above background, where 30 miRNAs were only detected in exosome samples, 139 miRNAs were only detected in parental cell samples, and 124 miRNAs were detected in both samples. Of all miRNAs detected above background, 96 were more abundant in the exosome samples and 197 miRNAs were either equally or more abundant in parental cell samples.
A heatmap visualized clustering between the samples and the miRNAs (Fig. 2D). Within this heatmap, clear division in the pattern of miRNA expression is evident between the samples such that all of the replicates cluster together. The abundance of miRNAs among the samples clusters similarly as the distributions shown in panel B. A cluster at the top of the heatmap represents those miRNAs that are equally distributed between parental cell and exosomes, a cluster in the middle of the heatmap represents those miRNAs with low abundance in the exosomes but higher abundance in the cell samples, a cluster at the bottom of the heatmap represents those miRNAs with high abundance in the exosomes and low abundance in the cell samples with a small set of miRNAs that are highly enriched in exosomes and nearly absent in the parental cells. This most highly enriched group included miRNAs: mmu-miR-711, mmu-miR-1187, and members of the miR-466 family (mmu-miR-466j, mmu-miR-466f-5p, mmu-miR-466f-3p, mmu-miR-466f, mmu-miR-466g, mmu-miR-466c-5p, and mmu-miR-466i-3p). While these miRNAs exhibited high exosomal enrichment, they were not the most abundant miRNAs in exosome samples. The twenty most abundant miRNAs and mRNAs found in the B16F0 exosome samples are listed in Table 1. Not surprising, the most abundant mRNA in exosomes was melanoma antigen (Mela). Interestingly, the most abundant miRNA, by a factor of 2.2 over the second most abundant miRNA, was mmu-miR-709, which has been reported to activate Wnt/β-catenin signaling by targeting GSK3β [22]. In comparing the results for probe sets that recognize the precursor hairpin versus mature miRNA, miRNAs in the exosomes were predominantly mature.
Table 1:
Twenty of the most highly abundant miRNAs and mRNAs in B16F0 exosomes.
| miRNAs | mRNAs | ||||
|---|---|---|---|---|---|
| Mature ID | Abundance (log2 (s.d.2)) | DABG* (p-value) | Gene Symbol | Abundance (log2 (s.d.2)) | DABG (p-value) |
| miR.709 | 13.951 (0.081) | 1.61E-12 | Mela | 13.989 (0.057) | 1.01E-11 |
| miR.2137 | 12.565 (0.121) | 3.36E-10 | Wsb2 | 13.869 (0.053) | 4.45E-12 |
| miR.2861 | 12.413 (0.057) | 7.82E-16 | Eif4ebp2 | 13.706 (0.041) | 5.50E-14 |
| miR.1195 | 12.221 (0.020) | 5.22E-101 | Fam168b | 13.419 (0.047) | 1.09E-12 |
| miR.762 | 12.079 (0.067) | 8.07E-14 | 0610007L01Rik | 13.359 (0.041) | 7.93E-14 |
| let.7c | 12.077 (0.035) | 2.59E-30 | Fth1 | 13.151 (0.026) | 6.54E-21 |
| let.7b | 12.011 (0.013) | 2.53E-292 | Tcf20 | 13.107 (0.062) | 3.41E-11 |
| miR.690 | 11.785 (0.091) | 2.28E-11 | Mlec | 13.042 (0.032) | 1.31E-16 |
| miR.1224 | 11.780 (0.021) | 2.87E-86 | Eef2 | 13.014 (0.034) | 1.27E-15 |
| miR.23a | 11.516 (0.132) | 1.11E-09 | Pabpc1 | 12.955 (0.028) | 7.00E-19 |
| let.7d | 11.262 (0.190) | 1.27E-08 | Cbx5 | 12.954 (0.052) | 5.89E-12 |
| miR.1894.5p | 11.250 (0.169) | 6.63E-09 | Scd2 | 12.859 (0.030) | 1.16E-17 |
| miR.24 | 11.150 (0.194) | 1.50E-08 | Ccnd2 | 12.825 (0.037) | 1.23E-14 |
| miR.17 | 11.124 (0.348) | 2.57E-07 | Cmtm4 | 12.750 (0.037) | 8.93E-15 |
| miR.1892 | 11.105 (0.207) | 2.17E-08 | Slc38a1 | 12.686 (0.024) | 8.10E-23 |
| miR.711 | 11.084 (0.111) | 2.90E-10 | Gnb1 | 12.652 (0.018) | 6.29E-36 |
| miR.466f.5p | 11.036 (0.091) | 3.17E-11 | Rcc2 | 12.641 (0.041) | 1.63E-13 |
| miR.705 | 11.021 (0.101) | 1.14E-10 | Mt2 | 12.639 (0.063) | 5.61E-11 |
| let.7a | 11.015 (0.397) | 4.73E-07 | Ptpn14 | 12.576 (0.058) | 2.44E-11 |
| miR.466f | 11.004 (0.258) | 6.93E-08 | Gnb2l1 | 12.552 (0.054) | 1.11E-11 |
Detected above background
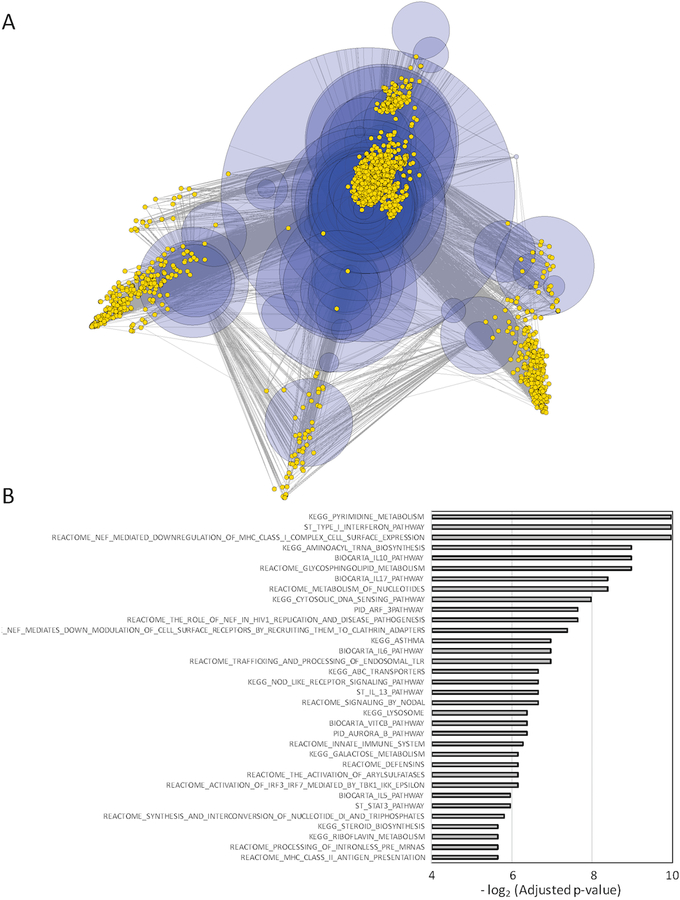
A subset of mRNAs that exhibited differential and similar abundance between exosome and whole cell samples were subject to semi-qualitative RT-PCR that confirmed that these transcripts were intact full-length open reading frames (see Fig. 4 in [20]) and subject to quantitative RT-PCR that confirmed the relative abundance predicted by the microarray results (Fig. 3A). A correlation coefficient of 0.90 between the abundance ratio observed by microarray compared to that observed by qRT-PCR suggests a strong positive correlation between these two measurements. Similarly, a subset of miRNAs present in both exosome and parental cell samples were subject to quantitative RT-PCR to confirm the relative abundance predicted by the microarray results (Fig. 3B). While the value was lower than for mRNA, a correlation coefficient of 0.57 still suggests a strong positive correlation between these two measurements of miRNA differential abundance. Interestingly, miRNAs in the exosomes overlap in their targeting of specific mRNA transcripts and are predicted to impact multiple pathways (Fig. 4 and Supplemental Table 1). Two of the top three most significantly enriched pathways were associated with cell-mediated immunity. These pathways were the Type I Interferon Pathway and NEF-mediated downregulation of MHC Class I complex cell surface expression. Targeted transcripts of the Type I Interferon Pathway included the Janus Kinases JAK1 and TYK2 while transcripts targeted in the NEF-mediated path-way included multiple components of the adaptor protein complex AP-1 (AP1G1, AP1M1, AP1M2, AP1S1, AP1S2). Collectively, the results suggest that B16F0 exosomes contain a payload of both coding and non-coding RNAs whose abundance does not reflect the abundance in the parental cell and can exert a concerted impact on the transcriptional landscape in recipient cells.
Figure 4: Exosomal miRNA overlap in targeting specific mRNA transcripts and collectively target multiple pathways.
(A) The targeting of specific mRNA transcripts (yellow nodes) by miRNAs present in B16 exosomes (blue nodes) is summarized by a multi-dimensionally scaled network diagram. The size of the miRNA nodes were scaled by relative abundance in exosomes. Nodes with less than two edges were pruned from the network to improve clarity. (B) Possible pathways targeted by exosomal miRNAs was determined by enrichment of miRNA to gene to pathway relationships. Using the miRNApath package, significance of pathway enrichment relative to random chance was assessed by random permutation where an adjusted p-value of less than 0.02 was considered significant.
Signature associated with mRNA but not miRNA exosomal payload is observed in cytotoxic T cell transcriptome
Given the importance of exosomes as a mode of intercellular communication and the emerging importance of tumor-derived exosomes in shaping the tumor microenvironment, we wanted to assess the impact of tumor-derived exosomes on the transcriptome of cytotoxic T lymphocytes, an important effector cell in controlling tumor growth. As a model system, we exposed CTLL2 T cells, an immortalized cytotoxic T cell line, to exosomes derived from B16F0 cells and monitored the transcriptome as a function of time using RNA sequencing. Specifically, mRNA isolated from the starting population of CTLL2 cells (0 hr) and from treated and untreated cells at 4 different time points (0.5, 2, 4, and 8 hour) were subject to RNA-Seq analysis. Using three biological replicates for each experimental condition, samples were randomly assigned to one of two HiSeq1500 batch runs. On average, 25.5 million raw reads (± 5.8 million reads) were generated for each sample and 22.5 million of these reads (± 4.7 million reads) were mappable to the host reference genomes. From the mappable reads, 2.3 million reads (± 0.6 million reads) were aligned to multiple positions in the reference genomes and were excluded. The resulting uniquely mapped reads, which averaged 19.5 million reads (± 4.0 with a range between 7.5 and 30.0 million reads), were retained for further analysis. In addition to controlling for batch effects, DESeq2 analysis of the uniquely mapped reads was used to identify differentially expressed (DE) transcripts [23], whereby we parsed the analysis into two comparisons. First, we identified transcripts that changed with time in the untreated samples. Second, we identified DE transcripts that were changed in exosome-treated samples at specific time points. These analyses are summarized in Fig. 5.
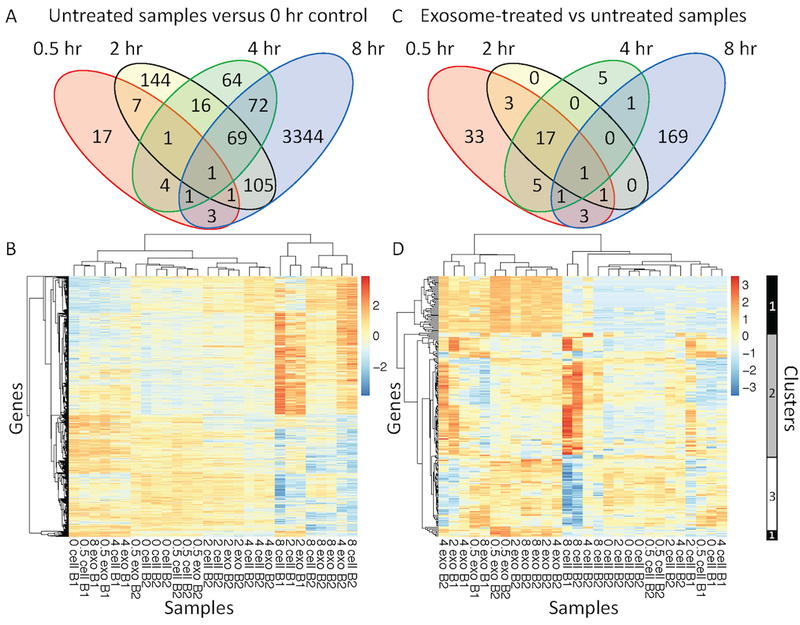
Figure 5: Exosome treatment initiated a different transcriptomic response in CTLL2 cells compared with the transcriptomic changes observed as a function of time.
Venn diagrams of the number of differentially expressed transcripts observed at different time points (A) in untreated CTLL2 cells compared to the 0 hour time point and (C) in exosome-treated versus untreated samples at each time point. Using p-values calculated using the DESeq2 package, a more stringent p-value cutoff was used at the 8 hour time point (p-value < 0.04 ) compared with the 0.5, 2, and 4 hour time points (p-value < 0.1). (B and D) Hierarchical clustering of collection of differentially expressed transcripts detected by RNA sequencing from 28 samples distributed across five time points (0, 0.5, 2, 4, and 8 hour), two experimental conditions (untreated versus exosome treated), and two RNA sequencing batches (B1 and B2). Heatmaps in panels B and D focus on the differentially expressed transcripts summarized in the Venn diagrams shown in panels A and C, respectively. Three clusters of dynamic transcript expression profiles are highlighted on the right margin of panel D. Cluster 1 corresponds to transcripts increased at all time points in exosome-treated samples. Cluster 2 corresponds to transcripts predominantly upregulated in untreated cells compared to exosome-treated cells at 8 hours. Cluster 3 corresponds to transcripts predominantly downregulated in untreated cells and upregulated in exosome-treated cells at 8 hours.
In untreated cells, the number of DE transcripts relative to the 0 hour sample generally increased with time (Fig. 5A). Even with reducing the p-value from 0.1 to 0.04, the 8 hour time point had greater than 10 times more DE transcripts than any other time point. In the exploratory data analysis, a reduced p-value was used so that the 8 hour time point did not swamp the information contained in the samples obtained at other time points. As illustrated by the heatmap (Fig. 5B), no clear trends emerged in the untreated cells, while a residual batch effect can be seen in clustering samples. In comparing exosome-treated to untreated samples at each time point, we also found that the number of DE transcripts was greatest at the 8 hour time point but by a lower margin than in the time-course analysis (Fig. 5C). Interestingly, there was no overlap in the set of DE transcripts identified in the time-course analysis versus the set of DE genes in exosome-treated samples. In addition, a set of 17 DE transcripts were shared across the 0.5, 2, and 4 hour time points, which corresponded to the majority of DE genes at 2 and 4 hours. DE genes identified at the 0.5, 2, and 4 hour times points were solely increased in exosome-treated samples with no genes exhibited a decrease following exosome treatment. In analyzing the set of DE genes across exosome-treated samples, samples tended to cluster based on time point rather than by batch. Overall, the number of DE genes is expected to increase with time as replating CTLL2 cells or treating with exosomes can elicit a primary response in gene expression that then propagates through genetic regulatory networks to elicit secondary changes in gene expression. To explore the biological significance of the clustering results, we focused on three distinct clusters of DE genes associated with exosome treatment, which are highlighted to the right of the heatmap in Fig. 5D. Cluster 1 is composed of transcripts that were increased within all exosome-treated samples. In contrast, transcripts within cluster 2 were upregulated at 8 hours in untreated CTLL2 cells compared to exosome-treated cells while cluster 3 includes transcripts that are downregulated at 8 hours in untreated samples relative to exosome-treated CTLL2 cells.
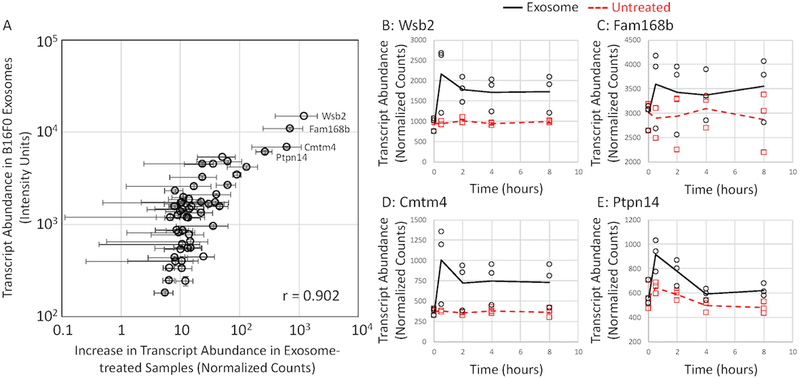
Given that genes associated with cluster 1 seemed to increase in abundance immediately upon exosome treatment, we compared the change in normalized transcript counts upon exosome treatment to mRNA abundance in B16 exosome samples assayed by Affymetrix microarray for genes observed by both of these platforms (Fig. 6). The comparison between the RNA-seq analysis of exosome-treated CTLL2 cells and the Affymetrix microarray analysis of exosomal mRNA payload revealed a linear trend, whereby a correlation coefficient of 0.90 suggested a strong positive correlation. This trend becomes more defined with highly abundant transcripts while low abundant transcripts seemed to deviate from linearity. Such a trend may be due to observational bias as microarray data can have a broader dynamic range while sequencing data is biased towards highly abundant transcripts. Low abundant transcripts that trend towards the mean may have too low of a frequency to be observed at the stated sequencing depth. To illustrate the overall trend for individual genes associated with cluster 1, the abundance in exosome-treated and untreated samples of four of the most highly abundant transcripts are plotted over time (Fig. 6B–E). Noticeable differences can be seen across all time points when examining all four genes. Greatest differences among all of the plots is visible at the 0.5 hour time point, indicating these genes are all initially upregulated following the exosome treatment and remain upregulated at the other time points. We also noted that not all highly abundant exosomal mRNAs that are listed in Table 1, such as Eif4ebp2, Fth1, Tcf20, and Mlec, were associated with cluster 1. High background levels of expression in CTLL2 cells of these transcripts coupled with the technical noise associated with RNA-seq analysis rendered the difference between untreated and exosome-treated CTLLs as not significant. Mela was an exception as it was not observed in any of the RNA-seq samples.
Figure 6: An increase in transcript abundance observed by RNAseq following exosome treatment correlates with mRNA payload contained within B16F0 exosomes as assayed by cDNA microarray.
(A) For transcripts detected above background using cDNA microarray, an increase in transcript abundance in exosome-treated cells over untreated cells after 0.5 hours (mean ± SD, n = 3) was plotted against the average fluorescence intensity for the corresponding transcript probeset assayed by cDNA microarray (mean ± SD, n = 4). A linear relationship was quantified by a correlation coefficient of 0.902. While highlighted in panel A, RNAseq results for four of the most abundance transcripts across experimental conditions (exosome treated: circles and black solid lines, untreated: squares and red dashed lines) are shown separately for Wsb2 (B), Fam168b (C), Cmtm4 (D) and Ptpn14 (E).
Of note, cluster 1 is associated with transcripts that increase with exosome-treated samples almost immediately. In contrast, clusters 2 and 3 develop over the course of 8 hours. We also noted that there were no transcripts that decrease in direct response to exosome treatment. This lack of a response has implications for the miRNA exosomal payload. There are three main functions associated with miRNA [24]. First, miRNAs may bind to the 3’ utr complementrary sequence of their target mRNA, thus inducing gene silencing and prohibiting the translation of proteins from that segment. Second, miRNAs may promote translational repression by cleaving and degrading mRNA sequences. Third, miRNAs have been shown to deadenylate and degrade mRNAs upon attachment. While all three functions of miRNA reduce protein abundance by interfering with translation, only the second two functions alter mRNA abundance. Given that there were no transcripts that were significantly decreased at the 0.5 and 2 hour time points, we concluded that transcript downregulation through miRNA targeting the 3’ utr for degradation is not a primary effect of exosome treatment. We note however that a decrease in mRNA target abundance by miRNA becomes more apparent when the miRNA is over-expressed, while exosomal delivery is closer to physiology.
Long non-coding RNA (lncRNA) can also regulate gene expression in cells [25, 26]. As the Affymetrix microarray platform does not assay lncRNAs, we used the RNA-seq data to identify lncRNA that were upregulated in CTLL2 cells upon stimulation with B16F0 exosomes. Given the concordance between mRNAs assayed in exosomes and their corresponding rapid increase in CTLL2 cells, we assumed that the rapid increase in lncRNA observed in exosome-treated CTLL2 cells originated from the B16 exosomes. There were two lncRNAs that were significantly increased in exosome-treated CTLL2 cells: Gm26809 and Gm26982. Gm26809 (ENSMUSG00000097815) was upregulated at all exosome treated time points (0.5hr untreated: 53 ± 11 counts, exosome-treated: 94 ± 14 counts, and p-value < 0.025). This is a lncRNA that targets LSM8, which was observed to be decreased at the 8 hour time point: (untreated: 988 ± 137 counts, exosome-treated: 719 ± 49, and p-value < 0.006). Gm26982 is an antisense lncRNA (ENSMUSG00000097994) that was significantly increased in exosome-treated samples at 0.5 hours (untreated: 57 ± 4 counts, exosome-treated: 93 ± 19 counts, and p-value < 0.05). Gm26982 overlaps with the protein-coding locus of ghrelin (Ghrl) on the opposite strand, although there was no significant difference in ghrelin transcript abundance associated with exosome treatment at any time point.
Exosome-elicited transcriptomic signature in cytotoxic T cells becomes apparent after 8 hours
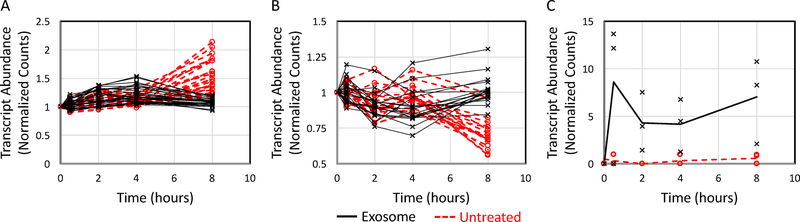
As the changes in gene expression associated with cluster 1 appears to be a direct result of exosome treatment, DE genes associated with clusters 2 and 3 characterize the secondary transcriptomic response to delivery of the exosomal payload, as both take hours to develop. Comparisons of overall gene expression of clusters 2 and 3 as a function of time between exosome-treated and untreated samples are illustrated by Fig. 7A–B, respectively. For genes associated with cluster 2, there was a significant increase in transcript abundance in untreated cells at the 8 hr time point while transcripts exosome-treated cells seemed to peak earlier at 4 hours and were not increased at 8 hours. The opposite effect was observed for cluster 3, whereby genes in untreated cells within cluster 3 undergo a reduction in transcript abundance at the 8 hour time point while these same genes in exosome-treated cells tended to decrease at the 2 and 4 hour time points and be upregulated at the 8 hour time point relative to untreated cells. Using the genes associated with either cluster 2 or 3, we performed gene enrichment analysis to identify cellular functions and pathways affected by the exosomal payload. As listed in Table 2, DE genes associated with clusters 2 and 3 correspond to pathways essential to cellular function and phenotypic maintenance.
Figure 7: Transcripts of genes associated with functional annotation of clusters 2 and 3 displayed different dynamic profiles.
Average transcript abundance for exosome-treated (solid black lines) and untreated (dotted red lines) for DE genes associated with the functional annotation of cluster 2 (panel A) and cluster 3 (panel B) are shown as a function of time. Transcript counts were normalized to the 0 hour time points. (C) Normalized counts for E2F transcription factor 2 (E2f2) across experimental conditions (exosome treated: X and black solid lines, untreated: circles and red dashed lines) and time.
Table 2:
Gene lists enriched in dynamic transcription clusters 2 and 3.
| Annotation Term | Identifier | Adjusted P-value | Enriched Genes |
|---|---|---|---|
| Cluster 2 | |||
| respiratory electron transport chain | GO:0022904 | 4.94E-06 | Atp5b Ndufb6 Ndufb11 Atp5e Ndufs4 Etfb Sdhd Ndufv2 |
| electron transport chain | GO:0022900 | 4.94E-06 | Atp5b Ndufb6 Ndufb11 Atp5e Ndufs4 Etfb Sdhd Ndufv2 |
| mitochondrial transport | GO:0006839 | 5.37E-05 | Chchd4 Atp5b Atp5e Timm22 Mrpl12 Bid Tomm5 |
| energy coupled proton transport, down electro-chemical gradient | GO:0015985 | 5.83E-03 | Atp5b Atp5e Atp5g3 |
| ATP synthesis coupled proton transport | GO:0015986 | 5.83E-03 | Atp5b Atp5e Atp5g3 |
| mitochondrial membrane part | GO:0044455 | 9.60E-08 | Atp5b Ndufb6 Ndufb11 Atp5e Ndufs4 Atp5g3 Sdhd Ndufv2 Bid Tomm5 |
| mitochondrion | GO:0005739 | 2.74E-06 | Oxnad1 Uqcc2 Bola1 Mrpl27 Mrpl14 Etfb Sdhd Mrpl12 Chchd4 Nt5c Mrpl41 Atp5b Fdx1 Ndufs4 Suclg1 Ndufv2 Bid Tomm5 Ccdc51 Tmem14c |
| mitochondrial membrane | GO:0031966 | 3.82E-05 | Atp5b Ndufb6 Ndufb11 Uqcc2 Ndufs4 Suclg1 Timm22 Sdhd Ndufv2 Bid Mrpl12 |
| mitochondrial matrix | GO:0005759 | 3.82E-05 | Atp5b Dtymk Uqcc2 Atp5e Fdx1 Suclg1 Etfb Fdx1l |
| organellar large ribosomal subunit | GO:0000315 | 3.33E-04 | Mrpl41 Mrpl27 Mrpl12 |
| Cluster 3 | |||
| positive regulation of gene expression, epigenetic | GO:0045815 | 8.87E-03 | Kmt2b Padi2 Arid1a |
| chromatin modification | GO:0016568 | 2.15E-03 | Rere Kmt2d Crebbp Kdm2b Sap130 Tet3 Kmt2b Padi2 Arid1a Bcorl1 |
| histone H3-K4 methylation | GO:0051568 | 1.21E-02 | Kmt2d Kmt2b Tet3 |
| histone modification | GO:0016570 | 8.87E-03 | Kmt2d Crebbp Kdm2b Kmt2b Sap130 Tet3 Padi2 |
| covalent chromatin modification | GO:0016569 | 8.87E-03 | Kmt2d Crebbp Kdm2b Kmt2b Sap130 Tet3 Padi2 |
| E2f2 LOF in MMTV-myc mouse, down | GDS4094 | 4.24E-03 | Ncor2 Zfhx3 Crebbp Creb3l2 Wasl Luc7l2 |
| Delta-Notch Signaling Pathway Mus musculus | WP265 | 8.97E-03 | Ncor2 Notch1 Furin Zfpm1 |
| Notch Signaling Pathway Mus musculus | WP29 | 1.01E-02 | Ncor2 Crebbp Notch1 |
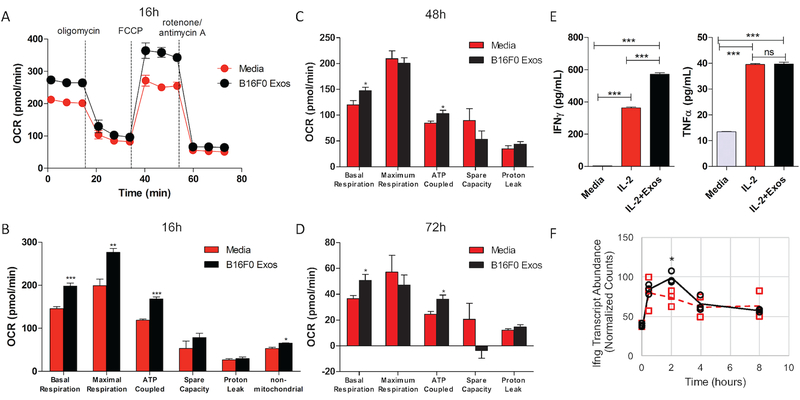
Specifically, cluster 2 is enriched with transcripts related to cellular energetics and mitochondrial function. The response of these genes with time was non-linear with an increase in transcript abundance in exosome-treated CTLL2 cells at 4 hours relative to the 0 hour time point and to untreated cells at the 4 hour time point (p-value < 0.001). In exosome-treated CTLL2 cells, the abundance of these transcripts returned to baseline by the 8 hour time point. In contrast, untreated CTLL2 cells increased expression of these genes over time suggesting that CTLL2 cells would increase their mitochondrial activity; yet, the timing between changes in gene expression and functional changes is unclear. Given the values at 8 hours, one might conclude that untreated cells would have increased mitochondrial activity as evidenced by annotation terms associated with this enriched pathway as many directly relate to ATP production. To validate this functional annotation results, we assessed mitochondrial respiration in CTLL2 cells pre-treated with B16F0 exosomes for 16 hours and in untreated CTLL2 cells using a Seahorse XFe96 Analyzer (Fig. 8A–D). Following the sequential addition of four chemical inhibitors, the measured oxygen consumption rate was analyzed to obtain the basal respiration, maximal respiration, ATP-coupled respiration, spare capacity, proton leak, and non-mitochondrial respiration for these two experimental groups. Compared to untreated CTLL2 cells, basal respiration, maximal respiration, and ATP-coupled respiration were significantly increased upon exosome treatment. Non-mitochondrial respiration was also slightly increased in exosome-treated cells. In contrast, spare capacity and proton leak were not significantly different between groups. Generally, these changes in basal and ATP-coupled respiration persisted at 48 and 72 hours, although the increase in maximal respiration diminished with time. Similar results were obtained when we increased the efficiency of exosomal payload delivery using the EV-Entry system. These functional results suggest that the observed increase in gene expression in exosome-treated CTLL2 cells at 4 hours translates to an increase in mitochondrial respiration.
Figure 8: Exosomes increased aspects of mitochondrial respiration and IFN-γ production in CTLL2 cells.
(A) Oxygen consumption rate (OCR) in CTLL2 cells treated with culture media (red circles) or media containing B16F0 exosomes (black circles) was measured after 16 hour culture while the indicated chemical inhibitors of the respiratory chain were sequentially added. As described in the methods, metrics associated with mitochondrial respiration were inferred from the trace of the OCR after 16 hours (B), 48 hours (C) and 72 hours (D). Significance associated with the difference in basal respiration, maximal respiration, ATP-coupled respiration, non-mitochondrial respiration, space capacity and proton leak in exosome treated (black bars) compared to untreated cells (red bars) were assessed. (E) IFN-γ and TNF-α were assayed in CTLL2 conditioned media by cytometric bead array following the indicated treatments. (F) RNA-seq results for IFN-γ mRNA are shown for comparison. Results representative of two independent experiments that each contained at least four biological replicates, where ***, **, and * correspond to p-values calculated using an unpaired t-test of < 0.001, < 0.01, and < 0.05, respectively.
Cluster 3 genes are related to the regulation of gene expression and DNA remodeling, including histone modification, histone methylation, and chromatin modification. Covalent modifications to both histones and DNA regulate transcription patterns within cells through mechanisms that alter the state of the nucleosome and influence the ability of proteins to access DNA. Such modifications can silence genes. Alternatively, a decrease in expression of genes that regulate the nucleosome suggests that the epigenetic state of DNA is less regulated with time in untreated cells and that some of the genes may no longer be effectively silenced. In contrast, epigenetic modification of gene expression seems to increase in exosome-treated cells upon prolonged tissue culture. In addition, a significant gene signature associated with cluster 3 is the down-regulation of genes, including Ncor2 and Crebbp that are shared with the Notch signaling pathway, upon the loss-of-function of the transcription factor E2f2. Instead of the loss-of-function, transcripts for E2f2 were observed to be significantly increased upon exosomal treatment (Fig. 7C), which suggests that the exosomal payload activated the Notch pathway in CTLL2 cells.
In contrast to intrinsic benefits to malignant cells [27–29], the impact on oncogenesis of activating Notch signaling in cytotoxic T cells by tumor cells is less clear. One body of literature suggests that activating Notch signaling in cytotoxic T cells enhances anti-tumor cytotoxicity. For instance, activated cytotoxic T cells lacking both Notch-1 and Notch-2 receptors have a reduced proliferation and impaired production of IFN-γ, TNF-α and granzyme B [30, 31]. By activating Notch through transgenic expression of the intracellular domain of Notch-1, antigen-specific cytotoxic T cells resist the immunosuppressive effect of tumor-induced MDSC and achieve higher reduction of 3LL-OVA tumor growth [30]. Notch signaling is also essential for differentiating short-lived effector cytotoxic T cells but is dispensable for generating memory precursor cells [31, 32]. This body of literature implies that an increase in Notch signaling would increase production of IFN-γ and TNF. Functionally, we observed that exosome treatment increased IFN-γ production while TNF-α was not increased over stimulating with IL-2 alone (Fig. 8E–F). In addition, CTLL2 cells did not produce IL-6, IL-10, IL-12p70, or MCP-1 under the conditions tested. Similar results were also obtained when we increased the efficiency of exosomal payload delivery using the EV-Entry system.
While most of these studies blocked Notch receptors or genetically modified their expression in cytotoxic T cells, the specific immune response depends on whether Notch signaling is triggered by either Delta-like or Jagged ligands. Interestingly, delivery of anti-Jagged1 antibody or Delta1-Fc fusion protein exacerbates experimental autoimmune encephalomyelitis in mice, whereas anti-Delta1 antibody or Jagged1-Fc fusion protein ameliorate disease progression [33]. These opposing results were attributed to differential regulation of T helper cells. Jagged1-Fc increases IL-10 producing T helper cells and reduces Th1 polarization, while Delta1-Fc has the opposite effect [33]. In the context of antigen presentation, ectopic expression of Delta1 or Delta4 in APC promotes Th1 differentiation while Jagged1 expression polarizes towards Th2 [34, 35]. In vivo, injecting a soluble Jagged1-encoding plasmid reduces the disease severity in an experimental arthritis model through the inhibition of cytotoxic T cell proliferation and effector functions [36]. Therefore, a potential increase of Notch signal activation in cytotoxic T cells by tumor exosomes could lead to activation or suppression of this population depending on subtleties of the transcriptional response, as similarly elicited by the Jagged and Delta-like ligands. While an increase in IFN-γ production and the absence of IL-10 are suggestive, finer resolution of the specific transcriptional response would be required to identify the specific impact on cytotoxic T cells by Notch signaling activation.
In summary, B16F0 exosomes seem to modify the epigenetic landscape and mitochondrial respiration within cytotoxic T cells. In vivo, metabolic changes in tumor infiltrating lmyphocytes reflect the collective effect of metabolic substrate deprivation, oxygen deprivation, and changes induced by tumor-immune cell crosstalk via a variety of mechanisms, including exosome transfer. As these factors are confounded in vivo, we have parsed the contribution of tumor-derived exosomes on immune cell function from metabolic changes elicited in cytotoxic T cells by the tumor microenvironment by delivering tumor-derived exosomes to cytotoxic T lymphocyte in vitro. While additional experiments may help clarify the longer-term effects of exosome transfer on anti-tumor immunity, these results suggest that B16 tumors produce exosomes that can alter mitochondrial respiration of tumor-infiltrating cytotoxic T lymphocytes independent of substrate and oxygen deprivation. Parsing how these mechanisms help condition the tumor microenvironment is the first step in developing therapeutic strategies that can enhance the functional persistence and efficacy of tumor-infiltrating cytotoxic T lymphocytes to control and eliminate malignancy.
Materials and Methods
Reagents and cell line culture
Cytokines, drugs, kits, and pharmacological inhibitors were obtained from commercial sources and used according to the suppliers’ recommendations unless otherwise indicated. The mouse melanoma cell line, B16F0, and a cytotoxic T lymphocyte cell line, CTLL2, were acquired from American Type Culture Collection. Base media for CTLL2 cells included RPMI 1640 (Cellgro/Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS-HI, Hyclone), 2.5% Glucose, 0.15% sodium bicarbonate, 10 mM HEPES, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin and 100 µg/ml streptomycin (Pen/Strep, Gibco), and 0.4% beta-mercaptoethanol. Complete media for CTLL2 cells comprised base media supplemented with10% T-STIM without ConA (Corning). B16F0 complete media was DMEM (Cellgro/Corning) with 10% FBS-HI, Pen/Strep, and 2 mM L-glutamine (Sigma). Extra-cellular vesicle (EV)-cleared media was prepared by removing EVs from the FBS-HI via ultracentrifugation, as described in the next paragraph, prior to making complete media.
Exosome isolation, electron microscopy (EM) imaging and Western blot analysis
Fresh extracellular vesicles were isolated from serum-free media conditioned by B16F0 cells for 24 hours and imaged with scanning electron microscopy (SEM) as described previously [18, 21]. Briefly, EVs were isolated from cell-conditioned media by differential centrifugation as follows: 300xg for 10 minutes to remove cells, 2,600xg for 10 minutes to remove residual cells and debris, 10,000xg for 60 minutes to remove microvesicles, and 100,000xg for 2 hours to collect nano-scaled vesicles in pellets. The resulting pellet was resuspended, washed once in DPBS, and re-pelleted at 100,000xg for 2 hours. Once isolated, nano-scaled vesicles were resuspended in DPBS and kept on ice. EVs were then imaged using SEM. The abundance of proteins contained in B16F0 EVs were compared against B16F0 whole cell lysate by Western blot analysis using methods described previously [20]. Specifically, membranes were probed with rabbit anti-CD9, CD63, CD81, and Hsp70 antibodies (System Biosciences, Mountain View, CA), and mouse anti-β-actin and β-tubulin antibodies (Santa Cruz Biotechnology, Dallas, Texas). Proteins were transferred to Bio Trace PVDF membrane (PALL Life Sciences, Pensacola, FL) and detected using Pierce ECL Western Blotting Substrate (Life Technologies).
Cell stimulation with exosomes and RNA analysis
As described in [20], GFP was delivered to CTLL2 cells using fresh exosomes isolated aseptically from B16F0-XPgfp cells, which were created with a lentivirus encoding an N-terminal XPack-GFP fusion protein by following the manufacturer’s instructions (XPAK530PA-1, System Biosciences, Inc., Mountain View, CA). CTLL2 cells were co-cultured with B16F0-XPgfp exosomes for the indicated times at a concentration of 1.0 mg/ml, and imaged using a Zeiss Axiovert 40 CFL fluorescent microscope using a standard GFP filter set. The efficiency of exosome payload delivery was increased using the EV-Entry System (System Biosciences, Inc., Mountain View, CA). After 24 hours, treated CTLL2 cells were also washed three times with DPBS and imaged. Total RNA was isolated from B16F0 exosomes and cells by TRIzol reagent extraction (Thermo Fisher Scientific) or RNAeasy (Qiagen), quantified using Nanodrop, and analyzed by on-chip-electrophoresis using the Agilent Bioanalyzer 2100. Message RNA isolated from B16F0 exosomes and cells were characterized using Mouse GeneChip 1.0 ST Exon Arrays (Affymetrix), as described previously [20]. For miRNA analysis, four hundred nanograms of each miRNA sample was biotin labeled using the Genisphere Flash-Tag HSR Kit according to the manufacturer’s instructions. The labeling of RNA was confirmed by Enzyme Linked Oligosorbent Assay according Genisphere FlashTag HSR protocol. The 21.5 µl of biotin-labeled RNA with added hybridization controls was hybridized to the GeneChip miRNA 2.0 Arrays (Affymetrix) at 48C and 60 rpm for 16 hours in GeneChip Hybridization Oven 640 (Affymetrix). GeneChip miRNA 2.0 Arrays were stained using FS 450 0003 protocol in Affymetrix GeneChip Fluidics Station 450. Briefly, biotin-labeled RNA was reacted using washes with a solution containing a streptavidin-phycoerythrin complex, with an intermediate treatment of biotin-labeled anti-streptadvidin antibody to amplify the signal. Phycoerythrin labeling was detected within the Affymetrix GeneChip Scanner 3000 7G plus using 532 nm light and detected by a photomultiplier tube. A miRNA QC Tool software (Affymetrix) was used to check quality controls of hybridized chips. All chips that passed quality controls were RMA normalized using miRNA QC Tool. Microarray data are accessible through GEO SuperSeries accession number GSE102951.
For RNA-seq analyses, CTLL2 cells were grown in complete media for 24 hrs, and then stimulated with fresh B16F0 exosomes resuspended in PBS, to a final exosome concentration of 0.2 mg/ml. Following RNA isolation by TRIzol extraction and Nanodrop quantification, RNA quality and integrity was determined using the Agilent Bioanalyzer 2100. Only RNA with RIN values >8.0 were used for RNA-Seq library preparation. RNA-Seq libraries were built with Illumina’s TruSeq RNA Kit V2 (RS-122–2001) as per manufacturer’s protocol. All libraries were indexed in such a way as to ensure only one index/lane on the HiSeq. Finished libraries were quality checked with a High Sensitivity DNA chip on the Bioanalyzer to determine average size and quantified on a Qubit with high sensitivity DNA reagents. The libraries were then sent to the Marshall University Genomics Core Facility where they were pooled in equimolar ratios and run on three lanes of an Illumina HiSeq1500 in a 2 × 50 base paired end design yielding a minimum of 49.1 million reads per sample. Samples included three independent biological replicates for each condition and four independent biological replicates for the 0 hr time point. Demultiplexing of samples and conversion from Illumina’s bcl format to standard fastq format was performed using CASAVA 1.8.4 (Illumina). The average transcriptomic alignment percentage was 86.0 ± 1.0 %. RNA-seq data are also accessible through GEO SuperSeries accession number GSE 102951.
Quantitative reverse-transcription PCR (q-RT-PCR) was used to validate the mRNA and miRNA microarray results. The results for mRNA analysis are described previously [20], where proprietary primer mixtures were purchased from Qiagen for the corresponding genes: Kpnb1 (QT00153419), Rnf14 (QT00157241), Rnd2 (QT00314216), Ptp4a3 (QT00138243), Eif2c2 (QT01757833), Hipk2 (QT00197890), Eif4ebp2 (QT00144606), Dnmt3a (QT00106519), Wsb2 (QT01747739). For miRNA validation, RNA samples were purified using an RNeasy mini kit (Qiagen) and reversely transcribed with TaqMan MicroRNA Reverse Transcription kit (Invitrogen 4366596). Quantitative PCR was carried out on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using Taqman Universal Master Mix II with UNG (Invitrogen 4440038). TaqMan Primer mixtures were purchased from Invitrogen for the indicated miRNA targets. Data was analyzed using the delta/delta CT method and the Sequence Detector Software version 2.2 (Applied Biosystems).
Mitochondrial respiration and other functional assay
Oxygen consumption rate (OCR) was measured at 37°C using a Seahorse XFe96 Analyzer (Agilent, Santa Clara, CA) according to the manufacturer’s instructions. Briefly, CTLL2 cells (100,000/well) were seeded into Seahorse Bioscience XFe96 cell culture plates previously coated with Cell-Tak (Corning, Inc., Corning, NY). Fresh B16F0 exosomes were added to a final concentration of 0.2 mg/mL. After overnight culture in the 37°C humidified incubator with 5% CO2, the media was changed to Mito-Stress Test Assay Media, which is DMEM containing 2 mM GlutaMax, 1 mM sodium pyruvate, and 25 mM glucose (media is un-buffered, pH 7.4), and incubated at 37oC without CO2 1 hr prior to the start of the extracellular flux assay. Oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), rotenone and antimycin A (all from Sigma, St. Louis, MO) of 10x compound dilutions were prepared for the assay as chemical inhibitors, and loaded into the assigned ports. A sensor cartridge was calibrated 24 hr prior to the assay, and following calibration, the cell plate was placed in the Bioanalyzer and the Mito-Stress Test assay protocol was completed on the samples. By adding the chemical inhibitors sequentially, this protocol determines basal respiration, ATP production, maximal respiration, spare respiratory capacity, proton leak, and non-mitochondrial respiration. Statistical analysis was done with unpaired t-test using GraphPad Prism 5 (GraphPad Software, San Diego, CA).
Cytokines produced by CTLL2 cells were measured in culture supernatants using a Cytometric Bead Array Mouse Inflammation kit and a BD LSRFortessa flow cytometer (BD Biosciences, Sparks, MD), according to the manufacturer’s instructions. Viable cell numbers were quantified using an ATPlite assay (PerkinElmer, Shelton, CT), performed according to the manufacturer’s instructions. For these assays, CTLL2 cells (20,000/well) were cultured for 48h in 100 µL of base CTLL2 media supplemented with 10 U/mL of recombinant murine IL-2 (eBioscience, ThermoFisher Scientific). Fresh B16F0-derived exosomes were added to a final concentration of 0.2 mg/mL. Conditioned media was then saved for cytometric bead analysis. For the ATPlite assay, the cells were treated with 50 µL/well of mammalian cell lysis solution, followed by 5 min. shaking. The substrate solution was added (50 µL/well), the plate was shaked another 5 min., and incubated 10 min. at room temperature protected from light. Finally, the luminescence was measured using a modulus microplate multimode reader (Turner BioSystems). CTLL2 cells cultured in base media supplemented with IL-2 but in the absence of B16F0 exosomes, as well as CTLL2 cells maintained in base media free of IL-2 and tumor exosomes, were included as controls. Statistically significant differences were detected with one way ANOVA and Tukey’s multiple comparison test using GraphPad Prism 5 (GraphPad Software).
Computational transcriptome analysis and statistics
Using the WVU Spruce Knob High-Performance Computing (HPC) cluster, fastq-formatted files were combined based on both lane designation and forward/reverse orientation. Quality control analysis was performed using the FastQC program to determine that trimming was not necessary prior to completing further downstream processing of the files. The Mus musculus FASTA file and a corresponding gene set annotation (GTF) file were downloaded (GRCm38.p2 from ensemble.org) to compile a genome index using STAR Aligner software (v2.4.2). Genome annotation files were comprehensive in that they included mappings for both exon and intron segments for the full selected genome. RNA sequence reads were then aligned to the index with STAR Aligner tools, converted to .bam files, sorted, and indexed. To determine the number of reads mapped to each genomic position or exon, the featureCounts program (v1.5.0) within the Subread package was used.
The remaining portion of the analysis was performed using R software platform (v3.3.2) along with Bioconductor packages. Using featureCounts files, the DESeq2 package [23] was used to determine the top differentially expressed genes by comparing the exosome-treated versus untreated samples at each time point and untreated samples at each time point relative to the 0 hour time point. A p-value adjusted for multiple hypothesis testing of less than 0.1 at the 0.5, 2, and 4 hour time points and of less than 0.04 at the 8 hour time point was considered significant. Transcript abundance was reported as variance stabilized normalized counts, whereby summary statistics were generated by averaging across all samples and standard deviation was used to represent uncertainty. Functional annotation of the RNAseq data was performed on the temporal clusters of differentially expressed genes associated with exosome-treated samples using the Enrichr algorithm [37, 38]. Additional microarray data containing both mRNA and miRNA samples was analyzed using R software. For microarray analyses, mRNA and miRNA abundance was estimated based on the average expression of all core probesets for a target measured by Affymetrix microarray. The probability of a mRNA/miRNA being detected above background (DABG) by random chance was estimated using the negative control probesets and a Welch’s t-test, where the degrees of freedom were calculated using the Welch-Satterthwaite equation. An adjusted p-value of less than 0.05 was considered as statistically significant. Functional annotation of exosomal miRNAs was determined using a multi-step approach in R. First, results from the Affymetrix GeneChip miRNA 2.0 microarrays were filtered based on the expression of Mus musculus miRNAs in exosomal samples detected above background. Next, the miR-NAtap package (v1.10.0) was used to relate specific miRNA with targeted mRNA transcripts based on aggregate predictions from at least 3 databases, which include DIANA, Miranda, PicTar, TargetScan, and miRDB [39]. Gene sets associated with canonical pathways was obtained from the Molecular Signatures Database v6.0 at the Broad Institute [40]. Finally, the miRNApath package (v1.36.0) was used to identify enriched pathways associated with the miRNA expression data, where the number of permutations was set to 1000 [41].
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation (CAREER 1053490 to D.J.K.) and the National Cancer Institute (NCI) (R15CA123123 and R01CA193473 to D.J.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI, the National Institutes of Health, or the National Science Foundation. CLB, CBH, AF, WD, SLR, and DJK declare that they have no competing interests. The authors also thank Aniello Infante for assistance with bioinformatic analyses, Kathleen Brundage for assistance with cytometric bead array analysis, the WVU and Marshall University Genomics core, and the Super Computing System (Spruce Knob) at WVU, which is funded in part by the National Science Foundation EPSCoR Research Infrastructure Improvement Cooperative Agreement #1003907, the state of West Virginia (WVEPSCoR via the Higher Education Policy Commission) and WVU. The cytometric bead analysis used the WVU Flow Cytometry & Single Cell core, which was supported by the National Institutes of Health (GM103488/RR032138, GM104942, GM103434, and OD016165).
Abbreviations:
- APC
antigen presenting cells
- ATP
adenosine triphosphate
- DABG
detected above background
- DE
differentially expressed
- DPBS
Dulbecco’s phosphate buffered saline
- EV
extracellular vesicle
- FBS-HI
fetal bovine serum-heat inactivated
- FCCP
carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone
- GEO
gene expression omnibus
- GFP
green fluorescent protein
- HEPES
4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid
- IL-2
Interleukin-2
- IL-6
Interleukin-6
- IL-10
Interleukin-10
- IL-12
Interleukin-12
- iNOS
inducible nitric oxide synthase
- lncRNA
long non-coding RNA
- MCP-1
monocyte chemotactic protein 1
- MDSC
myeloid-derived suppressor cells
- NADPH
nicotinamide adenine dinucleotide phosphate
- OCR
oxygen consumption rate
- PVDF
polyvinylidene fluoride
- QC
quality control
- RIN
RNA integrity number
- RT-PCR
reverse transcriptase polymerase chain reaction
- SEM
scanning electron microscopy
- TAM
tumor associated macrophages
- TAN
tumor associated neutrophils
- TGF-β
tumor growth factor beta
- TNF
tumor necrosis factor
- Tregs
regulatory T cells
Footnotes
Database: gene expression data are available in the GEO database under the accession SuperSeries number GSE102951.
Supporting Information
• Supplemental Table 1. The miRNA-Gene-Pathway enrichment results determined using the miRNApath package in R based on exosomal miRNAs. Pathways with an adjusted p-value < 0.02 are shown.
References
- [1].Vesely MD, Kershaw MH, Schreiber RD, and Smyth MJ. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol, 29:235–271, 2011. [DOI] [PubMed] [Google Scholar]
- [2].Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, Delgado A, Correa P, Brayer J, Sotomayor EM, Antonia S, Ochoa JB, and Ochoa AC. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res, 64(16):5839–5849, August 2004. [DOI] [PubMed] [Google Scholar]
- [3].Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, and Albelda SM. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell, 16(3):183–194, September 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gabrilovich DI, Ostrand-Rosenberg S, and Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol, 12(4):253–268, March 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ugel S, De Sanctis F, Mandruzzato S, and Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J. Clin. Invest, 125(9):3365–3376, September 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Joshi NS, Akama-Garren EH, Lu Y, Lee DY, Chang GP, Li A, DuPage M, Tammela T, Kerper NR, Farago AF, Robbins R, Crowley DM, Bronson RT, and Jacks T. Regulatory T Cells in Tumor-Associated Tertiary Lymphoid Structures Suppress Anti-tumor T Cell Responses. Immunity, 43(3):579–590, September 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tanaka A and Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res, 27(1):109–118, January 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Srivastava MK, Sinha P, Clements VK, Rodriguez P, and Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res, 70(1):68–77, January 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, Padhya T, McCaffrey TV, McCaffrey JC, and Gabrilovich DI. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J. Immunol, 182(9):5693–5701, May 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nagaraj S, Schrum AG, Cho HI, Celis E, and Gabrilovich DI. Mechanism of T cell tolerance induced by myeloid-derived suppressor cells. J. Immunol, 184(6): 3106–3116, March 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Torroella-Kouri M, Silvera R, Rodriguez D, Caso R, Shatry A, Opiela S, Ilkovitch D, Schwendener RA, Iragavarapu-Charyulu V, Cardentey Y, Strbo N, and Lopez DM. Identification of a subpopulation of macrophages in mammary tumor-bearing mice that are neither M1 nor M2 and are less differentiated. Cancer Res, 69(11):4800–4809, June 2009. [DOI] [PubMed] [Google Scholar]
- [12].Calcinotto A, Filipazzi P, Grioni M, Iero M, De Milito A, Ricupito A, Cova A, Canese R, Jachetti E, Rossetti M, Huber V, Parmiani G, Generoso L, Santinami M, Borghi M, Fais S, Bellone M, and Rivoltini L. Modulation of microenvironment acidity reverses anergy in human and murine tumor-infiltrating T lymphocytes. Cancer Res, 72(11):2746–2756, June 2012. [DOI] [PubMed] [Google Scholar]
- [13].Bellone M and Calcinotto A. Ways to enhance lymphocyte trafficking into tumors and fitness of tumor infiltrating lymphocytes. Front Oncol, 3:231, September 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, Tonc E, Schreiber RD, Pearce EJ, and Pearce EL. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell, 162(6):1229–1241, September 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, and Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ, 1:e201, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, and Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med, 183 (3):1161–1172, March 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, and Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med, 4(5):594–600, May 1998. [DOI] [PubMed] [Google Scholar]
- [18].Klinke DJ, Kulkarni YM, Wu Y, and Byrne-Hoffman C. Inferring alterations in cell-to-cell communication in HER2+ breast cancer using secretome profiling of three cell models. Biotechnol. Bioeng, 111(9):1853–1863, September 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, and Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med, 18(6):883–891, June 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wu Y, Deng W, McGinley EC, and Klinke DJ. Melanoma exosomes deliver a complex biological payload that upregulates PTPN11 to suppress T lymphocyte function. Pigment Cell Melanoma Res, 30(2):203–218, March 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu Y, Deng W, and Klinke DJ. Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst, 140(19):6631–6642, October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen H, Mo D, Li M, Zhang Y, Chen L, Zhang X, Li M, Zhou X, and Chen Y. miR-709 inhibits 3T3-L1 cell differentiation by targeting GSK3β of Wnt/β-catenin signaling. Cell. Signal, 26(11):2583–2589, November 2014. [DOI] [PubMed] [Google Scholar]
- [23].Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12):550, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guo H, Ingolia NT, Weissman JS, and Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature, 466(7308):835–840, August 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ponting CP, Oliver PL, and Reik W. Evolution and functions of long noncoding RNAs. Cell, 136(4):629–641, February 2009. [DOI] [PubMed] [Google Scholar]
- [26].Mercer TR and Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol, 20(3):300–307, March 2013. [DOI] [PubMed] [Google Scholar]
- [27].Zavadil J, Cermak L, Soto-Nieves N, and Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J, 23(5):1155–1165, March 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sahlgren C, Gustafsson MV, Jin S, Poellinger L, and Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. U.S.A, 105(17):6392–6397, April 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, Maru DM, Hawke DH, Rak J, Mani SA, Zweidler-McKay P, and Ellis LM. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell, 23(2):171–185, February 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sierra RA, Thevenot P, Raber PL, Cui Y, Parsons C, Ochoa AC, Trillo-Tinoco J, Del Valle L, and Rodriguez PC. Rescue of notch-1 signaling in antigen-specific CD8+ T cells overcomes tumor-induced T-cell suppression and enhances immunotherapy in cancer. Cancer Immunol Res, 2(8):800–811, August 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Backer RA, Helbig C, Gentek R, Kent A, Laidlaw BJ, Dominguez CX, de Souza YS, van Trierum SE, van Beek R, Rimmelzwaan GF, ten Brinke A, Willemsen AM, van Kampen AH, Kaech SM, Blander JM, van Gisbergen K, and Amsen D. A central role for Notch in effector CD8(+) T cell differentiation. Nat. Immunol, 15(12):1143–1151, December 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mathieu M, Duval F, Daudelin JF, and Labrecque N. The Notch signaling pathway controls short-lived effector CD8+ T cell differentiation but is dispensable for memory generation. J. Immunol, 194(12):5654–5662, June 2015. [DOI] [PubMed] [Google Scholar]
- [33].Elyaman W, Bradshaw EM, Wang Y, Oukka M, Kivisakk P, Chiba S, Yagita H, and Khoury SJ. JAGGED1 and delta1 differentially regulate the outcome of experimental autoimmune encephalomyelitis. J. Immunol, 179(9):5990–5998, November 2007. [DOI] [PubMed] [Google Scholar]
- [34].Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, and Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell, 117(4):515–526, May 2004. [DOI] [PubMed] [Google Scholar]
- [35].Sun J, Krawczyk CJ, and Pearce EJ. Suppression of Th2 cell development by Notch ligands Delta1 and Delta4. J. Immunol, 180(3):1655–1661, February 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kijima M, Iwata A, Maekawa Y, Uehara H, Izumi K, Kitamura A, Yagita H, Chiba S, Shiota H, and Yasutomo K. Jagged1 suppresses collagen-induced arthritis by indirectly providing a negative signal in CD8+ T cells. J. Immunol, 182(6):3566–3572, March 2009. [DOI] [PubMed] [Google Scholar]
- [37].Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14:128, April 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, and Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res, 44(W1):W90–97, July 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pajak M and Simpson TI. miRNAtap: microRNA Targets - Aggregated Predictions, 2016.
- [40].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A, 102(43):15545–15550, October 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, and Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis, 14(1):27–41, May 2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.