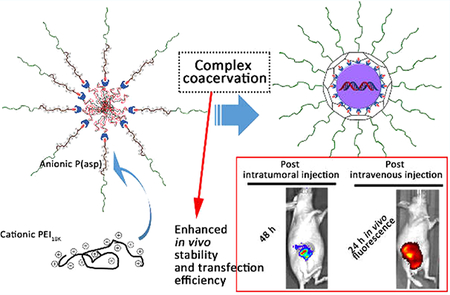
Abstract
Many polycation-based gene delivery vehicles have limited in vivo transfection efficiency because of their excessive exterior positive charges and/or PEGylation, both of which could result in premature dissociation and poor cellular uptake and trafficking. Here, we reported novel hybrid PEGylated nanoparticles (HNPs) that are composed of (a) poly(ethylene glycol)-b-poly(aspartate)-adamantane (PEG-P(asp)-Ad) constituting the outer PEG layer to provide colloidal stability; (b) poly(ethylenimine)10K (PEI10K) forming complex coacervate with P(asp) as the cross-linked cage preventing premature dissociation; (c) cyclodextrin-decorated PEI10K (PEI10K-CD) forming the core with reporter plasmid DNA (pDNA). These HNPs exhibited an increased stability and higher in vitro transfection efficiency compared to traditional PEGylated nanoparticles (PEG-NP). Intratumoral injections further demonstrated that HNPs were able to successfully deliver pDNAs into tumors, while PEG-NP and PEI25K had only negligible delivery efficiencies. Moreover, HNPs’ in vivo stability and pDNA delivery capability post intravenous injection were also confirmed by live animal bioluminescence and fluorescence image analysis. It is likely that the coacervation integration at the interface of PEI10K-CD/pDNA core and the PEG shell attributed to the significantly improved in vivo transfection efficiency of HNPs over PEG-NP and PEI25K. This study suggests that the HNP has the potential for in vivo gene delivery applications with significantly improved gene transfection efficiency.
Keywords: complex coacervation, PEGylated, transfection efficiency, plasmid DNA, in vivo delivery
Graphical Abstract
1. INTRODUCTION
One major obstacle for effective DNA delivery is the low in vivo transfection efficiency, despite many studies using polycations have claimed to have high in vitro transfection efficiencies. The high toxicity and instability of polycations in the presence of serum have been the major restriction for their in vivo applications.1,2 Polycations such as poly(ethylenimine) (PEI), which has been favored as an efficient nonviral gene carrier in vitro, also encounter the same problem.3,4 Indeed, polycations tend to bind and condense DNA effectively by electrostatic attractions to form nanoparticles, but nanoparticles with excessive positive charges are more likely to interact with negatively charged serum proteins as well as tissue components. This ultimately leads to aggregates in the physiological saline environment and low transfection efficiency in vivo.5,6 It has been shown that the in vivo transfection efficiency of DNA using high molecular weight polycations was very low.6,7 Even a direct intratumoral injection of the PEI/plasmid DNA (pDNA) complex expressing luciferase was unable to transfect efficiently.8 To reduce the nonspecific interactions with serum proteins or tissue components, nanocarriers containing poly(ethylene glycol) (PEG) and polycation blocks have been designed.6 However, likely due to the strong hydrophilicity of PEG,6 PEG-polycation/DNA complexes were prone to dissociation in the physiological environments,9,10 leading to unexpected release behavior and DNA payload degradation by nucleases. Additionally, PEG could also reduce the binding and uptake by target cells.6,11,12 While polycations as well as PEG-polycations have their own unique advantages, PEG-polycations do not significantly outperform polycations for in vivo transfection efficiency.13,14 Despite nonviral gene therapy has had great progress during the past 30 years, DNA-based drugs inherently pose greater delivery challenges than other nucleic acid therapeutics due to their large molecular sizes, the difficulty of entry into the nucleus, and the risk of mutagenesis.4
The host–guest effect combines two basic concepts of supramolecular chemistry: self-assembly and molecular recognition, offering a convenient tool for the construction of nanoparticles based on molecular building blocks.15,16 For instance, the surface of particles formed from cyclodextrin (CD)-grafted polycations and nucleic acids could be decorated with adamantane group (Ad) terminated PEG chains through the formation of Ad/β-CD inclusion complexes.17–20 However, using these kinds of PEGylated nanoparticles to accomplish gene delivery still remains a major challenge because of the reduction in transfection efficiency. This is likely due to the reduced cellular uptake and trafficking of these kinds of PEGylated nanoparticles.11,12,19 Thus, the PEG dilemma of polycations between avoiding nonspecific interaction and transfection efficiency has to be addressed for successful gene delivery in vivo.21,22
A recent study reported the complex coacervation phenomenon between PEI and P(glu) or P(asp) in a saline environment.23 More recently, some systems of complex coacervation have been explored for gene delivery.24–26 Due to the release of counter-ions, polyelectrolyte coacervation is entropy-driven and demonstrated very good characteristics of rigidity and viscoelasticity with the existence of salt.23,26 This suggests the nanoparticles integrated with complex coacervation modality could demonstrate a good stability in a physiological saline environment. Meanwhile, while considering the proton sponge effect of PEI, the stabilization from PEGylation, and the ease of condensation into sphere nanoparticle with the payload genes, we herein tried to introduce a poly(ethylenimine)10K (PEI10K)/P(asp) complex coacervation into the aforementioned PEGylated CD-containing polycations to construct a hybrid complex coacervation nanoparticle (HNP, Figure 1a) aiming for more efficient in vivo gene delivery.
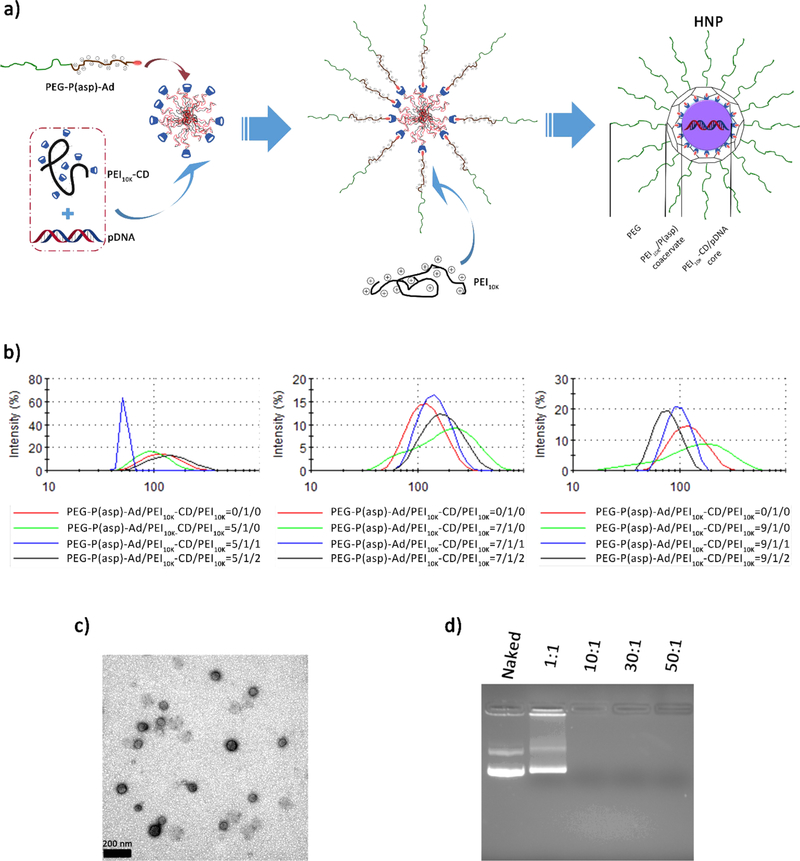
Figure 1.
Schematic illustration of the preparation and physicochemical characterizations of HNPs. (a) PEG-P(asp)-Ad binds to the PEI10K-CD/pDNA nanoparticle generating a PEG outer layer, while PEI10K interacts with P(asp) generating a coacervate for the PEI10K-CD/pDNA nanoparticles. Uniquely, the PEG-P(asp)-Ad conjugate combines with PEI10K to constrain and cage the nanoparticle core through the crosslinked network and also confers PEG corona to the nanoparticles. (b) Size distribution histograms of HNPs with different amounts of PEG coating and PEI10K addition. (c) A representative TEM image of HNPs (the mole ratio of PEG-P(asp)-Ad/PEI10K-CD/PEI10K = 9/1/2, N/P ratio = 30/1), scale bar = 200 nm. (d) A representative image of the gel-retardation experiment at different N/P ratios as indicated in the figure.
The uniqueness of our design is the use of a molecular building block, poly(ethylene glycol)-b-poly(aspartate)-adamantane (PEG-P(asp)-Ad), combining with PEI10K to compact and cage the cyclodextrin-decorated PEI10K (PEI10K-CD)/gene core and conferring a PEG corona. The Ad could form inclusion complexes with CD and thus provide a PEG shell protecting the payload by avoiding interactions with the surrounding biological environment. The caging of HNPs with PEI10K/P(asp) coacervate could not only provide lateral stabilization and prevent premature dissociation of the nanoparticles but also facilitate endosome escape after entering the target cells. The optimum densities of PEG and coacervate for HNPs were determined by in vitro stability studies. Furthermore, using the optimized formulation, the in vitro gene transfection efficiency of the HNPs was evaluated by monitoring the expression of enhanced green fluorescent protein (EGFP) and firefly luciferase 2 (Luc2) from HNP-loaded plasmid DNAs (pDNAs) using a fluorescence microscope and a dual luciferase reporter assay, respectively. The HNP in vivo distribution study post intravenous injection was determined using the IVIS live animal fluorescence imaging system. Finally, the in vivo gene transfection efficiency and stability of HNPs were investigated by monitoring the activity of Luc2 in mouse mammary xenograft tumor tissues from intratumoral and intravenous injection of HNP-loaded pDNA (pGL4.5-Luc2) using the IVIS live animal bioluminescence imaging system.
2. EXPERIMENTAL SECTION
2.1. Materials and Cell Culture.
PEI25K was purchased from Polysciences, Inc. (Monticello, IN). The pDNAs encoding for EGFP (pSM4-H1p-EGFP), Luc2 (pGL4.5-Luc2), and Renilla (pRL-TK) were obtained from Addgene (Cambridge, MA) and Promega (Madison, WI), respectively. Human breast cancer MDA-MB-231 and SUM159 cells were obtained and cultured as described in our recent publication.27 The LM2 cell, a derivative of MDA-MB-231 cells that was selected for its strong ability to metastasize to lung in vivo,28 was kindly provided by Dr. Joan Massagué (Memorial Sloan-Kettering Cancer Center, New York). The LM2 cells were cultured using the same medium as for MDA-MB-231 cells. All cells were cultured at 37 °C in a humidified 5% CO2 atmosphere.
2.2. Preparation and Characterization of HNPs To Identify Optimum Density of Coacervate.
PEG-anchored PEI10K-CD/pDNA nanoparticles (PEG-NP) were freshly prepared according to the previous reported method.19 HNPs loaded with pDNAs were freshly prepared before use. First, PEI10K-CD was mixed with pDNAs and then incubated at room temperature for 1–5 min to ensure complex formation. Second, PEG-P(asp)-Ad and PEI10K were added into the system in sequence and then incubated for 10 min. The amounts of PEI10K-CD and PEI10K were calculated on the basis of various N/P ratios. Here, N represents the total number of amine groups on PEI10K-CD and PEI10K, and P represents the number of anionic phosphate groups in pDNAs. The nanoparticles were suspended in a 0.9% (wt/vol) saline solution for the biological tests.
To evaluate the capability of HNPs to compress pDNAs, an agarose gel retardation assay was performed. Briefly, HNPs at different N/P ratios containing 500 ng of pDNAs were loaded into 1% agarose gel in TAE buffer and run at 100 V for 40 min. The final location pattern of pDNAs was visualized under UV irradiation and photographed using Gel Doc XR+ System (Bio-Rad).
The HNP size (diameter, nm) and surface charge (ζ-potential, mV) were obtained from three measurements using a Malvern Instruments Zetasizer ZS-90 instrument. HNPs at different polymer concentrations were prepared using double distilled water or 0.9% NaCl (wt/vol) saline solution. Z-average sizes of three sequential measurements were collected and analyzed, and ζ-potentials following three sequential measurements were also collected and analyzed. All measurements were made in triplicates, and results were expressed as mean ± SD. The sizes of PEI10K-CD and PEG-NP were determined by a dynamic light microscope (DLS) method as well.
HNPs were explored for their morphologies using a transmission electron microscope (TEM, JEOL 100CX II). Briefly, the aqueous solution of HNPs was applied dropwise onto a 400-mesh copper grid coated with carbon, stained with 1% uranyl acetate solution, observed and photographed using a TEM.
2.3. HNP in vitro Stability Study in the Presence of Bovine Serum Albumin (BSA) To Identify Optimum PEG Density.
HNPs with different formulations (PEG-P(asp)-Ad/PEI10K-CD/PEI10K = 5/1/1, 5/1/2, 7/1/1, 7/1/2, 9/1/1, and 9/1/2) were formed at an initial N/P ratio of 30:1 and at a pDNA concentration of 20 μg/mL in saline. HNPs were incubated with 0.005% (w/v) BSA, and an aliquot of HNP solutions was collected at each time point (0, 24, 48, and 72 h) to measure HNP size using the method described in Section 2.4.
2.4. Determination of the HNP-Loaded pDNA in vitro Stability in the Presence of Fetal Bovine Serum (FBS).
Serum stabilities of both naked pDNA and the pDNA loaded into HNPs (equivalent to 500 ng of pDNA in 15 μL of solution) were investigated by incubating each formulation at 37 °C with 20% volume addition of FBS. Samples at specified time points (1, 4, 24, and 48 h) were loaded into 1% agarose gel to determine the amount of remaining intact pDNAs. Naked pDNA, PEI25K/pDNA complex (N/P ratio = 30/1), and PEG-NP incubated in 20% FBS served as controls. Forced release of pDNAs from the nanoparticles was achieved by adding 5 mg/mL Heparin and mixing immediately before gel loading, as addition of excess Heparin could displace pDNAs from HNPs. The pDNA bands were visualized by ethidium bromide staining and photographed using the Gel Doc XR+ System (Bio-Rad).
2.5. HNP Cytotoxicity Analysis.
Cytotoxicities of HNPs and PEI25K without pDNA being loaded were evaluated using MDA-MB- 231, SUM159, and LM2 cells by the tetrazolium dye colorimetric test (MTT assay) as described previously.27 Briefly, cells were seeded into 96-well plates (3–5 × 103 cells/well in 100 μL of complete culture medium). Different concentrations of HNPs and PEI25K were added into the wells 24 h after cell seeding and incubated with cells for 48 h. The HNP concentrations were calculated on the basis of the concentration of PEI10K (the amount of PEI10K-CD was converted into that of PEI10K as well). At the end of incubation, 50 μL of the MTT reagent (5 mg/mL) was added to each well and incubated for 4 h. Then, 200 μL of dimethyl sulfoxide (DMSO) was added to each well and incubated for another hour. The plate was read using a microplate reader (SpectraMAX Plus, Molecular Devices, Sunnyvale, CA) at a wavelength of 570 nm. The relative cell viability was determined by the following formula:
2.6. HNP in vitro Gene Transfection Efficiency Determination.
HNPs were used to complex two different pDNAs encoding EGFP or Luc2, respectively, for in vitro gene transfection efficiency studies using two types of cell lines (SUM159 and MDA-MB-231). SUM159 cells were seeded into 6-well plates at a density of 3 × 104 cells per well and allowed to attach for 24 h at 37 °C. Transfections were carried out in complete growth media containing 5% FBS with pSM4-H1p-EGFP pDNA-uploaded PEI25k (N/P 10/1), PEI25k (N/P 30/1), PEG-NP, and HNP at a pDNA concentration of 3 μg per well. The nanoparticles were incubated with SUM159 cells for 48 h at 37 °C. The GFP expression was viewed and photographed with a Nikon Eclipse TE2000-U reverse fluorescence microscope (Nikon, Inc., Melville, NY) at 24 and 48 h time points. The in vitro transfection efficiencies of HNPs with different amounts of PEI10K (PEG-P(asp)-Ad/PEI10K-CD/PEI10K = 9/1/0, 9/1/1, and 9/1/2) at N/P ratios of 30/1 and 50/1 were also investigated using the same method.
In addition, the in vitro gene delivery efficiency of HNPs was also determined using a luciferase reporter assay as described previously.27 Briefly, SUM159 and MDA-MB-231 cells were seeded into 6-well plates at a density of 3 × 104 cells per well and allowed to attach for 24 h at 37 °C before luciferase reporter expressing plasmid transfection. The prepared PEI25K polyplexes and HNPs containing 3 μg of Luc2 expressing plasmid (pGL4.5-Luc2) and 90 ng of Renilla luciferase expressing plasmid (pRL-TK) at a N/P ratio of 30/1 were diluted with l00 μL of 0.9% NaCl solution, followed by incubation with SUM159 and MDA-MB-231 cells for 48 h. The relative luciferase reporter activity was calculated as relative light unit (RLU) per milligram (mg) cell protein.
2.7. HNP in vivo Gene Delivery Efficiency Study after Intratumoral Injection.
The in vivo gene transfection efficiency of HNPs was determined by directly injecting pGL4.5-Luc2-uploaded HNPs into nude mouse mammary xenograft tumors, and the activity of Luc2 was monitored using an IVIS live animal bioluminescence imaging system (PerkinElmer, Waltham, MA). All aspects of the animal studies were performed in accordance with the guidelines defined by the Institutional Animal Care and Use Committee of Michigan State University. Briefly, for the subcutaneous tumor model, 7 × 106 human breast cancer MDA-MB-231 cells suspended in 50 μL of blank media (no FBS) and 50 μL of growth factor-reduced Matrigel (BD Biosciences) were injected into the right flank close to the second mammary fat pad of 7-week old female nu/nu nude mice (Charles River Laboratories) to produce mammary xenograft tumors. The MDA-MB-231 orthotopic tumor model was established by our previously reported method.27 Briefly, we inoculated 5 × 106 human breast cancer MDA-MB-231 cells suspended in 50 μL of blank media (no FBS) and 50 μL of growth factor-reduced Matrigel (BD Biosciences) orthotopically in a fourth mammary fat pad of 7-week old female nu/nu nude mice (Charles River Laboratories) that were surgically exposed while the mice were anesthetized. When the subcutaneous and orthotopic tumors reached 300 and l000 mm3, respectively, a single dose of HNPs uploaded with 25 μg of pGL4.5-Luc2 pDNA was directly injected into tumor tissues and PEI25k/ pDNA complex (N/P ratio = 30/l) with the same dose serving as a control. The mice were intraperitoneally injected with the substrate D-luciferin (5 mg/kg mouse body weight) and then anesthetized by isoflurane inhalation at 24 and 48 h after the intratumoral injection. The activity of Luc2 in tumor tissue was monitored by determining the intensity of chemiluminescence signals 5 min after D-luciferin injection using an IVIS live animal bioluminescence imaging system. The mouse chemiluminescence images were taken every 2 min for 18 min consecutively, using the IVIS live animal imaging system (PerkinElmer, Waltham, MA). The spatial distribution of photon counts within the mouse was illustrated by the resulting pseudo color image.
2.8. HNP in vivo Stability Study Post Intravenous Injection.
The pDNA expression study post intravenous injection of HNPs was performed in mice bearing MDA-MB-231 orthotopic mammary tumors. Tumors were established as described in Section 2.7. When the tumor size reached l000 mm3, HNPs consisting of 50 μg dose of pGL4.5-Luc2 pDNA were administered via tail vein injection into the mice and the activity of Luc2 was monitored on day l, 3, 5 post injection by the same method described in the Section 2.7. At day 5 post injection, a second dose of HNPs (50 μg, pGL4.5-Luc2 pDNA) was administered and the activity of Luc2 was monitored on days l and 2 post injection.
2.9. HNP in vivo Distribution Study Post Intravenous Injection.
The in vivo distribution study of HNPs was performed in mice (n = 3) bearing MDA-MB-231 orthotopic mammary tumors. Tumors were established as described in Section 2.7. HNPs labeled with near-infrared (NIR) fluorophore-cyanine 7.5 (cy7.5) were administered via tail vein (50 μg cy7.5-labeled PEI10K per mouse), and fluorescence imaging (λex/λem= 745 nm/82O nm) was performed using the IVIS live animal imaging system (PerkinElmer, Waltham, MA) at 24 h postinjection. To verify the finding of in vivo imaging, tumors and lymph nodes were imaged ex vivo as well. The cy7.5-labeled HNPs were prepared by the same method except for using cy7.5-labeled PEI10K instead of PEI10K. More information about the synthesis of cy7.5-labeled PEI10K could be found in the Supporting Information.
2.10. Statistical Analysis.
The numerical data are expressed as mean ± SD, and the differences of effects among different treatment groups were tested via analysis of variance (ANOVA) using a general linear model. The differences between treatment groups were determined using a two sample t-test. A p value of <0.05 was considered statistically significant.
3. RESULTS
3.1. Preparation and Characterization of HNPs To Identify Optimum Density of Coacervation.
Two of the three molecular building blocks-PEG-P(asp)-Ad and PEI10K-CD were first prepared and characterized by lH NMR (Figures S1–S4). Herein, we put forward a convenient and modular construction method for HNPs (Figure 1a). The specially designed PEG-P(asp)-Ad was then combined with PEI10K-CD through host-guest complexation and then self-assembled with oppositely charged PEI10K to produce HNPs. By tuning the mixing ratios among the three molecular building blocks (PEG-P(asp)-Ad/PEI10K-CD/PEI10K) in 0.9% NaCl saline aqueous solution, the equilibrium between the coacervation/aggregation and capping/solvation of the three components could be altered in order to find the best HNP formulation.
To examine how the mixing ratios between PEG-P(asp)-Ad and PEI10K-CD affected the sizes of the resulting HNPs, we utilized DLS measurements to analyze the freshly prepared HNPs. In the absence of PEG-P(asp)-Ad, direct mixing of pDNAs and PEI10K-CD resulted in aggregation and precipitation. Thus, HNPs with various ratios of PEG-P(asp)-Ad were tested. In this case, PEG-P(asp)-Ad was slowly added into the mixtures containing PEI10K-CD and pDNAs at the N/P ratio of 30/1. As shown in Figure 1b, we were able to obtain a collection of water-soluble HNPs with variable sizes of around 100 nm. However, with increased addition of PEI10K, the sizes differentiated greatly among the groups with different amounts of PEG-P(asp)-Ad added. By adjusting the PEG-P(asp)-Ad/PEI10K-CD/PEI10K mole ratio of 9/1/2, we were able to obtain HNPs with a relatively smaller size (84.2 ± 2.9 nm) and better polydispersity index (0.26 ± 0.03) under similar self-assembly conditions. Moreover, the morphology and size of HNPs were examined using a TEM. Under a TEM, the HNP cores were observed by positive-staining with uranyl acetate, as their PEG shell could not be observed due to the lower electron density compared to the stained core. As shown in Figure 1c, TEM images suggested that the HNPs exhibit the spherical shapes and narrow size distributions, which are consistent to those observed by DLS (Figure 1b). The molecular building block PEG-AD was synthesized and characterized as well (Figure S5). At the same time, the particle size of PEG-NP in 0.9% saline solution was compared to that of the PEI10K-CD/pDNA and HNP as demonstrated in Figure S6. Due to the steric stabilization of PEG chains, PEG-NP has a relatively smaller size (222.3 ± 5.4 nm) than PEI10K-CD/pDNA complexes (446.1 ± 18.1 nm), despite the existence of exterior PEG layer in the physiological saline environment. This suggests that PEG-NP has good stability in a physiological saline solution and easily condenses genes into spherical particles compared to that of using PEG-polycation conjugates.19,20 However, HNPs with the extra coacervation layer did not show an increased size (102.5 ± 2.0 nm) compared with PEG-NP due to the further compacting effect from PEI10K/P(asp) coacervation in the saline environment.
Furthermore, the pDNA condensing and binding capability of HNPs was investigated by examining the stability of pDNAs loaded into HNPs (Figure 1d). It was found that, with the increase of N/P ratios, the pDNA condensing capability of HNPs was also greatly enhanced. The pDNA binding capability of HNPs was assured at the N/P ratio ranging from 10/1 to 30/1 (Figure 1d).
3.2. Stability Study of HNPs in the Presence of BSA To Identify Optimum Density of PEG.
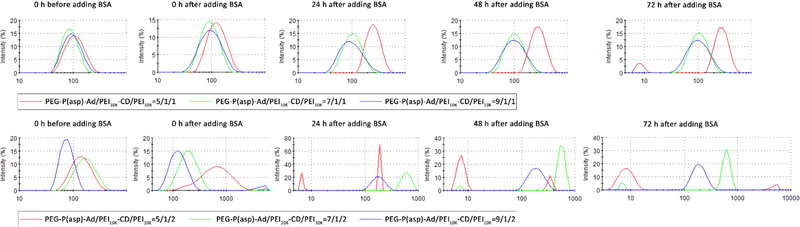
The application of a supramolecular approach conferred dynamic characteristics to the self-assembled HNPs. To understand the dynamic stability of HNPs, we employed DLS measurements to assess the size variation of HNPs loaded with pDNAs (N/P = 30/1) at different PEG capping (PEG-P(asp)-Ad/PEI10K-CD = 5/1, 7/1, and 9/1) and PEI10K caging (PEI10K-CD/PEI10K = 1/0, 1/1, 1/2) conditions in physiological ionic strength media (0.9% NaCl saline solution).
As shown in Figure 2, the addition of PEI10K, which cross-linked the negatively charged polypeptides on the PEG-P(asp)-Ad and compacted the nanoparticle, substantially increased the stability of the complexes in the presence of BSA. Although PEI10K molecules interact with surface exposed P(asp) via charge-to-charge interactions, they could not form an ionic cross-link between the complex particles because they are presumably shielded by PEG chains. The BSA stabilization was reversed upon the addition of excess PEI10K, indicating that it depends on an equilibrium between positive charges on PEI10K and negative charges on P(asp), which is consistent with the finding from a previous study.23 Despite that HNPs at the ratio of 5/1/1 displayed a very good distribution pattern as shown in Figure 1b, they failed to keep their integrity after 72 h of incubation with BSA (Figure 2). In contrast, HNPs at the ratio of 9/1/2 did not aggregate for at least 3 days in the presence of 0.005% (w/v) BSA (Figure 2). Taken together, these results suggest that the coacervate-caging of nanoparticles could stabilize their pDNA complexes in a serum-protein containing environment.
Figure 2.
Size distribution curves of HNPs with different formulations (PEG-P(asp)-Ad/PEI10K-CD/PEI10K = 5/1/1, 5/1/2, 7/1/1, 7/1/2, 9/1/1, and 9/1/2, N/P ratio = 30:1) in the presence of 0.005% (w/v) BSA. The histograms were recorded at the incubation time of 0, 24, 48, and 72 h, respectively.
During the whole 72 h incubation with BSA, the sizes of HNPs with the formulation of 9/1/1 were almost unchanged while the sizes of HNPs of 9/1/2 slightly increased (Figure 2). Although HNPs of 9/1/1 demonstrated better resistance to BSA than those of 9/1/2, HNPs of 9/1/1 had much poorer transfection efficiency as shown in later transfection experiments (Figure 6). In addition, as demonstrated in Figure S7, HNPs of 9/1/2 kept the structural integrity and monodistribution patterns with diameters under 1000 nm. This prompted us to choose P(asp)-Ad/PEI10K-CD/PEI10K of 9/1/2 as the final formulation of HNPs.
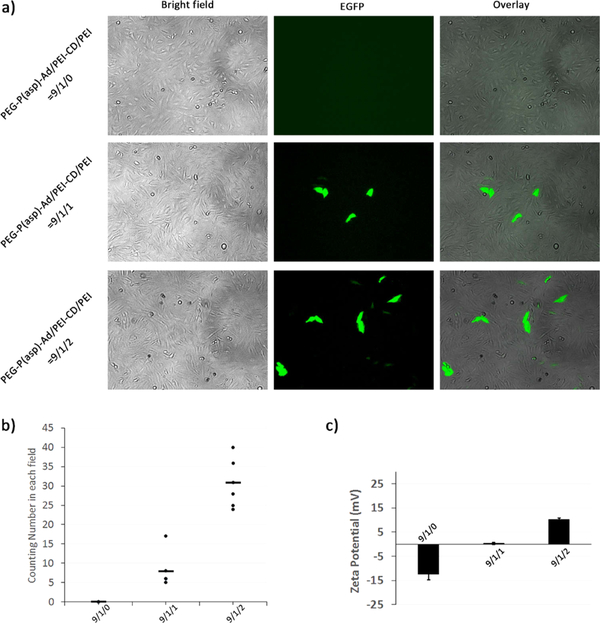
Figure 6.
Effect of different amounts of added-PEI10K on in vitro gene transfection efficiency of HNPs. SUM159 cells were transfected with GFP expressing pDNA-loaded HNPs formulated with different PEG-P(asp)-Ad/PEI10K-CD/PEI10K ratios under a N/P ratio of 30/1. The pDNA transfections were performed at a dose of 3 μg of GFP expressing pDNA. Cells were incubated in the transfection solution for 48 h and then viewed and photographed under a fluorescent microscope. (a) The representative images (100×) of cells transfected with HNPs formulated with different amounts of PEI10K under a N/P ratio of 30/1. (b) The numbers of GFP expression cells per field of view (POV) counted under a fluorescent microscope. Results are from two independent experiments (n = 6), and three randomly selected fields were counted in each experiment. (c) The ζ-potentials of HNPs formulated with different amounts of PEI10K.
3.3. Stability of pDNAs Loaded into HNPs in the Presence of FBS.
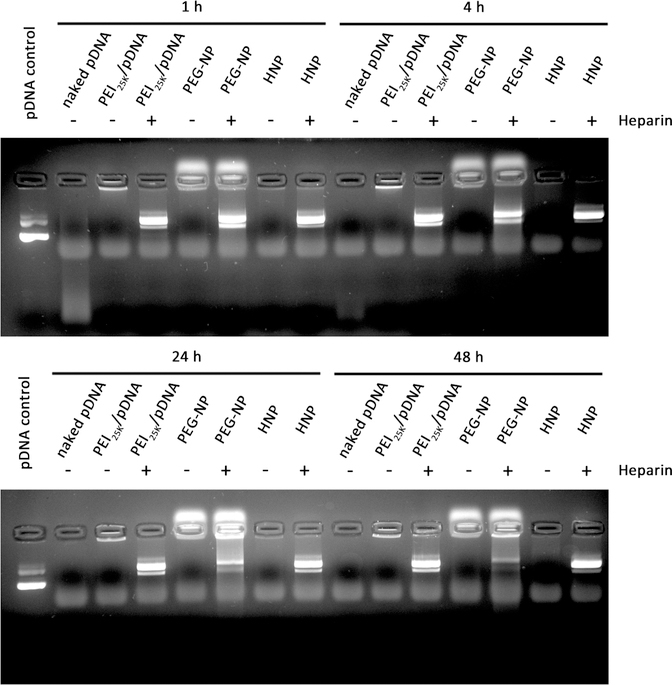
The stabilization of gene agents loaded into nanocarriers in serum is essential for in vivo gene delivery. In support of this, we found that the degradation of naked pDNAs was observed 30 min after the incubation, and naked pDNAs were fully degraded after 4 h of incubation in the presence of 20% FBS (Figure 3). The pDNAs in PEG-NP were partially degraded as well after 24 h of incubation, which was probably caused by premature dissociation and could lead to decreased transfection efficiency. In sharp contrast, our HNPs and PEI25K effectively stabilized pDNAs in the presence of 20% FBS for the entire duration of the experiment (48 h, Figure 3), which demonstrated that higher-density polyelectrolytes could compact pDNAs more effectively. These results indicate that integration of coacervate into HNPs provides much better rigidity than PEG-NP and could prevent genes from degradation in serum.
Figure 3.
Stability of naked pDNA (500 ng) and the pDNA (500 ng) uploaded into PEI25K, PEG-NP, and HNPs in the presence of FBS. They were incubated at 37 °C with 20% volume addition of FBS for 1, 4, 24, and 48 h. At the end of each determined time point, an aliquot of HNPs incubated with 20% FBS was added to 5 mg/mL Heparin to forcedly release the pDNA from HNPs to serve as controls. The released pDNAs were then visualized by agarose gel electrophoresis, followed by ethidium bromide staining and photographing using a Gel Doc XR+ System. The photos are representative pDNA images after agarose gel electrophoresis and ethidium bromide staining.
Moreover, HNPs with the PEG-P(asp)-Ad/PEI10K-CD ratio of 5/1 showed much worse size distribution after 0.005% (w/v) BSA was added than that of HNPs with the ratio of 9/1 (Figure 2). Furthermore, even after 72 h of incubation with 20% (v/v) FBS, the HNP with a PEG-P(asp)-Ad/PEI10K-CD/PEI10K ratio of 9/1/2 still kept its integrity and monodistribution pattern within the size range of 1000 nm (Figure S7). These findings are consistent with the report showing that a higher surface PEG density rendered nanoparticles more resistant to interactions with serum proteins.29
3.4. Cytotoxicity of HNPs.
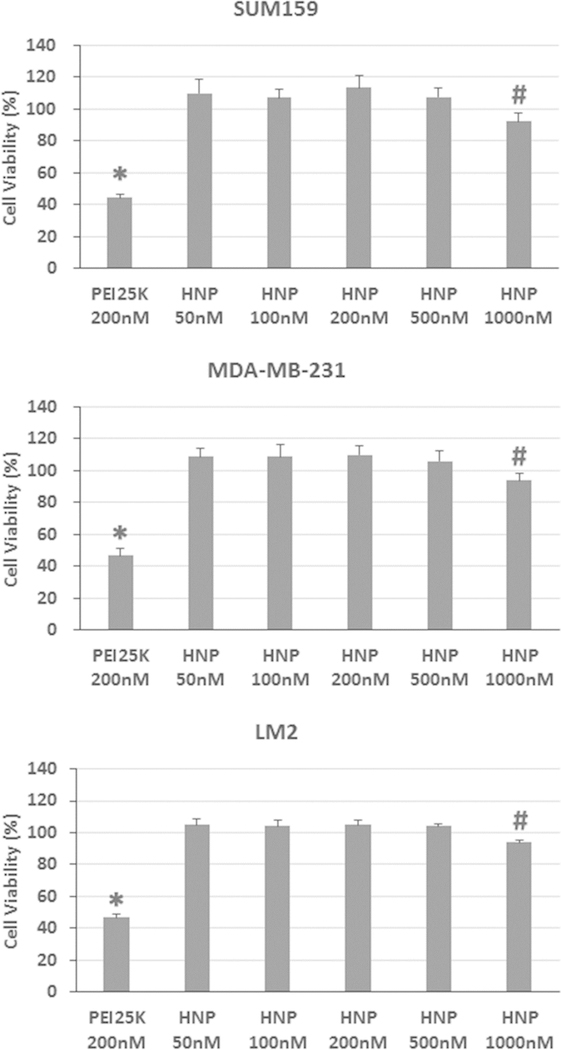
Cytotoxicity is an important factor to be considered when designing gene nanocarriers, so the cytotoxicity of blank HNPs at concentrations ranging from 50 to 1000 nM was evaluated with SUM159, MDA-MB-231, and LM2 cells using the standard MTT assay and compared with the cytotoxicity of PEI25K. Due to the strong interactions of cationic PEI25K with cell membranes through ionic or hydrophobic interactions, PEI25K usually causes severe damage to the cell membrane at certain concentrations and is thus highly cytotoxic.3 Indeed, as shown in Figure 4, the treatment with 200 nM of PEI25K was highly cytotoxic to all three tested model cell lines and reduced the cell viability to about 45%.
Figure 4.
Cytotoxicity analysis of PEI25K and blank HNPs in SUM159, MDA-MB-231, and LM2 cells by the MTT assay. Cells were cultured and treated with PEI25K and various concentrations of HNPs as described in the Experimental Section. The HNP concentrations were calculated on the basis of the concentration of PEI10K. (The amount of PEI10K-CD was converted into that of PEI10K as well.) Results are expressed as mean ± standard deviation (n = 8). *: p < 0.05, compared to 50, 100, 200, 500, and 1000 nM of HNP-treated groups; #: p < 0.05, compared to 50, 100, 200, and 500 nM of HNP-treated groups.
In striking contrast, negligible cytotoxicities were observed in all three tested model cell lines treated with various concentrations of HNPs as no cytotoxic effects were detected in all cells treated with 50 to 500 nM of HNPs (Figure 4). Moreover, although a very high concentration (1000 nM) of HNP treatment statistically significantly reduced the cell viability, it only decreased cell viability by about 7% compared to that of vehicle control-treated cells (Figure 4). This drastic reduction of the cytotoxic effect of HNPs is likely due to the reduced charge density by complex coacervation and the presence of surface anchored PEG chains.
3.5. in vitro pDNA Transfection Efficiency of HNPs.
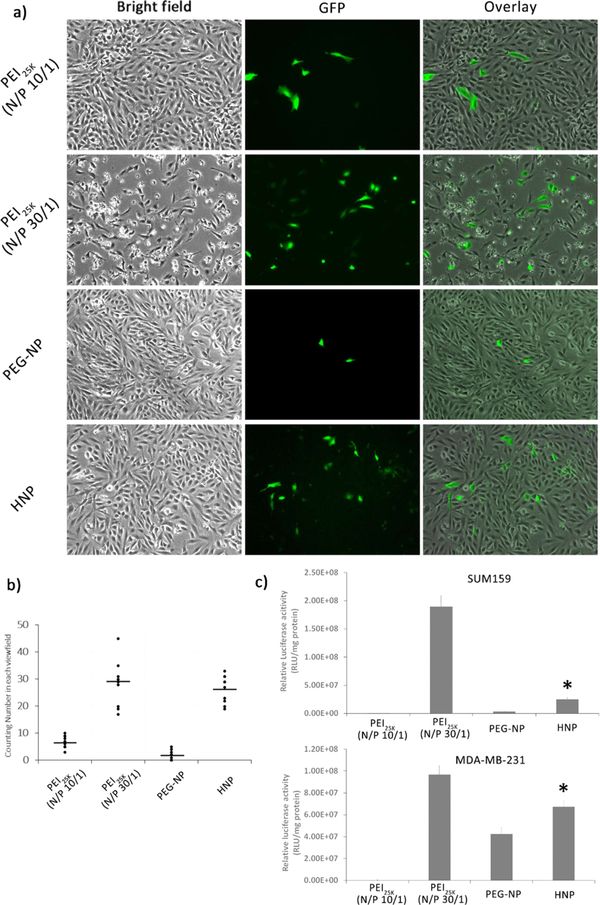
Having demonstrated that HNPs and PEI25K displayed extremely high stability compared to PEG-NP, we next set to determine the gene transfection efficiency of HNPs. The in vitro gene transfection capability of HNPs was investigated in two types of human breast cancer cell lines (SUM159 and MDA-MB-231) with two kinds of pDNAs, which express the reporter genes EGFP and Luc2, respectively. As shown in Figure 5a,b, HNPs efficiently delivered the EGFP expression plasmid into SUM159 cells as evidenced by observing a good number of GFP-expressing cells under a reverse fluorescence microscope. Moreover, HNPs exhibited a very similar EGFP expressing plasmid transfection efficiency in SUM159 cells to that of PEI25K (N/P 30/1) but displayed a much higher transfection efficiency than PEI25K (N/P 10/1) and PEG-NP (Figure 5a,b). In addition, the in vitro gene transfection efficiency of HNPs was further investigated with another pDNA expressing Luc2 by a dual luciferase reporter assay. It was found again that HNPs displayed a significantly higher gene transfection efficiency than PEI25K (N/P 10/1) and PEG-NP in SUM159 and MDA-MB-231 cells, despite showing a lower transfection efficiency than PEI25K (N/P 30/1) (Figure 5c). Even though PEI25K is claimed to show an optimal transfection efficiency in vitro at N/P ratio of 10/1 in serum-free medium in many studies,30 PEI25K (N/P 10/1) did not outperform our HNPs when using the 0.9% NaCl (153 mM) to prepare the complex and incubating with 5% FBS containing medium. Together, these results indicate that HNPs not only have a significantly lower cytotoxicity but also possess a high efficiency in delivering pDNAs into cultured cells, which is comparable to the gene transfection efficiency of the highly cytotoxic PEI25K.
Figure 5.
A comparison of in vitro gene transfection efficiencies of PEI25K (N/P = 10/1 and 30/1), PEG-NP, and HNPs by detecting the expression of a fluorescent protein GFP or Luc2 in SUM159 and MDA-MB-231 cells. HNPs loaded with the GFP or Luc2 expressing pDNA were prepared at a N/P ratio of 30/1, and the PEI25K/DNA complex was also prepared in the same way. The pDNA transfections were performed at a dose of 3 μg of the GFP or Luc2 expressing pDNA. Cells were incubated in the transfection solution for 48 h. (a) Representative images of GFP expression in cells transfected with the GFP expressing pDNA-loaded PEI25K or HNPs (100×). (b) The numbers of GFP expression cells per field of view (POV) counted under a fluorescent microscope. Results are from two independent experiments (n = 10), and five randomly selected fields were counted in each experiment. (c) The pDNA transfections were performed at a dose of 3 μg of Luc2 expressing pDNA and 30 ng of Renilla luciferase expressing pDNA. The activity of Luc2 in cells was determined using a dual luciferase reporter assay as described in the Experimental Section. Results are expressed as the relative luciferase reporter activity calculated as the ratio of firefly luciferase (Luc2) activity divided by the Renilla luciferase activity (mean ± SD, n = 3). *: p < 0.05, compared to the PEI25K (N/P = 10/1) and PEG-NP-transfected group.
To explore the potential mechanism by which HNPs exhibit high in vitro gene transfection efficiency, we determined the effect of the amount of added-PEI. It was found that, under an N/P ratio of 30/1 (Figure 6a,b) or 50/1 (Figure S8), cells transfected with HNPs at a PEG-P(asp)-Ad/PEI10K-CD/PEI10K ratio of 9/1/0 showed no detectable GFP expression 48 h after transfection indicating a poor gene delivery efficiency. With the addition of PEI10K, an increased number of GFP-positive cells were observed indicating an increase in gene delivery efficiency (Figure 6a,b). Moreover, it was found that HNPs at a PEG-P(asp)-Ad/PEI10K-CD/PEI10K ratio of 9/1/2 displayed a significantly higher gene transfection efficiency than HNPs at a PEG-P(asp)-Ad/PEI10K-CD/PEI10K ratio of 9/1/1. The GFP images for pDNA-loaded HNPs with an N/P ratio of either 30/1 or 50/1 validated the same trend (Figures 6a and S8, respectively). Further analysis with ζ-potential revealed that HNPs formulated with 0 equiv of PEI10K addition (compared to PEI10K-CD) showed an average surface charge of −12.4 ± 2.4 mV, while HNPs formulated with 1 equiv of PEI10K addition showed a higher surface charge of 0.3 ± 0.4 mV (Figure 6c). Moreover, an even higher surface charge of 10.2 ± 0.6 mV was observed in HNPs formulated with 2 equiv of PEI10K addition (Figure 6c). Therefore, a cationic nanoparticle surface has strong interactions with the cell membranes by ionic interactions or hydrophobic interactions. The increased surface positive charge of HNPs with the addition of 2 equiv of PEI10K may facilitate the transport of the complexes across the cell membrane and thus significantly improve the gene transfection efficiency.
3.6. in vivo pDNA Delivery Efficiency of HNPs Post Intratumoral Injection.
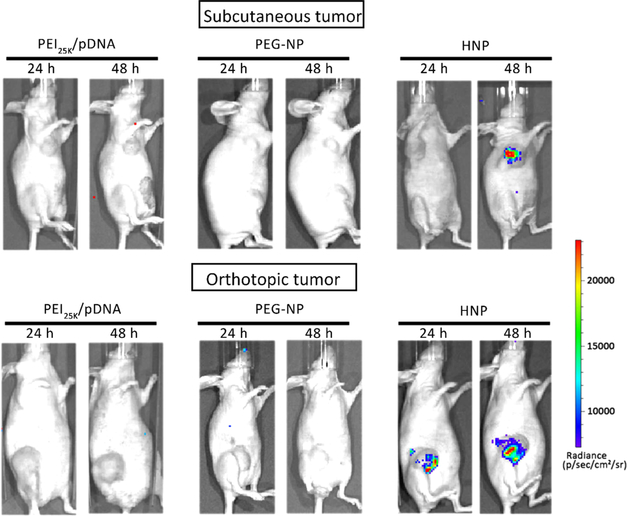
We further investigated the in vivo gene transfection efficiency of HNPs as it is an important component of evaluating nonviral gene carriers.31 Previous studies have done a direct intratumoral injection of therapeutic siRNA delivery complexes into various mouse xenograft models, which results in decreased expression levels of the targeting genes.32 However, we used the injection of a luciferase reporter gene because the usage of reporter genes for in vivo study could help separate the effects of the nanoparticles from cargo therapeutics it carries. By measuring the level of tumor tissue chemiluminescence, we were able to test the in vivo gene delivery efficiency of our HNPs uploaded with reporter pDNA (pGL4.5-Luc2).
To achieve efficient pDNA delivery in vivo, the carriers must be able to protect the payloads from degradation. The popular way to study the in vivo transfection efficiency of nanocarriers is blocking the expression of luciferase reporter gene existing in the xenograft tumor cells through siRNA,33 probably due to naked or poorly protected pDNA with a larger size are more easily degraded by nuclease compared to siRNA. To the best of our knowledge, few studies have used this direct transfection approach of pDNAs to investigate the in vivo gene transfection efficiency of nanocarriers. As shown in Figure 7, chemiluminescence signals in mouse mammary xenograft tumor tissue were detected 24 and 48 h after intratumoral injection of HNPs loaded with 25 μg of pDNA (pGL4.5-Luc2), which indicates the high activity or expression level of Luc2 in the tumor tissue. In contrast, negligible chemiluminescence signals could be observed from the tumors of the mice treated by the PEI25K/pDNA (pGL4.5-Luc2) complex and PEG-NP. The luciferase expression level was found to be time dependent, which was detectable in mice at 24 h postinjection, and reached significantly higher levels after approximately 48 h postinjection. These are consistent with the in vitro results. These results suggest that HNP-entrapped-pDNA could penetrate through tumor tissue and be internalized by the cells and released intracellularly displaying a high bioactivity.
Figure 7.
HNPs maintains its gene delivery capability in mouse mammary xenograft tumor tissues while PEG-NP and PEI25k lost that. The mouse mammary tumors were produced as described in the Experimental Section. A single dose of HNPs loaded with 25 μg of Luc2 expressing pDNA was introduced into tumor tissues through an intratumoral injection. Twenty-four and forty-eight hours later, the animal was given D-luciferin by intraperitoneal injection, and the activity of tumor tissue-expressed Luc2 was determined by measuring the intensity of chemiluminescence signals using an IVIS live animal bioluminescence imaging system as described in the Experimental Section. The photo is a representative image showing strong chemiluminescence signals in mouse mammary tumor tissue (n = 3).
3.7. in vivo Stability and Distribution Behaviors of HNPs Post Intravenous Injection.
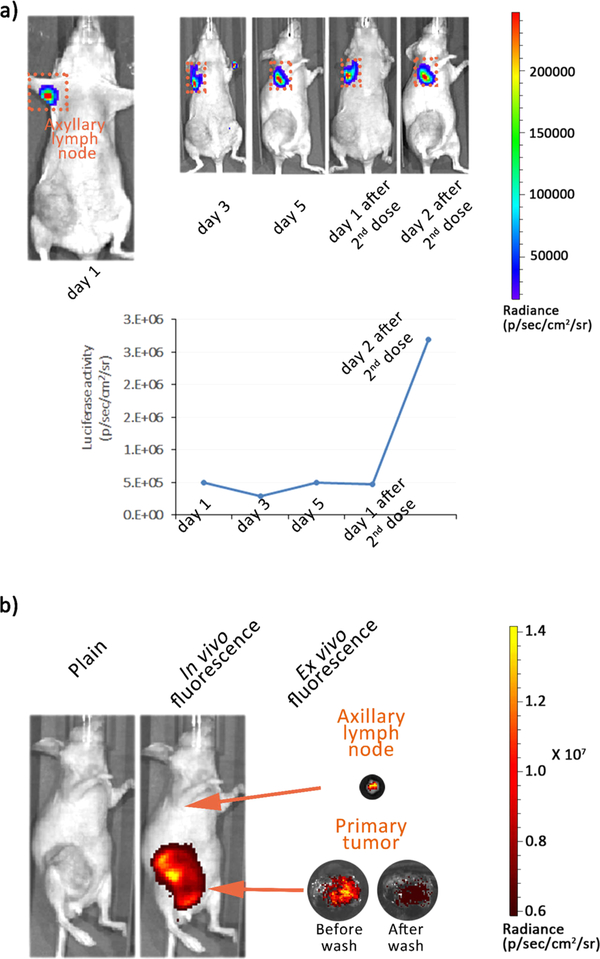
For in vivo gene delivery, intratumoral and intravenous injections are two frequently used methods. However, the behavior of nanoparticles post intravenous injection is significantly more complex than that post intratumoral injection. Intravenous administration of nanoparticles is usually associated with lower in vivo transfection efficiency due to clearance from circulation. Hence, the in vivo pDNA delivery efficiency of HNPs was further investigated following systemic injection. Live animal in vivo imaging can provide information about the time course of HNPs accumulation and bioavailability in target organs. On days 1, 3, and 5 post injection, high levels of chemiluminescence were detected in the positions of tumor-draining axillary lymph nodes (Figure 8a). To further validate that the signals are due to HNPs’ accumulation at the axillary lymph node, a second dose of HNPs was administered on day 5 post first injection and more than a 5-fold increase of Luc2 activities was observed on day 2 post second injection.
Figure 8.
HNPs demonstrate good in vivo stability post intravenous injection. (a) Bioluminescence images of MDA-MB-231-tumor-bearing mouse postinjection of HNPs loaded with 50 μg of pGL4.5-Luc2 pDNA. (b) in vivo and ex vivo fluorescent images of a MDA-MB-231-tumor-bearing mouse at 24 h postinjection of cy7.5-labeled HNPs (n = 3).
Although quantitative analysis from repeated administration indicated a significantly higher uptake of HNPs by lymph nodes, interestingly, negligible chemiluminescence signals were observed at the primary tumor upon HNP injections. To better understand this observation, we conjugated near IR dye cy7.5 with PEI10K and incorporated it into the HNPs. Cy7.5-labeled HNPs were injected intravenously into MDA-MB-231-tumor-bearing mice (n = 3) and imaged via fluorescence. The in vivo image at 24 h postinjection showed significantly higher cy7.5 fluorescence signals in tumor sites compared to other organs indicating selective HNP accumulation in tumor tissues (Figure 8a); the ex vivo images also revealed that HNPs were successfully delivered into lymph node and primary tumor (Figure 8b). HNPs’ uptake by axillary lymph node was well consistent with the biofluorescence result (Figure 8b). Moreover, the primary tumor was extracted from the mice and washed with PBS, which resulted in a reduced fluorescence intensity (Figure 8b) and demonstrated the poor penetration probably attributed to the negligible expression of pDNAs at the tumor site in Figure 8a.
4. DISCUSSION
The main objective of our study was to demonstrate the feasibility of in vivo delivering the pDNA through a novel form of hybrid PEGylated nanoparticles for transgene expression with high efficiency. We constructed HNPs using a self-assembling supramolecular method, and the pDNA was packaged into HNPs that have an average diameter of about 80–150 nm using PEGylated complex inclusion and coacervate matrix techniques. Although conventional nonviral vectors work very well in vitro, they have poor in vivo transfection efficiency for pDNAs. This is likely due to their nonspecific interactions with serum proteins, undesirable cells, or the extracellular matrix.8,34 Presumably, the instability of the formed nanocarrier/pDNA complexes during the transfection process is the major hurdle for the in vivo use of current delivery systems. Hence, an ideal gene-therapy nanocarrier should deliver intact pDNAs efficiently to the target cells. Indeed, in vivo delivery for pDNAs still remains a major challenge despite the similarity to siRNA formulation and delivery processes, including condensation, protection, and endosomal escape.4,35 Some nucleotides on the short 21–23 bp siRNAs can be chemically modified to improve stability and alter the duration of the therapeutic effect, so there are reports on the efficacy of naked siRNA upon direct injection into tumors in mice,36 which is particularly difficult for pDNAs with a size of several thousand bps using nonviral nanocarriers.4,37 To the best of our knowledge, our work is the first to show that luciferase activity images from mouse mammary xenograft tumors could be obtained by injecting HNP intratumorally, which indicated pGL4.5-Luc2 loaded in HNPs has been successfully transfected into the tumor tissue. This was probably attributed to the following three reasons.
First, the grafted PEG chains were likely to form a protective surface corona while PEI10K/P(asp)-coated PEI10K-CD/DNA served as a cross-linked shield of the internal core (Figure 1a). The capability to protect pDNA from degradation in the extracellular environment is crucial to successful delivery to a cell as well as to high transfection efficiency. The decreased particle—particle and particle—protein interactions resulting from steric effects of the PEG layer attributes to our HNPs’ stability. The PEI10K/P(asp) coacervate caging could also contribute significantly to increased pDNA stability through the prevention of an unfavorable dissociation of the formed nanoparticles, which is more condensed than PEG-NP (Figures 3 and S6). Additionally, the extra positive charges of PEI10K are capable of further promoting complex compression with the polyanionic nucleic acids of pDNA.
Second, the rapid escape of delivered-DNA from endosome to cytoplasm is necessary for efficient gene transfection, and PEI10K was used to achieve that purpose. This could induce an early escape from endosomal compartments by destabilizing the endosomes. From the PEG-P(asp)-Ad/PEI10K-CD/PEI10K mol ratio of 9/1/0 to 9/1/2, the gene transfection efficiencies of HNPs were significantly enhanced with an increasing amount of PEI10K (Figures 6a and S8). PEI is known to be a favorable polycation gene carrier with high pH-buffering capacity, which could enhance nanoplex unpacking in the cytosol and/or endosomes and eventually improve transfection efficiency.38 In response to a high acidic environment such as endosomal acidity (pH 4–5), the proton sponge effect of our HNPs due to the addition of positively charged PEI10K could strengthen the ability of these HNPs to escape from the endosomes.
Third, the surface charge of our produced HNP complex was negative at the PEG-P(asp)-Ad/PEI10K-CD/PEI10K mol ratio of 9/1/0, rose to zero at that of 9/1/1, and became positive at that of 9/1/2 (Figures 6c and S9). The ζ-potential change of HNPs was attributed to the presence of more PEI10K. The surface charge of a nanoparticle can dramatically influence the way it interacts with the target cell.39 A positively charged nanoparticle can facilitate uptake by associating with the negatively charged cellular membrane, while the negatively surface-charged nanoparticles presumably bind weakly with cells due to charge—charge repulsion with the cell surface. Therefore, the variant surface charges could be another important reason for the lack of the transfection ability of HNPs with low PEI10K loading.
As a whole, the PEI10K/P(asp) coacervate structure in HNPs could have advantages for interacting with cell membrane and facilitating the escape of the pDNA from endosomes into the cytoplasm compared to previously reported PEGylated nanocarriers. The presence of PEI10K in our HNP formulation is critical. PEI10K may partially detach from HNPs extracellularly, which could help enhance the interactions of HNPs with negatively charged cell membranes. Alternatively, PEI10K could dissociate from HNP intracellularly and enhance the escape of pDNA from endosomes/lysosomes for improved transfection efficiency through the proton sponge effect of PEI10K. The formation of polyelectrolyte complexes is determined by the anion/cation ratio, regardless of the total polymer concentration or molecular weight of the polyelectrolytes. According to our preliminary experiment (Figure S9), with the addition of more PEI10K, the surface charges of nanoparticles loaded with pDNA varied less dramatically compared to those of nanoparticles without pDNA. This suggests that cations of PEI10K were well matched with anions from pDNA and P(asp), which helped PEI10K to be well integrated into HNPs preventing the spontaneous release of PEI10K. Figures 2 and S7 also provide very strong evidence for the rigidity of HNP in the presence of serum proteins. Therefore, the enhancement of transfection efficiency by HNP most likely resulted from the proton sponge effect from intracellular coacervate. Further experiments are needed to firmly establish the mechanisms of enhanced HNP transfection efficiency in vitro and in vivo. According to our experimental data, more PEI10K addition can further improve the transfection efficiency but may increase the cytotoxicity at the same time. Therefore, a proper balance between transfection efficiency and cytotoxicity is needed during the formulation design in order to achieve efficient and safe in vivo gene delivery.
Strong chemiluminescence signals were detected at the site of axillary lymph node, which affirmed the in vivo transfection efficiency of HNP post intravenous injection (Figure 8a). The signals close to lymph nodes are probably attributed to that abnormal draining axillary lymph nodes associated with triple negative breast cancer metastases.40 After systemic administration, long-circulating nanocarriers with the size of 10–300 nm in diameter typically accumulate in lymph nodes, spleen, liver, and bone marrow, which contain high amounts of macrophages.41 The coacervate integration prevents the unexpected dissociation of HNPs, while exterior-anchored PEG could extend their persistence in the circulation and accumulation at the lymph node site. Abnormalities in nodal architecture or in lymph flow caused by metastases can lead to accumulation of nanoparticles, which are clinically detectable by MRI.42
The observed fluorescence signals at the primary tumor suggest that the HNPs could reach the tumor tissues presumably due to the enhanced permeability and retention (EPR) effect. The fact that PBS wash of the extracted tumor reduced the fluorescence intensities indicated that the HNPs were present extracellularly and did not penetrate deeply into the tumor and thus were easily removable by buffer washes. The extracellular distribution could also help explain the lack of chemiluminescence signals at the primary tumor despite the accumulation of HNPs. The low chemiluminescence at the primary tumor site upon systemic administration of HNPs is consistent with a literature report, where magnetic nanoparticles incorporating miRNA were injected intravenously into MDA-MB-231 tumor bearing mice.43 The miRNA delivered successfully arrested metastatic growth in the lymph nodes while growth of the primary tumor was still maintained, despite the extensive distribution of nanoparticles in both lymph nodes and primary tumor sites. This exact mechanism is unknown. The strategy of installing novel peptide ligands (e.g., iRGD) to deliver nanoparticles deeply into the tumor has been reported recently,44,45 which can potentially enhance penetration and transfection efficiencies of HNPs at the primary tumor site and will be adopted in the future.
5. CONCLUSION
In this study, we integrated the complex coacervate modularity with a previously reported PEGylated inclusion complex to construct a novel form of hybrid complex coacervation nanoparticles. Due to the incorporation of P(asp)/PEI10K coacervate into the PEGylated inclusion complex, the resulting HNPs had not only increased stability and reduced cytotoxicity but also significantly enhanced in vivo gene transfection efficiency compared to that of traditional PEGylated NP and PEI25K, a gold standard of in vitro transfection. The findings from this study suggest that the proposed HNPs have the potential to serve as a suitable nanoscale-based gene agent delivery vehicle. While exterior PEGylation protected HNPs from nonspecific interaction with serum protein, the integration of complex coacervation at the interface of PEI10K-CD/pDNA core and the PEG shell could prevent HNPs’ premature dissociation and increase the cellular uptake and trafficking. Hence, this integrative polycation coacervate concept could be extended into the design of new PEGylated gene delivery systems in the future.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by a Research Scholar Grant (RGS-15–026-01-CSM) from American Cancer Society to C.Y.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.6b10306.
The synthesis and characterization for PEG-P(asp)-Ad and PEI10K-CD and PEG-Ad; the particle size comparison among PEI10K-CD/pDNA, PEG-NP, and HNP in 0.9% saline solution; size distribution curves of HNPs before/after incubation with 20% (v/v) FBS at 37 °C; the representative images of cells transfected with HNPs formulated with different amounts of PEI10K (N/P ratio 50/1); the zeta-potential comparisons of the HNPs formulated with different amounts of PEI10K in water (PDF)
REFERENCES
- (1).Long X; Zhang Z; Han S; Tang M; Zhou J; Zhang J; Xue Z; Li Y; Zhang R; Deng L; et al. Structural Mediation on Polycation Nanoparticles by Sulfadiazine to Enhance DNA Transfection Efficiency and Reduce Toxicity. ACS Appl. Mater. Interfaces 2015, 7 (14), 7542–7551. [DOI] [PubMed] [Google Scholar]
- (2).Zheng N; Yin L; Song Z; Ma L; Tang H; Gabrielson NP; Lu H; Cheng J Maximizing Gene Delivery Efficiencies of Cationic Helical Polypeptides via Balanced Membrane Penetration and Cellular Targeting. Biomaterials 2014, 35 (4), 1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Moghimi SM; Symonds P; Murray JC; Hunter AC; Debska G; Szewczyk A A Two-stage Poly(ethylenimine)-Mediated Cytotoxicity: Implications for Gene Transfer/therapy. Mol. Ther. 2005, 11 (6), 990–995. [DOI] [PubMed] [Google Scholar]
- (4).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15 (8), 541–555. [DOI] [PubMed] [Google Scholar]
- (5).Jin L; Zeng X; Liu M; Deng Y; He N Current Progress in Gene Delivery Technology Based on Chemical Methods and Nano-Carriers. Theranostics 2014, 4 (3), 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gary DJ; Puri N; Won Y-Y Polymer-Based siRNA Delivery: Perspectives on the Fundamental and Phenomenological Distinctions from Polymer-Based DNA Delivery. J. Controlled Release 2007, 121 (1–2), 64–73. [DOI] [PubMed] [Google Scholar]
- (7).Abdallah B; Hassan A; Benoist C; Goula D; Behr JP; Demeneix BA A Powerful Nonviral Vector for in Vivo Gene Transfer into the Adult Mammalian Brain: Polyethylenimine. Hum. Gene Ther. 1996, 7 (16), 1947–1954. [DOI] [PubMed] [Google Scholar]
- (8).Coll J-L; Chollet P; Brambilla E; Desplanques D; Behr J-P; Favrot M In Vivo Delivery to Tumors of DNA Complexed with Linear Polyethylenimine. Hum. Gene Ther. 1999, 10 (10), 1659–1666. [DOI] [PubMed] [Google Scholar]
- (9).Oupicky D; Carlisle R; Seymour L Triggered Intracellular Activation of Disulfide Crosslinked Polyelectrolyte Gene Delivery Complexes with Extended Systemic Circulation in Vivo. Gene Ther. 2001, 8 (9), 713–725. [DOI] [PubMed] [Google Scholar]
- (10).Mullen PM; Lollo CP; Phan Q-C; Amini A; Banaszczyk MG; Fabrycki JM; Wu D; Carlo AT; Pezzoli P; Coffin CC; et al. Strength of Conjugate Binding to Plasmid DNA Affects Degradation Rate and Expression Level in Vivo. Biochim. Biophys. Acta, Gen. Subj 2000, 1523 (1), 103–110. [DOI] [PubMed] [Google Scholar]
- (11).Taratula O; Garbuzenko OB; Kirkpatrick P; Pandya I; Savla R; Pozharov VP; He H; Minko T Surface-Engineered Targeted PPI Dendrimer for Efficient Intracellular and Intratumoral siRNA Delivery. J. Controlled Release 2009, 140 (3), 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Pack DW; Hoffman AS; Pun S; Stayton PS Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discovery 2005, 4 (7), 581–593. [DOI] [PubMed] [Google Scholar]
- (13).Veiman K-L; Kunnapuu K; Lehto T; Kiisholts K; Pärn K; Langel Ü; Kurrikoff K PEG Shielded MMP Sensitive CPPs for Efficient and Tumor Specific Gene Delivery in Vivo. J. Controlled Release 2015, 209, 238–247. [DOI] [PubMed] [Google Scholar]
- (14).Yousefi A; Storm G; Schiffelers R; Mastrobattista E Trends in Polymeric Delivery of Nucleic Acids to Tumors. J. Controlled Release 2013, 170 (2), 209–218. [DOI] [PubMed] [Google Scholar]
- (15).Harada A; Takashima Y; Yamaguchi H Cyclodextrin-Based Supramolecular Polymers. Chem. Soc. Rev. 2009, 38 (4), 875–882. [DOI] [PubMed] [Google Scholar]
- (16).Yang J; Zhang Q; Chang H; Cheng Y Surface-Engineered Dendrimers in Gene Delivery. Chem. Rev. 2015, 115 (11), 5274–5300. [DOI] [PubMed] [Google Scholar]
- (17).Pun SH; Davis ME Development of a Nonviral Gene Delivery Vehicle for Systemic Application. Bioconjugate Chem. 2002, 13 (3), 630–639. [DOI] [PubMed] [Google Scholar]
- (18).Pun SH; Tack F; Bellocq NC; Cheng J; Grubbs BH; Jensen GS; Davis ME; Brewster M; Janicot M; Janssens B; et al. Targeted Delivery of RNA-Cleaving DNA Enzyme (DNAzyme) to Tumor Tissue by Transferrin-Modified, Cyclodextrin-Based Particles. Cancer Biol. Ther. 2004, 3 (7), 641–650. [DOI] [PubMed] [Google Scholar]
- (19).Pun SH; Bellocq NC; Liu A; Jensen G; Machemer T; Quijano E; Schluep T; Wen S; Engler H; Heidel J; et al. Cyclodextrin-Modified Polyethylenimine Polymers for Gene Delivery. Bioconjugate Chem. 2004, 15 (4), 831–840. [DOI] [PubMed] [Google Scholar]
- (20).Davis M; Pun S; Bellocq N; Reineke T; Popielarski S; Mishra S; Heidel J Self-Assembling Nucleic Acid Delivery Vehicles via Linear, Water-soluble, Cyclodextrin-Containing Polymers. Curr. Med. Chem. 2004, 11 (2), 179–197. [DOI] [PubMed] [Google Scholar]
- (21).Hatakeyama H; Akita H; Harashima H A Multifunctional Envelope Type Nano Device (MEND) for Gene Delivery to Tumours Based on the EPR Effect: A Strategy for Overcoming the PEG Dilemma. Adv. Drug Delivery Rev. 2011, 63 (3), 152–160. [DOI] [PubMed] [Google Scholar]
- (22).Majzoub RN; Chan C-L; Ewert KK; Silva BFB; Liang KS; Jacovetty EL; Carragher B; Potter CS; Safinya CR Üptake and Transfection Efficiency of PEGylated Cationic Liposome–DNA Complexes with and without RGD-tagging. Biomaterials 2014, 35 (18), 4996–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Priftis D; Megley K; Laugel N; Tirrell M Complex Coacervation of Poly (ethylene-imine)/polypeptide Aqueous Solutions: Thermodynamic and Rheological Characterization. J. Colloid Interface Sci 2013, 398, 39–50. [DOI] [PubMed] [Google Scholar]
- (24).Kuo C-H; Leon L; Chung EJ; Huang R-T; Sontag TJ; Reardon CA; Getz GS; Tirrell M; Fang Y Inhibition of Atherosclerosis-Promoting MicroRNAs via Targeted Polyelectrolyte Complex Micelles. J. Mater. Chem. B 2014, 2, 8142–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jessel N; Oulad-Abdelghani M; Meyer F; Lavalle P; Haikel Y; Schaaf P; Voegel J-C Multiple and Time-Scheduled in Situ DNA Delivery Mediated by β-Cyclodextrin Embedded in a Polyelectrolyte Multilayer. Proc. Natl. Acad. Sci U. S. A. 2006, 103 (23), 8618–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Priftis D; Leon L; Song Z; Perry SL; Margossian KO; Tropnikova A; Cheng J; Tirrell M Self-Assembly of α - Helical Polypeptides Driven by Complex Coacervation. Angew. Chem. 2015, 127 (38), 11280–11284. [DOI] [PubMed] [Google Scholar]
- (27).Humphries B; Wang Z; Oom AL; Fisher T; Tan D; Cui Y; Jiang Y; Yang C MicroRNA-200b Targets Protein Kinase Cα and Suppresses Triple-Negative Breast Cancer Metastasis. Carcinogenesis 2014, 35 (10), 2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Minn AJ; Gupta GP; Siegel PM; Bos PD; Shu W; Giri DD; Viale A; Olshen AB; Gerald WL; Massagué J Genes that Mediate Breast Cancer Metastasis to Lung. Nature 2005, 436 (7050), 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Du X-J; Wang J-L; Liu W-W; Yang J-X; Sun C-Y; Sun R; Li H-J; Shen S; Luo Y-L; Ye X-D; et al. Regulating the Surface Poly (ethylene glycol) Density of Polymeric Nanoparticles and Evaluating its Role in Drug Delivery in Vivo. Biomaterials 2015, 69, 1–11. [DOI] [PubMed] [Google Scholar]
- (30).Deng R; Yue Y; Jin F; Chen Y; Kung H-F; Lin MC; Wu C Revisit the Complexation of PEI and DNA—How to Make Low Cytotoxic and Highly Efficient PEI Gene Transfection non-Viral Vectors with a Controllable Chain Length and Structure? J. Controlled Release 2009, 140 (1), 40–46. [DOI] [PubMed] [Google Scholar]
- (31).Yang F; Huang W; Li Y; Liu S; Jin M; Wang Y; Jia L; Gao Z Anti-Tumor Effects in Mice Induced by Survivin-Targeted siRNA Delivered through Polysaccharide Nanoparticles. Biomaterials 2013, 34 (22), 5689–5699. [DOI] [PubMed] [Google Scholar]
- (32).Grzelinski M; Urban-Klein B; Martens T; Lamszus K; Bakowsky U; Hobel S; Czubayko F; Aigner A RNA Interference-Mediated Gene Silencing of Pleiotrophin through Polyethylenimine-Complexed Small Interfering RNAs in Vivo Exerts Antitumoral Effects in Glioblastoma Xenografts. Hum. Gene Ther. 2006, 17 (7), 751–766. [DOI] [PubMed] [Google Scholar]
- (33).Bartlett DW; Su H; Hildebrandt IJ; Weber WA; Davis ME Impact of Tumor-Specific Targeting on the Biodistribution and Efficacy of siRNA Nanoparticles Measured by Multimodality in Vivo Imaging. Proc. Natl. Acad. Sci U. S. A. 2007, 104 (39), 15549–15554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Glover DJ; Lipps HJ; Jans DA Towards Safe, Non-Viral Therapeutic Gene Expression in Humans. Nat. Rev. Genet. 2005, 6 (4), 299–310. [DOI] [PubMed] [Google Scholar]
- (35).Scholz C; Wagner E Therapeutic Plasmid DNA versus siRNA Delivery: Common and Different Tasks for Synthetic Carriers. J. Controlled Release 2012, 161 (2), 554–565. [DOI] [PubMed] [Google Scholar]
- (36).NIU XY; PENG ZL; DUAN WQ; Wang H; Wang P Inhibition of HPV 16 E6 Oncogene Expression by RNA Interference in vitro and in Vivo. Int. J. Gynecol. Cancer 2006, 16 (2), 743–751. [DOI] [PubMed] [Google Scholar]
- (37).Fernandes AR; Chari DM Part I: Minicircle Vector Technology Limits DNA Size Restrictions on ex Vivo Gene Delivery Using Nanoparticle Vectors: Overcoming a Translational Barrier in Neural Stem Cell Therapy. J. Controlled Release 2016, 238, 289. [DOI] [PubMed] [Google Scholar]
- (38).Gonçalves C; Akhter S; Pichon C; Midoux P Intra-Cellular Availability of pDNA and mRNA after Transfection: A Comparative Study in between Polyplexes, Lipoplexes and Lipopolyplexes. Mol. Pharmaceutics 2016, 13, 3153. [DOI] [PubMed] [Google Scholar]
- (39).Whitehead KA; Langer R; Anderson DG Knocking down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discovery 2009, 8 (2), 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Skobe M; Hawighorst T; Jackson DG; Prevo R; Janes L; Velasco P; Riccardi L; Alitalo K; Claffey K; Detmar M Induction of Tumor Lymphangiogenesis by VEGF-C Promotes Breast Cancer Metastasis. Nat. Med. 2001, 7 (2), 192–198. [DOI] [PubMed] [Google Scholar]
- (41).Weissleder R; Nahrendorf M; Pittet MJ Imaging Macrophages with Nanoparticles. Nat. Mater. 2014, 13 (2), 125–138. [DOI] [PubMed] [Google Scholar]
- (42).Harisinghani MG; Barentsz J; Hahn PF; Deserno WM; Tabatabaei S; van de Kaa CH; de la Rosette J; Weissleder R Noninvasive Detection of Clinically Occult Lymph-Node Metastases in Prostate Cancer. N. Engl. J. Med. 2003, 348 (25), 2491–2499. [DOI] [PubMed] [Google Scholar]
- (43).Yigit M; Ghosh S; Kumar M; Petkova V; Kavishwar A; Moore A; Medarova Z Context-Dependent Differences in miR-10b Breast Oncogenesis can be Targeted for the Prevention and Arrest of Lymph Node Metastasis. Oncogene 2013, 32 (12), 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sugahara KN; Teesalu T; Karmali PP; Kotamraju VR; Agemy L; Girard OM; Hanahan D; Mattrey RF; Ruoslahti E Tissue-Penetrating Delivery of Compounds and Nanoparticles into Tumors. Cancer Cell 2009, 16 (6), 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wang J; Wang H; Li J; Liu Z; Xie H; Wei X; Lu D; Zhuang R; Xu X; Zheng S iRGD-Decorated Polymeric Nanoparticles for the Efficient Delivery of Vandetanib to Hepatocellular Carcinoma: Preparation and in vitro and in Vivo Evaluation. ACS Appl Mater. Interfaces 2016, 8 (30), 19228–19237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.