Abstract
1. Syndromic surveillance is an incipient approach to early wildlife disease detection. Consequently, systematic assessments are needed for methodology validation in wildlife populations.
2. We evaluated the sensitivity of a syndromic surveillance protocol for respiratory disease detection among chimpanzees in Gombe National Park, Tanzania. Empirical health, behavioural and demographic data were integrated with an agent-based, network model to simulate disease transmission and surveillance.
3. Surveillance sensitivity was estimated as 66% (95% Confidence Interval: 63.1, 68.8%) and 59.5% (95% Confidence Interval: 56.5%, 62.4%) for two monitoring methods (weekly count and prevalence thresholds, respectively), but differences among calendar quarters in outbreak size and surveillance sensitivity suggest seasonal effects.
4. We determined that a weekly detection threshold of ≥2 chimpanzees with clinical respiratory disease leading to outbreak response protocols (enhanced observation and biological sampling) is an optimal algorithm for outbreak detection in this population.
5. Synthesis and applications. This is the first quantitative assessment of syndromic surveillance in wildlife, providing a model approach to detecting disease emergence. Coupling syndromic surveillance with targeted diagnostic sampling in the midst of suspected outbreaks will provide a powerful system for detecting disease transmission and understanding population impacts.
Keywords: agent-based modelling, apes, disease ecology, disease transmission, network model, respiratory disease, syndromic surveillance, wildlife surveillance
Introduction
With progressing wildlife habitat loss, a growing human population, and increasing contact among human, domestic animal and wildlife populations, concern about emerging infectious diseases is intensifying. The public health threat is apparent, with approximately 60% of emerging infectious diseases resulting from zoonotic transmission, but negative consequences of disease emergence are not unique to humans. Numerous examples exist of disease spillover from humans and domestic animals into wildlife, with long-term and devastating consequences (Cleaveland et al., 2005; Epstein & Price, 2009; Kock et al., 1999; Michel et al., 2009; Nituch, Bowman, Beauclerc, & Schulte-Hostedde, 2011; Nugent, 2011; Roelke-Parker et al., 1996). Primates are recognized as an important taxon in emerging disease events, given close phylogenetic relatedness to humans, serving as sentinels and sources of infections in humans, while themselves suffering from human-associated pathogens (Calvignac-Spencer, Leendertz, Gillespie, & Leendertz, 2012; Epstein & Price, 2009; Jones et al., 2008; Wolfe, Dunavan, & Diamond, 2007; Woolhouse & Gowtage-Sequeria, 2005). Primates, particularly great apes, have also gained growing conservation interest. Validated disease surveillance systems in these populations would contribute to both conservation and “One Health” efforts.
Syndromic surveillance, the collection of pre-diagnosis health data, was implemented in a well-studied population of wild, human-habituated chimpanzees (Pan troglodytes schweinfurthii) in Gombe National Park, Tanzania (EV Lonsdorf, Travis, Pusey, & Goodall, 2006; Travis, Lonsdorf, Mlengeya, & Raphael, 2008). Gombe is home to three chimpanzee communities: Mitumba in the park’s north, Kasekela centrally, and Kalande in the park’s south. Mitumba and Kasekela chimpanzees are habituated to humans, individually identifiable by researchers, and have been targets of behavioural research since mid-1990s and 1960s, respectively. The population has declined from as many as 120–150 in the 1960s [Pusey et al., 2007] to 96–100 in early 2013. Disease appears to have played a role, with clinical illness associated with >60% of known chimpanzee deaths in Kasekela and decline of the Kalande community (Williams et al., 2008). Syndromic surveillance began in 2004 to collect observational data on observable clinical signs associated with specific disease syndromes: respiratory, dermatologic and gastrointestinal disease, wasting, and trauma (EV Lonsdorf et al., 2006; Travis et al., 2008). The goal was to better understand baseline disease levels and improve outbreak detection. However, until recently (Elizabeth V. Lonsdorf et al., 2016), surveillance data were not systematically analysed or optimized for real-time outbreak detection.
Syndromic surveillance can provide an early “signal” of infectious disease outbreaks (Henning, 2004). This surveillance targets general disease types, or “syndromes” (e.g. fever or flu-like illness), rather than confirmatory diagnostic data. It is sensitive to changes in disease trends and has been effective for early outbreak detection (Dórea, Sanchez, & Revie, 2011). It is hypothesized that these methods could provide early warning for conservation managers of the emergence/re-emergence of infectious diseases in wildlife, particularly in situations where access to laboratory diagnostic methods are limited.
Reports on syndromic surveillance in wildlife, particularly in primate populations outside of Gombe, are sparse (Arenas et al., 2002; Oleaga et al., 2011; Rodríguez-Prieto et al., 2014; Warns-Petit, Morignat, Artois, Calavas, & Warns-petit, 2010). Formal system evaluations are even more limited. To help fill this gap, we began a systematic evaluation of the Gombe chimpanzee syndromic surveillance system (E. V. Lonsdorf et al. 2016; T. W. Wolf, unpublished data). In doing so, we compiled an epidemiological profile of syndromic disease (E. V. Lonsdorf et al., 2016) and a profile of baseline respiratory disease (coughing, sneezing, and/or rhinorrhoea) with development of two algorithms to be used for respiratory outbreak detection (T. W. Wolf, unpublished data). Both algorithms were developed to identify respiratory disease levels exceeding normal variation within each habituated chimpanzee community by using a weekly threshold of ≥2 chimpanzees observed with respiratory signs and prevalence thresholds (weekly proportion of observed chimpanzees with clinical respiratory disease) of ≥16.7% (Kasekela) and ≥33.3% (Mitumba). These simple, community-specific thresholds were designed for real-time implementation to alert park managers early to potential outbreaks.
An important component in formal surveillance system evaluation is the assessment of outbreak detection sensitivity (probability that a disease outbreak will be detected by the system) and specificity (resistance of the system to false alarms) (Rodríguez-Prieto et al., 2014; Stoto et al., 2006). Traditionally in syndromic surveillance, diagnostic laboratory data is used to confirm a potential outbreak, enabling estimation of surveillance specificity against a gold standard. Surveillance sensitivity can be estimated by simulated and/or empirical data (Dórea et al., 2013; Jackson, Baer, Painter, & Duchin, 2007; Warns-Petit et al., 2010). In such a process, simulated outbreaks, often injected into empirical baseline data, are used to determine the probability of detection by the surveillance algorithm. Together, specificity and sensitivity measures are used to optimize the system for the highest possible outbreak detection probability, while minimizing false alarms. In Gombe, we lack diagnostic data for confirming potential respiratory outbreaks and calculating surveillance specificity. However, here we describe an approach to surveillance sensitivity estimation using empirical field data combined with disease simulation modelling.
Theoretical disease models offer opportunities to evaluate disease transmission through a simplified representation of complex processes that are not easily observed or understood (Anderson & May, 1979). When combined with empirical population data, theoretical disease models become powerful epidemiological tools, enabling us to answer the “what if” questions related to pathogen transmission and quantitatively evaluate mitigation strategies (e.g. vaccination, treatment) before implementation. Agent-based models combine infectious disease epidemiology with individual behaviour. Instead of modelling the transition of groups or populations through disease states, agent-based models simulate the transition of individuals through disease states, explicitly integrating individual characteristics that influence disease transmission. This approach is well-suited for small, isolated populations such as the Gombe chimpanzees (Perez & Dragicevic, 2009). Social network modelling is useful for studying disease transmission among highly social species as it captures the heterogeneity of a population’s social structure (Carne, Semple, Morrogh-Bernard, Zuberbühler, & Lehmann, 2013; M. E. Craft & Caillaud, 2011; M. Craft & Volz, 2011; Rushmore et al., 2013). This is useful for modelling disease transmission in chimpanzee societies, which have fission-fusion dynamics in which members of a community form temporary subgroups or ‘parties’ (Goodall, 1986). Given these advantages, we combined these approaches in a model simulating disease transmission and surveillance.
To date, primate demographic and behavioural data have been utilized in transmission models, but empirical disease data have been lacking (Carne et al., 2013; Nunn, Thrall, Stewart, & Harcourt, 2007; Rushmore, Bisanzio, & Gillespie, 2017; Rushmore et al., 2014, 2013). Gombe databases include demographic, behavioural and disease/health data, providing a unique opportunity to create fully-informed disease transmission models. The primary objectives of this study were to 1) build a disease model - informed by empirical datasets - to couple respiratory disease transmission and syndromic surveillance simulation and 2) estimate the syndromic surveillance sensitivity for respiratory disease outbreak detection. Further, Gombe has marked seasonal fluctuations in climate based on rainfall: early wet (November-February), late wet (March-April), early dry (May-July), and late dry (August-October) seasons (Elizabeth V Lonsdorf et al., 2011). Given reported seasonal differences in chimpanzee behaviour (e.g. party size) (Murray, Eberly, & Pusey, 2006; Wrangham, 1977), activity budgets (Lodwick, Borries, Pusey, Goodall, & McGrew, 2004), and body mass ( a. E. Pusey, Oehlert, Williams, & Goodall, 2005), which may influence contact and disease susceptibility or researchers’ ability to find chimpanzees or clearly observe clinical signs, we stratified simulations by calendar quarter to approximate seasonal changes in population social structure and surveillance rates.
Materials and Methods
Empirical Data
Empirical data collected from March 2004 to December 2012 on the Kasekela chimpanzee community, which ranged in size from 56–68 individuals, was utilized for model parameterization (Table 1). Field research was conducted with the permission of the Tanzania Commission for Science and Technology, Tanzania Wildlife Research Institute, and Tanzania National Parks Association. Syndromic surveillance in Gombe gathers health data on community members through a variety of means, as described previously and in Supporting Information (EV Lonsdorf et al., 2006; Elizabeth V. Lonsdorf et al., 2016; Travis et al., 2008), but primarily consists of the collection of standardized health data on the presence or absence of clinical signs of specific disease syndromes. The Daily Health Record (DHR) captures detailed information on individuals, whether healthy or unhealthy, that are subjects of full-day behavioural studies (A. E. Pusey, Wilson, & Anthony Collins, 2008), as well as contact individuals that are perceived as unhealthy. Two disease cluster algorithms were recently developed to screen DHR data accumulated over the course of a week for respiratory disease levels exceeding baseline variation in cumulative case number and prevalence (the latter accounting for the weekly variation in number of chimpanzees observed) (T. W. Wolf, unpublished data). The thresholds for outbreak detection were a weekly case count of ≥2 chimpanzees with respiratory signs and prevalence (the proportion of chimpanzees observed each week with respiratory signs) of ≥16.7%.
Table 1.
Parameters included in the respiratory disease transmission model. All values used as rates in the model were Poisson distributed.
| Parameter | Value | Data source |
|---|---|---|
| Agent | Chimpanzees, n=60 | 2007 Kasekela community data, Jane Goodall Institute Research Center at Duke University |
| Party network | Quarter-specific, 2007 pairwise Dyadic Association Indices |
Jane Goodall Institute Research Center at Duke University |
| Grooming network | Quarter-specific, 2007 pairwise Grooming Association Indices |
Jane Goodall Institute Research Center at Duke University |
| Grooming contact rate (per day) |
0.535 | Model calibrated |
| Party-level contact rate (per day) |
0.436 | Model calibrated |
| Grooming transmission probability |
0.529 | Model calibrated |
| Party-level transmission probability |
0.085 | Model calibrated |
| Incubation period (days) | 2 | Model calibrated |
| Infectious period (days) | 4 | Model calibrated |
| Mean surveillance probabilities (s.e.)† |
2007 Gombe syndromic surveillance data | |
| Quarter 1 | 0.036 (± 0.024) | |
| Quarter 2 | 0.035 (± 0.024) | |
| Quarter 3 | 0.032 (± 0.023) | |
| Quarter 4 | 0.033 (± 0.024) |
Denoted here are community aggregated estimates of surveillance rates for each quarter; actual model inputs were individual surveillance rates for each chimpanzee for the specified quarter.
To test the sensitivity of the thresholds for outbreak detection, we created a disease transmission model to simulate DHR surveillance and used model output to estimate the number of outbreaks detected using the established weekly detection thresholds. To do this, we generated individual rates of observation by DHR surveillance (i.e. surveillance rates) to parameterize surveillance in the model. We used generalized estimating equations to determine that individual surveillance rates were optimally aggregated by quarter and year for parameterization (Supporting Information). Accordingly, we used the proportion of days in each quarter that an individual was observed by surveillance as quarter-specific, individual probabilities of surveillance.
We created two contact networks over which to simulate disease transmission from behavioural data of pairwise associations between Kasekela chimpanzees: 1) a party-level network, representing loose pair affiliations within a large chimpanzee group and 2) a grooming network, representing more intimate contact of grooming chimpanzees. A dyadic association index (DAI) was used in the creation of the party-level network and was estimated for each chimpanzee pair based on amount of time spent in the same party relative to the total amount of time each was observed (Supporting Information). A grooming association index (GAI), used to create a grooming network, was also calculated from behavioural data as the time a pair of chimpanzees spent grooming while within the same party (Supporting Information). Matrices of DAI and GAI data were integrated into the model as individual chimpanzee parameters to create pairwise connections between each community member.
Respiratory disease data from a major outbreak (≥20% community morbidity (Elizabeth V Lonsdorf et al., 2011; Williams et al., 2008)) in Kasekela community in 2007 was used to calibrate epidemiologic parameters (Table 1). This outbreak lasted 29 days and resulted in a cumulative case incidence of 25 (SI Figure 1) and 38% morbidity. While population managers in 2007 presumed this event an infectious disease outbreak, an etiologic agent was not confirmed with diagnostic data.
Model Overview
An agent-based, network model was constructed using the Researcher/Educational edition of AnyLogic software, version 7.1.2 (The AnyLogic Company, Chicago, Illinois, USA), a tool for creating models in the Java language. A stochastic Susceptible-Exposed-Infectious-Recovered (SEIR) disease model was created for a closed population (SI Figure 2), where individuals progressed through states of infection using contact parameters and transmission probabilities [Anderson and May, 1979] to simulate empirically observed respiratory disease outbreaks in ape populations: outbreaks of short duration (weeks) with a finite transmission period and variable morbidity [Kaur et al., 2008; Köndgen et al., 2008; Lonsdorf et al., 2011; Palacios et al., 2011; Williams et al., 2008]. An individual’s transition from Susceptible to Exposed state was dependent on its connection with an Infectious individual within its network, and defined by:
| (Equation 1) |
where βI is a daily contact rate with the Infectious connected individual and γ is a probability of disease transmission. The product of these two rates is the effective contact rate, or the rate at which contact between an Infectious and Susceptible individual results in a successful transmission event (Vynnycky and White 2010). Unique β and γ parameters were calibrated for grooming and party-level networks (using August 2007 outbreak data; see Supporting Information) to capture the heterogeneity of contact and associated disease transmission probability (Table 1). For example, grooming animals will have extended periods of direct contact, whereas animals within a party may or may not come into direct contact or even be in proximity with each other.
The remaining transitions between infection states were a function of the duration of time spent in a state. Transition from the Exposed state into the Infectious state was a rate set by the reciprocal of the number of days of latent infection or the latent period. Transition from the Infectious state to the Recovered state was a rate set as the reciprocal of the infectious period. All rates were Poisson distributed. Once recovered, individuals did not return to the susceptible state and were assumed to be immune for the remaining duration of simulation. This assumption was made based on empirical observations of clusters of respiratory disease, where clusters lasted for a finite period and persistence of clinical signs or recurrence of clinical signs were not a regular occurrence. Since the outbreak periods modelled were of short duration, a closed population model was constructed, excluding birth and mortality events. All epidemiologic parameters were calibrated in the model using August 2007 outbreak data, and none were varied between simulations.
At the start of each simulation a new population of agents (i.e. chimpanzees) was created from an Excel file containing a single year’s data of all individuals in the Kasekela community, their individual surveillance rates, and network matrices (DAI and GAI). DAI and GAI estimates were used as a probability for connection between pairs in the model. The networks were reset every six hours of model time, such that all connections were dissolved and re-established based on each pair’s connection probability, to mimic the fission-fusion social structure of chimpanzee communities (Goodall, 1986). Input data was stratified by quarter so that prior to a simulation run, the quarter was specified and corresponding data uploaded into the model. This design allows flexibility in modelling different populations and network structures.
Syndromic surveillance was simulated at each time step (daily) by testing whether each individual was observed by syndromic surveillance and logging if they were ill. In this process, two Boolean values were logged for each individual, where a value of true was set if a randomly generated number fell within the individual’s surveillance probability and true if the individual was in the Infectious state. As the simulation progressed, the weekly number of individuals that were 1) Infectious, 2) observed by surveillance, and 3) Infectious AND observed by surveillance were tabulated for output. Individuals in the Infectious state (and no other state) were assumed to express clinical signs, thus detection by surveillance while in the Infectious state was equivalent to observation of illness. Variations in clinical sign expression or surveillance rates by disease states were not modelled. In summary, individual-level empirical data were used to populate the model with chimpanzees having unique pairwise network connections with others in the community along with unique surveillance rates, both essential components of the agent-based model; whereas disease transmission parameters were estimated by fitting 2007 respiratory outbreak data to the model (Supporting Information).
Monte Carlo simulation methods were employed to obtain the relevant outbreak data for testing surveillance sensitivity, including outbreak duration, cumulative incidence, and weekly numbers of Infectious chimpanzees, chimpanzees observed by surveillance (sick and healthy), and sick chimpanzees detected by surveillance. Experiments were performed for each quarter of 2007 (consistent with the year from which calibration data originated), and each experiment was performed over 500 simulations with the introduction of a single, random Infectious individual. Each simulation was set to end when there were no additional Infectious agents or after 90 days of simulation. Model documentation can be retrieved from the Data Repository for University of Minnesota (https://doi.org/10.13020/D64X2Q).
Scenario Analyses
Three additional experiments were conducted to evaluate how empirical data choice for model parameterization impacted surveillance sensitivity estimates. Specifically, we wanted to know how the selection of surveillance rates and year modelled impacted results. To evaluate the impact of surveillance rate inputs on sensitivity estimation, a Monte Carlo experiment was performed in Quarter 3 (July – September) of the same year modelled in previous experiments, but surveillance rates were aggregated as an individual’s mean rate over its age-class for that quarter (Scenario 1, Supporting Information). To evaluate the effect of modelling multiple years versus a single year’s empirical data on sensitivity estimation, a series of experiments were performed in Quarter 3 with data from 2005–2012 (Scenario 2, Supplemental Information). We also wanted to determine if increased surveillance efforts by researchers could improve surveillance sensitivity. For this experiment, the surveillance probability was doubled to simulate a doubling of efforts (Scenario 3, Supporting Information). All disease transmission parameters were maintained across experiments. The resulting surveillance sensitivity estimates and 95% confidence intervals from each experiment were compared to the original quarter estimate (Vynnycky & White, 2010).
We did not conduct scenario analyses by varying the surveillance thresholds (count or prevalence) to determine if surveillance sensitivity could be improved with lower thresholds. The thresholds were established based on the normal variation of baseline respiratory disease in the study population, which was low (T. W. Wolf, unpublished data). We expect, based on observations of false alarms produced by screening retrospective data using the current prevalence threshold (T. W. Wolf, unpublished data), that lowering either of these (count to <2 chimps per week and prevalence to <16.7%) would result in a large number of false alarms.
Statistical Analyses and Sensitivity Estimation
Simulation results were aggregated by quarter to estimate the number of outbreaks produced (where at least one transmission occurred), mean cumulative incidence, mean weekly number of Infectious chimpanzees, mean outbreak duration, and associated 95% Probability Intervals. Surveillance output was aggregated to obtain the mean weekly number of chimpanzees observed by surveillance and mean weekly number detected sick by quarter. A weekly apparent prevalence was calculated as the number of Infectious chimpanzees observed by surveillance (apparently sick) out of the total number of chimpanzees observed by surveillance. This measure is comparable to the estimation of apparent prevalence from diagnostic testing (Dohoo, Martin, & Stryhn, 2009):
| (Equation 2) |
where in this case, T+ is the number of animals observed in the Infectious state and T- is the number of animals observed in any other disease state. We used maximum weekly apparent prevalence and maximum weekly number of apparently sick from each simulation to determine if our outbreak detection algorithms (i.e. weekly count and prevalence thresholds) would detect the simulated outbreak. So, if the maximum weekly apparent prevalence was ≥16.7%% or the maximum number of apparently sick was ≥2, then an outbreak was classified as “detected.” Surveillance sensitivity was calculated for each quarter as the number of outbreaks detected out of the total number of outbreaks produced.
Associations between outbreak size, quarter, and outbreak detection were examined by logistic regression modelling. For these analyses, a categorical value for outbreak size was generated, where outbreaks of ≥20% community morbidity were classified as major and those with less were classified as small. Outbreak detection was included as a binomial variable. Univariate logistic regression models were performed to examine associations between quarter and outbreak size, quarter and outbreak detection, and outbreak size and detection. A final multivariate model was generated with outbreak detection as the dependent variable and quarter and outbreak size as predictor variables; odds of outbreak detection and 95% confidence intervals were estimated by outbreak size and quarter. Logistic regression models were performed in SAS, Version 9.4 for Windows (SAS Institute Inc., Cary, NC, USA).
Results
Out of 2000 simulations (500 per calendar quarter), where a single chimpanzee was seeded with infection, 1064 iterations resulted in at least one secondary infection being produced. Average outbreak duration was 5 weeks (95% Probability Interval: 1, 9) (Figure 1) with an average cumulative incidence of 36 cases (95% PI: 2, 55) (Figure 2a). The mean weekly number of Infectious chimpanzees was 9 (95% PI: 1, 18). The mean weekly number of chimpanzees observed by surveillance was 12 (95% PI: 9, 15) and an average of 8.2% (95% PI: 0, 27%) of Infectious chimpanzees were detected weekly by surveillance (Figure 2b). Out of 1064 outbreaks, 702 were detected using the weekly count threshold for outbreak detection and 633 were detected using the weekly prevalence threshold, yielding a surveillance sensitivity for outbreak detection of 66% (95% Confidence Interval: 63.1, 68.8%) and 59.5% (95% Confidence Interval: 56.5%, 62.4%), respectively. The count threshold was consistently more sensitive in outbreak detection than the prevalence threshold (Table 2), with highest sensitivity in Quarter 1 (81%; 95% Confidence Interval: 77%, 86%) and Quarter 2 (79%; 95% Confidence Interval: 74%, 84%) and lowest sensitivity in Quarter 3 (30%; 95% Confidence Interval: 24%, 36%).
Figure 1.
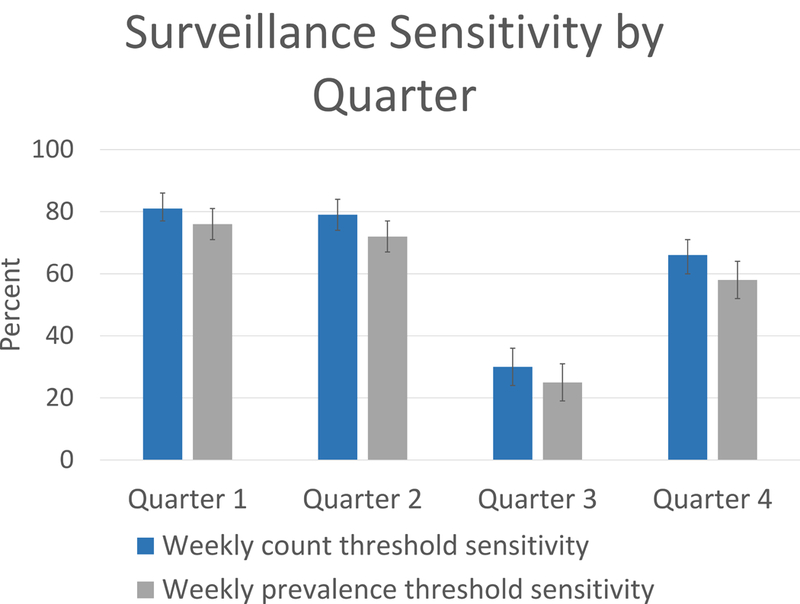
Estimated surveillance sensitivity and 95% confidence intervals for the two weekly outbreak detection thresholds. Sensitivity estimates were generated from model output for each quarter as the percent outbreaks detected by the weekly count and prevalence outbreak detection algorithms. Column bars represent the 95% confidence interval.
Figure 2.
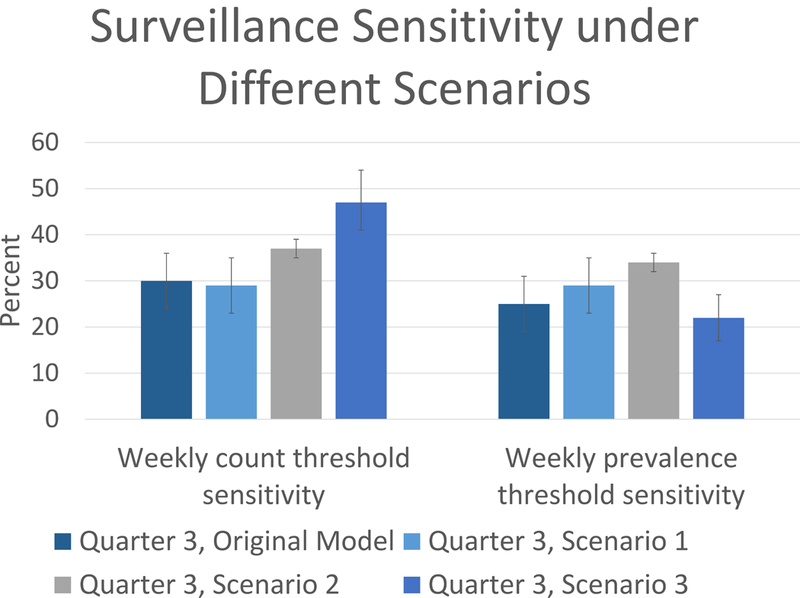
Estimated surveillance sensitivity and 95% confidence intervals from Quarter 3 scenario analyses for the two weekly outbreak detection thresholds. Compared to the estimated sensitivity for Quarter 3 from the original model (column 1) are scenario analyses of Quarter 3 using age-class-aggregated surveillance rates (Scenario 1, column 2), simulations across all years (2005–2012) (Scenario 2, column 3), and doubling of surveillance efforts (Scenario 3, column 4). Column bars represent the 95% confidence interval.
Table 2.
Summary of simulated outbreak characteristics and surveillance by quarter. Displayed are model results for experiments over all quarters from the primary model, as well as results from scenario analyses of Quarter 3 using age-class-aggregated surveillance rates (Scenario 1), simulations across all years (2005–2012) (Scenario 2), and doubling of surveillance efforts (Scenario 3).
| Model output | Quarter 1 mean (95%PI) |
Quarter 2 mean (95%PI) |
Quarter 3 mean (95%PI) |
Quarter 4 mean (95%PI) |
Quarter 3 Scenario 1, mean (95%PI) |
Quarter 3 Scenario 2, mean (95%PI) |
Quarter 3 Scenario 3, mean (95%PI) |
|---|---|---|---|---|---|---|---|
| Total outbreaks | 278 | 287 | 221 | 278 | 232 | 1702 | 237 |
| No. major outbreaks† | 246 | 245 | 90 | 220 | 113 | 1028 | 115 |
| Outbreak duration (weeks) | 5 (1, 9) |
5 (1, 9) |
4 (1, 9) |
5 (1, 9) |
4 (1, 9) |
5 (1, 9) |
4 (1,9) |
| Cumulative incidence | 46 (2, 56) |
42 (2, 54) |
13 (2, 40) |
37 (2, 53) |
16 (2, 41) |
27 (2, 53) |
16 (2, 42) |
| Mean weekly Infectious | 12 (1, 19) |
11 (2, 18) |
4 (1, 10) |
9 (2, 16) |
4 (1, 11) |
7 (1, 14) |
4 (1,11) |
| Mean weekly number observed by surveillance |
13 (10, 16) |
12 (9, 15) |
11 (7, 15) |
11 (9, 14) |
11 (8, 14) |
11 (6, 18) |
20 (16,24) |
| Mean weekly proportion detected sick | 0.09 (0, 0.25) |
0.08 (0, 0.22) |
0.08 (0, 0.33) |
0.08 (0, 0.23) |
0.08 (0, 0.36) |
0.06 (0, 0.28) |
0.14 (0, 0.50) |
Major outbreaks are those with ≥20% morbidity.
Out of 1064 outbreaks, 801 were classified as major outbreaks based on our criteria. Outbreak size was significantly associated with calendar quarter. Quarter 1 had significantly higher odds of a major outbreak and Quarter 3 had significantly lower odds of a major outbreak, as compared to Quarter 4 (Table 3). Outbreak detection using the count threshold was significantly associated with quarter, with Quarters 1 and 2 having significantly higher odds of outbreak detection than Quarter 4, and Quarter 3 having significantly lower odds of detection. Outbreak detection was significantly associated with outbreak size, with small outbreaks having significantly lower odds of detection than major outbreaks (OR: 0.003; 95% Confidence Interval: 0.001, 0.007). Since outbreak size was significantly associated with both outbreak detection and quarter, we examined its role as a confounder. A multivariate logistic regression model including outbreak size and quarter confirmed this, resulting in >10% change in the log odds estimate of the outbreak detection-quarter association. After controlling for outbreak size, we confirmed the differences observed in surveillance sensitivity by quarter.
Table 3.
Odds ratios (OR) for associations between quarter and outbreak detection and outbreak size. Quarter 4 was the reference for all models. The multivariate model included outbreak size as a predictor variable to control for confounding.
| OR estimate (95% Confidence Interval) |
||||
|---|---|---|---|---|
| Dependent variable | Model structure | Quarter 1 | Quarter 2 | Quarter 3 |
| Outbreak size | univariate | 2.0 (1.3, 3.2) |
1.5 (0.99, 2.4) |
0.18 (0.1, 0.3) |
| Outbreak detection | univariate | 2.3 (1.5, 3.3) |
2.0 (1.4, 2.9) |
0.22 (0.2, 0.3) |
| Outbreak detection | multivariate | 2.2 (1.3, 3.9) |
2.4 (1.4, 4.2) |
0.54 (0.3, 0.9) |
Scenario analyses
In scenario 1, surveillance rates averaged over chimpanzees’ age classes were generally lower than their surveillance rates in the primary experiments, although not significantly different. For example, average surveillance rates for Quarter 3 in primary experiments was 0.032 (s.e. ± 0.025), whereas surveillance rates for the same quarter, aggregated over chimpanzees’ age class averaged 0.029 (s.e. ± 0.015). Surveillance sensitivity was similar, with 29% (95% Confidence Interval: 23, 35%) of outbreaks detected by both thresholds (Table 2).
In scenario 2, experiments were conducted over all years of empirical data (2005–2012) during Quarter 3, where community members, contact networks and individual chimpanzee surveillance rates were varied by year. Out of 4000 simulations of Quarter 3 from 2005–2012, 1702 outbreaks were produced, resulting in overall sensitivity estimates of 37% (95% Confidence Interval: 35, 39%) and 34% (95% Confidence Interval: 32, 36%) by the count and prevalence thresholds, respectively. While the sensitivity estimate generated for Quarter 3 from the primary experiment was within the range of sensitivity estimates (two years resulted in even lower sensitivity estimates), it was low and contained more variation around the estimate as compared the multi-year analysis (Table 2). While long-term data sets are challenging to come by, this scenario demonstrates their value in such analyses.
Doubling each individual’s surveillance rate (Scenario 3) resulted in an increase in the average weekly number of animals detected by surveillance to 20 (95% PI: 16, 24). Quarter 3 surveillance sensitivity by the count threshold increased significantly to 47% (95% Confidence Interval: 41, 54%), but there was no change by the prevalence threshold, (estimate: 22%; 95% Confidence Interval: 17, 27%). Notably, among outbreaks with a cumulative incidence of 5 or more cases in Quarter 3, the sensitivity of outbreak detection by the count method increased from 53% (95% Confidence Interval: 44, 62%) to 74% (95% Confidence Interval: 67, 81%) with a doubling of surveillance efforts.
Discussion
We report the first quantitative assessment of syndromic surveillance sensitivity for respiratory outbreak detection in a population of free-living apes, and possibly the first such surveillance assessment for any wildlife population. We built an agent-based, network model to simulate disease transmission and syndromic surveillance in different calendar quarters, capturing seasonal variability in chimpanzee behaviour and surveillance. Using model output, we examined the sensitivity of two weekly outbreak detection thresholds, a weekly count threshold (i.e. absolute number ill) and a weekly prevalence threshold (i.e. proportion ill of those seen), and show consistently better performance by the count threshold. We show further that increasing surveillance effort (i.e. observing more chimpanzees each week) improves sensitivity of the count threshold. Given its simplicity, we expect easy implementation of this threshold for real-time detection of respiratory outbreaks by park managers and field researchers.
In comparison to a limited number of assessments of syndromic surveillance sensitivity (from the broader veterinary and public health research communities), our overall estimates of surveillance sensitivity fall within the reported range (50–98%) of different detection algorithms [Buckeridge et al. 2008; Dórea et al. 2013; Jackson et al. 2007]. Dórea et al. [2013] demonstrate how performance of different detection algorithms can vary under different scenarios. Our system’s sensitivity estimates are likely influenced by the challenge of observing disease in wildlife. However, if surveillance efforts were differentially increased (e.g. targeting “high risk” individuals), there may be improvement in outbreak detection with minimal change in observer effort. For example, high ranking females and their offspring are central in the social network and potentially important for disease transmission (Rushmore et al., 2013); these individuals tend also to be more easily observed in the Gombe communities. Although not tested in this model, focusing efforts on such individuals has the potential to improve outbreak detection.
Surveillance sensitivity varied by quarter, with the highest sensitivity in the first and second quarters and lowest in the third. In Tanzania , the first and second quarters coincide with the rainy season (January-March) and transition to the long dry season (April-June); whereas the third quarter coincides with the end of the long dry season (July – September). The latter is a period where food may be less abundant and chimpanzees more scattered in the park (Markham et al., 2013; a. E. Pusey et al., 2005). Our model indicates that seasonal dynamics of social structure may play a role in disease transmission and surveillance. In particular, fewer and smaller scale outbreaks were produced in the third quarter. This was the only quarter where the number of small outbreaks exceeded major outbreaks, although sensitivity remained significantly lower even after controlling for outbreak size. Seasonal changes in the environment may influence researchers’ ability to find and/or clearly observe chimpanzees; however, no seasonal differences have been observed in community coverage by surveillance (T. W. Wolf, unpublished data). Importantly, these findings raise awareness of seasonal differences in outbreak epidemiology (i.e. smaller outbreaks, with fewer weekly illnesses) that make disease detection more challenging. Further, although network analyses were not an objective of this study, our findings demonstrate a need to examine the seasonal dynamics of the social network in association with patterns of respiratory disease.
Model stochasticity resulted in outbreaks of similar incidence and duration of empirical respiratory outbreaks as intended, but additional output was validated by empirical data. For instance, the maximum number of chimpanzees identified as sick by surveillance (i.e. the observed cumulative incidence) in model simulations was ten (mean: 2.4, s.e. ± 1.8), even with high outbreak incidence. This aligned fairly well with empirical data, where during three respiratory outbreaks, 6, 10, and 15 sick animals were documented in the DHR; the full extent (i.e. cumulative incidence) of those outbreaks was captured by other data sources. There was no function in the model that adjusted surveillance probabilities after illness was initially detected, and since respiratory disease was an infrequent occurrence during the study period (Elizabeth V. Lonsdorf et al., 2016), it is reasonable to expect that recognition of disease in several individuals might alert researchers to the presence of contagious disease and trigger increased efforts to collect data on sick community members.
Ideally, a full quantitative surveillance assessment would include an estimate of specificity (i.e. a system’s tendency away from false alarms) with system optimization using both specificity and sensitivity estimates. As veterinary interventions have generally not been undertaken in Gombe and non-invasive sampling methodologies are relatively new for respiratory pathogen detection in primates, confirmatory diagnostic data were unavailable for specificity estimation during the study period. As our detection thresholds were low, we expect a higher sensitivity at the cost of lower specificity (i.e. high rate of false alarms). However, given the infrequent occurrence of respiratory diseases in the system (Elizabeth V. Lonsdorf et al., 2016) and ongoing concerns for their impact on great ape conservation, park officials prefer to err on investigating early and often.
Conclusions
Syndromic surveillance is a valuable, non-invasive tool for emerging infectious disease detection in wildlife. However, these systems must be evaluated and optimized. This study helps fill this gap and identifies key areas where the Gombe system could be improved. Notably, the integration of non-invasive, diagnostic data for respiratory pathogen detection (Gillespie, Nunn, & Leendertz, 2008; Kondgen, Schenk, Pauli, Hoesch, & Leendertz, 2010; Leendertz et al., 2006) with existing syndromic surveillance would allow the estimation of surveillance specificity. Refinement of the outbreak detection thresholds could be achieved through receiver operator characteristic (ROC) analysis, where sensitive outbreak detection (high sensitivity) and few false alarms (high specificity) can be optimized. Further, the model was developed with the flexibility to create a population of chimpanzees and their social network from empirical data at the start of simulation, enabling future exploration of population size, demographic, and social network structure impacts on disease transmission and surveillance. With growing concerns of emerging infectious disease in wildlife, advancement on this front is imperative.
Supplementary Material
Acknowledgements
Many thanks to Jane Goodall and the Jane Goodall Institute for supporting long-term chimpanzee research at Gombe National Park, the Gombe Stream Research Center staff collecting daily behavioural and health observations, and the remainder of the Gombe Ecosystem Health Project investigators (Carson Murray, Karen Terio, Beatrice Hahn) whose combined efforts ensure ongoing maintenance of syndromic surveillance in the park. We are grateful to the Tanzania Commission for Science and Technology, Tanzania Wildlife Research Institute, and Tanzania National Parks Association for their continued support and approval of this research. Funding support for collection and analysis of syndromic surveillance data comes from the National Institute of Health (R01 AI058715, R01 AI120810 and R00 HD057992), National Science Foundation (LTREB-1052693), Arcus Foundation, USFWS Great Ape Conservation Fund, Morris Animal Foundation (D10ZO-902), University of Minnesota Consortium on Law and Values in Health, Environment, and the Life Sciences, University of Minnesota Doctoral Dissertation Fellowship and Lincoln Park Zoo. Collection, digitization and analysis of behavioural data were supported by grants from the National Institutes of Health (R00 HD057992) and the Leo S. Guthman Foundation.
Footnotes
Data Availability
Data available through the Data Repository for University of Minnesota (DRUM) https://doi.org/10.13020/D64X2Q (Wolf et al., 2018).
References
- Anderson R, & May R (1979). Population biology of infectious diseases: Part I. Nature, 280, 361–367. doi:0028-0836/79/310361 [DOI] [PubMed] [Google Scholar]
- Arenas AJ, Gómez F, Salas R, Carrasco P, Borge C, Maldonado A, … Martinez-Moreno FJ (2002). An evaluation of the application of infrared thermal imaging to the tele-diagnosis of sarcoptic mange in the Spanish ibex ( Capra pyrenaica ). Veterinary Parasitology, 109(1), 111–117. [DOI] [PubMed] [Google Scholar]
- Buckeridge DL, Okhmatovskaia A, Tu S, O’Connor M, Nyulas C, & Musen MA (2008). Predicting outbreak detection in public health surveillance: quantitative analysis to enable evidence-based method selection. AMIA ... Annual Symposium Proceedings / AMIA Symposium. AMIA Symposium, 76–80. Retrieved from http://primo.lib.umn.edu/primo_library/libweb/action/PushToAction.do?indx=1&doc=TN_scopus2-s2.0-73949098160&recId=TN_scopus2-s2.0-73949098160&docs=TN_scopus2-s2.0-73949098160&pushToType=Mendeley&fromEshelf=false&vid=TWINCITIES&fromMendeley=true&fromSitemap=1 [PMC free article] [PubMed] [Google Scholar]
- Calvignac-Spencer S, Leendertz S, Gillespie TR, & Leendertz FH (2012). Wild great apes as sentinels and sources of infectious disease. Clinical Microbiology and Infection : The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases, 18(6), 521–7. doi: 10.1111/j.1469-0691.2012.03816.x [DOI] [PubMed] [Google Scholar]
- Carne C, Semple S, Morrogh-Bernard H, Zuberbühler K, & Lehmann J (2013). Predicting the vulnerability of great apes to disease: The role of superspreaders and their potential vaccination. PLoS ONE, 8(12), e84642 Retrieved from 10.1371/journal.pone.0084642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Mlengeya T, Kazwala RR, Michel a, Kaare MT, Jones SL, … Packer C (2005). Tuberculosis in Tanzanian wildlife. Journal of Wildlife Diseases, 41(2), 446–53. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/16107683 [DOI] [PubMed] [Google Scholar]
- Craft ME, & Caillaud D (2011). Network models: an underutilized tool in wildlife epidemiology? Interdisciplinary Perspectives on Infectious Diseases, 2011, 676949. doi: 10.1155/2011/676949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft M, & Volz E (2011). Disease transmission in territorial populations: the small-world network of Serengeti lions. Journal of the Royal Society, Interface, 8, 776–786. Retrieved from http://rsif.royalsocietypublishing.org/content/8/59/776.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohoo IR, Martin W, & Stryhn H (2009). Veterinary Epidemiologic Research (2nd ed.). Charlotte, P.E.I.: VER, Inc. [Google Scholar]
- Dórea FC, Mcewen BJ, Mcnab WB, Revie CW, Sanchez J, & Do FC (2013). Syndromic surveillance using veterinary laboratory data: data pre-processing and algorithm performance evaluation. Journal of the Royal Society, Interface, 10(April), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dórea FC, Sanchez J, & Revie CW (2011). Veterinary syndromic surveillance: Current initiatives and potential for development. Preventive Veterinary Medicine doi: 10.1016/j.prevetmed.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Epstein JH, & Price JT (2009). The significant but understudied impact of pathogen transmission from humans to animals. Mount Sinai Journal of Medicine, 76, 448–455. doi: 10.1002/MSJ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie TR, Nunn CL, & Leendertz FH (2008). Integrative approaches to the study of primate infectious disease: Implications for biodiversity conservation and global health. American Journal of Physical Anthropology, Suppl 47, 53–69. doi: 10.1002/ajpa.20949 [DOI] [PubMed] [Google Scholar]
- Goodall J (1986). The chimpanzees of Gombe: Patterns of behavior Cambridge, Massachusetts: Belknap Press of Harvard University Press. [Google Scholar]
- Henning K (2004). Overview of syndromic surveillance: What is syndromic surveillance? MMWR, 53(Suppl)(September 24, 2004), 5–11. Retrieved from http://www.cdc.gov/mmwr/preview/mmwrhtml/su5301a3.htm#top [PubMed] [Google Scholar]
- Jackson ML, Baer A, Painter I, & Duchin J (2007). A simulation study comparing aberration detection algorithms for syndromic surveillance. BMC Medical Informatics and Decision Making, 7(6). doi: 10.1186/1472-6947-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, & Daszak P (2008). Global trends in emerging infectious diseases. Nature, 451(7181), 990–3. doi: 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Singh J, Tong S, Humphrey C, Clevenger D, Tan W, … Nishida T (2008). Descriptive epidemiology of fatal respiratory outbreaks and detection of a human-related metapneumovirus in wild chimpanzees (Pan troglodytes) at Mahale Mountains National Park, Western Tanzania. American Journal of Primatology, 70(8), 755–65. doi: 10.1002/ajp.20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock RA, Wambua JM, Mwanzia J, Wamwayi H, Ndungu EK, Barrett T, … Rossiter PB (1999). Rinderpest epidemic in wild ruminants in Kenya 1993–97. The Veterinary Record, 145(10), 275–283. [DOI] [PubMed] [Google Scholar]
- Köndgen S, Kühl H, N’Goran PK, Walsh PD, Schenk S, Ernst N, … Leendertz FH (2008). Pandemic human viruses cause decline of endangered great apes. Current Biology, 18(4), 260–264. doi: 10.1016/j.cub.2008.01.012 [DOI] [PubMed] [Google Scholar]
- Kondgen S, Schenk S, Pauli G, Hoesch C, & Leendertz F (2010). Noninvasive monitoring of respiratory viruses in wild chimpanzees. EcoHealth, 7(3), 332–341. [DOI] [PubMed] [Google Scholar]
- Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, … Christophe B (2006). Pathogens as drivers of population declines: The importance of systematic monitoring in great apes and other threatened mammals. Biological Conservation, 131(2), 325–337. doi: 10.1016/j.biocon.2006.05.002 [DOI] [Google Scholar]
- Lodwick J, Borries C, Pusey A, Goodall J, & McGrew W (2004). From nest to nest: influence of ecology and reproduction on the active period of adult Gombe chimpanzees. American Journal of Primatology, 64, 249–260. [DOI] [PubMed] [Google Scholar]
- Lonsdorf E, Travis D, Pusey A, & Goodall J (2006). Using retrospective health data from the Gombe chimpanzee study to inform future monitoring efforts. American Journal of Primatology, 908(August 2005), 897–908. doi: 10.1002/ajp [DOI] [PubMed] [Google Scholar]
- Lonsdorf EV, Gillespie TR, Wolf TM, Lipende I, Raphael J, Bakuza J, … Travis DA (2016). Socioecological correlates of clinical signs in two communities of wild chimpanzees (Pan troglodytes) at Gombe National Park, Tanzania. American Journal of Primatology doi: 10.1002/ajp.22562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdorf EV, Gillespie TR, Wolf TM, Lipende I, Raphael J, Bakuza J, … Travis DA (2016). Socioecological correlates of clinical signs in two communities of wild chimpanzees ( Pan troglodytes ) at Gombe National Park, Tanzania. American Journal of Primatology, (August 2015), 1–20. doi: 10.1002/ajp.22562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdorf EV, Murray CM, Lonsdorf EV, Travis DA, Gilby IC, Chosy J, … Pusey AE (2011). A retrospective analysis of factors correlated to chimpanzee (Pan troglodytes schweinfurthii) respiratory health at Gombe National Park, Tanzania. EcoHealth, 8(1), 26–35. doi: 10.1007/s10393-011-0683-0 [DOI] [PubMed] [Google Scholar]
- Markham a. C., Santymire RM, Lonsdorf EV, Heintz MR, Lipende I, & Murray CM (2013). Rank effects on social stress in lactating chimpanzees. Animal Behaviour, 87, 195–202. doi: 10.1016/j.anbehav.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A, Coetzee ML, Keet DF, Maré L, Warren R, Cooper D, … van Helden P (2009). Molecular epidemiology of Mycobacterium bovis isolates from free-ranging wildlife in South African game reserves. Veterinary Microbiology, 133(4), 335–43. doi: 10.1016/j.vetmic.2008.07.023 [DOI] [PubMed] [Google Scholar]
- Murray CM, Eberly LE, & Pusey AE (2006). Foraging strategies as a function of season and rank among wild female chimpanzees (Pantroglodytes). Behavioral Ecology, 17(6), 1020–1028. doi: 10.1093/beheco/arl042 [DOI] [Google Scholar]
- Nituch LA, Bowman J, Beauclerc KB, & Schulte-Hostedde AI (2011). Mink farms predict Aleutian disease exposure in wild American mink. PLoS ONE, 6(7), e21693. doi: 10.1371/journal.pone.0021693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent G (2011). Maintenance, spillover and spillback transmission of bovine tuberculosis in multi-host wildlife complexes: A New Zealand case study. Veterinary Microbiology, 151(1–2), 34–42. doi: 10.1016/j.vetmic.2011.02.023 [DOI] [PubMed] [Google Scholar]
- Nunn CL, Thrall PH, Stewart K, & Harcourt AH (2007). Emerging infectious diseases and animal social systems. Evolutionary Ecology, 22(4), 519–543. doi: 10.1007/s10682-007-9180-x [DOI] [Google Scholar]
- Oleaga A, Casais R, Balseiro A, Espí A, Llaneza L, Hartasánchez A, & Gortázar C (2011). New techniques for an old disease: Sarcoptic mange in the Iberian wolf. Veterinary Parasitology, 181(2–4), 255–66. doi: 10.1016/j.vetpar.2011.04.036 [DOI] [PubMed] [Google Scholar]
- Palacios G, Lowenstine L, Cranfield MR, Gilardi KVK, Spelman L, Lukasik-Braum M, … Ian L (2011). Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerging Infectious Diseases, 17(4), 711–713. doi: 10.3201/eid1704100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L, & Dragicevic S (2009). An agent-based approach for modeling dynamics of contagious disease spread. International Journal of Health Geographics, 8, 50. doi: 10.1186/1476-072X-8-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey AE, Oehlert GW, Williams JM, & Goodall J (2005). Influence of ecological and social factors on body mass of wild chimpanzees. International Journal of Primatology, 26(1), 3–31. doi: 10.1007/s10764-005-0721-2 [DOI] [Google Scholar]
- Pusey AE, Wilson ML, & Anthony Collins D (2008). Human impacts, disease risk, and population dynamics in the chimpanzees of Gombe National Park, Tanzania. American Journal of Primatology, 70(8), 738–744. doi: 10.1002/ajp.20567 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Prieto V, Vicente-Rubiano M, Sánchez-Matamoros A, Rubio-Guerri C, Melero M, Martínez-López B, … Sánchez-Vizcaíno, JM (2014). Systematic review of surveillance systems and methods for early detection of exotic, new and re-emerging diseases in animal populations. Epidemiology and Infection, 1–25. doi: 10.1017/S095026881400212X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke-Parker ME, Munson L, Packer C, Kock R, Cleaveland S, Carpenter M, … Appel MJG (1996). A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature, 379(6564), 441–445. Retrieved from 10.1038/379441a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushmore J, Bisanzio D, & Gillespie TR (2017). Making New Connections: Insights from Primate-Parasite Networks. Trends in Parasitology, xx, 1–14. doi: 10.1016/j.pt.2017.01.013 [DOI] [PubMed] [Google Scholar]
- Rushmore J, Caillaud D, Hall RJ, Stumpf RM, Meyers LA, & Altizer S (2014). Network-based vaccination improves prospects for disease control in wild chimpanzees. Journal of the Royal Society, Interface, 11(97), 20140349. doi: 10.1098/rsif.2014.0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushmore J, Caillaud D, Matamba L, Stumpf RM, Borgatti SP, & Altizer S (2013). Social network analysis of wild chimpanzees provides insights for predicting infectious disease risk. The Journal of Animal Ecology, 82(5), 976–86. doi: 10.1111/1365-2656.12088 [DOI] [PubMed] [Google Scholar]
- Stoto MA, Fricker RD, Jain A, Diamond A, Davies-Cole JO, Glymph C, … Yuan C (2006). Evaluating statistical methods for syndromic surveillance. In Wilson A, Wilson G, & Olwell D (Eds.), Statistical Methods in Counterterrorism: Game Theory, Modeling, Syndromic Surveillance, and Biometric Authentication (pp. 141–172). New York, NY: Sprnger Science + Business Media, LLC. doi: 10.1007/0-387-35209-0_9 [DOI] [Google Scholar]
- Travis DA, Lonsdorf EV, Mlengeya T, & Raphael J (2008). A science-based approach to managing disease risks for ape conservation. American Journal of Primatology, 70(8), 745–50. doi: 10.1002/ajp.20566 [DOI] [PubMed] [Google Scholar]
- Vynnycky E, & White R (2010). An Introduction to Infectious Disease Modelling Oxford, New York: Oxford University Press. [Google Scholar]
- Warns-Petit E, Morignat E, Artois M, Calavas D, & Warns-petit E (2010). Unsupervised clustering of wildlife necropsy data for syndromic surveillance. BMC Veterinary Research, 6, 56. doi: 10.1186/1746-6148-6-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Lonsdorf E, Wilson M, Schumacher-Stankey J, Goodall J, & Pusey A (2008). Causes of death in the Kasekela chimpanzees of Gombe National Park, Tanzania. American Journal of Primatology, 70(8), 766–77. doi: 10.1002/ajp.20573 [DOI] [PubMed] [Google Scholar]
- Wolf TM, Wang WA, Lonsdorf EV, Gillespie T, Pusey A, Gilby I, … Singer R (2018). Data, Model Documentation, and Output Supporting “Optimizing syndromic health surveillance in free ranging great apes: the case of Gombe National Park.” Data Repository for the University of Minnesota,. doi: 10.13020/D64X2Q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, & Diamond J (2007). Origins of major human infectious diseases. Nature, 447(7142), 279–83. doi: 10.1038/nature05775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, & Gowtage-Sequeria S (2005). Host range and emerging and reemerging pathogens. Emerging Infectious Diseases, 11(12), 1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangham R (1977). Feeding behavior of chimpanzees in Gombe National Park Tanzania. In Clutton-Brock T (Ed.), Primate Ecology (pp. 504–538). London, U.K.: Academic Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


