Abstract
Cancer immunotherapy has been flourishing in recent years with remarkable clinical success. But as more patients are treated, a shadow is emerging that has haunted other cancer therapies: tumors develop resistance. Resistance is often caused by defects in the MHC class I (MHC I) antigen presentation pathway critical for CD8 T cell-mediated tumor clearance. TAP and tapasin, both key players in the pathway, are frequently downregulated in human cancers, correlating with poor patient survival. Reduced dependence on these factors may promote vaccine efficiency by limiting immune evasion. Here, we demonstrate that PMEL209-217, a promising phase 3 trial-tested anti-melanoma vaccine candidate, is robustly presented by various TAP- and/or tapasin-deficient cell lines. This striking characteristic may underlie its potency as a vaccine. Surprisingly, cytosolic proteasomes generate the peptide even for TAP-independent presentation, while tripeptidyl peptidase 2 (TPP2) efficiently degrades the epitope. Consequently, inhibiting TPP2 substantially boosts PMEL209-217 presentation, suggesting a possible strategy to improve the therapeutic efficacy of the vaccine.
Keywords: TAP, HLA-A2, MHC, tapasin, PMEL, antigen presentation, proteasome, TPP2
INTRODUCTION
Cancer is the second leading cause of death in the United States with more than 600,000 Americans projected to die in 2018 alone. Cancer immunotherapy activates and mobilizes the patients’ immune system against their tumor and has recently emerged as a forefront strategy to fight the disease. In particular, monoclonal antibodies targeting the immune checkpoints PD-1 or CTLA-4 induce long-term regression of metastatic tumors in a significant fraction of patients and can dramatically prolong survival (reviewed in (1, 2)). Patients not responding to treatment often bear tumors with a lower pre-existing T cell infiltrate or which lack an immune-active tumor microenvironment (3, 4). Such patients might benefit from combination therapies utilizing cancer vaccines whose efficacy would subsequently be boosted by immune checkpoint inhibitors.
Cancer vaccines are based on the in vivo activation of CD8+ cytolytic T lymphocytes (CTL). If CTLs recognize MHC I-presented antigenic peptides at the surface of cancer cells, they will kill these targets, thereby eradicating the tumor. A large number of tumor-expressed T cell epitopes have been identified (5), some of which are currently used as vaccines in immunotherapeutic trials. A better understanding of the expression profiles, processing and presentation pathways of the respective antigens will be crucial to select the most promising targets for the next generation of immunotherapies. To this end, it is important to determine which antigenic peptides are generated in tumors and which vaccines would most efficiently limit the development of immune escape variants.
Peptides recognized by CTLs typically originate from the degradation of intracellular proteins by the proteasome, which comprises three catalytic subunits: β1, β2, and β5 (2). In immune cells or cells stimulated with interferon-γ (IFN-γ), three other catalytic subunits are induced – LMP2 , MECL-1, and LMP7 – which replace the subunits β1, β2, and β5, respectively, to form immunoproteasomes. Intermediate proteasomes containing only one or two immunosubunits (β1, β2, LMP7 and LMP2, β2, LMP7) also exist. Standard, intermediate, and immunoproteasomes all have unique cleavage specificities, generating different sets of peptides from the same protein precursors (6, 7). Other cytosolic proteases such as tripeptidyl peptidase 2 (TPP2), insulysin, or nardilysin acting independently or in concert with the proteasome are also involved in the production of some antigenic peptides (8).
Peptides produced in the cytosol are subsequently transported into the lumen of the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP). TAP is a heterodimer composed of two subunits, TAP1 and TAP2, both of which are essential for peptide translocation (8). Inside the ER, the chaperone tapasin binds the N-terminal domains (N-domains) of the TAP chains (9) and bridges TAP to MHC I via its lumenal domain. Moreover, tapasin recruits two additional proteins: the thiol oxidoreductase ERp57, captured via a disulfide bond, and the chaperone calreticulin, which binds simultaneous to ERp57 and MHC I (8). The resulting complex consisting of one TAP heterodimer, two tapasin-ERp57 conjugates, and one or two MHC I-calreticulin units is known as the peptide-loading complex (PLC) (10, 11). Within the PLC, antigenic peptides are inserted into the binding groove of MHC I with tapasin acting as a crucial editor skewing the peptide repertoire towards high affinity ligands (12). Some peptides require N-terminal trimming via ER-associated aminopeptidases ERAP1 and/or ERAP2 (13-15). Once a ligand with sufficient affinity is captured, MHC I dissociates from the PLC and migrates to the plasma membrane. Various quality control pathways act along the secretory route (16-18), further ensuring that only optimally loaded MHC I molecules reach the cell surface. At the plasma membrane, peptide antigens are presented to CTLs.
Immunotherapeutic treatment can lead to the development of tumor escape variants no longer recognized by the immune system. This frequently occurs through loss of IFNγ signaling in the tumor (19, 20) leading to a reduction in antigen presentation by impairing the coordinated upregulation of the antigen processing machinery (21). Alternatively, components of the MHC I antigen processing and presentation machinery, such as β2-microglobulin (β2m), are often found directly mutated in non-responder or relapsing patients treated by immunotherapy (20, 22, 23).
The central role of the classical MHC I antigen presentation pathway in tumor clearance is also highlighted by the large number of cancers in which TAP (24-32) and/or tapasin (33-35) are downregulated or not expressed at all. Both molecules are key players in the process and their loss or downmodulation in tumors causes immune evasion and is frequently associated with a poor prognosis. While reduced expression of components in the MHC I antigen presentation pathway is often reversible by IFNγ treatment (21), structural defects that cannot be corrected by cytokine application have been observed for both TAP (36) and tapasin (37, 38). Interestingly, loss of TAP not only reduces MHC I antigen presentation but is associated with the presentation of an altered repertoire of self-epitopes, some of which are not presented by TAP-sufficient cells (39, 40). This reflects a growing number of non-classical mechanisms of MHC I antigen presentation, which have been described in recent years, partially compensating defects in the classical pathway (41). Targeting tumor antigens that are presented both in the presence and in the absence of TAP and/or tapasin (i.e. via classical and non-classical routes) provides a rationale for the development of novel immunotherapeutic vaccines designed to limit the possibility of tumor escape variants. However, to our knowledge, only three tumor-associated T cell epitopes have been shown to be presented in a TAP-independent manner in human cancers (39, 42, 43) and dependence on tapasin has only rarely been investigated (44, 45).
Among known melanoma antigens, PMEL209-217 has been shown to be one of the most potent peptides when used as a vaccine (typically in an anchor residue-modified form to enhance HLA-A2 binding) (46). It is derived from the pigmentation-associated melanocyte differentiation factor PMEL (also called gp100 or Pmel17), which is expressed in > 90% of metastatic melanomas. A recent phase 3 trial demonstrated a significant increase in objective clinical responses in stage ΙV or locally advanced stage ΙΙΙ melanoma patients, when the vaccine was combined with high-dose IL-2 compared to IL-2 alone (47). This trial also found a statistically significant positive effect of PMEL209-217 on progression-free survival and a strong trend towards increased overall survival (47). Even though there have been setbacks with this vaccine (48), a recent study suggested that PMEL209-217 is superior when compared against other melanoma peptide vaccines (46). However, the underlying mechanism is unclear. We hypothesized its potency may be explained if the peptide were less dependent on the classical MHC I pathway, making it less likely that tumors can evade its presentation. To this end, we investigated whether and to what extent PMEL209-217 presentation in melanoma cells can circumvent TAP or tapasin.
MATERIALS AND METHODS
Cell lines and cell culture
Buf1280 is a human melanoma cell line lacking TAP1- and HLA-A2 expression (36). LG2-MEL-220 (Mel220) is a human PMEL-deficient, HLA-A2-negative melanoma cell line positive for TAP1 expression (49, 50). M553 is a tapasin-deficient, HLA-A2-negative human melanoma cell line and its tapasin-CFP transfectant has been described (51). EB81-MEL2.7 is a human HLA-A2-positive melanoma cell line autologous to CTL7 and was obtained by limiting dilution cloning of EB81-MEL.2 cells (52). T2 is an HLA-A2-positive lymphoblastoid cell line lacking both TAP genes (53). CTL clones EB81-CGE 606 C/2.1 (CTL7) (54), as well as CTL IVSB and CTL 210/9 (55) have been described. Buf1280, Mel220, M553, EB81-MEL2.7, and T2 cells were cultured in IMDM (Sigma) / 10% FCS (HyClone) containing non-essential amino acids (Gibco), GlutaMax (Gibco), and penicillin/streptomycin (Gibco). Transfectants were grown in medium additionally containing 2 mg/ml G418 and/or 1.5 μg/ml puromycin. Cell lines lentivirally transduced with shRNAs were cloned by limiting dilution.
Treatment with pharmacological drugs
Unless otherwise noted, for pharmacological assays target cells were subjected to the following treatments at 37 °C for 2.5 hrs before measuring their recognition by CTLs: 20 μM lactacystin, 20 μM AAF-CMK (Biomol), 5 μM epoxomicin, 100 μM cycloheximide, 10 μg/ml brefeldin A, 100 μM butabindide (Tocris), 10 μM PAQ-22, 250 μM primaquine, and 2 nM concanamycin B. Drugs were purchased from Sigma unless otherwise specified.
Antibodies
Mouse monoclonal antibodies HC-A2 (HLA-A2) (56), BB7.2 (folded HLA-A2) (57), and 148.3 (TAP1) (58) have been described. Mouse monoclonal antibody ab54685 (TPP1) was purchased from Abcam. Rabbit polyclonal antibody Pep13h (PMEL C-terminus) (59) has been described. Rat monoclonal antibody 9G10 (Grp94, SPA-850) was purchased from Enzo Life Sciences. Rabbit polyclonal antibodies ab3329 (LMP7) (Abcam), 14120–1-AP (TPP2) (Proteintech), ab93341 (Derlin-1) (Abcam), ab15038 (Sec61β) (Abcam), AP2184A (Hrd1) (Abgent), 4108 (LC3A/B) (Cell Signaling Technology), and PA3–900 (calreticulin) (ThermoFisher) were purchased from the indicated suppliers. FITC-anti-HLA-A2 (551285), FITC-anti-CD107a (555800), FITC-anti-CD107b (555804), and APC-anti-CD8 (555369) antibodies were purchased from BD Biosciences.
Vector constructs
PMEL, IR-wt, and PMEL∆190–208 in pBMN-IRES-neo (50), as well as TAP1 in pLPCX (10) have been described. HLA-A2 in pBMN-IRES-neo was generated by cloning the HLA-A2 cDNA into pBMN-IRES-neo via EcoRI-sites. Quikchange mutagenesis was employed to clone HLA-A2 mutant T134K using HLA-A2 in pBMN-IRES-neo as template in combination with primer pair 5’- GGACCTGCGCTCTTGGAAGGCGGCGGACATGG C-3’/5’-GCCATGTCCGCCGCCTTCCAAGAGCGCAGGTCC-3’. Quikchange mutagenesis was employed to clone the PMEL209-217 minigene construct (pep209) using PMEL in pBMN-IRES-neo as a template in combination with primer pair 5’-GGAATTCCA CCATGATTACTGACCAGGTGCCTTTCTCCGTCTGAGCGGCG-3’/5’-CGCCGCTCAGA CGGAGAAAGGCACCTGGTCAGTAATCATGGTGGAATTCC-3’. PMEL-ΔSS was cloned into the expression vector as an EcoRI-EcoRI fragment after PCR-amplifying PMEL with primer pair 5’-AATGAATTCCACCATGGCTACAAAAGTACCCAG-3’/5’-TGAATTCGCCGCTCAGACCTGCTGCCC-3’. An HLA-A2-IRES-ICP47 construct to simultaneously render Mel220 cells HLA-A2 positive and TAP-inhibited was cloned by an overlap PCR using the following two fragments generated by a standard Pfu-driven reaction: (1) a 1810 bp fragment generated with primer pair 5’-CG GATCCCAGTGTGGTGGTAGG-3’/5’-CCATTTCCAGGGCCCACGACATGGTATTATCA TCGTGTTTTTCAAAGG-3’ and HLA-A2 in pBMN-IRES-neo as template and a (2) 310 bp fragment generated with primer pair 5’-CCTTTGAAAAACACGATGATAATACCATGT CGTGGGCCCTGGAAATGG-3’/5’-TATCGATGCGAACCCCAGAGTCCCGCTCAACGGGTTACCGGATTACGGG-3’ and ICP47 in pLPCX as template. The resulting overlap fragment was cloned into a pBMN vector as BamHI/ClaI fragment. LMP7 was cloned from IFNγ-stimulated buf1280 cells using primer pair 5’-ATCTCTGGGTGCTGGGCGGTC-3’/5’-GCTGCCACCACCACCATTATTGATTG-3’ in a standard RT-PCR and transferred into EcoRI-cleaved pBMN-IRES-puro as an MfeI/MfeI fragment amplified using primer pair : 5’-TTCAATTGCTCTGGGTGCTGGGCGGTCAT-3’/5’-AACAATTGGCTGCCACCACCACCATTATTG-3’. Lentiviral pLKO1.puro-based shRNA constructs targeting TPP2 were purchased from Sigma (shRNA construct #1 targets sequence 5’-CGCCTTAAAGACCTTCCATTT-3’; shRNA construct #5 targets sequence 5’-GCTGGATTCTAGTGACATTTA-3’). Lentiviral pLKO1.puro-based shRNA constructs targeting TPP1 were purchased from Sigma (shRNA construct #3 targets sequence 5’- CCTCGTCTGTTAAGTGTGAAT-3’; shRNA construct #5 targets sequence 5’- CCCTCTGTGATCCGTAAGCGA-3’). Lentiviral pLKO1.puro-based shRNA constructs specific for Hrd1 (construct #1), Derlin-1 (construct #2), and Sec61β (construct #3) were constructed to target sequences 5’-GAGACAGTTTCAGATGATT-3’, 5’-GCCAGCAGACTATTTATTCAT-3’, 5’-CCCAACATTTCTTGGACCAAA-3’, respectively. All vectors were sequenced before retroviral transduction (60) into Mel220, T2, or buf1280 cells and selected with appropriate antibiotics.
RT-PCR
Reverse transcription (RT) was performed on total RNA. Expression of PMEL, tyrosinase, and Melan-A was measured by a standard Taq-driven PCR. Primer pairs specific for tyrosinase, Melan-A, and PMEL were 5 ‘-GGATAGCGGATGCCTCTCAAAG-3’/5’-CCCAAGGAGCCATGACCAGAT-3’, 5 ‘-CTGACCCTACAAGATGCCAAGAG-3’/5’-ATCATGCATTGCAACATTTATTGATGGAG-3’, and 5’-ATGTGGAACAGGCAGCTGTAT-3’/5’-TTCAAGGGAAGATGCAGGCATCG-3’. PCR products were analyzed on an ethidium bromide-stained 1% agarose gel.
T cell assays
To measure T cell activation via degranulation, 60000 to 250000 target cells were co-cultured with the 50000 CTLs for 1h in the presence of FITC-labeled anti-CD107a and anti-CD107b antibodies (both at 1:50) in a total of 200 μl per well of a 96- or 48-well plate. Subsequently, BFA (Sigma) was added (10 μg/ml) and cells were incubated for another 3.5h. Finally, cells were harvested, CTLs were stained with APC-labeled anti-CD8 and T cell activation was assessed by flow cytometry measuring the surface exposure of CD107 caused by degranulation. To this end, propidium iodide (Sigma) was added for life-gating before 10000 CD8-positive, propidium iodide-negative events were acquired using a FACSCalibur or FACSVerse flow cytometer (BD Biosciences). Within this population, the percentage of CD107a/b-positive cells (activated CTLs post degranulation) was determined. To measure target cell lysis via europium release, the Europium Kit (Perkin Elmer) was used following the manufacturer’s instructions. The IFNγ ELISA to measure T cell activation was carried out as described (44).
Western blotting
Western blotting was carried out as described (61). Briefly, 5 × 106 cells were lysed for 1h in 500 μl PBS / 1% Triton-X100 containing Complete protease inhibitor cocktail (Roche) on ice. Lysates separated by SDS-PAGE were blotted onto Immobilon-P PVDF Transfer Membrane (Millipore). Immunoblots were probed with indicated antibodies.
RESULTS
PMEL209-217 can be presented by HLA-A2 in a TAP-independent manner
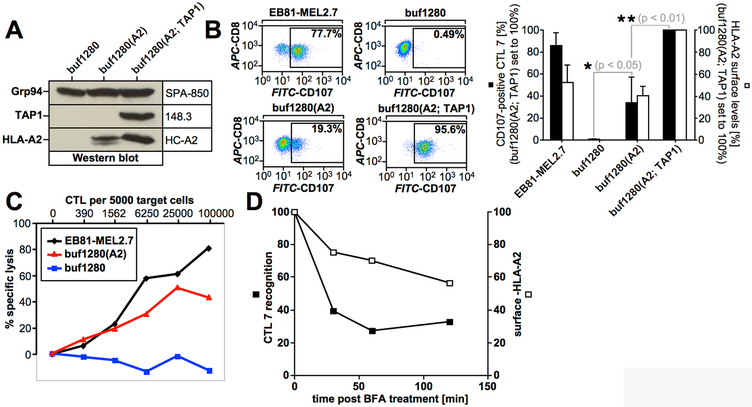
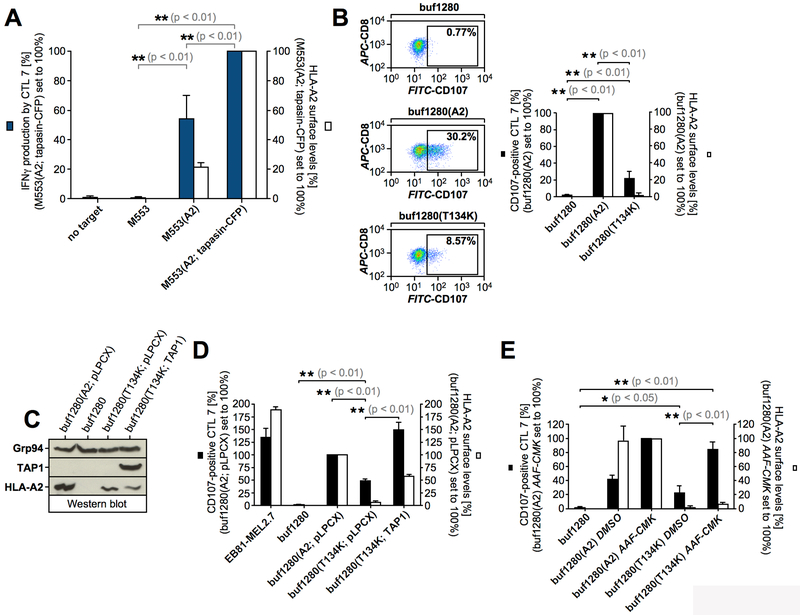
The human melanoma cell line buf1280 expresses PMEL (50), but lacks the expression of TAP1- and HLA-A2, because of frameshift mutations in both genes (36) (Table I). Since TAP transporter activity requires both TAP1 and TAP2 (8) and TAP2 stability requires physical presence of TAP1 (62, 63), neither TAP subunit is detectable at steady state and TAP is completely inactive in buf1280 cells (36). Therefore, buf1280 cells are a powerful tumor cell model system to investigate TAP-independent antigen presentation. To assess whether the HLA-A2-restricted epitope PMEL209-217 is a TAP-independent antigen, we analyzed its presentation in buf1280 derivative cell lines using the PMEL209-217-specific CTL clone EB81-CGE 606 C/2.1 (CTL7) (54). The buf1280-derived target cells examined in these experiments were stably transduced with either HLA-A2 alone or with both HLA-A2 and TAP1 (Fig. 1A, Table I). As expected, CTL7 efficiently recognized autologous tumor cells (EB81-MEL2.7) (Table I) but did not recognize untransduced, HLA-A2-negative buf1280 cells (Fig. 1B). However, CTL7 recognized TAP-deficient, HLA-A2-transduced buf1280(A2) cells in both T cell activation (Fig. 1B) and target cell lysis (Fig. 1C) assays, demonstrating that PMEL209-217 can be presented by HLA-A2 independently of TAP. Transduction of buf1280(A2) cells with TAP1, which restores TAP activity (36), further enhanced T cell recognition (Fig. 1B). This suggests that PMEL209-217 can be presented through both TAP-dependent and TAP-independent pathways.
Table I, Selected target cell lines used in T cell assays.
M553, Mel220, buf1280, and EB81-MEL2.7 are human melanoma cell lines. T2 is a human lymphoblastoid cell line.
| HLA-A2 | TAP | tapasin | PMEL | |
|---|---|---|---|---|
| buf1280 | − | − | + | + |
| buf1280(A2) | transduced | − | + | + |
| buf1280(A2; TAP1) | transduced | transduced | + | + |
| M553 | − | + | − | + |
| M553(A2) | transduced | + | − | + |
| M553(A2; tapasin-CFP) | transduced | + | transduced | + |
| Mel220 | − | + | + | − |
| Mel220(PMEL; A2) | transduced | + | + | transduced |
| Mel220(PMEL; A2; ICP47) | transduced | inhibited | + | transduced |
| EB81-MEL2.7 | + | + | + | + |
| T2 | + | − | + | − |
| T2(∆SS) | + | − | + | transduced* |
| T2(pep209) | +/+ | −/− | +/+ | transduced** |
cytosolic signal-sequence-deficient PMEL,
cytosolic PMEL209-217 minigene
Figure 1, TAP-independent presentation of PMEL209-217.
(A) Western blot analysis of buf1280 transfectants. (B) CTL7 recognition of target cells. Degranulated CTLs (CD107a/b+CD8+propidium iodide−) are shown in the gate and their percentage among total CTLs (CD8+propidium iodide−) is depicted in each dot plot (left panels). Average CTL activation in four independent degranulation assays (black bars) are shown in the bar diagram (right panel). For each experiment, HLA-A2 surface levels on target cells were determined (white bars). Error bars represent the standard deviation from the mean. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. (C) CTL7-mediated target cell lysis measured using a europium release assay. (D) buf1280(A2) cells were treated with BFA (5 μg/ml) for 30 min, 1h, or 2h before they were used as targets in a T cell activation assay as in Fig. 1B. CTL7 degranulation (black squares) and HLA-A2 surface levels (white squares) are depicted.
The stability of cell surface-exposed HLA-A2-PMEL209-217 complexes was measured in Brefeldin-A (BFA) experiments (17) using buf1280(A2) cells. This revealed a short surface half-life consistent with PMEL209-217 being a low affinity epitope (64, 65) (Fig. 1D). Sensitivity to BFA also suggests that TAP-independent presentation of PMEL209-217 requires functional secretion.
TAP-independent presentation of PMEL209-217 requires cytosolic processing
Residues 209-217 are part of the PMEL luminal domain exposed to potentially protease-rich secretory and endocytic environments while the molecule traffics to melanosomes (66). Moreover, these residues are part of the melanosomal PMEL amyloid core (67). Thus, proteolytic liberation of the epitope during early trafficking or during breakdown of aging PMEL fibrils could potentially underlie TAP-independent presentation of PMEL209-217. We therefore addressed requirements within the PMEL protein itself necessary for TAP-independent PMEL209-217 presentation.
To this end, we employed the human PMEL-free melanoma cell line Mel220 (49) stably transduced with HLA-A2 and the potent viral TAP-inhibitor ICP47 (68) (Table Ι). As expected, Mel220(A2; ICP47) cells were only recognized by CTL7 if they were also transfected with PMEL (Suppl. Fig. S1A). Since ICP47 was highly active in these cells (Suppl. Fig. S1B), these results established TAP-independent PMEL209-217 presentation in a second human melanoma system and also confirmed antigen specificity of the CTL clone. A previously described PMEL mutant, which fails to target to melanosomes but instead forms non-fibrillar aggregates in lysosomes (IR-wt) (50) also efficiently delivered the epitope in this system, suggesting that melanosomal degradation of PMEL amyloid fibrils is not the source of the TAP-independent epitope (Suppl. Fig. S1A/B). Strikingly, even a completely ER-retained PMEL mutant (Δ190–208) (50) gave rise to the epitope, strongly suggesting that TAP-independent presentation of PMEL209-217 does not require access of the protein to secretory or endocytic compartments (Suppl. Fig. S1C/D). Therefore, endocytic processing and/or recycling are unlikely to explain TAP-independent presentation of PMEL209-217.
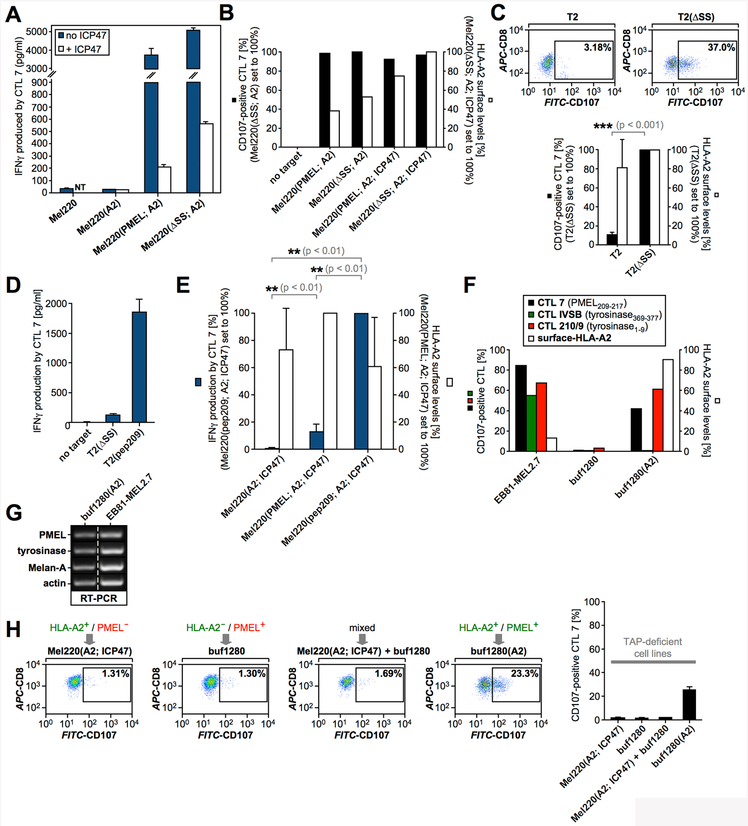
Next, we tested whether PMEL insertion into ER membranes was required for TAP-independent presentation of PMEL209-217. To this end, we examined whether Mel220(A2; ICP47) cells transduced with a cytosolically expressed PMEL mutant lacking the signal sequence (ΔSS) (Suppl. Fig. S1E) would be recognized by CTL7. These experiments demonstrated that co-translational translocation of PMEL into the ER membrane is not necessary for TAP-independent PMEL209-217 presentation (Fig. 2A/B). This result was further confirmed in T2 cells, in which both TAP1 and TAP2 genes are fully deleted (Table I), which additionally ruled out that presentation of PMEL209-217 might depend on residual low TAP activity (Fig. 2C). Taken together, this strongly suggests that even TAP-independent presentation of PMEL209-217 utilizes peptide that has been processed in the cytosol. To confirm that TAP-independent presentation of PMEL209-217 occurs when the peptide is produced in the cytosol, we transduced a cytosolic minigene encoding the antigenic peptide (preceded by a start methionine (pep209)) into T2 cells or TAP-deficient Mel220(A2; ICP47) cells. In both cell systems, PMEL209-217 was presented extremely efficiently, demonstrating that cytosolic production of the peptide drives its TAP-independent presentation (Fig. 2D/E).
Figure 2, Cytosolic processing of PMEL209-217.
(A) CTL7 activation measured by IFNγ ELISA in response to TAP-sufficient (blue bars) or TAP-inhibited (white bars) target cells (NT, not tested). (B, C) CTL7 degranulation measured in response to the indicated cell lines is shown as in Fig. 1B. CTL7 activation (black bars) and HLA-A2 surface levels on target cells (white bars) are depicted in bar diagrams. The experiment in Fig. 2C was repeated three times. Error bars represent the standard deviation from the mean of these three independent experiments. A paired two-tailed t-test was performed for statistical evaluation. Surface-HLA-A2 levels were measured in only two of these three experiments (white bars). (D, E) CTL7 activation measured by IFNγ ELISA (dark blue bars). The experiment in Fig. 2E was repeated four times. Error bars represent the standard deviation from the mean of these four independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Surface-HLA-A2 levels were measured by flow cytometry in only two of these four experiments (white bars). Relatively low T cell recognition of T2(∆SS) cells in Fig. 2D reflects consistently lower sensitivity of the IFNγ ELISA compared to LAMP degranulation (Fig. 2C). (F) T cell activation of clone CTL7 (black bars), tyrosinase1-9-specific clone CTL 210/9 (red bars) and tyrosinase369-377-specific clone CTL IVSB (green bars) measured using the degranulation assay in response to the indicated cell lines. HLA-A2 surface levels are shown in white bars. (G) Expression of melanocyte differentiation antigens analyzed by RT-PCR. The dashed line indicates where an irrelevant lane was removed. (H) CTL7 activation measured in response to the indicated cell lines or cell line mixtures. Degranulation is quantified in the rightmost panel. Error bars represent the standard deviation from the mean of two independent experiments.
TAP-independent presentation was observed for PMEL209-217 and another well-characterized TAP-independent epitope (tyrosinase1–9) but not for the classically TAP-dependent epitope tyrosinase369–377 (43) (Fig. 2F/G). This confirmed that TAP-independent presentation was a specific property of the PMEL-derived epitope, thus ruling out systematic leakage of all peptides into the ER. Moreover, experiments with mixed target cell cultures, in which half of the cells expressed HLA-A2 but not PMEL and the other half expressed PMEL but not HLA-A2, showed that the process was cell-autonomous and did not involve loading of antigens released from dying cells (Fig. 2H). Altogether, these results demonstrate that PMEL209-217 destined for TAP-independent presentation is generated in the cytosol and, hence, must cross a cellular membrane by a mechanism independent of TAP in order to gain access to HLA-A2.
The proteasome generates PMEL209-217 for TAP-independent presentation
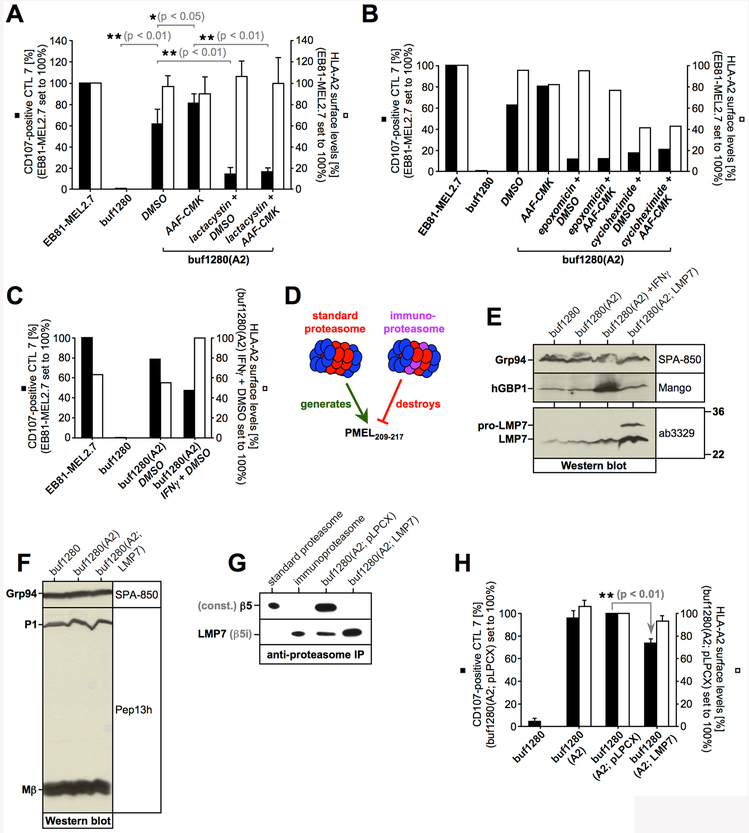
Proteasomal generation of PMEL209-217 has been extensively characterized. While the standard proteasome promotes the formation of the epitope, the immunoproteasome subunit LMP7 can cleave inside the peptide, leading to its destruction (6, 7). To assess whether TAP-independent presentation of PMEL209-217 requires proteasomal activity, we treated buf1280(A2) cells with proteasome inhibitors lactacystin or epoxomicin and used them as target cells in T cell assays. Both treatments dramatically reduced T cell recognition (Fig. 3A/B), suggesting that the proteasome generates the PMEL209-217 peptide in TAP-deficient cells for MHC I-mediated presentation. TAP-independent presentation of PMEL209-217 was also sensitive to treatment with cycloheximide, indicating that the process requires protein synthesis (Fig. 3B). In line with an involvement of the proteasome, stimulation of buf1280(A2) cells with interferon-γ (IFNγ), which induces the immunoproteasome, reduced PMEL209-217 presentation (Fig. 3C/D/E). Furthermore, overexpression of the immunoproteasome subunit LMP7 (Fig. 3E/F), which forces its incorporation into the proteasome at the cost of the standard subunit β5 (Fig. 3G), significantly reduced TAP-independent PMEL209-217 presentation (Fig. 3D/H).
Figure 3, PMEL209-217 destined for TAP-independent presentation is generated by the standard proteasome.
(A, B, C, H) CTL7 degranulation is shown as in Fig. 1B. CTL7 activation (black bars) and HLA-A2 surface levels on target cells (white bars) are depicted in bar diagrams. The experiment in Fig. 3A was repeated three times. Error bars represent the standard deviation from the mean of these three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Data from this experiment was also included in Fig. 3C and 4D. The experiment in Fig. 3H was repeated four times (pLPCX, empty vector). Error bars represent the standard deviation from the mean of these four independent experiments (the control cell line buf1280(A2) was included only three times). A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Data from this experiment was also included in Fig. 5D and 6C. (D) The standard proteasome promotes the generation of epitope PMEL209-217, while the immunoproteasome cleavage-destroys it. (E) Expression of the immunoproteasome subunit LMP7 in cells stably transduced with LMP7 or empty vector (pLPCX) analyzed by Western blotting. (F) Western blotting demonstrates that LMP7 expression does not affect endogenous PMEL expression. The PMEL ER form (P1) and the PMEL-Mβ fragment are labeled. (G) Proteasomes immunoisolated with antibody MCP21 from cells stably transduced with LMP7 or empty vector (pLPCX). Western blots were probed with antibodies against LMP7 or the standard proteasome subunit β5.
Tripeptidyl peptidase 2 degrades PMEL209-217 in the cytosol
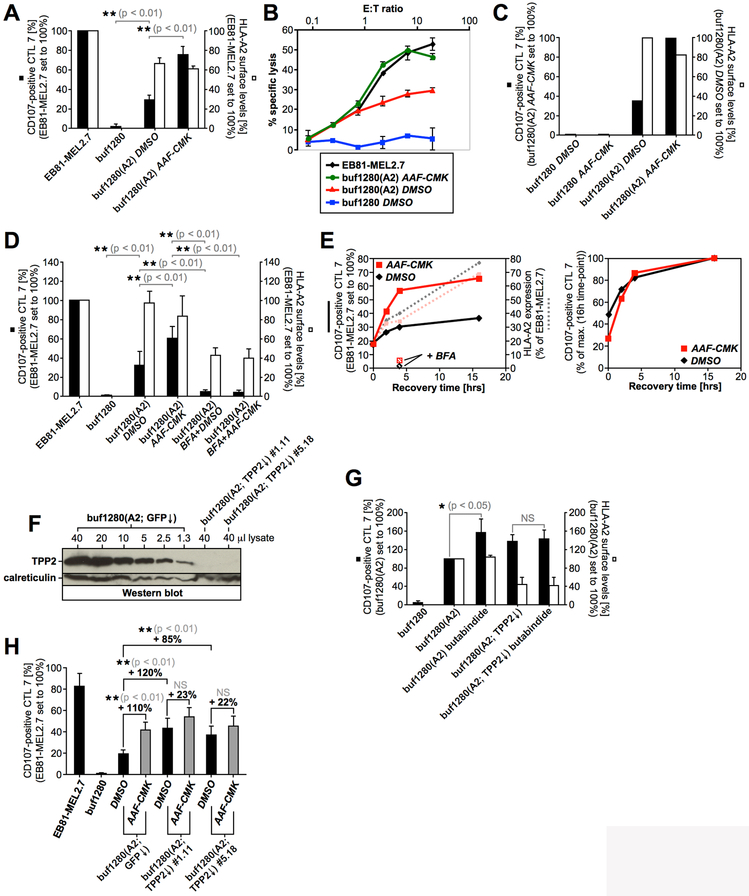
Tripeptidyl peptidase 2 (TPP2) is a cytosolic protease involved in MHC class I antigen processing. TPP2 promotes the formation of some epitopes (69, 70) while limiting others, but its global effect on MHC class I-presented antigens is likely negative (71, 72). AAF-CMK inhibits the activity of various proteases including TPP2, TPP1, and cytosolic puromycin-sensitive amino peptidase (PSAP) (73, 74). We found that in the presence of AAF-CMK, presentation of PMEL209-217 by buf1280(A2) cells was significantly enhanced without a major effect on global HLA-A2 levels (Fig. 4A/B). Not surprisingly, HLA-A2 expression by the target cells was required for the drug to increase T cell recognition (Fig. 4C). The effect of AAF-CMK was abrogated in the presence of proteasome or protein synthesis inhibition (Fig. 3A/B), indicating that the compound did not cause enhanced formation of the PMEL epitope but rather prevented its degradation. Moreover, for the drug to augment presentation of PMEL209-217, functional secretion was required (Fig. 4D). When surface-MHC class I molecules were removed by acid-wash (17), AAF-CMK-treated cells recovered higher T cell recognition, which was completely blocked by brefeldin A (BFA) (Fig. 4E, left panel), but the kinetics of recovery were similar in the absence and presence of the inhibitor (Fig. 4E, right panel).
Figure 4, Tripeptidyl peptidase 2 degrades PMEL209-217 in the cytosol.
(A, C, D, G) CTL7 degranulation is shown as in Fig. 1B. CTL7 activation (black bars) and HLA-A2 surface levels on target cells (white bars) are depicted in bar diagrams. The experiment in Fig. 4A was repeated four times. Error bars represent the standard deviation from the mean of these four independent experiments (surface-HLA-A2 levels (white bars) were measured in only three of these four experiments). A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. The experiment in Fig. 4D was repeated three times. Error bars represent the standard deviation from the mean of these three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Data from this experiment was also included in Fig. 3A and 5E. The experiment in Fig. 4G was repeated four times. Error bars represent the standard deviation from the mean of these four independent experiments. A Repeated Measures ANOVA with Dunnett’s post test was performed for statistical evaluation (NS = not significant). (B) CTL7-mediated target cell lysis of buf1280 transfectants treated or not with AAF-CMK (20 μM) was measured using a europium release assay. (E) buf1280(A2) cells were acid-washed to remove surface-MHC class I and incubated at 37 °C for the indicated times (0h, 2h, 4h, 16h) in the presence (red lines) or absence (black lines) of AAF-CMK (20 μM). HLA-A2 surface levels were determined (dotted lines) and cells were used as targets in a T cell activation assay as in Fig. 1B (full lines). Where indicated, samples were additionally treated with 10 μg/ml BFA. T cell recognition was normalized with respect to untreated autologous tumor cells (EB81-MEL2.7) (left panel), or maximal response (both 16h time-points set to 100%) (right panel). (F) Various amounts of a buf1280(A2; GFP↓) control lysate were loaded on an SDS-PAGE gel together with 40 μl of a lysate derived from TPP2-silenced buf1280(A2; TPP2↓) clones #1.11 (shRNA construct #1) and #5.18 (shRNA construct #5). (H) CTL7 degranulation measured in response to buf1280(A2) cell lines stably transduced with TPP2-specific shRNA constructs #1 (buf1280(A2; TPP2↓) #1.11) or #5 (buf1280(A2; TPP2↓) #5.18) or a GFP-specific control shRNA construct (buf1280(A2; GFP↓). Error bars represent the standard deviation from the mean of five independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation (NS = not significant).
The knockdown of TPP1 had no effect on TAP-independent PMEL209-217 presentation and the respective cells remained sensitive to AAF-CMK (Suppl. Fig. S2A/B). Treatment with the PSAP inhibitor PAQ-22 (75) also did not enhance PMEL209-217 presentation and AAF-CMK sensitivity was maintained (Suppl. Fig. S2C). This suggests that neither TPP1 nor PSAP play a major role in degrading the epitope. In contrast, and similar to AAF-CMK (Fig. 4A-D), the specific TPP2 inhibitor butabindide (76) significantly enhanced TAP-independent PMEL209-217 presentation, while TPP2-silenced cells lacked sensitivity to the drug (Fig. 4F/G and Suppl. Fig. S2D). Similarly, TPP2 knockdown cells, expressing either of two different TPP2-specific shRNAs, displayed enhanced PMEL209-217 presentation at steady state and the cells largely lost their sensitivity to AAF-CMK (Fig. 4H). Butabindide and AAF-CMK likely act on the same target, because they affected TAP-independent PMEL209-217 presentation to a similar extent and the two drugs produced no additive effect (Suppl. Fig. S2E). Altogether, this suggests that TPP2 is the protease that degrades PMEL209-217 in the cytosol.
Presentation of PMEL209-217 is independent of tapasin
To examine whether PMEL209-217 presentation depends on tapasin, we analyzed the recognition by CTL7 of the tapasin-deficient human melanoma cell line M553 transfected with HLA-A2 alone or co-transfected with HLA-A2 and tapasin (Table I). Both cell lines were strongly recognized by the CTL, but the presence of tapasin caused enhanced recognition (Fig. 5A). Thus, although both TAP and tapasin significantly promote PMEL209-217 presentation, neither factor is absolutely required for vigorous recognition of PMEL-expressing cells by CTL7.
Figure 5, TAP-independent PMEL209-217 presentation occurs in the absence of tapasin assistance.
(A) CTL7 activation measured by IFNγ ELISA (dark blue bars). The experiment was repeated three times and the error bars represent the standard deviation from the mean of these three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Surface-HLA-A2 levels were measured by flow cytometry (white bars). (B, D, E) CTL7 degranulation is shown as in Fig. 1B. CTL7 activation (black bars) and HLA-A2 surface levels on target cells (white bars) are depicted in bar diagrams. The experiment in Fig. 5B was repeated three times. Error bars represent the standard deviation from the mean of these three independent experiments (surface-HLA-A2 levels (white bars) were measured in only two of these three experiments). A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. The experiment in Fig. 5D was repeated three times (pLPCX, empty vector). Error bars represent the standard deviation from the mean of these three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Data from this experiment was also included in Fig. 3H and 6C. The experiment in Fig. 5E was repeated three times. Error bars represent the standard deviation from the mean of these three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation. Data from this experiment was also included in Fig. 4D. (C) Expression of HLA-A2 and TAP1 in cells stably transduced with wildtype HLA-A2 or HLA-A2 T134K and co-expressing either empty vector (pLPCX) or TAP1 was analyzed by Western blotting.
In a previous study, a subset of epitopes had been reported whose presentation survived the lack of TAP or the lack of tapasin individually, but not the loss of both factors simultaneously (77). Therefore, we tested whether PMEL209-217 presentation persists in cells in which neither TAP nor tapasin provide support for HLA-A2 loading. To this end, we analyzed the recognition of TAP-deficient buf1280 cells expressing the HLA-A2 mutant T134K, which does not associate with tapasin (78-81), by CTL7.
Surprisingly, although wildtype HLA-A2 is a rather TAP-independent MHC class I allele (82, 83) (Fig. 1A/B, white bars), its tapasin binding-deficient mutant T134K was dramatically reduced, in fact almost not detectable, at the plasma membrane of TAP-deficient buf1280(T134K) cells (Fig. 5B, white bars). However, HLA-A2 T134K reached significant surface levels when TAP1 was stably introduced into these cells, restoring TAP transporter activity (Fig. 5C/D, white bars). Thus, although global surface expression of HLA-A2 is not particularly dependent on either TAP or tapasin alone, simultaneous loss of support from both molecules severely collapses surface-HLA-A2.
Strikingly, even though only minute levels of HLA-A2 were detectable at the cell surface of buf1280(T134K) cells, PMEL209-217 continued to be recognized by CTL7 (Fig. 5B/D/E). Moreover, when these cells were treated with AAF-CMK, T cell activation even exceeded the recognition of tapasin-sufficient buf1280(A2) cells expressing wildtype HLA-A2 (Fig. 5E). This suggests that presentation of PMEL209-217 is extraordinarily resistant to simultaneous loss of TAP and tapasin. Such resistance may contribute to the superior qualities of this epitope as a vaccine (46).
PMEL209-217 presentation does not depend on Hrd1/Derlin-1-mediated ER-associated degradation (ERAD)
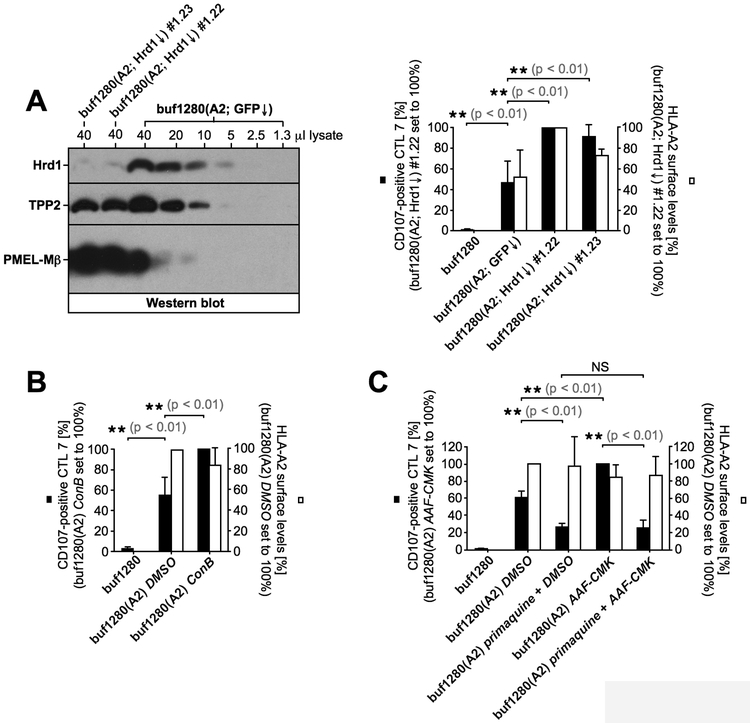
Cytosolically generated peptides, such as PMEL209-217 must cross a membrane in order to gain access to MHC I. How this occurs in TAP-deficient cells is poorly characterized, although some pathways have been proposed (41). One possibility is that an alternative peptide transporter exists in the ER membrane compensating for the loss of TAP. Obvious transporter candidates in such a scenario would be the molecules discussed as potential ERAD channels, Hrd1 (84) and Derlin-1 (85, 86), as well as the translocon Sec61. To investigate a possible role for these proteins in TAP-independent PMEL209-217 presentation, we generated buf1280(A2) cell clones with a stable knockdown of Hrd1 (Fig. 6A, left panel) or Derlin-1 (Suppl. Fig. S3A/B) and assessed their recognition by CTL7. These clones consistently displayed higher HLA-A2 surface levels, likely reflecting reduced degradation of this MHC class I allele as a consequence of reduced ERAD in the cell (white bars in Fig. 6A, right panel and Suppl. Fig. S3C). Concomitantly, these clones were better recognized by CTL7 indicating that neither Hrd1 nor Derlin-1 are necessary for TAP-independent PMEL209-217 presentation (black bars in Fig. 6A, right panel and Suppl. Fig. S3C). Interestingly, these results also argue against Hrd1/Derlin-1-mediated ERAD as a source for the generation of the PMEL209-217 peptide in buf1280(A2) cells. Unfortunately, cells are not viable without Sec61, which makes assessing its possible involvement in TAP-independent peptide transport a difficult task. However, the translocon component Sec61β is not essential, but involved in some, although not all functions of the channel (87). Nevertheless, a set of stable Sec61β knockdown clones showed no significant defect in PMEL209-217 presentation (Suppl. Fig. S3D-F, black bars), excluding at least an involvement of this particular Sec61 component in the process.
Figure 6, TAP-independent PMEL209-217 presentation requires endocytic recycling but not Hrd1-dependent ERAD.
(A) Various amounts of a buf1280(A2; GFP↓) control lysate were loaded on an SDS-PAGE gel together with 40 μl of a lysate derived from Hrd1-silenced buf1280(A2; Hrd1↓) clones #1.22 and #1.23 (shRNA construct #1) (left panel). CTL7 degranulation measured in response to buf1280(A2) cell lines stably transduced with Hrd1-specific shRNA construct #1 (buf1280(A2; Hrd1↓) #1.22 and buf1280(A2; Hrd1↓) #1.23) or a GFP-specific control shRNA construct (buf1280(A2; GFP↓). Error bars represent the standard deviation from the mean of three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation (right panel). (B, C) CTL7 degranulation is shown as in Fig. 1B. CTL7 activation (black bars) and HLA-A2 surface levels on target cells (white bars) are depicted in bar diagrams. Both experiments were repeated three times. Error bars represent the standard deviation from the mean of these three independent experiments. A One-way ANOVA with Dunnett’s post test was performed for statistical evaluation (NS = not significant). Some data shown in Fig. 6C was also included in Fig. 3A, 4D, and 5D.
PMEL209-217 presentation requires endocytic recycling
Autophagy has been discussed as a potential mechanism delivering cytosolic peptides into endosomes for capture by recycling MHC I in TAP-deficient cells (88). Interestingly, treatment of buf1280(A2) cells with concanamycin B (ConB), a specific inhibitor of vacuolar-type H+-ATPase, significantly augmented PMEL209-217 presentation (Fig. 6B, black bars) without affecting global surface-HLA-A2 levels (Fig. 6B, white bars). This is consistent with a scenario in which neutralization of endosomal pH protects the peptide antigen from degradation. We note that a similar phenomenon had been reported previously for a murine TAP-independent epitope (89). However, this scenario would predict that after capturing its peptide cargo, PMEL209-217-loaded HLA-A2 would employ endocytic recycling to present its antigenic ligand to CTL7. In line with such a mechanism, primaquine, a specific inhibitor of endocytic recycling, significantly suppressed PMEL209-217 presentation in buf1280(A2) cells (Fig. 6C, black bars) – again without affecting global surface-HLA-A2 (Fig. 6C, white bars). Interestingly, CTL7 recognition was comparably low in primaquine-treated cells irrespective of whether these cells had been co-treated or not with AAF-CMK (Fig. 6C). This indicates that peptide protection in the cytosol through inhibition of TPP2 drives higher PMEL209-217 presentation only when endocytic recycling is allowed to occur. Altogether, these data are consistent with a model in which cytosolic PMEL209-217 is delivered into endosomes by a yet uncharacterized mechanism and there gains access to recycling HLA-A2 leading to TAP-independent presentation.
Surprisingly, conventional macroautophagy, a potential candidate for such a mechanism, is unlikely to be involved in the process. Specifically, the autophagy inducer rapamycin did not augment CTL7 recognition of buf1280(A2) cells (Suppl. Fig. S4A). In fact, if anything, at very high concentrations rapamycin suppressed PMEL209-217 presentation (Suppl. Fig. S4B), although this might reflect toxicity of the drug at these levels. Moreover, the autophagy inhibitor 3-methyladenine (3-MA) did not affect PMEL209-217 presentation at any concentration tested (Suppl. Fig. S4C/D). In line with this, knockdown of the key autophagy gene ATG5 did not significantly affect PMEL209-217 presentation (Suppl. Fig. S4E/F, black bars). A number of possible mechanisms remain that could deliver the cytosolic PMEL209-217 peptide into endosomal compartments for HLA-A2 loading. These potential mechanisms include TAP-L-mediated import into lysosomes (90), ATG5-independent macroautophagy (91), microautophagy (92), and chaperone-assisted autophagy (93).
DISCUSSION
PMEL209-217 has long been known as a potential melanoma vaccine. Early work by Rosenberg and colleagues identified the epitope and demonstrated that TILs isolated from a subset of melanoma patients recognized the antigen (94). These groundbreaking studies also found a significant correlation between the reactivity of such TILs against PMEL and clinical responses in the context of adoptive T cell transfer therapy (94). Subsequently, a phase 2 trial using an anchor residue-optimized derivative of PMEL209-217 (PMEL209-217/T210M) in combination with high-dose IL-2 reported tumor regression in 42% of treated metastatic melanoma patients (95). This suggested a substantial potency of the peptide as a successful vaccine. Analyzing a large cohort of 684 metastatic melanoma cases treated at the Surgery Branch of the National Cancer Institute that had received either high-dose IL-2 alone or IL-2 in conjunction with various vaccines, further underscored this potential. In fact, the study identified PMEL209-217/T210M as a highly effective ingredient giving rise to almost twice the number of clinical objective responses than IL-2 alone or IL-2 in combination with other vaccines (46). This was corroborated in an extensive randomized phase 3 trial involving 185 patients, which demonstrated increased potency of the PMEL209-217/T210M/high dose IL-2 combination over treatment with IL-2 alone (47). Specifically, PMEL209-217/T210M significantly extended progression-free survival, displayed a trend towards increased overall survival, and almost tripled the number of clinical responses (16% versus 6%) (47). Although there have been setbacks with this vaccine as well (48), taken together, above studies support the view that PMEL209-217 is a particularly effective antigen. Our finding that the peptide is highly flexible with respect to its route of antigen presentation and is presented through TAP-dependent, tapasin-dependent, but also TAP/tapasin-independent pathways provides a possible explanation for its potency.
Strikingly, specific CTLs recognize PMEL209-217 even on target cells in which neither TAP nor tapasin provide support for MHC I loading (Fig. 5B/D/E, black bars). This is important, because various epitopes have been described that can bypass the need for TAP or tapasin individually but cannot overcome the lack of both factors simultaneously (77). In line with this, we show that the tapasin-binding mutant T134K is almost completely absent from the surface of TAP-deficient buf1280 cells (Fig. 5B/D/E, white bars), while HLA-A2-T134K efficiently reaches the plasma membrane in TAP-sufficient counterparts (Fig. 5D, white bars). In fact, surface levels of the T134K mutant in buf1280 cells are so drastically reduced that it is surprising that anything at all is presented in this situation (Fig. 5B/D/E, white bars). For this reason, we note that we included untransfected HLA-A2-negative buf1280 cells as unrecognized negative controls in all above experiments (Fig. 5B/D/E). Together this suggests that the vast majority of TAP-independent HLA-A2 epitopes is not presented without tapasin assistance, while the vast majority of tapasin-independent epitopes has no access to MHC I without TAP. PMEL209-217 appears to be exceptional in its ability to cause notable T cell recognition even in such an unfavorable environment. The behavior of HLA-A2 may at least in part reflect the interdependence of TAP and tapasin, because tapasin not only organizes the PLC and edits the peptide cargo loaded onto MHC I but also stabilizes and mediates TAP heterodimerization (12, 45, 63). Tumors frequently display coordinated downregulation of the antigen presentation machinery (21) or multiple independent defects in the MHC I pathway (38). Thus, simultaneous loss or downmodulation of TAP and tapasin – alongside other factors – is a scenario that is often encountered in cancer patients and should be taken into consideration for the design of future vaccines. We postulate that the success of PMEL209-217 as a vaccine is at least in part attributable to its high capacity to be presented not only via the classical, TAP/tapasin-dependent MHC I pathway, but also through non-classical routes.
Loss of IFNγ signaling is emerging as a major mechanism of resistance to modern checkpoint blockade immunotherapies (19, 20). Given that many components of the MHC I antigen processing machinery including TAP and tapasin are IFNγ-inducible, antigens exhibiting persistent presentation in TAPlow/tapasinlow cells may be particularly well suited for use as vaccines in combination with such therapies. In this context, it is disappointing that PMEL209-217 did not further improve the efficiency of anti-CTLA-4 treatment in a recent trial (48). The absence of activity in this particular study is baffling and unexplained at present and may reflect loss of antigen (96), lack of CD4+ T cell help (97), or choice of adjuvant (98, 99) among other factors. In principle, some of these potential pitfalls can be addressed by using multi-epitope vaccines and/or extended, longer peptides additionally containing MHC class II-epitopes. Moreover, higher efficacy of cancer vaccines might be achieved by changing the mode of delivery of the vaccine. Peptide and protein-based vaccines are often not optimal to induce strong CTL responses, while viral vector-based vaccines designed to express specific antigenic peptides can be highly immunogenic and induce strong CTL-mediated anti-tumor responses (100).
We believe PMEL209-217 remains a strong candidate for such therapies. Unlike fully personalized vaccines based exclusively on mutant neoepitopes (23, 101), PMEL expression is shared between a majority of patients (102), rendering the peptide a potential off-the-shelf drug with known reliable potency that might be added to personalized vaccines. Moreover, as an unmutated self-peptide, PMEL209-217 presentation is independent of the high mutagenicity caused, for instance, by mismatch repair deficiency that was recently shown to predict the response to PD-1 blockade therapy (103). In case of cancers that do not generate a lot of mutated neoepitopes, conserved peptides like PMEL209-217 may be the only available option.
Tumor cells may not only be equipped with proteases that can generate PMEL209-217 but may additionally express proteases that efficiently degrade the peptide, thereby limiting its presentation. In this context, we identify TPP2 as a cytosolic enzyme counteracting proteasome-mediated formation of the epitope. TPP2 inhibition (Fig. 4A/B/G and Suppl. Fig. S2E) or silencing (Fig. 4F/H) sharply increases CTL recognition, strongly suggesting that the protease degrades the peptide in the cytosol. Particularly, in a background mimicking a TAP/tapasin-defective environment the effect of TPP2 inhibition is dramatic (Fig. 5E, black bars), even though HLA-A2 surface levels remain minimal (Fig. 5E, white bars). Although TPP2 has been reported to productively generate a certain subset of epitopes (69, 70), its overall net effect on antigen presentation is believed to be negative (71, 72). Our findings are in line with this trend. Importantly, we also reveal a possible Achilles’ heel of PMEL209-217-mediated immunotherapy. Our results predict that upregulation of TPP2 in the tumor might substantially limit the presentation of PMEL209-217, potentially allowing the tumor to evade recognition by CTLs and continue to grow. In this context, it would be interesting to examine whether TPP2 levels are augmented in progressing metastases of PMEL209-217-treated patients. If this were to be the case and/or to impede basal TPP2 activity to boost the peptide’s presentation, blockade of TPP2 during PMEL209-217 therapy may be a strategy worth considering.
At present, we have little information about the nature of the non-classical pathway of PMEL209-217 presentation aside from the formation of the epitope in the cytosol (Fig. 2A-E) through the proteasome (Fig. 3A/B). As a charged molecule additionally containing several polar residues, the peptide is highly unlikely to passively cross the ER (or another cellular) membrane, as some strongly hydrophobic, TAP-independent antigens have been reported to do (77). Moreover, PMEL209-217 is not derived from a signal sequence (signal sequences are a common source for TAP-independent, HLA-A2-presented epitopes (39, 41, 43, 104)). Our observation that neutralization of endosomal pH vigorously boosts PMEL209-217 presentation (Fig. 6B) while inhibition of endocytic recycling blocks the process (Fig. 6C) suggests an involvement of endocytic compartments in the mechanism. Given our results, the most likely scenario is that some as yet uncharacterized process, likely not conventional macroautophagy (Suppl. Fig. S4A-F), imports cytosolic PMEL209-217 into the endosomal system where the peptide accesses recycling HLA-A2 leading to its subsequent presentation at the cell surface. Interestingly, the non-classical MHC I pathway employed by PMEL209-217 seems to possess a certain specificity with respect to the nature of its peptide ligands (Fig. 2F). Characterizing the associated machinery that drives the process and revealing its ligands may identify novel promising candidates for anti-tumor vaccines that share favorable features with PMEL209-217. Our current efforts are focused on this important goal.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. W. Yuan for providing the ICP47 construct in pLPCX, to Dr. S. Ferrone for providing melanoma cell lines, to Dr. D. Stepensky for providing M553 transfectants, and to Dr. M. Marks for providing the PMEL antibodies Pep13h and PMEL-N. PAQ-22 was a kind gift from Dr. Y. Hashimoto. The ATG5 shRNA construct was a kind gift from Dr. P. Agostinis. We thank Aline Depasse, Rui Cheng, and Susan Mitchell for excellent technical assistance.
This work was supported by a postdoctoral fellowship from the Cancer Research Institute (to RML) and by the NIH/NIAMS under award number R21-AR068518 (to RML). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study was further supported by a Marie Curie Outgoing International Fellowship (OIF) from the European Union (NV) and by grants from the Fonds National de la Recherche Scientifique (F.N.R.S), Belgium, from the Walloon excellence in life sciences and biotechnology (WELBIO), from the Fondation contre le Cancer, and the Fonds Maisin, Belgium (NV and BV). VF is supported by a fellowship from the Fonds National de la Recherche Scientifique, Belgium (FRIA grant No. 1.E091.14). The project received further support from the NIH/NIAID under award number R01-AI097206 (to PC) and through a Yale SPORE in Skin Cancer Grant 5P50 CA121974 (to PC).
Footnotes
DISCLOSURES
The authors declare no competing financial interests.
REFERENCES
- 1.Dimberu PM, and Leonhardt RM. 2011. Cancer immunotherapy takes a multi-faceted approach to kick the immune system into gear. The Yale journal of biology and medicine 84:371–380. [PMC free article] [PubMed] [Google Scholar]
- 2.Vigneron N 2015. Human Tumor Antigens and Cancer Immunotherapy. BioMed research international 2015:948501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, Jackson JR, and Shahabi V. 2012. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 61:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, and Ribas A. 2014. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vigneron N, Stroobant V, Van den Eynde BJ, and van der Bruggen P. 2013. Database of T cell-defined human tumor antigens: the 2013 update. Cancer immunity 13:15. [PMC free article] [PubMed] [Google Scholar]
- 6.Chapiro J, Claverol S, Piette F, Ma W, Stroobant V, Guillaume B, Gairin JE, Morel S, Burlet-Schiltz O, Monsarrat B, Boon T, and Van den Eynde BJ. 2006. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J Immunol 176:1053–1061. [DOI] [PubMed] [Google Scholar]
- 7.Guillaume B, Stroobant V, Bousquet-Dubouch MP, Colau D, Chapiro J, Parmentier N, Dalet A, and Van den Eynde BJ. 2012. Analysis of the processing of seven human tumor antigens by intermediate proteasomes. J Immunol 189:3538–3547. [DOI] [PubMed] [Google Scholar]
- 8.Saunders PM, and van Endert P. 2011. Running the gauntlet: from peptide generation to antigen presentation by MHC class I. Tissue antigens 78:161–170. [DOI] [PubMed] [Google Scholar]
- 9.Rufer E, Kagebein D, Leonhardt RM, and Knittler MR. 2015. Hydrophobic Interactions Are Key To Drive the Association of Tapasin with Peptide Transporter Subunit TAP2. J Immunol 195:5482–5494. [DOI] [PubMed] [Google Scholar]
- 10.Panter MS, Jain A, Leonhardt RM, Ha T, and Cresswell P. 2012. Dynamics of major histocompatibility complex class I association with the human peptide-loading complex. The Journal of biological chemistry 287:31172–31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rufer E, Leonhardt RM, and Knittler MR. 2007. Molecular architecture of the TAP-associated MHC class I peptide-loading complex. J Immunol 179:5717–5727. [DOI] [PubMed] [Google Scholar]
- 12.Williams AP, Peh CA, Purcell AW, McCluskey J, and Elliott T. 2002. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity 16:509–520. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Li L, Weimershaus M, Evnouchidou I, van Endert P, and Bouvier M. 2016. ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Scientific reports 6:28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, and van Endert PM. 2005. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nature immunology 6:689–697. [DOI] [PubMed] [Google Scholar]
- 15.Serwold T, Gonzalez F, Kim J, Jacob R, and Shastri N. 2002. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419:480–483. [DOI] [PubMed] [Google Scholar]
- 16.Howe C, Garstka M, Al-Balushi M, Ghanem E, Antoniou AN, Fritzsche S, Jankevicius G, Kontouli N, Schneeweiss C, Williams A, Elliott T, and Springer S. 2009. Calreticulin-dependent recycling in the early secretory pathway mediates optimal peptide loading of MHC class I molecules. The EMBO journal 28:3730–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonhardt RM, Fiegl D, Rufer E, Karger A, Bettin B, and Knittler MR. 2010. Post-endoplasmic reticulum rescue of unstable MHC class I requires proprotein convertase PC7. J Immunol 184:2985–2998. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Wearsch PA, Zhu Y, Leonhardt RM, and Cresswell P. 2011. A role for UDP-glucose glycoprotein glucosyltransferase in expression and quality control of MHC class I molecules. Proceedings of the National Academy of Sciences of the United States of America 108:4956–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, Chen PL, Hwu P, Allison JP, Futreal A, Wargo JA, and Sharma P. 2016. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 167:397–404 e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TN, Lo RS, and Ribas A. 2016. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. The New England journal of medicine 375:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero JM, Jimenez P, Cabrera T, Cozar JM, Pedrinaci S, Tallada M, Garrido F, and Ruiz-Cabello F. 2005. Coordinated downregulation of the antigen presentation machinery and HLA class I/beta2-microglobulin complex is responsible for HLA-ABC loss in bladder cancer. International journal of cancer 113:605–610. [DOI] [PubMed] [Google Scholar]
- 22.Restifo NP, Marincola FM, Kawakami Y, Taubenberger J, Yannelli JR, and Rosenberg SA. 1996. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. Journal of the National Cancer Institute 88:100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, Bukur V, Tadmor AD, Luxemburger U, Schrors B, Omokoko T, Vormehr M, Albrecht C, Paruzynski A, Kuhn AN, Buck J, Heesch S, Schreeb KH, Muller F, Ortseifer I, Vogler I, Godehardt E, Attig S, Rae R, Breitkreuz A, Tolliver C, Suchan M, Martic G, Hohberger A, Sorn P, Diekmann J, Ciesla J, Waksmann O, Bruck AK, Witt M, Zillgen M, Rothermel A, Kasemann B, Langer D, Bolte S, Diken M, Kreiter S, Nemecek R, Gebhardt C, Grabbe S, Holler C, Utikal J, Huber C, Loquai C, and Tureci O. 2017. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547:222–226. [DOI] [PubMed] [Google Scholar]
- 24.Cabrera CM, Jimenez P, Cabrera T, Esparza C, Ruiz-Cabello F, and Garrido F. 2003. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue antigens 61:211–219. [DOI] [PubMed] [Google Scholar]
- 25.Dissemond J, Gotte P, Mors J, Lindeke A, Goos M, Ferrone S, and Wagner SN. 2003. Association of TAP1 downregulation in human primary melanoma lesions with lack of spontaneous regression. Melanoma Res 13:253–258. [DOI] [PubMed] [Google Scholar]
- 26.Kageshita T, Hirai S, Ono T, Hicklin DJ, and Ferrone S. 1999. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. The American journal of pathology 154:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamarashev J, Ferrone S, Seifert B, Boni R, Nestle FO, Burg G, and Dummer R. 2001. TAP1 down-regulation in primary melanoma lesions: an independent marker of poor prognosis. International journal of cancer 95:23–28. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Hao C, Su P, and Shi J. 2009. Down-regulation of HLA class I antigen-processing machinery components in esophageal squamous cell carcinomas: association with disease progression. Scandinavian journal of gastroenterology 44:960–969. [DOI] [PubMed] [Google Scholar]
- 29.Meissner M, Reichert TE, Kunkel M, Gooding W, Whiteside TL, Ferrone S, and Seliger B. 2005. Defects in the human leukocyte antigen class I antigen processing machinery in head and neck squamous cell carcinoma: association with clinical outcome. Clin Cancer Res 11:2552–2560. [DOI] [PubMed] [Google Scholar]
- 30.Seliger B, Atkins D, Bock M, Ritz U, Ferrone S, Huber C, and Storkel S. 2003. Characterization of human lymphocyte antigen class I antigen-processing machinery defects in renal cell carcinoma lesions with special emphasis on transporter-associated with antigen-processing down-regulation. Clin Cancer Res 9:1721–1727. [PubMed] [Google Scholar]
- 31.Tao J, Li Y, Liu YQ, Li L, Liu J, Shen X, Shen GX, and Tu YT. 2008. Expression of transporters associated with antigen processing and human leucocyte antigen class I in malignant melanoma and its association with prognostic factors. The British journal of dermatology 158:88–94. [DOI] [PubMed] [Google Scholar]
- 32.Vitale M, Rezzani R, Rodella L, Zauli G, Grigolato P, Cadei M, Hicklin DJ, and Ferrone S. 1998. HLA class I antigen and transporter associated with antigen processing (TAP1 and TAP2) down-regulation in high-grade primary breast carcinoma lesions. Cancer research 58:737–742. [PubMed] [Google Scholar]
- 33.Jiang Q, Pan HY, Ye DX, Zhang P, Zhong LP, and Zhang ZY. 2010. Downregulation of tapasin expression in primary human oral squamous cell carcinoma: association with clinical outcome. Tumour Biol 31:451–459. [DOI] [PubMed] [Google Scholar]
- 34.Shionoya Y, Kanaseki T, Miyamoto S, Tokita S, Hongo A, Kikuchi Y, Kochin V, Watanabe K, Horibe R, Saijo H, Tsukahara T, Hirohashi Y, Takahashi H, Sato N, and Torigoe T. 2017. Loss of tapasin in human lung and colon cancer cells and escape from tumor-associated antigen-specific CTL recognition. Oncoimmunology 6:e1274476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokol L, Koelzer VH, Rau TT, Karamitopoulou E, Zlobec I, and Lugli A. 2015. Loss of tapasin correlates with diminished CD8(+) T-cell immunity and prognosis in colorectal cancer. Journal of translational medicine 13:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seliger B, Ritz U, Abele R, Bock M, Tampe R, Sutter G, Drexler I, Huber C, and Ferrone S. 2001. Immune escape of melanoma: first evidence of structural alterations in two distinct components of the MHC class I antigen processing pathway. Cancer research 61:8647–8650. [PubMed] [Google Scholar]
- 37.Belicha-Villanueva A, McEvoy S, Cycon K, Ferrone S, Gollnick SO, and Bangia N. 2008. Differential contribution of TAP and tapasin to HLA class I antigen expression. Immunology 124:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang CC, Pirozzi G, Wen SH, Chung IH, Chiu BL, Errico S, Luongo M, Lombardi ML, and Ferrone S. 2015. Multiple structural and epigenetic defects in the human leukocyte antigen class I antigen presentation pathway in a recurrent metastatic melanoma following immunotherapy. The Journal of biological chemistry 290:26562–26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durgeau A, El Hage F, Vergnon I, Validire P, de Montpreville V, Besse B, Soria JC, van Hall T, and Mami-Chouaib F. 2011. Different expression levels of the TAP peptide transporter lead to recognition of different antigenic peptides by tumor-specific CTL. J Immunol 187:5532–5539. [DOI] [PubMed] [Google Scholar]
- 40.van Hall T, Wolpert EZ, van Veelen P, Laban S, van der Veer M, Roseboom M, Bres S, Grufman P, de Ru A, Meiring H, de Jong A, Franken K, Teixeira A, Valentijn R, Drijfhout JW, Koning F, Camps M, Ossendorp F, Karre K, Ljunggren HG, Melief CJ, and Offringa R. 2006. Selective cytotoxic T-lymphocyte targeting of tumor immune escape variants. Nature medicine 12:417–424. [DOI] [PubMed] [Google Scholar]
- 41.Oliveira CC, and van Hall T. 2015. Alternative Antigen Processing for MHC Class I: Multiple Roads Lead to Rome. Frontiers in immunology 6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tykodi SS, Fujii N, Vigneron N, Lu SM, Mito JK, Miranda MX, Chou J, Voong LN, Thompson JA, Sandmaier BM, Cresswell P, Van den Eynde B, Riddell SR, and Warren EH. 2008. C19orf48 encodes a minor histocompatibility antigen recognized by CD8+ cytotoxic T cells from renal cell carcinoma patients. Clin Cancer Res 14:5260–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfel C, Drexler I, Van Pel A, Thres T, Leister N, Herr W, Sutter G, Huber C, and Wolfel T. 2000. Transporter (TAP)- and proteasome-independent presentation of a melanoma-associated tyrosinase epitope. International journal of cancer 88:432–438. [PubMed] [Google Scholar]
- 44.Stroobant V, Demotte N, Luiten RM, Leonhardt RM, Cresswell P, Bonehill A, Michaux A, Ma W, Mulder A, Van den Eynde BJ, van der Bruggen P, and Vigneron N. 2012. Inefficient exogenous loading of a tapasin-dependent peptide onto HLA-B*44:02 can be improved by acid treatment or fixation of target cells. European journal of immunology 42:1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vigneron N, Peaper DR, Leonhardt RM, and Cresswell P. 2009. Functional significance of tapasin membrane association and disulfide linkage to ERp57 in MHC class I presentation. European journal of immunology 39:2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith FO, Downey SG, Klapper JA, Yang JC, Sherry RM, Royal RE, Kammula US, Hughes MS, Restifo NP, Levy CL, White DE, Steinberg SM, and Rosenberg SA. 2008. Treatment of metastatic melanoma using interleukin-2 alone or in conjunction with vaccines. Clin Cancer Res 14:5610–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartzentruber DJ, Lawson DH, Richards JM, Conry RM, Miller DM, Treisman J, Gailani F, Riley L, Conlon K, Pockaj B, Kendra KL, White RL, Gonzalez R, Kuzel TM, Curti B, Leming PD, Whitman ED, Balkissoon J, Reintgen DS, Kaufman H, Marincola FM, Merino MJ, Rosenberg SA, Choyke P, Vena D, and Hwu P. 2011. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. The New England journal of medicine 364:2119–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbe C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, and Urba WJ. 2010. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine 363:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vigneron N, Ooms A, Morel S, Ma W, Degiovanni G, and Van den Eynde BJ. 2005. A peptide derived from melanocytic protein gp100 and presented by HLA-B35 is recognized by autologous cytolytic T lymphocytes on melanoma cells. Tissue antigens 65:156–162. [DOI] [PubMed] [Google Scholar]
- 50.Leonhardt RM, Vigneron N, Rahner C, Van den Eynde BJ, and Cresswell P. 2010. Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction. The Journal of biological chemistry 285:16166–16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stepensky D, Bangia N, and Cresswell P. 2007. Aggregate formation by ERp57-deficient MHC class I peptide-loading complexes. Traffic (Copenhagen, Denmark) 8:1530–1542. [DOI] [PubMed] [Google Scholar]
- 52.Ma W, Germeau C, Vigneron N, Maernoudt AS, Morel S, Boon T, Coulie PG, and Van den Eynde BJ. 2004. Two new tumor-specific antigenic peptides encoded by gene MAGE-C2 and presented to cytolytic T lymphocytes by HLA-A2. International journal of cancer 109:698–702. [DOI] [PubMed] [Google Scholar]
- 53.Salter RD, Howell DN, and Cresswell P. 1985. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235–246. [DOI] [PubMed] [Google Scholar]
- 54.Germeau C, Ma W, Schiavetti F, Lurquin C, Henry E, Vigneron N, Brasseur F, Lethe B, De Plaen E, Velu T, Boon T, and Coulie PG. 2005. High frequency of antitumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens. The Journal of experimental medicine 201:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfel T, Van Pel A, Brichard V, Schneider J, Seliger B, Meyer zum Buschenfelde KH, and Boon T. 1994. Two tyrosinase nonapeptides recognized on HLA-A2 melanomas by autologous cytolytic T lymphocytes. European journal of immunology 24:759–764. [DOI] [PubMed] [Google Scholar]
- 56.Stam NJ, Vroom TM, Peters PJ, Pastoors EB, and Ploegh HL. 1990. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. International immunology 2:113–125. [DOI] [PubMed] [Google Scholar]
- 57.Parham P, and Brodsky FM. 1981. Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Human immunology 3:277–299. [DOI] [PubMed] [Google Scholar]
- 58.Meyer TH, van Endert PM, Uebel S, Ehring B, and Tampe R. 1994. Functional expression and purification of the ABC transporter complex associated with antigen processing (TAP) in insect cells. FEBS letters 351:443–447. [DOI] [PubMed] [Google Scholar]
- 59.Berson JF, Harper DC, Tenza D, Raposo G, and Marks MS. 2001. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Molecular biology of the cell 12:3451–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leonhardt RM, Vigneron N, Hee JS, Graham M, and Cresswell P. 2013. Critical residues in the PMEL/Pmel17 N-terminus direct the hierarchical assembly of melanosomal fibrils. Molecular biology of the cell 24:964–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ehses S, Leonhardt RM, Hansen G, and Knittler MR. 2005. Functional role of C-terminal sequence elements in the transporter associated with antigen processing. J Immunol 174:328–339. [DOI] [PubMed] [Google Scholar]
- 62.Keusekotten K, Leonhardt RM, Ehses S, and Knittler MR. 2006. Biogenesis of functional antigenic peptide transporter TAP requires assembly of pre-existing TAP1 with newly synthesized TAP2. The Journal of biological chemistry 281:17545–17551. [DOI] [PubMed] [Google Scholar]
- 63.Leonhardt RM, Abrahimi P, Mitchell SM, and Cresswell P. 2014. Three tapasin docking sites in TAP cooperate to facilitate transporter stabilization and heterodimerization. J Immunol 192:2480–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parkhurst MR, Salgaller ML, Southwood S, Robbins PF, Sette A, Rosenberg SA, and Kawakami Y. 1996. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol 157:2539–2548. [PubMed] [Google Scholar]
- 65.Yu Z, Theoret MR, Touloukian CE, Surman DR, Garman SC, Feigenbaum L, Baxter TK, Baker BM, and Restifo NP. 2004. Poor immunogenicity of a self/tumor antigen derives from peptide-MHC-I instability and is independent of tolerance. The Journal of clinical investigation 114:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leonhardt RM, Vigneron N, Rahner C, and Cresswell P. 2011. Proprotein convertases process Pmel17 during secretion. The Journal of biological chemistry 286:9321–9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hee JS, Mitchell SM, Liu X, and Leonhardt RM. 2017. Melanosomal formation of PMEL core amyloid is driven by aromatic residues. Scientific reports 7:44064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, and Johnson D. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411–415. [DOI] [PubMed] [Google Scholar]
- 69.Guil S, Rodriguez-Castro M, Aguilar F, Villasevil EM, Anton LC, and Del Val M. 2006. Need for tripeptidyl-peptidase II in major histocompatibility complex class I viral antigen processing when proteasomes are detrimental. The Journal of biological chemistry 281:39925–39934. [DOI] [PubMed] [Google Scholar]
- 70.Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, Weinschenk T, Schild H, Laderach D, Galy A, Haas G, Kloetzel PM, Reiss Y, and Hosmalin A. 2003. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nature immunology 4:375–379. [DOI] [PubMed] [Google Scholar]
- 71.Firat E, Huai J, Saveanu L, Gaedicke S, Aichele P, Eichmann K, van Endert P, and Niedermann G. 2007. Analysis of direct and cross-presentation of antigens in TPPII knockout mice. J Immunol 179:8137–8145. [DOI] [PubMed] [Google Scholar]
- 72.Kawahara M, York IA, Hearn A, Farfan D, and Rock KL. 2009. Analysis of the role of tripeptidyl peptidase II in MHC class I antigen presentation in vivo. J Immunol 183:6069–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stoltze L, Schirle M, Schwarz G, Schroter C, Thompson MW, Hersh LB, Kalbacher H, Stevanovic S, Rammensee HG, and Schild H. 2000. Two new proteases in the MHC class I processing pathway. Nature immunology 1:413–418. [DOI] [PubMed] [Google Scholar]
- 74.Vines D, and Warburton MJ. 1998. Purification and characterisation of a tripeptidyl aminopeptidase I from rat spleen. Biochimica et biophysica acta 1384:233–242. [DOI] [PubMed] [Google Scholar]
- 75.Kakuta H, Tanatani A, Nagasawa K, and Hashimoto Y. 2003. Specific nonpeptide inhibitors of puromycin-sensitive aminopeptidase with a 2,4(1H,3H)-quinazolinedione skeleton. Chemical & pharmaceutical bulletin 51:1273–1282. [DOI] [PubMed] [Google Scholar]
- 76.Rose C, Vargas F, Facchinetti P, Bourgeat P, Bambal RB, Bishop PB, Chan SM, Moore AN, Ganellin CR, and Schwartz JC. 1996. Characterization and inhibition of a cholecystokinin-inactivating serine peptidase. Nature 380:403–409. [DOI] [PubMed] [Google Scholar]
- 77.Aladin F, Lautscham G, Humphries E, Coulson J, and Blake N. 2007. Targeting tumour cells with defects in the MHC Class I antigen processing pathway with CD8+ T cells specific for hydrophobic TAP- and Tapasin-independent peptides: the requirement for directed access into the ER. Cancer Immunol Immunother 56:1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hermann C, Strittmatter LM, Deane JE, and Boyle LH. 2013. The binding of TAPBPR and Tapasin to MHC class I is mutually exclusive. J Immunol 191:5743–5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson DR, and Mook-Kanamori B. 2000. Dependence of elevated human leukocyte antigen class I molecule expression on increased heavy chain, light chain (beta 2-microglobulin), transporter associated with antigen processing, tapasin, and peptide. The Journal of biological chemistry 275:16643–16649. [DOI] [PubMed] [Google Scholar]
- 80.Lewis JW, and Elliott T. 1998. Evidence for successive peptide binding and quality control stages during MHC class I assembly. Curr Biol 8:717–720. [DOI] [PubMed] [Google Scholar]
- 81.Lewis JW, Neisig A, Neefjes J, and Elliott T. 1996. Point mutations in the alpha 2 domain of HLA-A2.1 define a functionally relevant interaction with TAP. Curr Biol 6:873–883. [DOI] [PubMed] [Google Scholar]
- 82.Matsui M, Ikeda M, and Akatsuka T. 2001. High expression of HLA-A2 on an oral squamous cell carcinoma with down-regulated transporter for antigen presentation. Biochemical and biophysical research communications 280:1008–1014. [DOI] [PubMed] [Google Scholar]
- 83.Young NT, Mulder A, Cerundolo V, Claas FH, and Welsh KI. 1998. Expression of HLA class I antigens in transporter associated with antigen processing (TAP)-deficient mutant cell lines. Tissue antigens 52:368–373. [DOI] [PubMed] [Google Scholar]
- 84.Carvalho P, Stanley AM, and Rapoport TA. 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143:579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lilley BN, and Ploegh HL. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429:834–840. [DOI] [PubMed] [Google Scholar]
- 86.Mehnert M, Sommer T, and Jarosch E. 2014. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nature cell biology 16:77–86. [DOI] [PubMed] [Google Scholar]
- 87.Kalies KU, Rapoport TA, and Hartmann E. 1998. The beta subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. The Journal of cell biology 141:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tey SK, and Khanna R. 2012. Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway. Blood 120:994–1004. [DOI] [PubMed] [Google Scholar]
- 89.Oliveira CC, Sluijter M, Querido B, Ossendorp F, van der Burg SH, and van Hall T. 2014. Dominant contribution of the proteasome and metalloproteinases to TAP-independent MHC-I peptide repertoire. Molecular immunology 62:129–136. [DOI] [PubMed] [Google Scholar]
- 90.Zollmann T, Moiset G, Tumulka F, Tampe R, Poolman B, and Abele R. 2015. Single liposome analysis of peptide translocation by the ABC transporter TAPL. Proceedings of the National Academy of Sciences of the United States of America 112:2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, and Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. [DOI] [PubMed] [Google Scholar]
- 92.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, and Santambrogio L. 2011. Microautophagy of cytosolic proteins by late endosomes. Developmental cell 20:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orenstein SJ, and Cuervo AM. 2010. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Seminars in cell & developmental biology 21:719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins PF, Sette A, Appella E, and Rosenberg SA. 1995. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol 154:3961–3968. [PubMed] [Google Scholar]
- 95.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, Wunderlich JR, Parkhurst MR, Kawakami Y, Seipp CA, Einhorn JH, and White DE. 1998. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nature medicine 4:321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee KH, Panelli MC, Kim CJ, Riker AI, Bettinotti MP, Roden MM, Fetsch P, Abati A, Rosenberg SA, and Marincola FM. 1998. Functional dissociation between local and systemic immune response during anti-melanoma peptide vaccination. J Immunol 161:4183–4194. [PubMed] [Google Scholar]
- 97.Bijker MS, van den Eeden SJ, Franken KL, Melief CJ, Offringa R, and van der Burg SH. 2007. CD8+ CTL priming by exact peptide epitopes in incomplete Freund’s adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J Immunol 179:5033–5040. [DOI] [PubMed] [Google Scholar]
- 98.Rosenberg SA, Yang JC, Kammula US, Hughes MS, Restifo NP, Schwarz SL, Morton KE, Laurencot CM, and Sherry RM. 2010. Different adjuvanticity of incomplete freund’s adjuvant derived from beef or vegetable components in melanoma patients immunized with a peptide vaccine. J Immunother 33:626–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hailemichael Y, Woods A, Fu T, He Q, Nielsen MC, Hasan F, Roszik J, Xiao Z, Vianden C, Khong H, Singh M, Sharma M, Faak F, Moore D, Dai Z, Anthony SM, Schluns KS, Sharma P, Engelhard VH, and Overwijk WW. 2018. Cancer vaccine formulation dictates synergy with CTLA-4 and PD-L1 checkpoint blockade therapy. The Journal of clinical investigation 128:1338–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Naslund TI, Uyttenhove C, Nordstrom EK, Colau D, Warnier G, Jondal M, Van den Eynde BJ, and Liljestrom P. 2007. Comparative prime-boost vaccinations using Semliki Forest virus, adenovirus, and ALVAC vectors demonstrate differences in the generation of a protective central memory CTL response against the P815 tumor. J Immunol 178:6761–6769. [DOI] [PubMed] [Google Scholar]
- 101.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, Chen C, Olive O, Carter TA, Li S, Lieb DJ, Eisenhaure T, Gjini E, Stevens J, Lane WJ, Javeri I, Nellaiappan K, Salazar AM, Daley H, Seaman M, Buchbinder EI, Yoon CH, Harden M, Lennon N, Gabriel S, Rodig SJ, Barouch DH, Aster JC, Getz G, Wucherpfennig K, Neuberg D, Ritz J, Lander ES, Fritsch EF, Hacohen N, and Wu CJ. 2017. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boon T, Coulie PG, Van den Eynde BJ, and van der Bruggen P. 2006. Human T cell responses against melanoma. Annual review of immunology 24:175–208. [DOI] [PubMed] [Google Scholar]
- 103.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, and Diaz LA Jr. 2017. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (New York, N.Y 357:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weinzierl AO, Rudolf D, Hillen N, Tenzer S, van Endert P, Schild H, Rammensee HG, and Stevanovic S. 2008. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. European journal of immunology 38:1503–1510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.