Abstract
Multigene families of CKX genes encode cytokinin oxidase/dehydrogenase proteins (CKX), which regulate cytokinin content in organs of developing plants. It has already been documented that some of them play important roles in plant productivity. The presented research is the first step of comprehensive characterization of the bread wheat TaCKX gene family with the goal to select genes determining yield-related traits. The specificity of expression patterns of fifteen formerly annotated members of the TaCKX family was tested in different organs during wheat development. Based on this, the genes were assigned to four groups: TaCKX10, TaCKX5 and TaCKX4, highly specific to leaves; TaCKX3, TaCKX6 and TaCKX11, expressed in various levels through all the organs tested; TaCKX1, TaCKX2.3, TaCKX2.2, TaCKX2.1, TaCKX2.4 and TaCKX2.5 specific to developing spikes and inflorescences; TaCKX9, TaCKX8 and TaCKX7, highly specific to roots. Amplification products of tested genes were mapped to the chromosomes of the A, B or D genome using T. aestivum Ensembl Plants. Based on analysis of TaCKX transcripts as well as encoded amino acids in T. aestivum and Hordeum vulgare the number of CKX genes in wheat was limited to 11 and new numbering of selected TaCKX genes was proposed. Moreover, we found that there were developmental differences in expression of TaCKX in the first and the second spike and expression of some of the genes was daily time dependent. A very high and significant correlation was found between expression levels of TaCKX7 and TaCKX9, genes specific to seedling roots, TaCKX1, TaCKX2.1 and TaCKX2.2, specific to developing spikes, and the group of TaCKX3, 4, 5, 6, 10 and 11, highly expressed in leaves and other organs. The genes also co-operated among organs and were included in two groups representing younger or maturating stages of developing plants. The first group was represented by seedling roots, leaves from 4-week old plants, inflorescences and 0 DAP spikes; the second by developing spikes, 0 DAP, 7 DAP and 14 DAP. The key genes which might determine yield-related traits are indicated and their possible roles in breeding strategies are discussed.
Introduction
Bread/common wheat is a very important crop in global agriculture because its grains are a worldwide staple food. Its productivity is high, but rising consumption and changing climate indicate the need of further improvement in yield potential [1]. The genome of this allohexaploid species is complex (2n = 6x = 42), composed of three homologous, diploid genomes (AABBDD). It is very large (1.7 x 1010 bp/1C), which makes genetic and molecular research as well as breeding very challenging. Recent development of advanced biotechnology tools indicated the role of specific key genes in wheat and barley for plant productivity (reviewed in Nadolska-Orczyk et al. [2]). Among those listed are CKX genes. CKX genes form multigene families in different plant species. In allohexaploid bread wheat (Triticum aestivum L.) 11 TaCKX genes have been identified so far and numbered from TaCKX1 to TaCKX11 (NCBI databases) and five TaCKX2 (TaCKX2.1 to TaCKX2.5), which have undergone a Triticeae-specific gene-duplication event [3]. TaCKX1 and TaCKX5 have been cloned by Feng et al. [4] and by Lei et al. [5]. Both TaCKX2.1 (FJ648070) and TaCKX2.2 (GUO84177) were isolated and characterized by Zhang et al. [6], and TaCKX2.3, TaCKX2.4 and TaCKX2.5 by Mameaux et al. [3]. Cloning and preliminary characterization of TaCKX3 were done by Ma et al. [7]. The genes encode cytokinin oxidase/dehydrogenase proteins (CKX), which irreversibly degrade cytokinins [8–10], and thereby regulate cytokinin content in developing plants. Their expression is tissue-/organ- and developmentally-specific, suggesting genes’ specific functions [11, 12]. The detailed biological function of most of the TaCKX genes in wheat is not known.
Cytokinins (CKs) are known to be key regulators of seed yield in plants [13]. They act locally or alternatively they function as long distance signaling messengers [14] mediating and modulating sink strength [15–17]. The level of CKs in developing tissues/organs is regulated by balancing biosynthesis and degradation, and the CKX enzymes possibly play the principal role in this regulation [18–20]. The important role of selected CKX genes in cereal productivity has already been documented. In rice, loss-of-function mutation of OsCKX2 caused an elevated cytokinin level, leading to an increase in reproductive organ number and seeds [21]. The same was also evidenced by RNAi silencing of the gene expression. In barley, decrease of expression of HvCKX1 and HvCKX9 by RNAi gene silencing in developing kernels and seedling roots led to an increase of seed and spike number, as well as plant productivity [12, 22, 23]. There are also two known examples of TaCKX variation in wheat leading to higher productivity. In the first one the haplotype variant of TaCKX6-D1 was associated with a higher filling rate and grain size [24, 25]. In the second, the QTL found in recombinant inbred lines (RILs) containing a higher copy number of TaCKX4 was associated with higher chlorophyll content and grain size [26].
According to Zalewski et al. [12], expression patterns of HvCKX genes indicated their role in growth and reproductive development of barley. Thinking about CKX regulation in wheat we started with the same hypothesis, which had been positively verified for barley. The goal of this research was to select those TaCKX genes which might regulate yield-related traits in wheat.
This first-step study on the role of TaCKX genes in plant development and productivity of wheat presents results on the specificity of the genes’ expression in various organs during wheat plant development and their co-operation. We also tested daily time dependency and spike number dependency of expression levels of the tested genes. In our forthcoming papers we will present phenotypic consequences of the observed TaCKX expression patterns for yield-related traits in breeding materials as well as the detailed roles of TaCKX1 and TaCKX2.1/TaCKX2.2 based on RNAi silenced wheat lines.
Materials and methods
Plant material
The experimental tissue samples were collected from three cultivars of common wheat (Triticum aestivum L.): Kontesa, Ostka and Trappe. Ten seeds from each cultivar were germinated into Petri dishes for one day at 4°C and then five days at room temperature in the dark. Six out of ten seedlings from each Petri dish were replanted into pots with soil. The plants were grown in a growth chamber under controlled environmental conditions with 20°C/18°C day/night temperatures and a 16 h light/8 h dark photoperiod. The light intensity was 350 μmol· s-1·m-2. Plants were irrigated three times a week and fertilized once a week with Florovit according to the manufacturer’s instructions.
The following tissue samples in three biological replicates from each cultivar were collected: 5-day-old seedlings roots, well-developed leaves from 4-week-old plants (the longest leaf); 5–6 cm long inflorescences and spikes: 0 days after pollination (0 DAP), first spike 7 DAP (I), second spike 7 DAP (II) and 14 DAP. All these samples were collected at 9:00 am. Additionally, the first 7 DAP spikes were collected at three time points: 9 am, 12 pm and 3 pm. The collected material was frozen in liquid nitrogen and kept at -80°C until use.
RNA extraction and cDNA synthesis
Total RNA from all collected tissues was extracted using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s protocol. The purity and concentration of the isolated RNA were determined using a NanoDrop spectrophotometer (NanoDrop ND-1000) and the integrity was checked by electrophoresis on 1.5% (w/v) agarose gels. To remove the residual DNA the RNA samples were treated with DNase I, RNase-free (Thermo Fisher Scientific). Each time 1 μg of good quality RNA was used for cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. The obtained cDNA was diluted 20 times before use in RT-qPCR assays.
Quantitative RT-qPCR
RT-qPCR assays were performed for 15 target genes. Primer sequences designed for each gene are shown in S1 Table. All real-time reactions were performed in a Rotor-Gene Q (Qiagen) thermal cycler using 1x HOT FIREPol EvaGreen qPCR Mix Plus (no ROX) (Solis BioDyne), 0.2 μM of each primer, and 4 μl of cDNA in a total volume of 10 μl. Each reaction was carried out in 3 biological and 3 technical replicates at the following temperature profile: 95°C– 15 min initial denaturation and polymerase activation (95°C– 25 s, 62°C– 25 s, 72°C– 25 s) x 45 cycles, 72°C– 5 min, with melting curve at 72–99°C 5 s per step. The expression of TaCKX genes was calculated according to the two standard curves method using ADP-ribosylation factor as a normalizer.
Relative expression was related to mean expression of TaCKX3 measured in all organs and set as 1.00. Additionally, all data were counted in relation to the organ correction factor (OCR), which was the quotient of Ct number of the reference gene in the cDNA sample divided by the average Ct number for all samples.
Statistical analysis was performed using Statistica 13 (StatSoft) software. The Kruskal-Wallis test was used to verify the significance of the relative expression differences at the confidence level p<0.05. Correlations coefficients were determined using correlation matrices (Pearson test).
Sequence data and phylogenetic analysis
Homologous sequences were retrieved from Ensembl Plants database [27]. The alignments of TaCKX and HvCKX amino acid sequences was conducted using the MAFFT version 7 program [28, 29]. The maximum likelihood phylogenetic tree was done on alignments of full-length protein sequences using MEGA X performed with the JTT model and 1000 bootstrap replicates [30].
Results
The family of TaCKX genes in wheat
According to the NCBI database and references there are 15 TaCKX genes; 11 of them are numbered from TaCKX1 to TaCKX11 (Table 1). In the case of TaCKX2 five duplicates (TaCKX2.1 to TaCKX2.5) are identified [3, 6]. Specific primers for expression analysis were designed for all of them, including TaCKX2 duplicates. Based on T. aestivum IWGSC assembly (Ensembl Plants [27]), the three groups of transcripts: TaCKX2.1, TaCKX2.2, TaCKX2.4 and TaCKX6D1; TaCKX2.3, TaCKX2.5 and TaCKX6a02 as well as TaCKX7 and TaCKX8 were located to the same homeoparalogues. Suggested names are: TaCKX2.2 for the first group, TaCKX2.1 for the second and TaCKX7 for the third as indicated in the Table 1. The amplified regions of the genes were located to the following chromosomes: TaCKX1 to chromosomes 3A, 3B, 3D; TaCKX2.2, TaCKX2.1 and TaCKX4 to chromosomes 3B, 3D; TaCKX5 to chromosome 3B. The TaCKX6 amplification product is located to chromosomes 1A, 1B, 1D and these locations are different from the TaCKX6-D1b and a alleles, which is 3D [24] and from the TaCKX6a02 allele located to the 3DS [25]. There is no pairwise homology among them. Investigated by us TaCKX7 is located to chromosomes 6B and 6D, TaCKX9 to chromosomes 7A, 7B and 7D, and TaCKX11 to chromosome 2A. According to NCBI T. aestivum genome assembly the primers for TaCKX3 gene amplification hybridized with chromosome 7B and for TaCKX10 with 1A, 1B and 1C (not showed).
Table 1. Comparison of analysed wheat TaCKX gene family members published in NCBI with ensemble plants (IWGSC assembly) databases.
Primers for amplification were designed based on accession numbers of the genes in NCBI (in bold).
| Gene (suggested annotation in bold) | Former annotation | Accesion numer NCBI or Ensembl Plants |
Chrom. location | No. of exons/ coding exons | Protein (aa) | Amplicon length (bp) | Refer. (query cover/ ident. %) |
|---|---|---|---|---|---|---|---|
| TaCKX1 | JN128583 | - | - | 501 | 188 | [31] | |
| TraesCS3A02G109500 | 3A | 3 | 524 | 188 | (100/98) | ||
| TraesCS3B02G128700 | 3B | 3 | 524 | 188 | (100/98) | ||
| TraesCS3D02G111300 | 3D | 3 | 524 | 188 | (100/98) | ||
| TaCKX2.11 | TaCKX2.31 | JF293079 | - | - | 553 | 144 | [3] |
| TraesCS3A02G311000 | 3A | 3 | 567 | na | (87/91) | ||
| TraesCS3B02G161100 | 3B | 3 | 578 | 144 | (94/95) | ||
| TraesCS3D02G143600 | 3D | 3 | 551 | 144 | (97/100) | ||
| TaCKX2.51 | JN381556 | - | 3 | 545 | 147 | [3] | |
| TraesCS3A02G311000 | 3A | 3 | 567 | na | (41/91) | ||
| TraesCS3B02G161100 | 3B | 3 | 578 | na | (41/100) | ||
| TraesCS3D02G143600 | 3D | 3 | 551 | na | (41/97) | ||
| TaCKX6a021 | NI | 3D | - | - | na | [25] | |
| TraesCS3D02G143600 | 3D | 3 | 551 | na | (100/99) | ||
| TaCKX2.22 | TaCKX2.12 | FJ648070 | 3D* | - | 547 | 205 | [6] |
| TraesCS3A02G311100 | 3A | 3 | 552 | na | (91/92) | ||
| TraesCS3B02G161000 | 3B | 3 | 547 | 205 | (99/99) | ||
| TraesCS3D02G143300 | 3D | 3 | 547 | 221 | (96/95) | ||
| TraesCS3D02G143500 | 3D | 3 | 547 | 224 | (100/94) | ||
| TaCKX2.22 | GU084177 | 3D* | - | 547 | 175 | [6] | |
| TraesCS3A02G311100 | 3A | 3 | 552 | na | (96/92) | ||
| TraesCS3B02G161000 | 3B | 3 | 547 | 175 | (97/95) | ||
| TraesCS3D02G143300 | 3D | 3 | 547 | 175 | (100/99) | ||
| TraesCS3D02G143500 | 3D | 3 | 547 | 175 | (97/95) | ||
| TaCKX2.42 | JN381555 | - | 3 | 552 | 220 | [3] | |
| TraesCS3A02G311100 | 3A | 3 | 552 | na | (53/100) | ||
| TraesCS3B02G161000 | 3B | 3 | 547 | na | (53/94) | ||
| TraesCS3D02G143300 | 3D | 3 | 547 | na | (53/94) | ||
| TraesCS3D02G143500 | 3D | 3 | 547 | na | (53/94) | ||
| TaCKX6D12 | JQ797673 | 3D* | 3 | 545 | 155 | [24] | |
| TraesCS3A02G311100 | 3A | 3 | 552 | na | (44/94) | ||
| TraesCS3B02G161000 | 3B | 3 | 547 | na | (44/97) | ||
| TraesCS3D02G143300 | 3D | 3 | 547 | na | (44/99) | ||
| TraesCS3D02G143500 | 3D | 3 | 547 | na | (44/99) | ||
| TaCKX3 | JN128585 | - | - | - | 150 | [31] | |
| TraesCS7A02G536900 | 7A | 3 | 516 | na | (76/98) | ||
| TraesCS7B02G455000 | 7B | 4 | 516 | na | (98/99) | ||
| TaCKX4 | JN128586 | - | - | - | 112 | [31] | |
| TraesCS3B02G525300 | 3B | 5 | 525 | 111 | (99/99) | ||
| TraesCS3D02G475800 | 3D | 5 | 525 | 112 | (100/94) | ||
| TaCKX5 | NI | 3B | - | - | 150 | [5] | |
| TraesCS3A02G321100 | 3A | 5 | 530 | 144 | (95/95) | ||
| TraesCS3B02G344600 | 3B | 5 | 531 | 150 | (98/100) | ||
| TraesCS3D02G310200 | 3D | 5 | 531 | na | (90/96) | ||
| TaCKX6 | JN128587 | - | - | - | 182 | [31] | |
| TraesCS1A02G159600 | 1A | 5 | 522 | 182 | (99/97) | ||
| TraesCS1B02G176000 | 1B | 6/5 | 522 | 182 | (94/98) | ||
| TraesCS1D02G157000 | 1D | 5 | 522 | 182 | (100/99) | ||
| TaCKX73 | TaCKX73 | JN128588 | - | 1 | 534 | 144 | [31] |
| TraesCS6A02G185800 | 6A | 1 | 533 | na | (100/96) | ||
| TraesCS6B02G214700 | 6B | 1 | 533 | na | (100/96) | ||
| TraesCS6D02G172900 | 6D | 2 | 467 | na | (87/97) | ||
| TaCKX83 | JN128589 | - | 1 | 534 | 198 | [31] | |
| TraesCS6A02G185800 | 6A | 1 | 533 | na | (100/96) | ||
| TraesCS6B02G214700 | 6B | 1 | 533 | 198 | (100/99) | ||
| TraesCS6D02G172900 | 6D | 2 | 467 | 198 | (87/96) | ||
| TaCKX9 | JN128590 | - | 1 | 262 | 278 | [31] | |
| TraesCS7A02G363400 | 7A | 2 | 551 | 278 | (100/95) | ||
| TraesCS7B02G264400 | 7B | 2 | 540 | 278 | (100/99) | ||
| TraesCS7D02G359700 | 7D | 2 | 532 | 278 | (98/96) | ||
| TaCKX10 | JN128591 | - | - | - | 167 | [31] | |
| TraesCS1A02G234800 | 1A | 6/5 | 521 | na | (96/99) | ||
| TraesCS1B02G248700 | 1B | 5 | 521 | na | (96/99) | ||
| TraesCS1D02G237200 | 1D | 5 | 521 | na | (96/99) | ||
| TaCKX11 | JN128592 | - | - | 245 | 184 | [31] | |
| TraesCS2A02G378300 | 2A | 5 | 528 | 184 | (99/97) |
*—primary mapped
1,2,3 –located in the same gene respectively; NI—no identified;—no information; na–not amplified
The distance tree of pairwise comparison (Figs A-N in S1 File) revealed that homologues of TaCKX1 were located on chromosomes 3A, 3B, 3D (T. aestivum genome assembly) with identity of 98% of whole sequences (100% cover). The gene is highly homologous to Aegilops tauschii cytokinin dehydrogenase 1-like (99% cover/98% identity), Secale cereale ScCKX1 (45% cover/97% identity), Hordeum vulgare CKX1 (100% cover/94% identity) and Zea mays CKX1 (57% cover/84% identity). The TaCKX2.2 isolated by Zhang et al. [6] is proved to be located on 3D and their homologue on 3B has 95% identity and on 3A 92% identity. The gene is highly homologous to A. tauschii cytokinin dehydrogenase 2-like (95% cover/96% identity), H. vulgare CKX2.2 (93% cover/92% identity), Z. mays CKX5 (83% cover/77% identity), and Oryza sativa (85% cover/79% identity). Moreover, TaCKX2.2 is 91% identical (under 38% cover) to S. cereale ScCKX2.2. The third TaCKX2.1, for which primers were designed based on JF293079 [3] has highest cover/identity with 3D (97/100%) and lower with 3B (94/95%) and 3A (87/91%) T. aestivum Esembl Plants and is highly homologous to A. tauschii cytokinin dehydrogenase 2-like (96% cover/99% identity).
The primers for TaCKX3 were designed based on the JN128585 sequence isolated by Song et al. [31] hybridized with NCBI LS992099 from T. aestivum NCBI genome assembly (not showed) and showed 99% identity with TaCKX3 (GQ925404) isolated by Ma et al. [7]. The whole sequence located on chromosome 7B has 99% identity to the sequence from T. aestivum IWGSC assembly chromosome 7B and 98% identity with 76% cover with 7A. The closest orthologues among other cereal species are: A. tauschii cytokinin dehydrogenase 11 (96% cover/95% identity), S. cereale ScCKX11 (72% cover/95% identity), H. vulgare (64% cover/93% identity), Z. mays CKX10 (74% cover/86% identity), and O. sativa (75% cover/87% identity).
The primers for TaCKX4, isolated by Song et al. [31] and deposited in NCBI as an unverified accession, hybridized with the closest homologue found on chromosome 3B T. aestivum IWGSC assembly (99% cover/99% identity) and 3D (100% cover/94% identity). Close orthologues of the gene were A. tauschii CKX4-like showing 94% identity under 97% cover, H. vulgare CKX4 with 89% identity under 98% cover and Lolium perenne with 85% identity and under 82% cover.
Homologous copies of TaCKX5 isolated by Lei et al. [5] have been localised on chromosomes 3A, 3B and 3D, however primers hybridized with 3B and the only amplification product was 150 bp long. Closest orthologues were A. tauschii with 96% identity and 90% cover, S. cereale with 96% identity and 73% cover, O. sativa CKX5-like, Sorghum bicolor CKX5, Z. mays CKX4b, Panicum hallii CKX5 and Setaria italica CKX5, all with identity 77–78% and 78–80% cover.
TaCKX6 analysed in this paper was isolated by Song et al. [31] and based on T. aestivum IWGSC assembly mapped to chromosomes 1A, 1B and 1D. In Table 1 two additional, described in the literature, TaCKX6 genes were included: TaCKX6-D1b [24] and TaCKX6a02 [25]. However transcript of the first one was located to the TaCKX2.2 and the second to the TaCKX2.1 of chromosome 3D. The closest homologue of TaCKX6 was T. aestivum CKX8 sharing 98% identity under 89% cover of the sequences. Very close orthologues were: H. vulgare CKX3 with 94% identity, L. perenne CKX6 with 96% identity and Z. mays CKX6 sharing 81% identity under 83% cover. The closest homologues of TaCKX-D1b and a alleles was TaCKX2.2 sharing 98% identity under 43% cover. Their close orthologues were A. tauschii CKX2-like with 99% identity and 43% cover and H. vulgare CKX2.2 with 88% identity under 69% cover. The closest homologue of TaCKX6a02 was T. aestivum CKX2.1 with 99% identity under 100% cover.
The sequences of TaCKX7 and TaCKX8 isolated by Song et al. [31] were located to the same TaCKX7 gene and their primers hybridized with 6B and 6D, and shared 100% cover with chromosomes 6A and 6B, however with lower, 96% identity. Very close orthologues were: A. tauschii CKX6-like with 96% similarity under 100% cover, H. vulgare CKX7 with 99% cover and 90% identity, and Z. mays CKX7, S. bicolor CKX7, S. italica CKX7, P. halli CKX7, all with 77–78% identity under 93–95% cover.
Other TaCKX genes isolated by Song et al. [31] and analysed in this paper are TaCKX9, TaCKX10 and TaCKX11. Based on T. aestivum IWGSC assembly primers for the first one hybridized to chromosomes 7A, 7B and 7D. The second one was in silico amplified on chromosomes 1A, 1B and 1D, according to NCBI T. aestivum genome assemble (not showed) but not to IWGSC assembly. Close orthologues to TaCKX9 were: H. vulgare CKX10 showing 95% identity under 71% cover, A. tauschii cytokinin dehydrogenase 10-like (99% cover/97% identity), and Z. mays CKX10, S. italic CKX10 and P. halli CKX10 with 83–84% identity and under 95–96% cover. The TaCKX10 sequence had 100% cover and 99% identity to chromosomes 1A, 1B and 1D according to NCBI T. aestivum genome assemble but 99% cover and 96% identity according to IWGSC assembly. Close homologues to TaCKX10 were: A. tauschii cytokinin dehydrogenase 9 (96% cover/99% identity), H. vulgare CKX3 (96% cover/99% identity), H. vulgare CKX2 (98% cover/96% identity), which was further annotated as HvCKX9 [3], Brachypodium distachyon CKX9 (96% cover/90% identity), O. brachyantha CKX9 (96% cover/85% identity), O. sativa chromosome 5 (97% cover/86% identity), O. sativa CKX9 (96% cover/86% identity), Sorghum bicolor CKX9 (95% cover/83% identity), Setaria italica CKX9 (95% cover/84% identity).
The TaCKX11 sequence had 99% cover and 97% identity with the corresponding region of chromosome 2A. Very close orthologues of the gene were: A. tauschii CKX8-like with 95% identity, under 99% cover and H. vulgare CKX8 (81% cover/95% identity), B. distachyon CKX8-like (97% cover/86% identity), O. sativa CKX8 (100% cover/83% identity), P. hallii CKX8-like (95% cover/82% identity), S. italica CKX8 (97% cover/83% identity).
Phylogenetic comparison of proteins encoded by TaCKX and HvCKX
The proteins encoded by TaCKX2.3, TaCKX2.5 and TaCKX6a02 grouped together in close association with the protein encoded by HvCKX2.1 (S2 Table and S1 Fig). Hence the genes were assigned as TaCKX2.1. Another group composed of TaCKX2.1, TaCKX2.2, TaCKX2.4 and TaCKX6D1 was associated with HvCKX2.2. Based on this four wheat genes coding these proteins were assigned as TaCKX2.2. The proteins encoded by TaCKX7 and TaCKX8 grouped together in association with the protein encoded by HvCKX7. Hence the genes were assigned as TaCKX7.
The TaCKX genes are more or less specifically expressed in different tissues during plant development
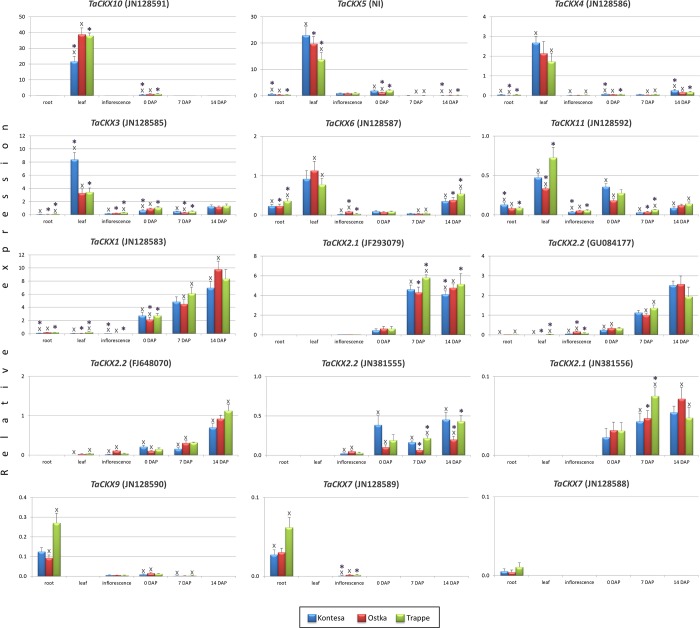
Specificity of TaCKX genes’ expression was tested in 5-day-old seedling roots, well-developed leaves from 4-week old plants, 5–6 cm long inflorescences, and 0 DAP, 7 DAP, 14 DAP spikes. The expression data are shown as relative values related to the mean expression of TaCKX3 in all organs tested designated as 1.0. The most highly and specifically expressed gene was TaCKX10 (Fig 1). The relative expression level of the gene ranging from 21.54 to 38.75 in well-developed leaves of 4-week-old plants depended on the cultivar tested. The differences in expression between cv. Kontesa and cvs. Ostka and Trappe were significant. The gene was also expressed in 0 DAP spikes with about a 40 times lower level compared to leaves (0.63–1.04).
Fig 1. Specificity of TaCKX family gene expression in developing tissues/organs.
The genes are annotated as suggested in Table 1 with the corresponding NCBI accession number in brackets. The relative expression of the genes was measured in: seedling root, well-developed leaf from 4-week-old plant, inflorescence, 0 DAP, 7 DAP and 14 DAP spikes in three cultivars, Kontesa, Ostka, Trappe. The level of expression was related to the mean expression of TaCKX3 in all organs tested set as 1.00. The order of the genes is from the highest to the lowest expression, taking into consideration the specificity of expression. *–significant differences at P<0.05.
The next gene in order of expression level was TaCKX5, which showed the highest expression in well-developed leaves (13.78–22.90) and then in 0 DAP spikes (1.19–1.90), inflorescence (0.88–0.96) and seedling roots (0.33–0.60) depending on the cultivar. There were significant differences in expression of the gene among cultivars in seedling root, leaf and 0 DAP spike.
TaCKX1 was highly and specifically expressed in developing spikes with the highest expression level in 14 DAP spikes ranging from 6.96 in cv. Kontesa to 9.79 in cv. Ostka through 4.49 to 6.11 in 7 DAP spikes and from 2.11 in cv. Ostka to 2.73 in cvs. Kontesa and Trappe in 0 DAP spikes. Very low expression, below 0.2, was found in seedling roots and leaves. There were significant differences in expression of TaCKX1 in developing spikes as well in seedling roots among the cultivars.
Interestingly, TaCKX3 was expressed in all organs tested. The highest expression level of the gene was in leaves, ranging from 3.28 to 3.43 in cvs. Ostka and Trappe and to more than twice as high (8.39) in cv. Kontesa. Similarly, the relative expression level was above 1.0 in 14 DAP spikes and there were no significant differences among cultivars, and in 0 DAP spikes of cv. Trappe. There was a lower expression level in 7 DAP spikes (0.31–0.49), inflorescence (0.19–0.31) and the lowest in the seedling roots (0.05–0.12). Significant differences in expression levels among cvs. Kontesa, Ostka and Trappe were found in all organs tested excluding 14 DAP.
TaCKX2.1 (JF293079) was highly and specifically expressed in developing spikes with the highest expression level in 7 DAP and 14 DAP spikes ranging from 4.11 in 14 DAP cv. Kontesa to 5.80 in 7 DAP cv. Trappe. There were significant differences in level of expression among the cultivars. Low expression (0.43–0.61) was found in 0 DAP spikes.
TaCKX4, TaCKX2.2(FJ648070), TaCKX2.2(GU084177), TaCKX2.2(JN381555), TaCKX2.1(JN381556), TaCKX6, TaCKX11, TaCKX9, TaCKX7(JN128589) and TaCKX7(JN128588) were low-expressed genes. The highest expression of the first one was in leaves (1.73–2.68). The gene was also expressed in other tested organs although at a very low level, from 0.01 in roots of cv. Ostka to 0.26 in 14 DAP spikes of cv. Kontesa. There were significant differences among the cultivars for almost all genes tested. TaCKX2.2(FJ648070), TaCKX2.2(GU084177), TaCKX2.2(JN381555) and TaCKX2.1(JN381556) were specifically expressed in generative organs: inflorescences and developing spikes, with much higher expression values in the second one. The relative expression of TaCKX2.2(FJ648070) in inflorescence ranged from 0.02 in cv. Kontesa to 0.11 in cv. Ostka, while for TaCKX2.2(GU084177) it was from 0.05 in cv. Kontesa to 0.16 in cv. Ostka. The expression patterns of TaCKX2.2(FJ648070) varied in 0 DAP spikes, showing the highest level in cv. Kontesa (0.21) and the lowest in cv. Ostka (0.10). Relative expression of the gene in 7 DAP spikes was about 3 times higher in cvs. Ostka and Trappe, but it was lower in cv. Kontesa. The highest expression of TaCKX2.2(FJ648070) observed in 14 DAP spikes ranged from 0.70 in cv. Kontesa to 1.12 in cv. Trappe. There were significant differences in the level of expression between cv. Kontesa and the two other cultivars.
The expression of TaCKX2.2(GU084177) in 0 DAP spikes was from 0.24 in cv. Kontesa up to 0.34 in cv. Ostka. In 7 DAP spikes the expression was about 4 times higher and in 14 DAP spikes about 8 times higher compared to 0 DAP spikes. There were significant differences in expression in all organs excluding 14 DAP among cultivars tested. The expression of TaCKX2.2(JN381555) and TaCKX2.1(JN381556) was very low, below 0.5 in developing spikes for the first one and below 0.1 for the second.
TaCKX6 was expressed in all organs tested, showing a higher expression level in leaves (0.77–1.13) and about twice as low in 14 DAP spikes (0. 35–0.54). Interestingly, expression level of the gene was relatively high in the roots, ranging from 0.23 in cvs. Kontesa and Ostka to 0.36 in cv. Trappe. There were significant differences of the gene expression among cultivars in all organs tested excluding 0 DAP.
Similar to TaCKX6, TaCKX11 was expressed in all organs tested, with expression levels ranging from 0.04 to 0.07 in inflorescences and 7 DAP spikes, about 0.1 in roots and 14 DAP spikes, 0.18 to 0.36 in 0 DAP spikes, up to 0.33–0.73 in leaves. There were significant differences of expression levels among cultivars in all organs.
TaCKX9, TaCKX7(JN128589) and TaCKX7(JN128588) were specifically expressed in roots showing low, from 0.09 in cv. Ostka to 0.27 in cv. Trappe and significant differences among cultivars for the first one; very low, from 0.03 to 0.06 for the second and minimal expression, below 0.012 for the third gene.
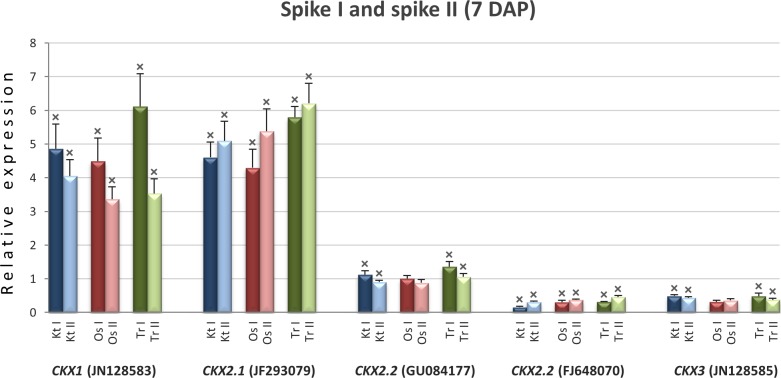
There were differences between TaCKX expression levels in the first and the second spike
The differences in expression levels of TaCKX were measured in the first (I) and the second (II) 7 DAP spikes of plants from the three cultivars tested (Fig 2). There were significant differences in expression levels of I and II spikes in the case of TaCKX1, TaCKX2.1(JF293079) and TaCKX2.2(FJ648070) in the three cultivars tested and TaCKX2.2(GU084177) and TaCKX3 in cvs. Kontesa and Trappe. Low expressing genes (TaCKX4, TaCKX5, TaCKX6, not showed) differ in expression in both spikes depending on the cultivar. In most cases expression levels of the genes in the I spikes were higher than in the II spikes. Opposite results were observed for TaCKX2.1(JF293079) and TaCKX2.2(FJ648070).
Fig 2.
TaCKX gene expression in the first (I) and the second (II) developed 7 DAP spike in three cultivars, Kontesa, Ostka, Trappe. The level of expression was related to the mean expression of TaCKX3 set as 1.00. *–significant differences at P<0.05.
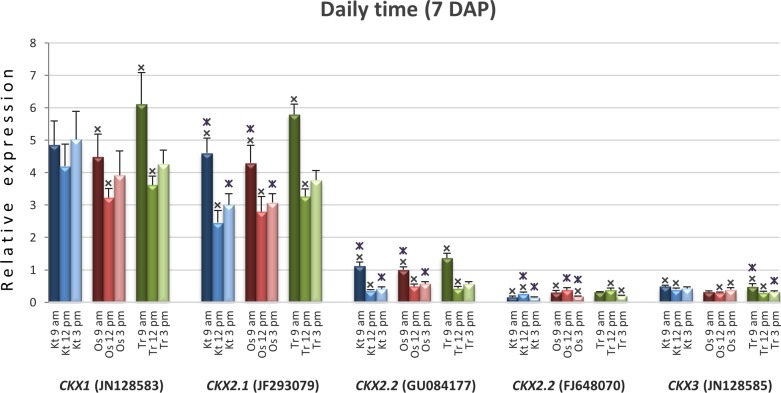
TaCKX level of expression was daily time dependent
Daily time dependence of expression level was measured in 7 DAP spikes I collected at 9:00 am, 12:00 pm and 3:00 pm in the three tested cultivars (Fig 3). There were significant differences among daily time expression for all tested TaCKX genes and in all three or two cultivars tested. No significant differences were observed for TaCKX1 expression in cv. Kontesa and TaCKX4 expression (not showed) in cv. Ostka.
Fig 3. Daily time expression of TaCKX family genes in 7 DAP spikes for relatively high expressing genes.
The first spikes were collected at 9:00 am, 12:00 pm and 3:00 pm from three cultivars, Kontesa, Ostka, Trappe. The level of expression was related to the mean expression of TaCKX3 set as 1.00. *–significant differences at P<0.05.
Co-operation of TaCKX genes among themselves and among organs
Correlation coefficients among TaCKX gene expression through investigated organs are presented in Table 2. The highest correlations were found between TaCKX9, TaCKX7(JN128589) and TaCKX7(JN128588) (0.99–1.0) and TaCKX4 and TaCKX5 (0.99). Very high coefficients were found in the group of TaCKX3, TaCKX4, TaCKX5 and TaCKX6 (0.79–0.93) and in the group of TaCKX1, TaCKX2.2(FJ648070) and TaCKX2.2(GU084177) (0.87–0.90) and between TaCKX2.1(FJ648070) and TaCKX 2.1(JN381556) (0.93). Slightly lower correlations from 0.55 to 0.83 were among TaCKX1and all clones of TaCKX2.1 and TaCKX2.2. Similar level of correlation coefficients ranging from 0.71 to 0.91 were observed among TaCKX11, TaCKX10 and TaCKX3, 4, 5, 6. Moreover, TaCKX6 correlated with TaCKX3, TaCKX4, TaCKX5 at the level of 0.79, 0.89 and 0.87 respectively. There were also significant negative correlation coefficients between TaCKX1 and TaCKX5 at the level of -0.43 and TaCKX1 and TaCKX10 at -0.39. Besides, TaCKX2.1(FJ648070) and TaCKX2.1(JN381556) negatively correlated with TaCKX5, 10 and 11 at the level of -0.38 to -0.49.
Table 2. Correlations coefficients among TaCKX genes through investigated organs in three cultivars (N = 27).
NCBI clones are in brackets.
|
CKX2.2 (FJ648070) |
CKX2.2 (GU084177) |
CKX2.1 (JF293079) |
CKX2.2 (JN381555) |
CKX2.1 (JN381556) |
CKX3 | CKX4 | CKX5 | CKX6 |
CKX7 (JN128588) |
CKX7 (JN128589) |
CKX9 | CKX10 | CKX11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CKX1 | 0.87** | 0.90** | 0.82** | 0.70** | 0.83** | no | -ns | -0.43* | no | -ns | -ns | -ns | -0.39* | -ns |
| CKX2.2 (FJ648070) | 0.88** | 0.70** | 0.72** | 0.71** | no | no | -ns | no | -ns | -ns | -ns | -ns | no | |
| CKX2.2 (GU084177) | 0.77** | 0.71** | 0.81** | no | no | -ns | no | -ns | -ns | -ns | -ns | -ns | ||
| CKX2.1 (JF293079) | 0.55** | 0.93** | -ns | -ns | -0.43* | -ns | -ns | -ns | -ns | -0.38* | -0.49* | |||
| CKX2.2 (JN381555) | 0.64** | no | -ns | -ns | no | -ns | -ns | -ns | -ns | no | ||||
| CKX2.1 (JN381556) | -ns | -ns | -0.43* | -ns | -ns | -0.39* | -ns | -0.40* | -0.39* | |||||
| CKX3 | 0.93** | 0.91** | 0.79** | no | no | no | 0.71** | 0.72** | ||||||
| CKX4 | 0.99** | 0.89** | no | no | no | 0.91** | 0.77** | |||||||
| CKX5 | 0.87** | no | no | no | 0.91** | 0.78** | ||||||||
| CKX6 | no | no | no | 0.85** | 0.71** | |||||||||
| CKX7 (JN128588) | 0.99** | 1.00** | no | no | ||||||||||
| CKX7 (JN128589) | 0.99** | no | no | |||||||||||
| CKX9 | no | no | ||||||||||||
| CKX10 | 0.81** |
no—lack of correlation; ns—low and not significant
*- significant at p<0.05
**- significant at p<0.01
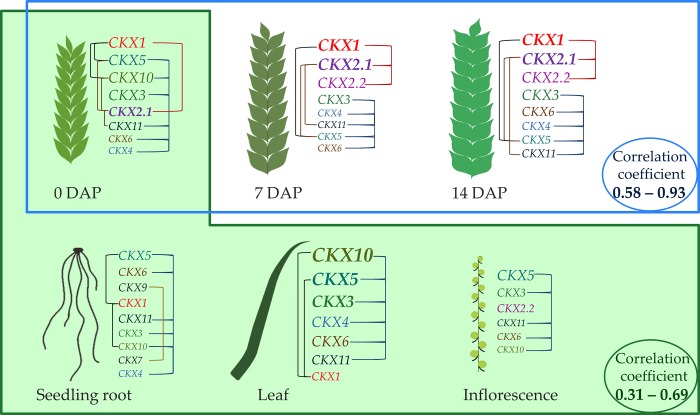
Correlation coefficients of TaCKX gene family expression among organs are presented in Table 3 and a graphic presentation of co-operation of TaCKX genes in the investigated organs is shown in Fig 4. There were two groups of developing tissues/organs showing high correlation of TaCKX gene expression. The first one groups seedling roots, leaves from 4-week-old plants, inflorescences and 0 DAP spikes in which correlations of gene expression ranged from 0.31 to 0. 69 for the three cultivars tested. The second groups developing spikes, 0 DAP, 7 DAP and 14 DAP. In this group the lowest correlation coefficient for TaCKX gene expression was between 0 DAP and 7 DAP spikes (0.58), higher between 0 DAP and 14 DAP (0.66), and the highest between 7 DAP and 14 DAP (0.84).
Table 3. Correlations coefficients of TaCKX gene family among organs in three cultivars (N = 45).
| Leaf | Inflorescence | 0 DAP | 7 DAP | 14 DAP | |
|---|---|---|---|---|---|
| Seedling root | 0.31* | 0.69** | 0.45** | no | no |
| Leaf | 0.43** | 0.35* | no | no | |
| Inflorescence | 0.44** | no | no | ||
| 0 DAP | 0.58** | 0.66** | |||
| 7 DAP | 0.93** |
no—lack of correlation
*- significant at p<0.05
**- significant at p<0.01
Fig 4. Graphic presentation of co-operation of TaCKX genes in the investigated organs in three cultivars tested.
The genes with the highest expression in tested tissue/organ are at the top of the lists. Correlations of TaCKX gene expression within an organ are marked by lines and among organs by green and white boxes and blue line.
Discussion
New nomenclature of selected TaCKX
The nucleotide sequences of NCBI accessions: FJ648070, GU084177, JN381555 and JQ797673 previously annotated as TaCKX2.1, TaCKX2.2, TaCKX2.4 and TaCKX6D1 respectively, were located to the same homeoparalogues in wheat A, B and D subgenomes: TraesCS3A02G311100, TraesCS3B02G161000 TraesCS3D02G143300 and TraesCS3D02G143500. Proteins encoded by the genes were phylogenetically closely associated with the protein encoded by HvCKX2.2 therefore our suggested name for these homeoparalogues is TaCKX2.2. Similarly, accessions JF293079 and JN381556 previously annotated as TaCKX2.3 and TaCKX2.5 were located, along with reported by [25] TaCKX6a02, to the same homeoparalogues TraesCS3A02G311000, TraesCS3B02G161100 and TraesCS3D02G143600. Proteins encoded by the genes were phylogenetically closely associated with the HvCKX2.1 encoded protein. Hence, the wheat homeoparalogues were assigned as TaCKX2.1. The accessions JN128588 and JN128589, previously assigned as TaCKX7 and TaCKX8, were highly similar to homeoparalogues TraesCS6A02G185800, TraesCS6B02G214700 and TraesCS6D02G172900. The genes-encoded proteins were phylogenetically close to HvCKX7, therefore our suggested name for the gene is TaCKX7. The similarities of proteins encoded by barley and wheat CKX genes as well as discussed below expression patterns and expression correlations of wheat CKX genes supported suggested by us numbering of CKX genes in wheat. The number of 11 CKX genes in wheat and barley are coherent for the both species.
Specificity of expression of TaCKX family genes and phylogenic analysis
Considering the specificity of expression, the TaCKX genes can be assigned to four groups: TaCKX10, TaCKX5 and TaCKX4 were highly specific to leaves, TaCKX1, TaCKX2.1 and TaCKX2.2 to developing spikes, TaCKX9 and TaCKX7 to roots, and TaCKX3, TaCKX6, TaCKX11 are more or less expressed through the all organs tested.
The first two genes from the group specific to leaves, TaCKX10 and TaCKX5 have the highest expression level among tested organs of developing wheat plants. Based on T. aestivum genome assembly data the amplification product of the TaCKX10 gene isolated by Song et al. [31] was proved to be located to chromosomes 1A, 1B, 1D and was one of few TaCKX, which was most likely expressed from three homologous genomes. Function of the gene is not characterized. A very close homolog of TaCKX10 found in databases is H. vulgare CKX9, formerly annotated as HvCKX2, and this gene has been previously characterized [12, 23, 32]. In the last report anti-HvCKX9 antibodies predominantly detected proteins in the leaf vasculature; however, according to Zalewski et al. [12] expression of HvCKX9 was highest in 14 DAP kernels, 30-fold higher than in other tissues of two barley cultivars. Overexpression of the gene under a constitutive promoter caused very slow growth and plants died without flowering [32]. In contrast, the phenotypic result of HvCKX9 silencing by stable, Agrobacterium-mediated RNAi was a higher number of seeds and higher grain yield [23]. Very close homology of TaCKX10 with HvCKX9 (96% identity under 98% cover) and the proteins encoded by these genes might suggest similar functions; however, expression patterns of these orthologues, although measured in the same organs during wheat and barley plant development, were completely different. The same function of TaCKX10 is expected to be different from the known function of HvCKX9.
The starters for TaCKX5 were found to hybridized to 3B and might be expressed only from this genome. The closest homologues of the investigated TaCKX5 were CKX5 orthologues belonging to several species: S. cereale, S. brachyantha, S. bicolor, S. italica, B. distachyon as well as CKX5-like O. sativa. None of them has been analysed. Nonetheless, TaCKX5 seemed to be a very important element of the family, also showing the highest expression among the others in seedling roots and inflorescences and very high, just after TaCKX1, in 0 DAP spikes.
The primers for the third gene specific to leaves, TaCKX4 [31], based on T. aestivum IWGSC assembly, hybridized to chromosome 3B and 3D. The gene was not homologous to TaCKX4-1 to 4–3 copies characterized by Chang et al. [26], which were located to chromosome 3A. However, TaCKX4 investigated by us was orthologous to A. tauschii CKX4-like with 94% identity, H. vulgare CKX4 with 89% identity and L. perenne CKX4 with 85% identity, suggesting that the number of the gene is correct. According to Song et al. [31], expression of TaCKX4 was very low during reproductive development, which was in agreement with our data. The authors also found a low mRNA level in flag leaves, which differs from our results, probably because the developmental stage of well-developed leaves from young plants used by us was not comparable with the developmental stage and role of flag leaves. The only research on the role of TaCKX4-1 to 4–3 was reported by Chang et al. [26], who merged copy number variation with grain weight and chlorophyll content in the RIL population. Unexpectedly, the two-copy locus of TaCKX4 corresponded to higher chlorophyll content and grain weight compared to null or three-copy genotypes. These data are difficult to interpret, since we know nothing about expression of these copies of the TaCKX4 gene. Higher copy number should correlate with higher expression and should increase CKX enzyme activity and irreversible cytokinin degradation. Since an increased level of CKs delays senescence and causes nutrient mobilization [33], which positively affects grain yield, one should expect that higher copy number should be negatively associated with chlorophyll content compared to the null phenotype. Anyway, TaCKX4 investigated in this study was not comparable with three-copied TaCKX4, since the genes were not homologous and were located on different genomes.
The second group of TaCKX genes which were highly and specifically expressed in developing spikes were represented by TaCKX1,TaCKX2.1and TaCKX2.2. Amplicon hybridization with T. aestivum IWGSC assembly data showed that the TaCKX1 was expressed from chromosomes 3A, 3B, 3D and both TaCKX2.1 and TaCKX2.2 from chromosomes 3B and 3D. Similar to our data, TaCKX1 was the most highly expressed gene in developing spikes among the six tested by Song et al. [31] and the only one with high expression in mature and senescing flag leaves. Expression measured by semi-quantitative RT-PCR proved that the gene was not expressed in root, sheath, and leaf [4]. Phylogenetic analysis showed that TaCKX1 shares high sequence similarity with other CKX1-type orthologues from A. tauschii CKX1-like, S. cereale ScCKX1, H. vulgare CKX1, and lower with Z. mays CKX1. Analysis of the transcript level of HvCKX1 in different organs of developing barley plants showed a similar pattern of expression to wheat [12]. The level of transcript was the highest in 14 DAP spikes and was lower in 7 DAP and 0 DAP spikes. Stable RNAi silencing of HvCKX1 expression was associated with decreased CKX enzyme activity in developing spikes leading to a higher number of seeds and higher plant productivity [22]. Taking into account the high similarity of CKX1 orthologues, especially HvCKX1, which additionally showed close similarity to TaCKX1 in wheat expression pattern in developing plants of barley, we might expect an important role of TaCKX1 in common wheat productivity.
Former annotated five duplicates of TaCKX2: TaCKX2.3, TaCKX2.2, TaCKX2.1, TaCKX2.4 and TaCKX2.5 showed 2, 4, 8, 20 and 100 times lower expression respectively than in the case of TaCKX1. Profiles of expression of investigated by us genes are close to the result of TaCKX2 and TaCKX1 presented by Song et al. [31], however the levels of expression are not comparable since expression pattern for former five duplicates was not investigated. Semi-quantitative analysis proved high expression of two TaCKX2 genes in young spikes and culms [6]. Moreover, there was a positive correlation between expression level of both genes and grain number per spike in 12 wheat varieties. As discussed for TaCKX4, we should expect a negative correlation between these traits. Phylogenetic analysis showed that TaCKX2.2 is most closely related to A. tauschii CKX2-like, H. vulgare CKX2.2 and S. cereale ScCKX2.2 as well as sharing very high sequence similarity to TaCKX2.2(JN381555). Proteins encoded by TaCKX2.2(FJ648070), TaCKX2.2(GU084177) were found to be most closely related to OsCKX2 and have been located in clustered clade I of monocots [6] and according to our results to HvCKX2.2. These data did not confirm the investigation by Mameaux et al. [3] showing that TaCKX2 has five duplicated copies, since all three clones are located in one TaCKX2.2 gene. The most similar sequence to TaCKX2.2 found in databases was the TaCKX6-D1b allele isolated and analyzed by Zhang et al. [24]. Their expression patterns were similar, however differ from TaCKX6 investigated in this research.
The TaCKX9, TaCKX7(JN128589) and TaCKX7(JN128588) expression was highly specific to seedling roots, but very low compared to other TaCKX in other organs tested including roots. Despite their very high correlation coefficient of expression (0.99–1.00), the genes were expressed from different chromosomes, 7A, 7B and 7D as well as 6B and 6D, respectively. Both, TaCKX9 and TaCKX7 were expressed from genome D, which was shown to play a crucial role in the increased lateral root number [34]. According to phylogenetic analysis the most closely related orthologues of TaCKX7 was H. vulgare CKX7 as well as protein encoded by the gene and for TaCKX9 A. tauschii, CKX10-like. None of these genes have been analysed.
TaCKX3, TaCKX6 and TaCKX11 were more or less expressed through all the organs tested. TaCKX3 used in our research was isolated by Song et al. [31] and expression of the gene investigated by the same group was reported as very low during both reproductive stage and flag leaf development. The data are comparable with ours for developing spikes, but are not comparable in the case of leaves, which differ in stages of plant development. Because TaCKX3 was expressed through the all organs tested, showing an average expression level among TaCKX genes, we set relative expression as 1.00 for the mean TaCKX3 expression measured in all organs. The highest relative expression of the gene, ranging from 3 to 8, was revealed in well-developed leaves and was around 1.0 in 14 DAP and 0 DAP spikes. There are notably large differences between the high expression level of the gene in leaves of the old cultivar Kontesa and the modern cultivars Ostka and Trappe. Highly homologous and mapped to the same chromosome 7B, TaCKX3 isolated by Ma et al. [7] had no signal peptide at the N terminus, which means that the gene functions in the cytoplasm. Very close orthologues of TaCKX3 were A. tauschii CKX11, S. cereale CKX11 and one clone of H. vulgare. 97% similarity was shown by O. sativa CKX11-like and Z. mays CKX10. Despite close similarity of TaCKX3 to CKX11 orthologues, TaCKX11 showed a different pattern of expression and chromosome localisation compared to TaCKX3. The closest orthologue to TaCKX11 was H. vulgare CKX8, sharing 95% similarity as well as protein encoded by the gene; less close was orthologue of Z. mays CKX12.
TaCKX6 investigated by us, isolated and shown as the third highly expressed TaCKX in developing spikes by Song et al. [31], in our case was the seventh one taking into account the level of expression, and showed higher expression in leaf, 14 DAP spikes and seedling roots than in other organs tested. These results are not comparable because of using different objects/cultivars, organs, reference genes, number of tested TaCKX genes and the way of counting results. The coding sequence of TaCKX6 used by us was located on chromosomes 1A, 1B and 1D, but it was not similar to TaCKX6-D1b or -D1a isolated and characterized by Zhang et al. [24]. Haplotype TaCKX6-D1a has an 18 nt deletion compared to TaCKX6-D1b, and its expression was negatively associated with higher grain weight. Performed by us transcript analysis revealed that TaCKX6-D1 is indeed the TaCKX2.2 and is located on chromosome 3. Another TaCKX6 allele associated with grain size, filling rate and weight, TaCKX6a02 [25] was located on chromosome 3DS, but again was not similar to the one investigated by us or TaCKX6-D1. Performed by us transcript analysis revealed that TaCKX66a02 is in fact the TaCKX2.1 which was located on chromosome 3D. Results of alignment proved that all three TaCKX6 genes differed in their close relationship with other TaCKX genes and their orthologues. The TaCKX6 located on 1D was the closest homologue of T. aestivum CKX8, sharing 98% identity under 89% cover of the sequences. The closest orthologue was H. vulgare CKX3 with 94% identity and L. perenne CKX6 with 96% identity. Z. mays CKX6 shared 81% identity under 83% coverage. The TaCKX-D1 located on 3D was the closest homologue of TaCKX2.2, sharing 98% identity, and TaCKX2.1 with 97% identity, and under 43% cover for both. Their close orthologues were A. tauschii CKX2-like with 99% identity and 43% cover and H. vulgare CKX2.2 (69% cover/88% identity). The closest homologue of TaCKX6a02 located on 3D was T. aestivum CKX2.1(JF293079) with 99% identity under 100% cover. Summarizing alignment and phylogenetic tree analysis, TaCKX6 investigated by us was among TaCKX3, 5, 4, 8, 9 and the closest to other CKX6 orthologues. Since TaCKX6 is not the same as TaCKX6-D1 characterized by Zhang et al. [24] as well as TaCKX6-D1a by Lu et al. [25], the genes probably would not share the same function.
Developmental and daily time dependence of TaCKX expression levels and their co-operation
The TaCKX1, TaCKX2.2 and TaCKX2.1 genes, which are specific to developing spikes, and unspecific but well expressed in this organ TaCKX3 were shown to be developmentally and daily time dependent. The level of expression of TaCKX1 and TaCKX3 was significantly higher in the first 7 DAP spikes comparing to the second 7 DAP spikes in two or three cultivars and the result was opposed to TaCKX2.1. Similar data were observed for daily time expression of TaCKX1, TaCKX2.1 and TaCKX3 in 7 DAP spikes, which was highest at 9:00 am, lower at 3:00 pm and lowest at 12:00 pm and various for TaCKX2.1. These differences in developmental and daily time expression of TaCKX2.1 and TaCKX2.2 as well as their different levels of expression indicate that detailed functions of the TaCKX2 genes might be different. The data of respectively high and specific expression of TaCKX1, TaCKX2.1 and TaCKX2.2 genes were compatible with their strong correlation coefficients of expression in investigated organs assuming their powerful cooperation in spike development. Daily time dependence of expression in 7 DAP spikes was also significant for TaCKX3 and low expressing genes. These data showed that developmental and daily time expression of most TaCKX genes was cultivar independent, but some of them are developmentally and day-time insensitive.
Besides the very high correlation coefficients of expression in the group of spike-specific genes the correlation was also very high among leaf-specific genes: TaCKX4, TaCKX5, TaCKX10 and highly expressed in this organ TaCKX3. Generally we showed that there were two groups of TaCKX genes which positively cooperate in developing wheat plants. The first groups genes expressed in organs from young plants: seedling roots, leaves from 4-week-old plants, inflorescences and 0 DAP spikes. The second group includes TaCKX genes expressed in developing spikes of maturing plants: 0 DAP, 7 DAP and 14 DAP, reaching a very high correlation coefficient.
Differential expression of the genotypes
Although the expression patterns of individual TaCKX genes are tissue- and developmentally-specific, their expression levels measured in individual organs in most cases significantly differ among three cultivars used in the experiments. These differences are more distinct in the larger group of breeding material (not published yet) and might be result of various alleles of the tested genes, indicating for possible selection of these alleles of interest for breeding.
Looking for TaCKX genes regulating yield-related traits
The goal of this research was to select those TaCKX genes which might regulate yield-related traits in wheat. To achieve the goal we hypothesized that expression patterns of TaCKX genes indicate their role in growth and reproductive development. The same hypothesis for HvCKX genes has been positively verified for barley [12]. The data with barley showed that RNAi silencing of HvCKX genes that are highly and specifically expressed in developing spikes led to higher productivity [22, 23]. This effect was the consequence of a decreased level of expression of selected HvCKX genes, which was associated with a decrease of CKX enzyme activity. Since CKX enzyme irreversibly degrades cytokinins, a decreased level of expression of the TaCKX genes in selected organs is expected to increase cytokinin content. High levels of CKs promote numerous developmental features, which in the case of rice [21] and barley spikes [22, 23] led to higher grain numbers. Assuming this, yield-related traits in wheat might be regulated most of all by those TaCKX genes which are specifically expressed in developing spikes. We found three of 11 newly numbered, TaCKX1, TaCKX2.2 and TaCKX2.1, highly specific to inflorescences and developing spikes and a fourth, TaCKX3, which is not specific but is relatively highly expressed in the same organs. As discussed above, a positive effect of these genes on yield-related traits might operate on similar mechanism as in barley or rice. We chose two ways to reach this goal and positively verify the hypothesis. The first is to decrease selected gene expression by direct silencing, as was done in barley or rice. The second one is to look for natural variation of expression of these genes in generative organs among wheat genotypes. Preliminary, not yet published results prove that both ways might lead to the goal. Moreover, the first way provides us well-characterized material for detailed, functional analysis of the silenced genes. The second way gives us an opportunity to select non-GMO breeding lines for breeding purposes.
Supporting information
(PDF)
(PDF)
Figs A-N. The distance tree of pairwise comparison of TaCKX1 (A), TaCKX2.1 (B), TaCKX2.2 (C), TaCKX3 (D), TaCKX4 (E), TaCKX5 (F), TaCKX6 (G), TaCKX6-D1 (H), TaCKX6a02 (I), TaCKX7 (J), TaCKX8 (K), TaCKX9 (L), TaCKX10 (M), TaCKX11 (N) with their homologues and orthologues (queries are highlighted by yellow).
(PDF)
(JPG)
Acknowledgments
This research was supported by the National Science Centre, grant UMO-2014/13/B/NZ9/02376, by the Ministry of Agriculture and Rural Development, grant No. 5 PBwPR and a statutory grant of PBAI-NRI.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by the National Science Centre, grant UMO-2014/13/B/NZ9/02376, by the Ministry of Agriculture and Rural Development, grant No. 5 PBwPR and a statutory grant of PBAI-NRI.
References
- 1.Curtis T, Halford NG. Food security: the challenge of increasing wheat yield and the importance of not compromising food safety. Ann Appl Biol. 2014;164(3):354–72. 10.1111/aab.12108 WOS:000334359200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nadolska-Orczyk A, Rajchel IK, Orczyk W, Gasparis S. Major genes determining yield-related traits in wheat and barley. Theor Appl Genet. 2017;130(6):1081–98. 10.1007/s00122-017-2880-x WOS:000401980500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mameaux S, Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, et al. Molecular, phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the Poaceae. Plant Biotechnol J. 2012;10(1):67–82. 10.1111/j.1467-7652.2011.00645.x WOS:000298094800008. [DOI] [PubMed] [Google Scholar]
- 4.Feng DS, Wang HG, Zhang XS, Kong LR, Tian JC, Li XF. Using an Inverse PCR Method to Clone the Wheat Cytokinin Oxidase/Dehydrogenase Gene TaCKX1. Plant Mol Biol Rep. 2008;26(3):143–55. 10.1007/s11105-008-0033-8 WOS:000259962600001. [DOI] [Google Scholar]
- 5.Lei Z, Baoshi Z, Ronghua Z. Isolation and chromosomal localization of cytokinin oxidase/dehydrogenase gene (TaCKX5) in wheat. Scientia Agricultura Sinica 2008;41:636–642. [Google Scholar]
- 6.Zhang JP, Liu WH, Yang XM, Gao AN, Li XQ, Wu XY, Li LH. Isolation and characterization of two putative cytokinin oxidase genes related to grain number per spike phenotype in wheat. Mol Biol Rep. 2011;38(4):2337–47. 10.1007/s11033-010-0367-9 WOS:000289257100017. [DOI] [PubMed] [Google Scholar]
- 7.Ma X, Feng DS, Wang HG, Li XF, Kong LR. Cloning and Expression Analysis of Wheat Cytokinin Oxidase/Dehydrogenase Gene TaCKX3. Plant Mol Biol Rep. 2011;29(1):98–105. 10.1007/s11105-010-0209-x WOS:000286674600011. [DOI] [Google Scholar]
- 8.Hare PD, Vanstaden J. Cytokinin oxidase—Biochemical Features and Physiological Significance. Physiol Plantarum. 1994;91(1):128–36. 10.1034/j.1399-3054.1994.910118.x WOS:A1994NN55600018. [DOI] [Google Scholar]
- 9.Jones RJ, Schreiber BMN. Role and function of cytokinin oxidase in plants. Plant Growth Regul. 1997;23(1–2):123–34. 10.1023/A:1005913311266 WOS:000071317800008. [DOI] [Google Scholar]
- 10.Mok DWS, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Phys. 2001;52:89–118. 10.1146/annurev.arplant.52.1.89 WOS:000169615600005. [DOI] [PubMed] [Google Scholar]
- 11.Werner T, Schmulling T. Cytokinin action in plant development. Curr Opin Plant Biol. 2009;12(5):527–38. 10.1016/j.pbi.2009.07.002 WOS:000271137200003. [DOI] [PubMed] [Google Scholar]
- 12.Zalewski W, Gasparis S, Boczkowska M, Rajchel IK, Kala M, Orczyk W, et al. Expression patterns of HvCKX genes indicate their role in growth and reproductive development of barley. Plos One. 2014;9(12):e115729 10.1371/journal.pone.0115729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J, Jiang L, Jameson PE. Identification and quantitative expression of cytokinin regulatory genes during seed and leaf development in wheat. Agronomy Society of New Zealand Special Publication No l3 / Grassland Research and Practice Series No 14. 2015. [Google Scholar]
- 14.Kudo T, Kiba T, Sakakibara H. Metabolism and Long-distance Translocation of Cytokinins. J Integr Plant Biol. 2010;52(1):53–60. 10.1111/j.1744-7909.2010.00898.x WOS:000274795500007. [DOI] [PubMed] [Google Scholar]
- 15.Kuiper D. Sink Strength—Established and Regulated by Plant-Growth Regulators. Plant Cell Environ. 1993;16(9):1025–6. 10.1111/j.1365-3040.1996.tb02052.x WOS:A1993MH66800006. [DOI] [Google Scholar]
- 16.Roitsch T, Ehness R. Regulation of source/sink relations by cytokinins. Plant Growth Regul. 2000;32(2–3):359–67. 10.1023/A:1010781500705 WOS:000167858500031. [DOI] [Google Scholar]
- 17.Werner T, Holst K, Pors Y, Guivarc'h A, Mustroph A, Chriqui D, et al. Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J Exp Bot. 2008;59(10):2659–72. 10.1093/jxb/ern134 WOS:000257401500008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmulling T, Werner T, Riefler M, Krupkova E, Bartrina y Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res. 2003;116(3):241–52. 10.1007/s10265-003-0096-4 WOS:000183664100009. [DOI] [PubMed] [Google Scholar]
- 19.Werner T, Motyka V, Strnad M, Schmulling T. Regulation of plant growth by cytokinin. P Natl Acad Sci USA. 2001;98(18):10487–92. 10.1073/pnas.171304098 WOS:000170738000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner T, Kollmer I, Bartrina I, Holst K, Schmulling T. New insights into the biology of cytokinin degradation. Plant Biology. 2006;8(3):371–81. 10.1055/s-2006-923928 WOS:000238359400013. [DOI] [PubMed] [Google Scholar]
- 21.Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–5. 10.1126/science.1113373 WOS:000230938200041. [DOI] [PubMed] [Google Scholar]
- 22.Zalewski W, Galuszka P, Gasparis S, Orczyk W, Nadolska-Orczyk A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J Exp Bot. 2010;61(6):1839–51. 10.1093/jxb/erq052 WOS:000276735300024. [DOI] [PubMed] [Google Scholar]
- 23.Zalewski W, Orczyk W, Gasparis S, Nadolska-Orczyk A. HvCKX2 gene silencing by biolistic or Agrobacterium-mediated transformation in barley leads to different phenotypes. Bmc Plant Biol. 2012;12:206 10.1186/1471-2229-12-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Zhao YL, Gao LF, Zhao GY, Zhou RH, Zhang BS, et al. TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 2012;195(3):574–84. 10.1111/j.1469-8137.2012.04194.x WOS:000306179200011. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Chang C, Zhang HP, Wang SX, Sun G, Xiao SH, et al. Identification of a Novel Allele of TaCKX6a02 Associated with Grain Size, Filling Rate and Weight of Common Wheat. Plos One. 2015;10(12):e0144765 10.1371/journal.pone.0144765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang C, Lu J, Zhang HP, Ma CX, Sun GL. Copy Number Variation of Cytokinin Oxidase Gene Tackx4 Associated with Grain Weight and Chlorophyll Content of Flag Leaf in Common Wheat. Plos One. 2015;10(12):15 10.1371/journal.pone.0145970 WOS:000367481900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, et al. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018;46(D1):D802–D8. 10.1093/nar/gkx1011 WOS:000419550700119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J, Jiang L, Jameson PE. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. Bmc Plant Biol. 2012;12:78 10.1186/1471-2229-12-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mrizova K, Jiskrova E, Vyroubalova S, Novak O, Ohnoutkova L, Pospisilova H, et al. Overexpression of cytokinin dehydrogenase genes in barley (Hordeum vulgare cv. Golden Promise) fundamentally affects morphology and fertility. Plos One. 2013;8(11):e79029 10.1371/journal.pone.0079029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lara MEB, Garcia MCG, Fatima T, Ehness R, Lee TK, Proels R, et al. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell. 2004;16(5):1276–87. 10.1105/tpc.018929 WOS:000221461400016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang HF, Hu ZR, Huang K, Han Y, Zhao AJ, Han HM, et al. Three genomes differentially contribute to the seedling lateral root number in allohexaploid wheat: evidence from phenotype evolution and gene expression. Plant J. 2018;95(6):976–87. 10.1111/tpj.14005 WOS:000443813300005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Figs A-N. The distance tree of pairwise comparison of TaCKX1 (A), TaCKX2.1 (B), TaCKX2.2 (C), TaCKX3 (D), TaCKX4 (E), TaCKX5 (F), TaCKX6 (G), TaCKX6-D1 (H), TaCKX6a02 (I), TaCKX7 (J), TaCKX8 (K), TaCKX9 (L), TaCKX10 (M), TaCKX11 (N) with their homologues and orthologues (queries are highlighted by yellow).
(PDF)
(JPG)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.