Abstract
Filoviruses, which include Ebola virus and Marburg virus, are negative-sense RNA viruses associated with sporadic outbreaks of severe viral hemorrhagic fever characterized by uncontrolled virus replication. The extreme virulence and emerging nature of these zoonotic pathogens make them a significant threat to human health. Replication of the filovirus genome and production of viral RNAs requires the function of a complex of four viral proteins, the nucleoprotein (NP), viral protein 35 (VP35), viral protein 30 (VP30) and large protein (L). The latter performs the enzymatic activities required for production of viral RNAs and capping of viral mRNAs. Although it has been recognized that interactions between the virus-encoded components of the EBOV RNA polymerase complex are required for viral RNA synthesis reactions, specific molecular details have, until recently, been lacking. New efforts have combined structural biology and molecular virology to reveal in great detail the molecular basis for critical protein-protein interactions (PPIs) necessary for viral RNA synthesis. These efforts include recent studies that have identified a range of host factors and in some instances demonstrated unique mechanisms by which they act. For a select number of these interactions, combined use of mutagenesis, over-expressing of peptides corresponding to PPI interfaces and identification of small molecules that disrupt PPIs have demonstrated the functional significance of virus-virus and virus-host PPIs and suggest several as potential targets for therapeutic intervention.
1. Significance of filoviruses as human pathogens.
Filoviruses are filamentous, enveloped viruses with non-segmented, negative-sense RNA genomes (Messaoudi et al., 2015). The family Filoviridae is comprised of the genus Ebolavirus includes Zaire ebolavirus (Ebola virus, EBOV) and five other species, the genus Marburgvirus, which includes Marburg virus (MARV), and the genus Cuevavirus (Afonso et al., 2016; Goldstein et al., 2018). Members of the Ebolavirus and Marburgvirus genera are zoonotic pathogens that have caused repeated outbreaks with substantial lethality in humans (Rougeron et al., 2015). The largest such outbreak on record was caused by EBOV and occurred in West Africa between 2013-2016. This resulted in upwards of 28,000 infections, more than 11,000 deaths, and the export of infected cases to the United States and Europe (Spengler et al.). In pregnant women, the fatality rate is estimated to be 70% (Hayden et al., 2017). The only treatments available for infected individuals were supportive care and experimental therapies, hampering patient treatment and leaving healthcare workers at risk. Survivors are known to exhibit persistent infections with virus residing in immune privileged sites, including the eye and testes (Jacobs et al., 2016; Uyeki et al., 2016; Yeh et al., 2015; Zeng et al., 2017). The West Africa epidemic reinforced the threat posed by filoviruses and resulted in a major push to provide potential therapeutics and vaccines for infected individuals. The results of a trial with vesicular stomatitis virus (VSV)-based vaccine in which the VSV glycoprotein is replaced by the Ebola virus glycoprotein (GP) were very promising (Henao-Restrepo et al., 2017). However, even if effective vaccines are developed, it is unlikely that universal vaccination will be implemented for EBOV or for other filoviruses. Therefore, there will remain a need for effective complimentary therapies, including small molecule drugs. This reinforces the need for ongoing development of new filovirus therapeutics and continued research into filovirus biology.
2. Overview of filovirus replication.
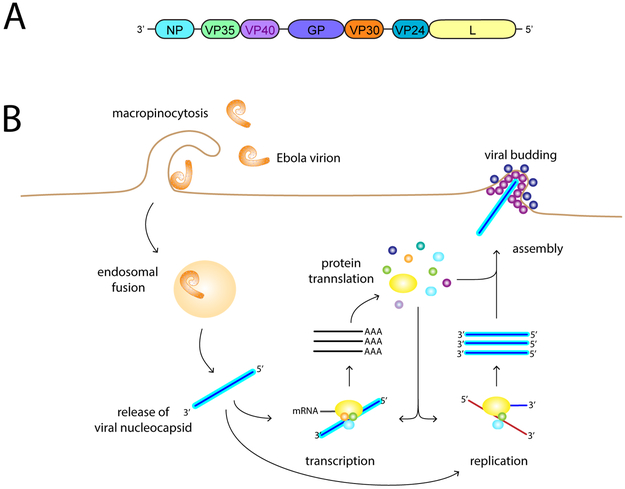
The filovirus genome is approximately 19 kilobases in length and encodes up to nine translation products from seven separate transcriptional units (Feldmann et al., 2015; Messaoudi et al., 2015) (Fig 1A). These genes encode the viral nucleoprotein (NP), viral protein of 35 kDa (VP35), VP40, a type I transmembrane glycoprotein (GP), VP30, VP24, and the large protein (L), which is the viral polymerase. Members of the Ebolavirus genus also produce at least two secreted forms of the GP protein (Messaoudi et al., 2015).
Figure 1. The filovirus genome and replication cycle.
A. A schematic diagram of the EBOV genome showing individual genes, named after the proteins they encode: NP, nucleoprotein; VP35, viral protein 35; VP40, viral protein 40; GP, glycoprotein/soluble glycoprotein; VP30, viral protein 30; VP24, viral protein 24; L, Large protein. The GP gene of members of the Ebolavirus and Cuevavirus genera also encode a soluble glycoprotein (sGP). B. Overview of the filovirus replication cycle. Proteins are indicated by circles and ovals with colors corresponding to those in panel A. Depicted are viral entry via micropinocytosis, release of the genomic RNA into the cytoplasm where the depicted viral mRNA and genome replication take place. Assembly and budding occur at the plasma membrane.
The filovirus replication cycle can be summarized as follows (Fig. 1B). Viral entry is mediated by GP which acts as an attachment factor and mediates fusion of viral and host cell membranes within an endosomal compartment (Davey et al., 2017). A prerequisite for fusion is processing of GP by host cell cathepsin proteases within endosomes, allowing cleaved GP to interact with an intra-endosomal receptor, Niemann-Pick disease, type C1 (NPC-1) (Carette et al., 2011; Chandran et al., 2005; Cote et al., 2011; Miller et al., 2012). The viral genome is released into the cytoplasm as a ribonucleoprotein complex. This complex serves as the template for the RNA synthesis reactions that replicate the viral genomic RNA and transcribe the mRNAs for viral gene expression. Replication of the viral genomic RNA requires NP, which associates with the viral genomic and antigenomic RNAs throughout the course of infection; VP35, a non-enzymatic cofactor for the viral RNA-dependent RNA polymerase that also serves as a potent suppressor of innate antiviral signaling pathways, and L. L possesses all the enzymatic activities required for viral transcription and genome replication, including RNA-dependent RNA polymerase activity, guanyltransferase, and methyltransferase activities (Muhlberger, 2007). Viral transcription (mRNA synthesis) involves the synthesis of distinct 5’-capped, 3’-polyadenylated mRNAs from each of the viral genes and requires, in addition to NP, VP35 and L, the VP30 protein (Fig. 2) (Muhlberger, 2007). In addition to the required viral proteins, host factors modulate viral RNA synthesis through interaction with viral factors. (Batra et al., 2018; Luthra et al., 2015; Luthra et al., 2013; Smith et al., 2010). However, a complete understanding as to how host factors contribute to viral RNA synthesis remains incomplete.
Figure 2. Schematic diagram of the Ebola virus RNA synthesis complex.
The template viral genome RNA is depicted as a blue line. The RNA is encapsidated by the bilobed NP. Viral transcription requires the presence of VP30 and the requirement for VP30 is due to the presence of a stem loop near the transcription start site in the NP gene. VP35 interacts with NP and L and brings L, which possesses the necessary enzymatic activities required for viral transcription and replication, to the template. The interaction of VP35-L with NP presumably exposes the template RNA to allow access to L.
Other viral functions include filovirus assembly and release (Kolesnikova et al., 2017). The VP40 matrix protein drives the membrane budding events that lead to release of new virus particles. GP is incorporated into the membrane of viral particles and enhances budding. Viral RNPs that contain genomic RNA, NP, VP35, VP30 and VP24 are recruited into the budding particles. In addition to playing roles in replication and assembly, several filovirus proteins counteract host innate antiviral defenses (Olejnik et al., 2017). The filovirus VP35 proteins block interferon (IFN)-α/β production and the VP24 proteins of Ebolavirus and Cuevavirus genera members and the VP40 proteins of the Marburgvirus genus block IFN-induced antiviral signaling (Basler et al., 2000; Cardenas et al., 2006; Leung et al., 2010; Mateo et al., 2010; Reid et al., 2006; Reid et al., 2007; Valmas and Basler, 2011; Valmas et al., 2010; Xu et al., 2014). The MARV VP24 protein also modulates host antioxidant response pathways through interaction with the host protein Kelch-like ECH-associated protein 1 (Keap1) (Edwards et al., 2014; Johnson et al., 2016; Page et al., 2014).
2. Evidence that EBOV RNA synthesis reactions are viable targets for therapeutics
Given their essential role for viral gene expression and viral genome RNA replication, targeting the viral RNA synthesis reactions is a logical strategy for therapeutic development. It is a strategy that also shows substantial promise. As examples, GS-5734 (Remdesivir) and BCX4430, are two nucleoside analogs that target viral RNA synthesis and have proven effective in non-human primate models (Warren et al., 2016; Warren et al., 2014). Other examples of small molecules with anti-EBOV activity include the nucleoside analog favipiravir that is effective in cell culture, can protect mice, reduce viral titers and showed some evidence of efficacy in EBOV- and MARV-infected macaques (Oestereich et al., 2014). Carbocyclic nucleosides, including 3-deazaneplanocin A have broad spectrum activity for non-segmented negative sense RNA viruses including EBOV and MARV (Bray et al., 2000; De Clercq, 1985; Huggins et al., 1999; Ye and Schneller, 2014). This antiviral activity is likely due to inhibition of cellular enzyme S-adenosylhomocysteine (SAH) hydrolase (SAHase), which breaks down the SAH produced from S-adenosylmethionine (SAM), the essential cofactor in macromolecular methylation reactions. Inhibition of SAHase increases intracellular levels of SAH, blocking cellular transmethylation reactions through feedback inhibition, causing diminished methylation of the 5′ cap on viral mRNAs, a reaction carried out by the EBOV L protein, and consequent loss of viral protein synthesis (De Clercq, 2004). 6-azauridine and azacytidine (5-aza-2′-deoxycytidine) have also been demonstrated to inhibit EBOV replication in cell culture (Edwards et al., 2015; Uebelhoer et al., 2014). In another example, hsp90 inhibitors, which likely act by destabilizing the EBOV L, and are inhibitory towards EBOV in cell culture (Oestereich et al., 2014; Smith et al., 2010; Smither et al., 2014; Warren et al., 2016).
In addition to targeting the enzymatic activities of the L protein or causing L protein degradation, compounds that target VP30 phosphorylation status, which influences viral transcription, or expression can also impair virus replication or RNA synthesis in cell culture. Okadaic acid (OA), an inhibitor of cellular phosphatases, can prevent dephosphorylation of VP30 and inhibit EBOV transcription (Modrof et al., 2002). This observation led to the identification of small molecules that promote VP30 phosphorylation, inhibit EBOV transcription and suppress EBOV replication in cell culture (Ilinykh et al., 2014). In a separate study, small molecule inhibitors of polyamine production were found to inhibit EBOV RNA synthesis measured by minigenome assay. siRNA knockdown of spermidine synthase (SRM) a critical enzyme in polyamine biosynthesis exerted an antiviral effect against an EBOV expressing green fluorescent protein (GFP) (Olsen et al., 2016). Spermidine is required for the hypusination of eukaryotic translation initiation factor 5A (eIF5A), which is modified in this manner at lysine 50. The compounds ciclopirox (CPX) and deferiprone (DEF), which block the enzyme deoxyhypusine hydroxylase (DOHH) required for hypusination, were demonstrated to inhibit minigenome activity and to inhibit both EBOV and MARV growth in cell culture. As eIF5A is a translation factor, the impact of the inhibitors on expression of the EBOV replication complex proteins was assessed in minigenome assays. The results point to inhibition of VP30 expression as an explanation for inhibition of viral replication (Olsen et al., 2016).
Finally, there are examples of host factors identified as modulating viral RNA synthesis that suggest avenues for therapeutic development. For example, genetic and small molecule screens identified inhibition of either host purine or pyrimidine biosynthesis as an effective means of impairing viral RNA synthesis (Edwards et al., 2015; Luthra et al., 2018; Martin et al., 2018). Host RNA binding Staufen was demonstrated to interact with EBOV RNA and with components of the viral replication machinery and to modulate viral RNA synthesis (Fang et al., 2018). DNA topoisomerase 1 was found to interact with and colocalize with EBOV L and its knockdown decrease EBOV replication and viral polymerase function (Takahashi et al., 2013).
3.1. Overview of interactions among EBOV encoded proteins required for viral RNA synthesis
Four viral proteins, NP, VP35, VP30 and L are required to carry out the full complement of EBOV RNA synthesis reactions, with VP30 being required for viral transcription but dispensable for viral genome RNA replication (Muhlberger et al., 1999). Using minigenome assays, which model filoviral RNA synthesis in transfected cells, VP30 was demonstrated to not be required for MARV transcription (Muhlberger et al., 1998). Each of the proteins of the RNA synthesis complex homo-oligomerizes and can interact with RNA. NP consists of an N-terminal domain, a potentially flexible bridge between the N- and C-terminal region that has low complexity sequences, and a C-terminal domain. NP spontaneously associates with ssRNA and oligomerizes (Kirchdoerfer et al., 2015; Leung et al., 2015; Liu et al., 2017a; Mavrakis et al., 2002; Su et al., 2018; Sugita et al., 2018; Wan et al., 2017; Zhu et al., 2017). VP35s homo-oligomerize through a region in the VP35 N-terminus; EBOV VP35 likely forms various multimers, including tetramers and trimers in solution, whereas the N-terminus of MARV VP35 is reported to form trimers (Figure 3A-B) (Bruhn et al., 2017; Edwards et al., 2016; Luthra et al., 2015; Moller et al., 2005; Reid et al., 2005)(Zinzula et al., 2018). It is not clear at present which forms are critical to mediate each VP35 function. VP35 also possesses a C-terminal domain that binds to dsRNA and is required for inhibition of signaling through retinoic acid-inducible gene I (RIG-I)-like receptor signaling and suppression of IFN and other innate antiviral responses (Bale et al., 2012; Bale et al., 2013; Cardenas et al., 2006; Kimberlin et al., 2010; Leung et al., 2009; Leung et al., 2010; Ramanan et al., 2012). VP30 is a Zn2+ binding, phosphoprotein that can also interact with RNA. Zn2+ and RNA binding are mediated by an N-terminal domain (John et al., 2007; Modrof et al., 2003). VP30 homo-oligomerization has been demonstrated and VP30-derived peptides that block oligomerization inhibit EBOV growth when introduced into infected cells (Hartlieb et al., 2003). The C-terminal domain of VP30 can dimerize when expressed alone. However, full-length VP30 forms a variety of multimeric complexes, including homohexamers via a domain in the N-terminus (Hartlieb et al., 2007). L polymerase carries out the capping and cap methylation reactions for viral mRNA synthesis as well as RNA-dependent RNA polymerase activities for viral mRNA synthesis and viral genome replication. Studies expressing the N-terminal region of L indicate that L can also homo-oligomerize (Trunschke et al., 2013).
Figure 3. Structural characterization of specific interactions that are important for modulating filoviral replication.
A. Coiled-coil motif of Reston VP35 N-terminal oligomerization domain arranged as a parallel tetramer (slate; PDB 6GBR). B. Coiled-coil motif of Ebola VP35 N-terminal oligomerization domain arranged as an antiparallel dimer of parallel trimers (molecule A, marine; molecule B, tv blue; PDB 6GBO). C. Ebola VP30 C-terminal domain (warm pink) bound to an Ebola NP peptide (pale green) (PDB 5VAP). D. CDRMs modulating Ebola NP-NP interactions (molecule A, limon; molecule B, split pea; PDB 6C54). E. RNA (yellow) bound Ebola NP (forest) (PDB 5Z9W). F. Ebola NP (chartreuse) bound to an Ebola VP35 peptide NPBP (sky blue) (PDB 4YPI). Structures are colored according to filoviral proteins. Different shades of a color are used to indicate different conformational arrangements. Simple named colors used are defined by PyMOL.
The filovirus RNA synthesis machinery assembles via several protein-protein interactions (PPIs) between different components (Becker et al., 1998). NP associates with and oligomerizes on the viral template RNAs, either negative-sense genomic RNA or positive-sense anti-genomic (complementary) RNA (Kirchdoerfer et al., 2015; Leung et al., 2015; Liu et al., 2017a; Mavrakis et al., 2002; Noda et al., 2010; Su et al., 2018; Sugita et al., 2018; Wan et al., 2017; Watanabe et al., 2006; Zhu et al., 2017). NP also interacts with VP35 and VP30 (Becker et al., 1998; Biedenkopf et al., 2013; Kirchdoerfer et al., 2015; Kirchdoerfer et al., 2016; Kruse et al., 2018; Leung et al., 2015; Liu et al., 2017a; Xu et al., 2017; Zhu et al., 2017). VP35 makes two distinct interactions with NP, one via a sequence in the VP35 N-terminus that interacts with NP near the hinge region between the N- and C-lobes (Kirchdoerfer et al., 2015; Leung et al., 2010; Liu et al., 2017a; Zhu et al., 2017), and one via the C-terminal VP35 IFN inhibitory domain (IID) that interacts with NP, although the exact region is unclear (Prins et al., 2010). VP35 also interacts with L, with an interaction site having been mapped to the first 380 N-terminal residues in L (Trunschke et al., 2013). This presumably brings L to the RNA template via the VP35-NP interaction. The unphosphorylated form of VP30 promotes EBOV transcription and, to a lesser extent MARV transcription (Modrof et al., 2002; Tigabu et al., 2018). VP30 interacts with NP and this occurs via a cleft in the C-terminal domain of VP30 and a proline-rich peptide near residues 610-612 of NP (Hartlieb et al., 2007; Kirchdoerfer et al., 2016; Xu et al., 2017). The interaction with NP, as detected by co-immunoprecipitation assay, is stronger when phosphomimetic mutants of VP30 are used (Biedenkopf et al., 2013). The mechanism by which VP30 phosphorylation impacts NP interaction remains to be clarified, however. VP30 has also been reported to interact with VP35 and L (Biedenkopf et al., 2013; Tigabu et al., 2018). The former interaction is influenced by the phosphorylation status of VP30 and is suggested to require the presence of RNA but the regions of VP30 and VP35 have not been defined. The VP30-L interaction, as well as evidence of an NP-VP30-L bridge have been demonstrated by co-localization assessed by indirect immunofluorescence studies performed on cells transfected with expression plasmids for individual or combinations of RESTV proteins (Groseth et al., 2009). The domains involved in the VP30-L interaction and the functional significance of the VP30-VP35 and VP30-L interaction remain to be defined.
Filovirus RNA synthesis takes place in cytoplasmic inclusions (Dolnik et al., 2015; Hoenen et al., 2012). These inclusions are also sites of the PPIs described above. NP expressed alone forms inclusion bodies and when co-expressed with NP, VP35, VP30 and L also co-localize in these inclusions (Becker et al., 1998; Nanbo et al., 2013). Further, the ratio of NP to VP35 can alter propensity of NP to form inclusions (Noda et al., 2011). Inclusions are associated with cellular membranes as well as with a variety of cellular factors including endosomal complex required for transport (ESCRT) associated proteins, autophagy-associated proteins such as microtubule associated protein 1 light remain to be defined. chain 3 alpha (LC3)and stress granule proteins (Dolnik et al., 2010; Dolnik et al., 2015; Nelson et al., 2016). Whether these proteins influence filoviral RNA synthesis and by which mechanisms remains to be determined.
4. Recent insights into the protein-protein interactions that modulate Ebola virus RNA synthesis.
Although it has long been known that interactions between NP, VP35, VP30 and L are required for filovirus RNA synthesis, detailed understanding of these interactions has been lacking. Further, several host-viral RDRP complex interactions have been defined. Recently, the combination of structural, biophysical and molecular biology approaches has revealed in substantial detail the molecular basis for protein-protein interactions (PPIs) and defined how specific interactions contribute to the function of the viral RDRP complex (Figure 3). In several cases, disruption of an interaction has been demonstrated to inhibit viral RNA synthesis and EBOV replication.
4.1. Structures of EBOV NP reveal structure and dynamics of NP-RNA and NP-protein interactions critical for viral RNA synthesis and genome RNA encapsidation
Three recent studies reported electron cryo-microscopy and -tomography derived structures of EBOV NP complexes. These data revealed, in great detail, interactions necessary for NP assembly and dynamics that allow NP to participate in EBOV RNA synthesis and nucleocapsid assembly (Su et al., 2018; Sugita et al., 2018; Wan et al., 2017). In one study, a structure of NP residues 1-450, a region sufficient to form α-helical structures and encapsidate RNA was solved, as were structures of an NP-VP35-VP24-VP40 nucleocapsid and nucleocapsids in EBOV and MARV particles. In a second study, NP nucleocapsids formed by the N-terminal domain of NP were examined by a combination of methods that included hydrogen-deuterium exchange mass spectrometry (HDX-MS) and functional assays coupled with cryoEM. In a third study, cryo-EM and single-particle analysis enabled determination and an atomic model of the NP 1-450 structure. While the studies and conclusion differ in some details, the overall picture that emerges is one where NP is a highly dynamic molecule and that these dynamics play critical roles in EBOV RNA synthesis and assembly. These studies yielded electronic density into which previously-determined EBOV NP X-ray crystal structures could be fitted. These studies therefore clarified how individual NP monomers assemble into helical structures in which rings of NP stack upon one another. Among the findings were the identification of α-helices within NP monomers that mediate NP-NP interactions and NP-RNA interactions. Of particular note is the binding pocket on NP where the VP35 NP-binding peptide (NPBP) binds. In the assembled NP structures, a-helices from nearby NP molecules occupies this groove.
Wan et al. also determined the nucleocapsid structures present in virus-like particles formed by co-expression of full-length NP–VP35, VP24 and VP40 as well as nucleocapsids present in EBOV and MARV particles. Comparative analysis of these structures suggested that NP oligomerization causes structural changes allowing an α-helix in NP to acts as a clamp that facilitates RNA binding in the NP RNA binding cleft. The RNA binding promotes stabilization of the NP assembly (Figure 3D). For the NP N-terminus alone, this is seen to result in a condensed structure. With full-length NP, condensation is incomplete until the nucleocapsid also associates with VP35 and VP24. In this model, NP oligomerization, RNA association and recruitment of other viral proteins to the nucleocapsid are interconnected processes. However, the hierarchy of these processes remains largely unexplored.
Su et al. began their analysis from the perspective of their previously described X-ray crystal structure of the NP N-terminus, where two α-helices at the C-terminus of the NP construct, α21 and α22, made NP-NP contacts. Mutagenesis and HDX-MS studies indicated that NP conformational changes occur due to NP-NP interactions and that these affect accessibility of the VP35-NPBP binding site. RNA binding was also demonstrated to be regulated by the first 24 amino acid residues of NP and the cooperation of this region with the α21 and α22 helices was described as a context-dependent regulatory module (CDRM). The CDRM appears to be a critical regulator of NP intermolecular interactions that couples NP oligomerization, RNA binding and access the viral RNA template for viral RNA synthesis reactions (Figure 3E).
Functional assays that utilized minigenome assays and either wild-type of mutated NPs indicated that viral RNA synthesis is dependent on NP oligomerization. Modeling NP dynamics through the use of Markov state modeling (MSM), which maps the conformational space adopted by dominant structural states, suggested that monomeric NP can sample conformations similar to those seen with the NP oligomer and that oligomerization stabilizes the structure. From these studies, the authors propose a similar model to that proposed by Wan et al. whereby NP assembles into a helical nucleocapsid structure through structural rearrangements in both the N- and the C-termini of the NP N-terminal domain. Interactions within the CDRM, including those involving the N-terminal 24 amino acids and more C-terminal sequences regulate NP oligomerization and through this, NP-ssRNA interaction. These dynamic interactions are likely to play important roles in the regulation of filoviral RNA synthesis. Based on their high resolution structure, Sugita et al. propose that peptidomimetics could be used to competitively inhibit or cross-link interactions within the NP oligomer to achieve antiviral activity (Sugita et al., 2018).
4.2. Interactions between NP and VP35 critical for viral RNA synthesis
Based on existing data and by analogy to other NNSVs, VP35 and NP interaction has been thought to serve at least 2 functions in the context of viral RNA synthesis. Interaction of VP35 with NP serves a chaperoning function, preventing NP from non-specifically associating with RNA while maintaining NP as a monomer. VP35-NP interaction is also expected to bring the L protein to the NP-encapsidated template viral genomic or antigenomic RNAs. Several studies that included structural, biophysical and functional analyses have now defined in detail how the chaperoning function is mediated by either EBOV VP35 or MARV VP35 (Kirchdoerfer et al., 2015; Leung et al., 2015; Liu et al., 2017a; Zhu et al., 2017).
Leung et al. and Kirchdoerfer et al., in separate studies, employed multi-disciplinary approaches to address the EBOV VP35 chaperoning function for EBOV NP. In each study, core domains of the NP N-terminus and truncations of VP35 were used to define short peptides capable of high affinity binding to the NP N-terminus (Kirchdoerfer et al., 2015; Leung et al., 2015). In the case of Leung et al., a VP35 peptide corresponding to residues 20-48 was sufficient to bind and was named NPBP. In solution, NPBP was found to be intrinsically disordered, but it exhibited a propensity to form helical structures in the presence of trifluoroethanol, suggesting that interaction with NP might occur in the form of an α helix. To further explain the functional consequences of the interaction, the X-ray crystal structure of VP35 NPBP in complex with the NP N-terminal domain (ΔNPNTD) was solved to 3.7 Å resolution (Figure 3F). The NPBP/ΔNPNTD complex formed double stacked rings with eight ΔNPNTD molecules per rung. ΔNPNTD was found to have two lobes, a head lobe and a foot lobe. The two C-terminal α-helices in the foot lobe (α21 and α 22) make contacts with adjacent ΔNPNTD molecules in a ring. In the structure, NPBP interacts with the foot lobe of ΔNPNTD as two orthogonal helices. The NPBP binding site was identified as critical for NP RNA binding and NPBP was able to compete with ssRNA for binding to ΔNPNTD providing evidence that this VP35-derived peptide is a key regulator of NP RNA binding. Further, when NPBP was added to ring structures formed by NP 1-457, the ring structures were disrupted. Size exclusion chromatography confirmed that NPBP dissociated the oligomeric forms of NP. Although approaches differed, Kirchdoerfer et al. similarly identified a peptide derived from the VP35 N-terminus capable of binding to the NP N-terminus. The NP N-terminal construct used was sufficient to bind RNA and to oligomerize. In the presence of the VP35 peptide, however, NP oligomerization was prevented. While oligomerized NP 1-450 binds RNA, the presence of VP35 peptide blocks RNA binding. Their approach to crystallization of the complex was to produce a protein containing VP35 residues 15–60 fused to NP residues 34–367. This also identified a bilobed structure for the NP N-terminus, demonstrated binding of VP35 peptide near the RNA binding site and identified mutations that disrupt VP35 peptide-NP interaction and impair viral RNA synthesis. Subsequent studies on MARV VP35-NP N-terminal domain interactions reached similar conclusions (Liu et al., 2017a; Zhu et al., 2017). Remaining to be clarified are the precise mechanisms by which the VP35-chaperoned NP, upon delivery to assembling viral nucleocapsid structures, is freed from VP35 allowing NP oligomerization and NP-RNA association. Also, the details by which the viral RNA template, which is tightly associated with oligomerized NP in the nucleocapsid, becomes accessible to the viral replication complex that contains L and VP35 proteins remain to be explained. It is tempting to speculate that NPBP can interact with NP to temporarily disengage the RNA from NP to allow it to be transcribed and replicated.
Data from minigenome assays using mutants that disrupt the NPBP-NP interaction suggest the NPBP-NP interface as an antiviral target. In support of this, Leung et al demonstrated that expression of the NPBP is sufficient to inhibit EBOV RNA synthesis, as assessed by minigenome assays (Leung et al., 2015). In order to take advantage of the mechanistic insights and to aid the development of potential therapies that target the NPBP binding site on EBOV NP, Liu et al. developed a sensitive fluorescence polarization assay (FPA) for high-throughput screening (HTS) of the EBOV NPNTD-NPBP interaction (Liu et al., 2017b). A pilot screen was then performed using a library of 640 FDA approved drugs. This identified Tolcapone, an FDA-approved drug that inhibits catechol-O-methyltransferase (COMT) and is used in the treatment of Parkinson’s Disease (Politi et al., 2018), as an inhibitor or NP:NPBP with an IC50 of 2 μM in the FPA. Tolcapone was further demonstrated to inhibit growth or EBOV in cell culture and, consistent with the role of NP:NPBP interaction for EBOV RNA synthesis, to inhibit the EBOV minigenome assay. Because the NPBP sequence is relatively well conserved across filoviruses, the binding of NPBP from SUDV, RESTV and MARV to EBOV NP was assessed and found to yield similar affinities. When Tolcapone was tested against the MARV NPBP interaction, inhibition was demonstrated (Liu et al., 2017b). These findings suggest that the NP:NPBP interaction has potential as a pan-filovirus target.
As noted above, VP35 not only chaperones NP but may mediate delivery of the viral polymerase to the NP-encapsidated template RNA for viral RNA synthesis. It is not clear that the NPBP-NP interaction fully explains this function. A second interaction between VP35 and NP that may serve this role has also been described (Kirchdoerfer et al., 2015; Prins et al., 2010). Present in the C-terminal IID of EBOV VP35 are two basic patches, the central basic patch, which mediates interaction with dsRNA, and the first basic patch which consists of VP35 residues K222, R225, K248 and K251 (Leung et al., 2010; Prins et al., 2010). Mutation of first basic patch residues to alanine disrupted viral RNA synthesis in minigenome assays (Prins et al., 2010). These same mutants exhibited impaired interaction with NP in co-immunoprecipitation experiments and when maltose binding protein-VP35 IID fusions were used to pull-down VP35 from transfected cell lysates (Prins et al., 2010). These data indicate that the IID can mediate an interaction between VP35 and NP. However, pulldowns performed in a separate study suggest that sequences outside of the IID can also contribute to this NP interaction (Kirchdoerfer et al., 2015).
As with the NPBP-NP interactions, mutations that disrupt the IID-NP interaction impair viral RNA synthesis, suggesting this interaction could be a viable target for therapeutics. In further support of this, two strategies that targeted the IID domain of VP35 were able to inhibit EBOV RNA synthesis. In one approach, the X-ray crystal structure of the EBOV VP35 IID was used as a target for in silico-based screening (Brown et al., 2014). Compounds were sought that could target the IID. Taking hits from the in silico screen and combining NMR, X-ray crystallography and medicinal chemistry, small molecules were identified that target a binding pocket adjacent to the IID first basic patch. NMR mapping experiments and high-resolution x-ray crystal structures showed that select small molecules bind to a region of VP35 IID that is important for replication complex formation through interactions with the viral nucleoprotein (NP). Select compounds from this series could impair VP35 IID-NP interaction in vitro, inhibit EBOV minigenome assay activity and replication of EBOV in tissue culture. These results confirm the ability of compounds identified in this study to inhibit VP35-NP interactions in vitro and to impair EBOV replication in cell culture (Brown et al., 2014). In another study, RNA aptamers that target the EBOV VP35 IID were selected for. These could compete with dsRNA for binding to eVP35 and disrupt VP25-NP interaction and select aptamers could also inhibit EBOV RNA synthesis (Binning et al., 2013).
4.3. VP35-L interactions.
It is generally accepted that VP35 interacts with L and via its interaction with NP, VP35 brings L to the NP-encapsidated template RNAs but the underlying molecular mechanisms remain to be characterized at high resolution. VP35 has been demonstrated to interact with the N-terminal region of L (Shabman et al., 2013; Trunschke et al., 2013). Studies found that co-expression of VP35 can substantially enhance expression of L residues 1-505 (L1-505), compared with when L1-505 is expressed alone, suggesting a possible role of VP35 as a chaperone for L, although further studies using full-length L are required to address this question (Shabman et al., 2013). Further analysis, defined L residues 280-370 as sufficient to mediate a weak VP35-L interaction and L residues 1-370 as sufficient for stronger interaction (Trunschke et al., 2013). Co-immunoprecipitation studies pointed to an L-L interaction domain within the N-terminal 450 amino acid residues, but this interaction was not altered by co-expression of VP35. Supporting a role for these interactions in EBOV RNA synthesis, expression of fragments of L sufficient to interact with VP35 inhibited minigenome assays (Trunschke et al., 2013).
4.4. VP35 interactions with host proteins that impact viral RNA synthesis
In addition to its interactions with viral factors, VP35 interactions with host factors can also modulate viral RNA synthesis. VP35 interaction with host protein 8 kilodalton dynein light chain (DLC8 or LC8) was initially identified based on a yeast two-hybrid screen for VP35 interactors. The interaction mapped to the amino-terminal half of VP35 and requires the motif SQTQT, which matches the LC8 consensus binding motif K/SxTQT (Kubota et al., 2009; Lo et al., 2001). In a separate study, this interaction was demonstrated to be direct and of high affinity with an equilibrium dissociation constant [KD] = 82.4 ± 18 nM. LC8 forms dimers and VP35 forms tetramers; in complex, two LC8 dimers interact with the VP35 tetramer. Functionally, the interaction was demonstrated to enhance VP35 stability. Mutations to the consensus binding site substantially decrease binding. Further, these mutations negatively impact viral RNA synthesis, as measured by minigenome activity, whereas LC8 over-expression enhances activity, with the main impact being on viral transcription (Luthra et al., 2015). These data suggest that blocking LC8-VP35 interaction would negatively impact EBOV RNA synthesis during infection.
VP35 was demonstrated to interact with the host protein known as protein activator of interferon induced protein kinase eIF2AK2 (PACT) (Luthra et al., 2013). PACT can stimulate innate antiviral defenses through activation of the IFN-induced, dsRNA activated protein kinase (PKR) kinase activity, which phosphorylates translation initiation factor eukaryotic translation initiation factor 2 alpha (eIF-2α) to suppress translation, and by facilitating activation of RIG-I, which leads to induction of type I IFN (IFN) responses (Kok et al., 2011; Patel and Sen, 1998). The interaction with PACT occurs through the C-terminal VP35 IID which is critical for potent suppression of IFN responses (Luthra et al., 2013). It was further demonstrated that interaction with VP35 binding prevent PACT binding to RIG-I to suppress innate immune responses. Because VP35 is also critical for EBOV RNA synthesis, the impact of PACT interaction on this function was also assessed. PACT was demonstrated to inhibit EBOV RNA synthesis in the minigenome assay. The inhibition was relieved when VP35 IID point mutants that lose interaction with PACT were used in place of wild-type VP35 in the same assay. Co-immunoprecipitation assays demonstrated that PACT can interfere with VP35 interaction with the N-terminus of L and that this effect was also lost with the VP35 IID mutant. Consistent with these findings, in EBOV infected cells over-expression of PACT increased IFN responses and decreased viral RNA production; whereas PACT knockdown decreased IFN responses and increased viral RNA synthesis (Luthra et al., 2013). These data provided evidence that host factors can negatively modulate EBOV replication by targeting components of the RNA synthesis machinery. These findings also suggest as therapeutic strategies the identification of molecules that target the VP35-L interaction in a manner similar to PACT or that increase the availability of PACT to target VP35.
Another host interaction with VP35 involves tripartite motif-containing protein 6 (TRIM6), a host cell E3-ubiquitin ligase demonstrated to produce free poly-ubiquitin chains that promote type I IFN responses (Bharaj et al., 2017). VP35 and TRIM6 were found to co-immunoprecipitate, and VP35 was demonstrated to be ubiquitinated in the IID on K309, which was enhanced by over-expression of TRIM6. VP35 was demonstrated to interact with polyubiquitin and to inhibit TRIM-6 induced IFN responses. Significantly, expression of a wild-type but not a catalytically-inactive TRIM6 enhanced EBOV RNA synthesis, as measured by minigenome assay, and TRIM6 knockout cells exhibited reduced EBOV replication. Therefore, while the mechanisms by which TRIM6 modulates EBOV RNA synthesis and propagation require further clarification, strategies that either reduce TRIM6 E3-ubiquitin ligase activity or disrupt VP35-TRIM6 interaction might effectively suppress EBOV growth.
Finally, double-stranded RNA-binding protein, 76 KD (DRBP76;also referred to as NFIL3, NF90 and other names) has been demonstrated to interact with the VP35 IID (Shabman et al., 2011). VP35-IID-DRBP76 interaction was detectable in the presence of exogenously added dsRNA, suggesting the possibility of RNA bridging the protein-protein interaction. However, full-length VP35 bound to DRBP76 when exogenous dsRNA was absent. Regardless of the basis for interaction, when DRBP76 was over-expressed it impaired EBOV minigenome activity, demonstrating a capacity to modulate the function of the EBOV RNA synthesis machinery (Shabman et al., 2011).
4.5. Interactions between VP30 and NP modulate viral RNA synthesis
The C-terminal domain of VP30 interacts with NP and this interaction is important for viral RNA synthesis (Hartlieb et al., 2007). Two studies that combined structural, biophysical and molecular approaches defined the basis and demonstrated the functional significance of VP30-NP interactions for EBOV (Kirchdoerfer et al., 2016; Xu et al., 2017). Xu et al. demonstrated that the C-terminal region of NP (residues 600-739) can bind to the C-terminal domain of VP30 (residue 110-272) and were able to generate co-crystals that revealed NP residues 600-612 in complex with a cleft on the surface of VP30 (110-272) (Figure 3C) (Xu et al., 2017). The interacting NP peptide contains a motif, PPxPxY, conserved across filoviral NPs. A combination of in vitro FPAs and cell-based co-immunoprecipitation assays identified interface loss of binding mutants. Because VP30 and NP each participate in EBOV RNA synthesis, the mutants were assessed in a minigenome assay. Several of the mutants exhibited loss of activity, demonstrating a role for the interaction in viral RNA synthesis. The loss of function of these mutants is related to the role of VP30 as a transcription initiation factor. This was demonstrated by introducing into the minigenome template RNAs that disrupt the stem-loop at the NP gene transcription start site. These render the minigenome assay VP30- dependent and restored function in the presence of the loss of function mutants (Xu et al., 2017).
Kirchdoerfer et al. similarly demonstrated binding of the C-terminus of NP to the C-terminal domain of VP30 and mapped an NP peptide, residues 600-617, sufficient for binding (Kirchdoerfer et al., 2016). For crystallization, to overcome relatively low NP peptide-VP30 affinity, the NP peptide was fused to the N-terminus of the VP30 C-terminal domain, with a short spacer sequence between. The conclusions based on the structure parallel those described above, and mutations designed to alter NP-VP30 affinities also yielded variable results but demonstrated an important role for the interaction for viral RNA synthesis. By comparing VP30-NP binding affinities of various mutants with activity in the minigenome assay, the authors proposed that there is an optimal binding affinity such that too high or too low binding affinities negatively impacts filoviral RNA synthesis (Kirchdoerfer et al., 2016). As both studies implicated the VP30-NP interaction in viral RNA synthesis, disrupting the interaction would be a potential therapeutic target. Consistent with this, Xu et al. demonstrated that expression of the NP peptide as a fusion with GFP impaired RNA synthesis measured by the minigenome assay (Xu et al., 2017).
4.6. An interaction between NP and host protein phosphatase regulatory subunit B56 mediates VP30 dephosphorylation.
VP30 function is modulated by phosphorylation, with the dephosphorylated state promoting viral mRNA synthesis. Protein phosphatases PP1, PP2A and PP2C have been implicated as enzymes that mediate VP30 dephosphorylation (Modrof et al., 2002). However, details as to how the VP30 phosphorylation state is regulated have been unclear. Significant light was shed on this when it was recognized the EBOV NP possesses an LxxIxE motif, a sequence that can serve as a binding site for the cellular PP2A-B56 protein phosphatase (Hertz et al., 2016). PP2A possesses three distinct subunits, one catalytic (PP2A-C), one which serves a scaffolding function (PP2A-A) and any of several B subunits which regulate activity (Shi, 2009). One B subunit family, B56, interacts with the LxxIxE motifs that were noted in filovirus NPs (Hertz et al., 2016). The NP LxxIxE motifs lie 18-28 amino acids, depending on which filovirus is examined, from the PPxPxY motif that mediates VP30-NP interaction (Kruse et al., 2018). It was demonstrated that NP recruits PP2A-B56 to the LxxIxE motif and that this interaction is needed for VP30 dephosphorylation and, hence, for viral transcription. Further, LxxIxE-containing peptides were developed and were demonstrated to inhibit EBOV RNA synthesis in a minigenome assay and to inhibit EBOV replication (Kruse et al., 2018). These data therefore define a critical NP-host interaction that modulates the balance of VP30 phosphorylation/dephosphorylation and demonstrates that disruption of NP-B56 interaction could serve as a therapeutic approach. A caveat to the approach tested is that the peptides would presumably target other B56 PPIs as well.
4.7. Comprehensive profiling of EBOV-host PPIs identifies novel modulators of EBOV RNA synthesis.
The increasing evidence that host factors engage and modulate filoviral RNA synthesis reactions, suggested that more systematic and comprehensive approaches to identify virus-host protein-protein interactions PPIs, was warranted. A four lab, four institution effort sought to systematically characterize Ebola virus-human PPIs using an affinity purification mass spectrometry (AP-MS) approach (Batra et al., 2018) (Shah et al., 2015). In order to build this interaction network, individual Ebola virus open reading frames for NP, VP35, VP40, GP, VP30, and VP24 were cloned into a mammalian expression vector fusing each viral protein to a 2×Strep affinity tag (Jager et al., 2011b). Tagged plasmids were transfected into HEK293T and Huh7 cells and EBOV-human protein complexes were purified by StrepTactin affinity purification and subjected to quantitative mass spectrometry analysis. Each purification was repeated multiple times in both cell lines and rigorous tests of statistical significance were applied (Verschueren et al., 2015), yielding, a network of 194 virus-host PPIs (Batra et al., 2018). This approach has proven successful in defining a number of virus-host networks, including networks for human immunodeficiency virus 1 (HIV-1), hepatitis C virus (HCV), Kaposi's sarcoma-associated herpesvirus (KSHV), dengue virus (DENV), human papillomavirus (HPV) and now EBOV (Batra et al., 2018; Davis et al., 2015; De Maio et al., 2016; Eckhardt et al., 2018; Jager et al., 2011a; Ramage et al., 2015)(Shah et al., Cell in press). The EBOV network recapitulates known interactions, such as between EBOV VP24 and human karyopherin alpha (KPNA; also knows as importin alpha), which mediates suppression of antiviral signaling by blocking nuclear translocation of tyrosine phosphorylated signal transducer and activator of transcription 1 (STAT1) (Mateo et al., 2010; Reid et al., 2006; Reid et al., 2007; Xu et al., 2014), and between VP35 and LC8 (Kubota et al., 2009; Luthra et al., 2015). The L protein was not included in the analysis because low L expression reduced confidence in hits obtained (data not shown). Therefore, strategies that effectively increase L expression are needed to address host interactions with this critical viral enzyme. Nonetheless, two components of the RNA replication complex, VP35 and VP30, each had very robust interactomes (Figure 4), suggesting the likelihood that host factors capable of influencing viral RNA synthesis reactions had been identified.
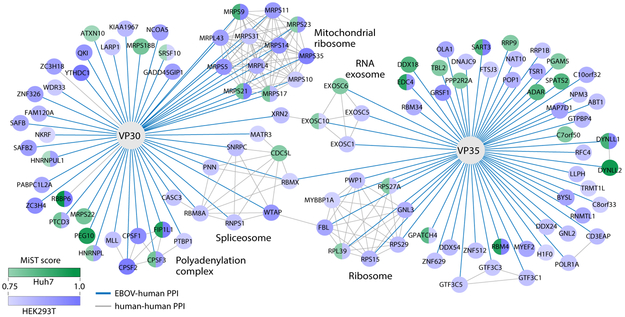
Figure 4. EBOV-human protein-protein interactions with VP30 and VP35.
EBOV-human protein-protein interactions are represented (blue lines) between VP30 and VP35 proteins and human proteins. Each human prey node is colored according to the cell line in which it was identified: blue indicates HEK293T cells, green indicates Huh7 cells, and blue-green bifurcation indicates the protein was identified as an interactor in both cell types. The shade of the prey node correlates with the MiST score of the interaction (scale at lower left). Grey lines correspond to human-human protein-protein interactions curated in the publicly available CORUM database and several human protein complexes are labeled.
For example, we found that VP30 was physically associated with components of the spliceosome and the polyadenylation machinery, two complexes which are functionally linked (Kornblihtt et al., 2004). These connections suggest that VP30 may be impinging on host transcriptional events, similar to what we have previously observed for DENV and host mRNA splicing (De Maio et al., 2016). Furthermore, further manipulation of the host transcriptional apparatus via VP35 may be occurring, as we identified physical associations with components of both TFIIIC, a transcription factor for RNA polymerase III (Pascali and Teichmann, 2013), as well as the RNA exosome (Morton et al., 2018), a complex involved in a myriad of processes, including RNA degradation during transcription. Interestingly, we recently found that RNA exosome physically associates with the influenza A virus (IAV), a connection required for efficient IAV RNA synthesis (Rialdi et al., 2017). Finally, we also uncovered a connection between VP30 and VPS35 with the mitochondrial and canonical ribosome, respectively, implying a direct connection between EBOV infection to host translational as well (Figure 4). Further work will be required to tease out the functional relevance of these and other physical interactions uncovered by our unbiased AP-MS analysis of the EBOV proteome.
4.8. Host protein RBBP6 interacts with VP30 to modulate EBOV RNA synthesis.
The first EBOV-host PPI derived from the network that was investigated in mechanistic detail was between VP30 and retinoblastoma binding protein 6 (RBBP6), an interaction that was among the highest confidence scores in the entire PPI network, both in HEK293T and Huh7 cells. The interaction could be confirmed in an immunoprecipitation (IP)-Western blot experiment, and IP-Western blotting of other filoviral VP30 proteins demonstrated that the interaction is conserved across all filoviruses tested (EBOV, Reston virus (RESTV), MARV, and Lloviu virus (LLOV)). Truncation mutants of EBOV VP30 and RBBP6 were used to identify specific domains required for the interaction in both cell-based co-immunoprecipitation and in vitro binding assays, the latter using purified VP30 and RBBP6 domains. These efforts mapped the interaction to the C-terminal domain of VP30 and to a proline rich motif within RBBP6, data that enabled the co-crystallization of the VP30 C-terminus with a RBBP6 peptide spanning its interaction domain and the structure of the complex was solved to 1.5 A resolution. Strikingly, the RBBP6 peptide was found to interact with the same cleft on VP30 that interacts with NP (Kirchdoerfer et al., 2016; Kruse et al., 2018; Xu et al., 2017). Quantitative binding measurements demonstrated that the RBBP6 peptide binds the eVP30 C-terminus with ~5 fold higher affinity than the NP-derived peptide. Examination of the NP and RBBP6 peptides that bind eVP30 identified the motif PPxPxY as being present in both. Notably, PPxPxY is conserved among filovirus NPs (Kirchdoerfer et al., 2015) and mutations within this motif can disrupt RBBP6 interaction with eVP30 (Batra et al., 2018).
Because the interaction with NP at this cleft on VP30 is important for EBOV RNA synthesis, experiments were performed to determine whether RBBP6 modulates viral RNA synthesis (Batra et al., 2018). Knockdown of RBBP6 with siRNA stimulated viral RNA synthesis, as measured by minigenome assay, while overexpression of RBBP6 or its VP30-interacting peptide, which was fused to green fluorescence protein (GFP), was strongly inhibitory. That these inhibitory activities are mediated through direct VP30-RBBP6 interaction was supported by the loss of inhibitory activity when RBBP6 or VP30 mutants that were unable to bind were tested. Mechanistically, the inhibition mediated by RBBP6 appears to occur by at least 2 mechanisms. RBBP6 was demonstrated to effectively compete with NP for binding to VP30 and, at least when over-expressed at high levels, to also promote VP30 degradation. Based on the studies examining VP30-NP interaction, loss of VP30-NP interaction should lead to inhibition of viral transcription because this would be expected to inhibit the eVP30 dephosphorylation which occurs on NP. Further study will be required to determine if this is the case.
4.9. The VP30 cleft that binds NP and RBBP6 as a site for therapeutic targeting.
To determine the impact of RBBP6 on actual EBOV replication a combination of knockdown and over-expression studies were performed (Batra et al., 2018). Knockdown of endogenous RBBP6 expression substantially increased infectivity as measured by GFP expression from an EBOV that encodes GFP (EBOV-GFP). Further, expression of RBBP6 fused to GFP significantly decreased EBOV infectivity compared to cells expressing GFP. These data demonstrated the relevance of the interaction for EBOV replication and suggest that methods, perhaps peptidomimetic or other small molecules, that target the VP30 cleft that binds either NP or RBBP6 could serve as antivirals. Given the conservation of the interaction across multiple filoviruses, such an approach could have pan-filovirus potential.
It is striking that human cells possess a protein that negatively impacts EBOV gene expression. It is possible that the VP30-RBBP6 interaction serves a beneficial, perhaps regulatory role for the virus. Further study will be required to develop insight into this question, to further define the mechanisms by which RBBP6 impacts the viral RNA synthesis machinery and to explore the VP30 cleft as a therapeutic target. Intriguingly, the host-EBOV PPI network contains 3 other VP30 interactors, hnRNP L, hnRNP UL1 and PEG10, possessing PPxPxY motifs (Batra et al., 2018). This raises the intriguing possibility that multiple host factors, potentially with different abundance and affinities for eVP30 might together regulate EBOV replication.
5. Conclusions and Future Directions
The efforts described above have provided substantial insights into the assembly and function of the filovirus RNA synthesis machinery and suggest strategies to interfere with critical viral functions. Continued study should further define in high resolution the basis for critical interactions between viral proteins and host proteins and identify the most promising therapeutic targets. At the same time, it is important to recognize that viral transcription and replication are dynamic processes, with specific interactions playing distinct roles on different aspects of these reactions. For example, the interaction between NP and the NPBP from VP35 are thought to chaperone free NP, preventing it from inappropriately binding to cellular RNAs and facilitating its appropriate oligomerization on viral genomic and antigenomic RNAs (Leung et al., 2015). As these dynamics are revealed in higher resolution, additional opportunities will arise to intervene in critical intra- and intermolecular movements in order to inhibit viral growth. The same should also hold true for virus-host PPIs, and it is likely that the field has only scratched the surface of those that modulate viral RNA synthesis. Finally, it should be recognized that, with the exception of VP30 phosphorylation, the impact of post-translational modifications on relevant PPIs and on viral RNA synthesis reactions has largely gone unaddressed. A full accounting of the host factors that interact with and impact filoviral RNA synthesis will require that post-translational modifications also be comprehensively defined.
Highlights.
Filoviruses remain important human pathogens for which therapeutics are needed.
The viral RNA synthesis machinery is appealing as a therapeutic target.
Multiple protein-protein interactions (PPIs) are required for viral RNA synthesis.
Targeting these PPIs impairs viral RNA synthesis and inhibits virus replication.
Acknowledgements.
This work was supported by NIH grants U19AI109664 (C.F.B. D.W.L., G.K.A.), U19AI109945 (C.F.B. D.W.L., G.K.A.), P01120943 (C.F.B. D.W.L., G.K.A.) and R01AI 125453 (C.F.B.). C.F.B. is a Georgia Research Alliance Eminent Scholar in Microbial Pathogenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, Formenty P, Fouchier RA, Fu Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiang D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liu L, Longdon B, Marton S, Maisner A, Muhlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Randall RE, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Shi M, Smither SJ, Stenglein MD, Stone DM, Takada A, Terregino C, Tesh RB, Tian JH, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Verbeek M, Volchkov VE, Wahl-Jensen V, Walsh JA, Walker PJ, Wang D, Wang LF, Wetzel T, Whitfield AE, Xie JT, Yuen KY, Zhang YZ, Kuhn JH, 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161, 2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Kimberlin CR, Halfmann P, Zandonatti MA, Kunert J, Kroon GJ, Kawaoka Y, MacRae IJ, Wilson IA, Saphire EO, 2012. Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNA for interferon antagonism. PLoS Pathog 8, e1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Krois AS, Wilson IA, Saphire EO, 2013. Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism. J Virol 87, 10385–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P, 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A 97, 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra J, Hultquist JF, Liu D, Shtanko O, Von Dollen J, Satkamp L, Jang GM, Luthra P, Schwarz TM, Small GI, Arnett E, Anantpadma M, Reyes A, Leung DW, Kaake R, Haas P, Schmidt CB, Schlesinger LS, LaCount DJ, Davey RA, Amarasinghe GK, Basler CF, 2018. Protein interaction mapping identifies RBBP6 as a negative regulator of Ebola virus replication. Cell in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Rinne C, Hofsass U, Klenk HD, Muhlberger E, 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249, 406–417. [DOI] [PubMed] [Google Scholar]
- Bharaj P, Atkins C, Luthra P, Giraldo MI, Dawes BE, Miorin L, Johnson JR, Krogan NJ, Basler CF, Freiberg AN, Rajsbaum R, 2017. The Host E3-Ubiquitin Ligase TRIM6 Ubiquitinates the Ebola Virus VP35 Protein and Promotes Virus Replication. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkopf N, Hartlieb B, Hoenen T, Becker S, 2013. Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: impact on viral transcription and replication. J Biol Chem 288, 11165–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binning JM, Wang T, Luthra P, Shabman RS, Borek DM, Liu G, Xu W, Leung DW, Basler CF, Amarasinghe GK, 2013. Development of RNA aptamers targeting Ebola virus VP35. Biochemistry 52, 8406–8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Driscoll J, Huggins JW, 2000. Treatment of lethal Ebola virus infection in mice with a single dose of an S-adenosyl-L-homocysteine hydrolase inhibitor. Antiviral Res 45, 135–147. [DOI] [PubMed] [Google Scholar]
- Brown CS, Lee MS, Leung DW, Wang T, Xu W, Luthra P, Anantpadma M, Shabman RS, Melito LM, MacMillan KS, Borek DM, Otwinowski Z, Ramanan P, Stubbs AJ, Peterson DS, Binning JM, Tonelli M, Olson MA, Davey RA, Ready JM, Basler CF, Amarasinghe GK, 2014. In silico derived small molecules bind the filovirus VP35 protein and inhibit its polymerase cofactor activity. J Mol Biol 426, 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn JF, Kirchdoerfer RN, Urata SM, Li S, Tickle IJ, Bricogne G, Saphire EO, 2017. Crystal Structure of the Marburg Virus VP35 Oligomerization Domain. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M Jr., Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF, 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 80, 5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR, 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM, 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J, 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey RA, Shtanko O, Anantpadma M, Sakurai Y, Chandran K, Maury W, 2017. Mechanisms of Filovirus Entry. Curr Top Microbiol Immunol 411, 323–352. [DOI] [PubMed] [Google Scholar]
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, Jager S, Shales M, Horner J, Hernandez RD, Krogan NJ, Glaunsinger BA, 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E, 1985. Antiviral and antimetabolic activities of neplanocins. Antimicrob Agents Chemother 28, 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E, 2004. Antivirals and antiviral strategies. Nat Rev Microbiol 2, 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, Mammi P, Mancini E, Yanovsky MJ, Andino R, Krogan N, Srebrow A, Gamarnik AV, 2016. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog 12, e1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnik O, Kolesnikova L, Stevermann L, Becker S, 2010. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol 84, 7847–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnik O, Stevermann L, Kolesnikova L, Becker S, 2015. Marburg virus inclusions: A virus-induced microcompartment and interface to multivesicular bodies and the late endosomal compartment. Eur J Cell Biol 94, 323–331. [DOI] [PubMed] [Google Scholar]
- Eckhardt M, Zhang W, Gross AM, Von Dollen J, Johnson JR, Franks-Skiba KE, Swaney DL, Johnson TL, Jang GM, Shah PS, Brand TM, Archambault J, Kreisberg JF, Grandis JR, Ideker T, Krogan NJ, 2018. Multiple Routes to Oncogenesis Are Promoted by the Human Papillomavirus-Host Protein Network. Cancer Discov 8, 1474–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF, 2014. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep 6, 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Liu G, Mire CE, Sureshchandra S, Luthra P, Yen B, Shabman RS, Leung DW, Messaoudi I, Geisbert TW, Amarasinghe GK, Basler CF, 2016. Differential Regulation of Interferon Responses by Ebola and Marburg Virus VP35 Proteins. Cell Rep 14, 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Pietzsch C, Vausselin T, Shaw ML, Bukreyev A, Basler CF, 2015. High-Throughput Minigenome System for Identifying Small-Molecule Inhibitors of Ebola Virus Replication. ACS Infect Dis 1, 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Pietzsch C, Ramanathan P, Santos RI, Ilinykh PA, Garcia-Blanco MA, Bukreyev A, Bradrick SS, 2018. Staufen1 Interacts with Multiple Components of the Ebola Virus Ribonucleoprotein and Enhances Viral RNA Synthesis. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Sanchez A, Geisbert TW, 2015. Filoviridae: Marburg and Ebola Viruses. Wolters Kluwer, Philadelphia: :. [Google Scholar]
- Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, Belaganahalli MN, Wells HL, Dhanota JK, Liang E, Grodus M, Jangra RK, DeJesus VA, Lasso G, Smith BR, Jambai A, Kamara BO, Kamara S, Bangura W, Monagin C, Shapira S, Johnson CK, Saylors K, Rubin EM, Chandran K, Lipkin WI, Mazet JAK, 2018. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 3, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Charton JE, Sauerborn M, Feldmann F, Jones SM, Hoenen T, Feldmann H, 2009. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res 140, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlieb B, Modrof J, Muhlberger E, Klenk HD, Becker S, 2003. Oligomerization of Ebola virus VP30 is essential for viral transcription and can be inhibited by a synthetic peptide. J Biol Chem 278, 41830–41836. [DOI] [PubMed] [Google Scholar]
- Hartlieb B, Muziol T, Weissenhorn W, Becker S, 2007. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc Natl Acad Sci U S A 104, 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG, Friede M, Bausch DG, 2017. Experimental Therapies for Ebola Virus Disease: What Have We Learned? J Infect Dis 215, 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Gsell PS, Hossmann S, Watle SV, Konde MK, Keita S, Kone S, Kuisma E, Levine MM, Mandal S, Mauget T, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Rottingen JA, Kieny MP, 2017. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 389, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz EPT, Kruse T, Davey NE, Lopez-Mendez B, Sigurethsson JO, Montoya G, Olsen JV, Nilsson J, 2016. A Conserved Motif Provides Binding Specificity to the PP2A-B56 Phosphatase. Mol Cell 63, 686–695. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Shabman RS, Groseth A, Herwig A, Weber M, Schudt G, Dolnik O, Basler CF, Becker S, Feldmann H, 2012. Inclusion bodies are a site of ebolavirus replication. J Virol 86, 11779–11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins J, Zhang ZX, Bray M, 1999. Antiviral drug therapy of filovirus infections: S-adenosylhomocysteine hydrolase inhibitors inhibit Ebola virus in vitro and in a lethal mouse model. J Infect Dis 179 Suppl 1, S240–247. [DOI] [PubMed] [Google Scholar]
- Ilinykh PA, Tigabu B, Ivanov A, Ammosova T, Obukhov Y, Garron T, Kumari N, Kovalskyy D, Platonov MO, Naumchik VS, Freiberg AN, Nekhai S, Bukreyev A, 2014. Role of protein phosphatase 1 in dephosphorylation of Ebola virus VP30 protein and its targeting for the inhibition of viral transcription. J Biol Chem 289, 22723–22738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Rodger A, Bell DJ, Bhagani S, Cropley I, Filipe A, Gifford RJ, Hopkins S, Hughes J, Jabeen F, Johannessen I, Karageorgopoulos D, Lackenby A, Lester R, Liu RS, MacConnachie A, Mahungu T, Martin D, Marshall N, Mepham S, Orton R, Palmarini M, Patel M, Perry C, Peters SE, Porter D, Ritchie D, Ritchie ND, Seaton RA, Sreenu VB, Templeton K, Warren S, Wilkie GS, Zambon M, Gopal R, Thomson EC, 2016. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 388, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ, 2011a. Global landscape of HIV-human protein complexes. Nature 481, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Gulbahce N, Cimermancic P, Kane J, He N, Chou S, D'Orso I, Fernandes J, Jang G, Frankel AD, Alber T, Zhou Q, Krogan NJ, 2011b. Purification and characterization of HIV-human protein complexes. Methods 53, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John SP, Wang T, Steffen S, Longhi S, Schmaljohn CS, Jonsson CB, 2007. Ebola virus VP30 is an RNA binding protein. J Virol 81, 8967–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B, Li J, Adhikari J, Edwards MR, Zhang H, Schwarz T, Leung DW, Basler CF, Gross ML, Amarasinghe GK, 2016. Dimerization Controls Marburg Virus VP24-dependent Modulation of Host Antioxidative Stress Responses. J Mol Biol 428, 3483–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin CR, Bornholdt ZA, Li S, Woods VL Jr., MacRae IJ, Saphire EO, 2010. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci U S A 107, 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Abelson DM, Li S, Wood MR, Saphire EO, 2015. Assembly of the Ebola Virus Nucleoprotein from a Chaperoned VP35 Complex. Cell Rep 12, 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Moyer CL, Abelson DM, Saphire EO, 2016. The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis. PLoS Pathog 12, e1005937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY, 2011. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9, 299–309. [DOI] [PubMed] [Google Scholar]
- Kolesnikova L, Nanbo A, Becker S, Kawaoka Y, 2017. Inside the Cell: Assembly of Filoviruses. Curr Top Microbiol Immunol 411, 353–380. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G, 2004. Multiple links between transcription and splicing. RNA 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Biedenkopf N, Hertz EPT, Dietzel E, Stalmann G, Lopez-Mendez B, Davey NE, Nilsson J, Becker S, 2018. The Ebola Virus Nucleoprotein Recruits the Host PP2A-B56 Phosphatase to Activate Transcriptional Support Activity of VP30. Mol Cell 69, 136–145 e136. [DOI] [PubMed] [Google Scholar]
- Kubota T, Matsuoka M, Chang TH, Bray M, Jones S, Tashiro M, Kato A, Ozato K, 2009. Ebolavirus VP35 interacts with the cytoplasmic dynein light chain 8. J Virol 83, 6952–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, Shabman RS, Chiu W, Davey RA, Otwinowski Z, Basler CF, Amarasinghe GK, 2015. An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions. Cell Rep 11, 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, Amarasinghe GK, 2009. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci U S A 106, 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, Basler CF, Amarasinghe GK, 2010. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol 17, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Dong S, Li G, Wang W, Liu X, Wang Y, Yang C, Rao Z, Guo Y, 2017a. Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Nash PJ, Johnson B, Pietzsch C, Ilagan MX, Bukreyev A, Basler CF, Bowlin TL, Moir DT, Leung DW, Amarasinghe GK, 2017b. A Sensitive in Vitro High-Throughput Screen To Identify Pan-filoviral Replication Inhibitors Targeting the VP35-NP Interface. ACS Infect Dis 3, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KW, Naisbitt S, Fan JS, Sheng M, Zhang M, 2001. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem 276, 14059–14066. [DOI] [PubMed] [Google Scholar]
- Luthra P, Jordan DS, Leung DW, Amarasinghe GK, Basler CF, 2015. Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis. J Virol 89, 5148–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Naidoo J, Pietzsch CA, De S, Khadka S, Anantpadma M, Williams CG, Edwards MR, Davey RA, Bukreyev A, Ready JM, Basler CF, 2018. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antiviral Res 158, 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF, 2013. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 14, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Chiramel AI, Schmidt ML, Chen YC, Whitt N, Watt A, Dunham EC, Shifflett K, Traeger S, Leske A, Buehler E, Martellaro C, Brandt J, Wendt L, Muller A, Peitsch S, Best SM, Stech J, Finke S, Romer-Oberdorfer A, Groseth A, Feldmann H, Hoenen T, 2018. A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med 10, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE, 2010. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol 84, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M, Kolesnikova L, Schoehn G, Becker S, Ruigrok RW, 2002. Morphology of Marburg virus NP-RNA. Virology 296, 300–307. [DOI] [PubMed] [Google Scholar]
- Messaoudi I, Amarasinghe GK, Basler CF, 2015. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Microbiol 13, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K, 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31, 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrof J, Becker S, Muhlberger E, 2003. Ebola virus transcription activator VP30 is a zinc-binding protein. J Virol 77, 3334–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrof J, Muhlberger E, Klenk HD, Becker S, 2002. Phosphorylation of VP30 impairs ebola virus transcription. J Biol Chem 277, 33099–33104. [DOI] [PubMed] [Google Scholar]
- Moller P, Pariente N, Klenk HD, Becker S, 2005. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J Virol 79, 14876–14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH, Fasken MB, 2018. The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberger E, 2007. Filovirus replication and transcription. Future Virol 2, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberger E, Lotfering B, Klenk HD, Becker S, 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol 72, 8756–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S, 1999. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol 73, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Watanabe S, Halfmann P, Kawaoka Y, 2013. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep 3, 1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EV, Schmidt KM, Deflube LR, Doganay S, Banadyga L, Olejnik J, Hume AJ, Ryabchikova E, Ebihara H, Kedersha N, Ha T, Muhlberger E, 2016. Ebola Virus Does Not Induce Stress Granule Formation during Infection and Sequesters Stress Granule Proteins within Viral Inclusions. J Virol 90, 7268–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Hagiwara K, Sagara H, Kawaoka Y, 2010. Characterization of the Ebola virus nucleoprotein-RNA complex. J Gen Virol 91, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kolesnikova L, Becker S, Kawaoka Y, 2011. The importance of the NP: VP35 ratio in Ebola virus nucleocapsid formation. J Infect Dis 204 Suppl 3, S878–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestereich L, Ludtke A, Wurr S, Rieger T, Munoz-Fontela C, Gunther S, 2014. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res 105, 17–21. [DOI] [PubMed] [Google Scholar]
- Olejnik J, Hume AJ, Leung DW, Amarasinghe GK, Basler CF, Muhlberger E, 2017. Filovirus Strategies to Escape Antiviral Responses. Curr Top Microbiol Immunol 411, 293–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen ME, Filone CM, Rozelle D, Mire CE, Agans KN, Hensley L, Connor JH, 2016. Polyamines and Hypusination Are Required for Ebolavirus Gene Expression and Replication. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A, Volchkova VA, Reid SP, Mateo M, Bagnaud-Baule A, Nemirov K, Shurtleff AC, Lawrence P, Reynard O, Ottmann M, Lotteau V, Biswal SS, Thimmulappa RK, Bavari S, Volchkov VE, 2014. Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep 6, 1026–1036. [DOI] [PubMed] [Google Scholar]
- Pascali C, Teichmann M, 2013. RNA polymerase III transcription - regulated by chromatin structure and regulator of nuclear chromatin organization. Subcell Biochem 61, 261–287. [DOI] [PubMed] [Google Scholar]
- Patel RC, Sen GC, 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J 17, 4379–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi C, Ciccacci C, Novelli G, Borgiani P, 2018. Genetics and Treatment Response in Parkinson's Disease: An Update on Pharmacogenetic Studies. Neuromolecular Med 20, 1–17. [DOI] [PubMed] [Google Scholar]
- Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF, 2010. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J Virol 84, 10581–10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, Ott M, 2015. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, Amarasinghe GK, 2012. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc Natl Acad Sci U S A 109, 20661–20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Cardenas WB, Basler CF, 2005. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology 341, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF, 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol 80, 5156–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF, 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol 81, 13469–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rialdi A, Hultquist J, Jimenez-Morales D, Peralta Z, Campisi L, Fenouil R, Moshkina N, Wang ZZ, Laffleur B, Kaake RM, McGregor MJ, Haas K, Pefanis E, Albrecht RA, Pache L, Chanda S, Jen J, Ochando J, Byun M, Basu U, Garcia-Sastre A, Krogan N, van Bakel H, Marazzi I, 2017. The RNA Exosome Syncs IAV-RNAPII Transcription to Promote Viral Ribogenesis and Infectivity. Cell 169, 679–692 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V, Feldmann H, Grard G, Becker S, Leroy EM, 2015. Ebola and Marburg haemorrhagic fever. J Clin Virol 64, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabman RS, Hoenen T, Groseth A, Jabado O, Binning JM, Amarasinghe GK, Feldmann H, Basler CF, 2013. An upstream open reading frame modulates ebola virus polymerase translation and virus replication. PLoS Pathog 9, e1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabman RS, Leung DW, Johnson J, Glennon N, Gulcicek EE, Stone KL, Leung L, Hensley L, Amarasinghe GK, Basler CF, 2011. DRBP76 associates with Ebola virus VP35 and suppresses viral polymerase function. J Infect Dis 204 Suppl 3, S911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PS, Wojcechowskyj JA, Eckhardt M, Krogan NJ, 2015. Comparative mapping of host-pathogen protein-protein interactions. Curr Opin Microbiol 27, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. [DOI] [PubMed] [Google Scholar]
- Smith DR, McCarthy S, Chrovian A, Olinger G, Stossel A, Geisbert TW, Hensley LE, Connor JH, 2010. Inhibition of heat-shock protein 90 reduces Ebola virus replication. Antiviral Res 87, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smither SJ, Eastaugh LS, Steward JA, Nelson M, Lenk RP, Lever MS, 2014. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antiviral Res 104, 153–155. [DOI] [PubMed] [Google Scholar]
- Spengler JR, Ervin ED, Towner JS, Rollin PE, Nichol ST, Perspectives on West Africa Ebola Virus Disease Outbreak, 2013-2016. Emerg Infect Dis 22, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wu C, Shi L, Luthra P, Pintilie GD, Johnson B, Porter JR, Ge P, Chen M, Liu G, Frederick TE, Binning JM, Bowman GR, Zhou ZH, Basler CF, Gross ML, Leung DW, Chiu W, Amarasinghe GK, 2018. Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly. Cell 172, 966–978 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita Y, Matsunami H, Kawaoka Y, Noda T, Wolf M, 2018. Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution. Nature. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Halfmann P, Oyama M, Kozuka-Hata H, Noda T, Kawaoka Y, 2013. DNA topoisomerase 1 facilitates the transcription and replication of the Ebola virus genome. J Virol 87, 8862–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigabu B, Ramanathan P, Ivanov A, Lin X, Ilinykh PA, Parry CS, Freiberg AN, Nekhai S, Bukreyev A, 2018. Phosphorylated Vp30 of Marburg Virus Is a Repressor of Transcription. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunschke M, Conrad D, Enterlein S, Olejnik J, Brauburger K, Muhlberger E, 2013. The L-VP35 and L-L interaction domains reside in the amino terminus of the Ebola virus L protein and are potential targets for antivirals. Virology 441, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebelhoer LS, Albarino CG, McMullan LK, Chakrabarti AK, Vincent JP, Nichol ST, Towner JS, 2014. High-throughput, luciferase-based reverse genetics systems for identifying inhibitors of Marburg and Ebola viruses. Antiviral Res 106, 86–94. [DOI] [PubMed] [Google Scholar]
- Uyeki TM, Erickson BR, Brown S, McElroy AK, Cannon D, Gibbons A, Sealy T, Kainulainen MH, Schuh AJ, Kraft CS, Mehta AK, Lyon GM 3rd, Varkey JB, Ribner BS, Ellison RT 3rd, Carmody E, Nau GJ, Spiropoulou C, Nichol ST, Stroher U, 2016. Ebola Virus Persistence in Semen of Male Survivors. Clin Infect Dis 62, 1552–1555. [DOI] [PubMed] [Google Scholar]
- Valmas C, Basler CF, 2011. Marburg virus VP40 antagonizes interferon signaling in a species-specific manner. J Virol 85, 4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basler CF, Muhlberger E, 2010. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog 6, e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren E, Von Dollen J, Cimermancic P, Gulbahce N, Sali A, Krogan NJ, 2015. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics 49, 8 19 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Kolesnikova L, Clarke M, Koehler A, Noda T, Becker S, Briggs JAG, 2017. Structure and assembly of the Ebola virus nucleocapsid. Nature 551, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, Flint M, McMullan LK, Chen SS, Fearns R, Swaminathan S, Mayers DL, Spiropoulou CF, Lee WA, Nichol ST, Cihlar T, Bavari S, 2016. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, Honnold S, Bantia S, Kotian P, Chen X, Taubenheim BR, Welch LS, Minning DM, Babu YS, Sheridan WP, Bavari S, 2014. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 508, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Noda T, Kawaoka Y, 2006. Functional mapping of the nucleoprotein of Ebola virus. J Virol 80, 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, Berry KN, Yen B, Hamilton J, Brett TJ, Pappu RV, Leung DW, Basler CF, Amarasinghe GK, 2014. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 16, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Luthra P, Wu C, Batra J, Leung DW, Basler CF, Amarasinghe GK, 2017. Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis. Nat Commun 8, 15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Schneller SW, 2014. The enantiomers of the 1',6'-isomer of neplanocin A: synthesis and antiviral properties. Bioorg Med Chem 22, 5315–5319. [DOI] [PubMed] [Google Scholar]
- Yeh S, Varkey JB, Crozier I, 2015. Persistent Ebola Virus in the Eye. N Engl J Med 373, 1982–1983. [DOI] [PubMed] [Google Scholar]
- Zeng X, Blancett CD, Koistinen KA, Schellhase CW, Bearss JJ, Radoshitzky SR, Honnold SP, Chance TB, Warren TK, Froude JW, Cashman KA, Dye JM, Bavari S, Palacios G, Kuhn JH, Sun MG, 2017. Identification and pathological characterization of persistent asymptomatic Ebola virus infection in rhesus monkeys. Nat Microbiol 2, 17113. [DOI] [PubMed] [Google Scholar]
- Zhu T, Song H, Peng R, Shi Y, Qi J, Gao GF, 2017. Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide Reveals a Conserved Drug Target for Filovirus. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L, Nagy I, Orsini M, Weyher-Stingl E, Bracher A, Baumeister W, 2018. Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species. Structure. [DOI] [PubMed] [Google Scholar]