VPS4 constricts ESCRT-III CHMP2A-CHMP3.
Abstract
Many cellular processes such as endosomal vesicle budding, virus budding, and cytokinesis require extensive membrane remodeling by the endosomal sorting complex required for transport III (ESCRT-III). ESCRT-III protein family members form spirals with variable diameters in vitro and in vivo inside tubular membrane structures, which need to be constricted to proceed to membrane fission. Here, we show, using high-speed atomic force microscopy and electron microscopy, that the AAA-type adenosine triphosphatase VPS4 constricts and cleaves ESCRT-III CHMP2A-CHMP3 helical filaments in vitro. Constriction starts asymmetrically and progressively decreases the diameter of CHMP2A-CHMP3 tubular structure, thereby coiling up the CHMP2A-CHMP3 filaments into dome-like end caps. Our results demonstrate that VPS4 actively constricts ESCRT-III filaments and cleaves them before their complete disassembly. We propose that the formation of ESCRT-III dome-like end caps by VPS4 within a membrane neck structure constricts the membrane to set the stage for membrane fission.
INTRODUCTION
The endosomal sorting complex required for transport (ESCRT) machinery catalyzes a wide range of membrane remodeling processes. ESCRTs are required for vesicle formation at endosomes, budding of some enveloped viruses, cytokinesis, nuclear envelope reformation, plasma membrane repair, exosome formation, and axon pruning (1–8). Common to all processes is the recruitment of ESCRT-III and the AAA-type adenosine triphosphatase (ATPase) VPS4 (vacuolar protein sorting 4), which have been suggested to constrict membrane necks to the point of membrane fission. However, how constriction of membrane necks is achieved by ESCRT-III is yet unknown.
Higher eukaryotes express 12 ESCRT-III proteins named CHMP1 (charged multivesicular body protein 1) to CHMP7 and IST1 (increased sodium tolerance 1 gene product) (3). Yeast ESCRT-III (Vps20, Snf7, Vps24, and Vps2) and their human homologs (CHMP6, CHMP4A or CHMP4B or CHMP4C, CHMP3 and CHMP2A or CHMP2B) are recruited sequentially in the indicated order to the membrane (9, 10). Once CHMP4 is recruited, it provides a platform for the recruitment of CHMP3 and CHMP2 (11), two ESCRT-III members that cap ESCRT-III assembly before recycling (9).
ESCRT-III proteins adopt a closed autoinhibited conformation in the cytosol (12–14), and release of autoinhibition (13, 15, 16) leads to polymerization into homomeric (17–21) or heteromeric spirals in vitro (13, 22–24) and in vivo (18, 25–28). Current understanding is that ESCRT-III spirals assemble within membrane neck structures, although spirals have yet been only imaged at the cytokinetic midbody under physiological conditions (28–30).
An early on-established role for the hexameric VPS4 ATPase in the regulation of ESCRT-III polymers is their disassembly, thereby recycling ESCRT-III proteins (31). VPS4 uses its “microtubule interacting and trafficking” (MIT) domain to interact with ESCRT-III proteins containing a C-terminal “MIT interaction motif” (MIM) (32, 33). Upon disassembly, ESCRT-III protein substrate passes via the central pore of dynamic VPS4 hexameric assemblies driven by adenosine 5′-triphosphate (ATP) hydrolysis (34). Efficient and fast VPS4-catalyzed ESCRT-III disassembly in vitro has been confirmed on CHMP3 filaments (35) and CHMP2A-CHMP3 polymers (22, 36). Recently, the role of VPS4 has been extended to promote the growth and turnover of ESCRT-III assemblies in vitro on flat membranes (29), consistent with a dynamic recruitment of ESCRT-III to endosomal membranes (37). The residence time of ESCRT-III on cellular membranes is rather short and complemented by a short transient recruitment of several VPS4 complexes required for completion of budding and cytokinesis (37–41).
CHMP2A and CHMP3 are engaged late in ESCRT-III assembly, recruit VPS4 (31, 42), and block Snf7 (CHMP4) polymerization (9, 42). Although only one member of CHMP4 and CHMP2 is essential for HIV-1 release (11), CHMP3 exerts a substantial synergistic effect on CHMP2A in HIV-1 budding (43). This is consistent with a strong dominant negative effect of truncated CHMP3 on HIV-1 budding (12), further suggesting an interdependence of CHMP2A and CHMP3. In vitro, CHMP2A and CHMP3 form helical tubular structures that bind VPS4 on the inside and membrane on the outside (22), with average diameters in the range of 47 to 51 nm (43). Few of the in vitro assembled CHMP2A-CHMP3 tubes have the filament of one tube end coiled up to form a dome-like structure (22). Because of the late final engagement of this complex, it has been suggested that such a structure could induce membrane neck constriction to a few nanometers, which would provide a favorable setting for membrane fission (44, 45), although other models have been suggested (46).
However, since it is not clear whether a dome-like end cap structure can form spontaneously in vivo at all and whether its formation is regulated, the central question that remained is how ESCRT-III polymers are constricted. Thus, we set out to analyze how CHMP2A-CHMP3 filamentous tubes might be constricted from 50 nm to a few nanometers to guide the membrane remodeling process toward membrane fission. Our results show that VPS4 actively remodels ESCRT-III CHMP2A-CHMP3 filaments, leading to constriction, cleavage, and the generation of dome-like end caps. We propose that the generation of these structures within a bud neck in vivo primes ESCRT-III for membrane fission.
RESULTS
Effect of VPS4 on CHMP2A-CHMP3 tubular polymers observed by high-speed atomic force microscopy
CHMP2A-CHMP3 tubes were assembled as described (22) and bound to a lipid bilayer composed of 60% dioleoylphosphatidylcholine (DOPC) and 40% dioleoylphosphatidylserine (DOPS). The CHMP2A-CHMP3 tubes were then imaged over a period of ~15 min using high-speed atomic force microscopy (HS-AFM; Fig. 1A and movie S1) (47). The maximal height of these tubes was 50 nm (Fig. 1A), and their surface appeared smooth in agreement with previous electron microscopy (EM) analysis (43). Next, we tested the effect of VPS4B on the tube without ATP. Upon addition of 10 μM VPS4B, the tube undergoes very small, local reductions of the tube radius (Fig. 1B and movie S2). The line profile in the last panel of Fig. 1B indicates multiple reductions of at least 3 nm in tube radius. No further changes in the tube surface topography were observed, not even after an extended imaging time (fig. S1).
Fig. 1. Local deformation of VPS4B-treated CHMP2A-CHMP3 tubes and disassembly upon ATP hydrolysis.
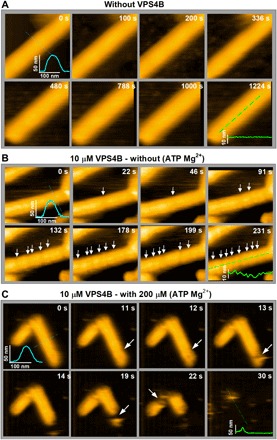
(A) Clips of HS-AFM images of CHMP2A-CHMP3 tubes captured at 0.5 frame/s in the absence of VPS4B, showing no significant change in roughness of the tube surface. The inset represents the cross section (axial or radial) along the corresponding lines. (B) Clips of HS-AFM images captured at 1 frame/s, showing that the 10 μM VPS4-treated CHMP2A-CHMP3 tubes undergo discrete reductions of the tube radius (indicated with white arrows) without further structural changes in the absence of ATP Mg2+. The axial cross section at 231 s shows a clear reduction of at least 3 nm in the tube radius. Because of tip convolution, the exact value is likely higher than the observed result. (C) Clips of HS-AFM images captured at 1 frame/s from 10 μM VPS4-treated CHMP2A-CHMP3 tubes in presence of 200 μM ATP Mg2+. Upon ATP hydrolysis, CHMP2A-CHMP3 tubes disassembled within less than 1 min. White arrows show disassembly sites. Average tube height is 48 ± 3 nm (n = 30). Scale bars, 100 nm.
Constriction and disassembly of CHMP2A-CHMP3 tubular polymers in the presence of VPS4 and ATP observed by HS-AFM
Adding ATP and Mg2+ to the sample led to rapid cleavage (completion of constriction) and disassembly of the CHMP2A-CHMP3 tube, as could be observed by imaging at 1 frame/s (Fig. 1C). A first cleavage occurs at a single position after 13 s, leading to partial tube disassembly at 14 s. A second cleavage takes place at approximately 19 s, which is followed by other cleavage and disassembly events and completion of tube disassembly at 31 s (Fig. 1C and movie S3). This behavior is in agreement with the rapid disassembly of CHMP2A-CHMP3 tubes revealed by a fluorescent-based assay in vitro (36). Reducing the VPS4B concentration to 5 μM slows down the process and reveals local constriction of the CHMP2A-CHMP3 tubes before cleavage and disassembly. Constriction typically starts asymmetrically, as shown in Fig. 2A (movie S4). Figure 2B depicts kymographs along the long axis of the tubes (green lines in Fig. 2A), representing the progress of constrictions over time. The height profiles at the constriction points over time (Fig. 2C) show a reduction of the CHMP2A-CHMP3 tube diameter from 45 to <10 nm with an average constriction time of 87 ± 20 s (fig. S2). Constriction was not due to the forces applied by the AFM tip, as control experiments without VPS4B or with VPS4 without ATP did not lead to constriction (Fig. 1, A and B, fig. S1, and movies S1 and S2).
Fig. 2. In presence of ATP Mg2+, VPS4B mediates local constriction (asymmetric) of the CHMP2A-CHMP3 tubes before disassembly.
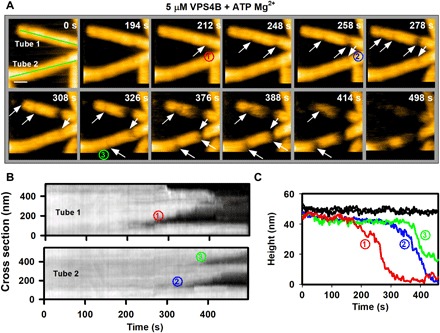
(A) Clips of HS-AFM images captured at 0.5 frame/s, showing that the 5 μM VPS4B-treated CHMP2A-CHMP3 tubes undergo local constriction before disassembly. The constriction normally appears asymmetric (white arrows), i.e., the deformation starts from one side of the tube. Numbers in circles identify three constriction sites in the two tubes. Scale bar, 100 nm. (B) Kymographs of both tubes [green lines in (A)]. The progress of the constrictions (numbers in circles) is identifiable from the kymograph. (C) The height profiles of the tubes in (A) at nonconstricted site (in black curves) and at the constriction site over time derived from (A) and (B). Each color code is similar to the constriction sites (numbers in circles) in (A) and (B).
To further corroborate that ATP hydrolysis and the correct positioning of the VPS4B MIT domain are required for CHMP2A-CHMP3 tube remodeling, we performed experiments with two different VPS4B mutants, which were inactive in a CHMP2A-CHMP3 tube disassembly assay (36). The catalytic site of the E235Q mutant is deficient in ATP hydrolysis, and the I124E mutation affects the conformation of the linker that connects the MIT domain to the ATPase domains but retains ATPase activity. HS-AFM imaging using 10 μM VPS4B E235Q and ATP revealed local deformations along the tube, as observed for VPS4B without ATP. This indicates the proper assembly of mutant VPS4B hexamers upon binding to ESCRT-III, which, however, does not lead to constriction or cleavage despite the presence of ATP (fig. S3 and movie S5). VPS4B I124E plus ATP does not even show local tube deformation nor constriction upon incubation of CHMP2A-CHMP3 tubes (fig. S3 and movie S6), suggesting that the linker conformation is essential for the functional assembly of VPS4B hexamers on ESCRT-III filaments. This thus corroborates that constriction and cleavage of CHMP2A-CHMP3 tubes by VPS4 are ATP dependent and require an optimal positioning of the VPS4B hexamer. The latter depends on the linker conformation and not only on the MIT domain interaction with ESCRT-III, which is consistent with the role of the linker region in VPS4 assembly and its activity (48).
The kinetics of ESCRT-III CHMP2A-CHMP3 remodeling depends on the VPS4 concentration
We next compared the effect of remodeling at decreasing VPS4B concentrations. As shown above, 10 μM VPS4B induces rapid cleavage and almost complete degradation within 20 s (Figs. 1C and 3A and movies S3 and S7). The kymograph in Fig. 3B corroborates how constriction and disassembly progress over the course of the experiment (see Materials and Methods). At a concentration of 5 μM VPS4B, cleavage and disassembly occurred significantly later, at time scales of hundreds of seconds (Fig. 3, C and D, and movies S4 and S8). Under 3 μM VPS4B conditions (Fig. 3E and movie S9), remodeling is further slowed down as indicated by a complete constriction at ~500 s (see kymograph in Fig. 3F) but without subsequent disassembly. One micromolar VPS4B induces only some minor asymmetric constriction after a long incubation (>2000 s) time without completion of constriction (Fig. 3, G and H, and movie S10). Furthermore, we observed that cleavage produces structures resembling dome-like end caps or straight ends (fig. S4). The probability of having these shapes varies over VPS4B concentration. These results indicate that the local concentration of VPS4B is crucial for the assembly of functional hexameric complexes. Achieving the critical concentration for hexamer assembly is likely limited by the necessity of VPS4B to diffuse into the interior of the tube. Figure 3I quantifies how the kinetics of CHMP2A-CHMP3 tube remodeling is dependent on the VPS4B concentration and shows that the time required for full constriction is significantly reduced at high VPS4B concentrations.
Fig. 3. Effect of VPS4 concentration on CHMP2A-CHMP3 tube remodeling in presence of ATP Mg2+.
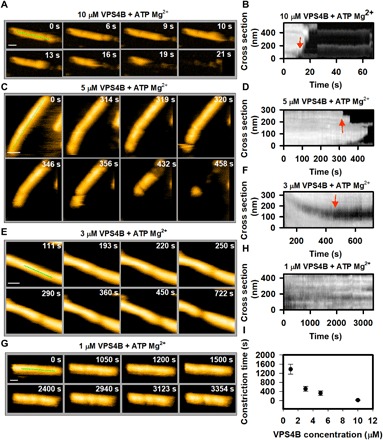
(A) Clips of HS-AFM images captured at 1 frame/s, showing a rapid disassembly of tube, treated with 10 μM VPS4B. (B) Kymograph taken from (A), showing the time and position of first constriction and/or disassembly (marked by red arrow). (C) Same as (A) but for 5 μM VPS4B-treated tubes. Frame rate, 1 frame/s. (D) Same as (B) but derived from (C). (E and F) Same as (C) and (D), respectively, but for 3 μM VPS4B-treated tubes. One can see that, at this concentration, the tube goes through constriction but not disassembly. (G and H) Same as (C) and (D), respectively, but for 1 μM VPS4B-treated tubes. Images in (G) show that at, 1 μM VPS4B concentration (with ATP Mg2+), the tube undergoes a partial constriction after a significant amount of time. Frame rate, 0.33 frame/s. (I) Effect of VPS4B concentration on the cleavage of CHMP2A-CHMP3 tubes. The cleavage time is considered from the moment of addition of 200 μM ATP Mg2+ until the first cleavage occurs (red arrows in the kymograph). For four different concentrations of VPS4B, more than 20 tubes were investigated. Scale bars, 100 nm.
EM analyses of CHMP2A-CHMP3 tube remodeling by VPS4
We next confirmed CHMP2A-CHMP3 tube remodeling by EM analyses. Untreated CHMP2A-CHMP3 generally forms straight tubes (fig. S5) with variable diameters ranging from 47 to 51 nm formed by one-start or multistart helices (43). Consistent with HS-AFM imaging (Fig. 1B), the presence of VPS4B inside the CHMP2A-CHMP3 tube was confirmed by cryo-EM, revealing extra density inside the tube in the presence of VPS4 (fig. S6). Furthermore, the presence of VPS4 does not induce major morphological changes (fig. S6), although small local reductions in diameter can be observed by HS-AFM imaging (Fig. 1B, fig. S1, and movie S2). On the basis of the HS-AFM imaging results, we used 5 μM VPS4B plus ATP and Mg2+ at various incubation times to image remodeling by negative-staining EM. This confirmed that constriction often starts asymmetrically (Fig. 4A and fig. S7). Complete constriction leads to the generation of at least one dome-like end cap (Fig. 4B), a structure that was shown to catalyze membrane constriction in silico (44). CHMP2A-CHMP3 tube constriction was further confirmed by cryo-EM, which revealed early stages of remodeling (Fig. 4C), asymmetric cleavage start sites (Fig. 4D), and the generation of dome-like end caps upon tube constriction (Fig. 4E). These results show that VPS4 remodels CHMP2A-CHMP3 tubes, constricts them, and cleaves them, thereby generating at least one dome-like end cap structure.
Fig. 4. EM imaging of CHMP2A-CHMP3 tube remodeling by VPS4B.
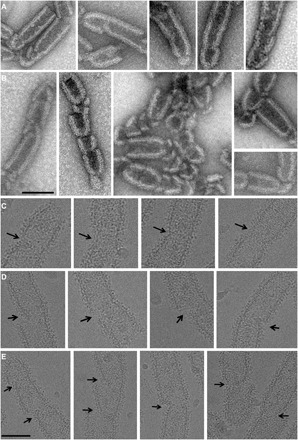
(A and B) Negative-stain images of CHMP2A-CHMP3 tubes incubated with 5 μM VPS4B and 200 μM ATP Mg2+, indicating (A) start sites of cleavage and (B) the generation of dome-like end caps. Scale bar, 100 nm. (C to E) Cryo-EM images of CHMP2A-CHMP3 tubes incubated with 5 μM VPS4B and 200 μM ATP Mg2+. (C) Images of early stages of constriction sites, (D) asymmetric constriction start sites, and (E) the generation of dome-like end caps. Scale bar, 100 nm.
DISCUSSION
ESCRT-III has been proposed to assemble inside the membrane neck formed at a late stage of a budding vesicle or an enveloped virus. The membrane neck needs to be constricted to proceed to membrane fission, thereby splitting the vesicle or virus from the cellular membrane (1, 11). In case of HIV-1 budding, it has been shown that ESCRT-III proteins are recruited once Gag assembly is completed, leading to the transient appearance of ESCRT-III and VPS4 at the bud neck (38, 39, 41). Super-resolution imaging revealed an average cluster size of 56 ± 12 nm for CHMP2A structures colocalizing with HIV-1 buds (40), indicating bud neck diameters of a similar range. Upon CHMP4 assembly at the budding site, which might itself already induce constriction of the neck, CHMP3 and CHMP2A are recruited during HIV-1 budding (11, 41). The preferred filament diameter of 47 to 51 nm observed in vitro (43) would thus fit well with the range of neck diameters produced upon HIV-1 Gag assembly. Here, we show that VPS4 can constrict and cleave CHMP2A-CHMP3 helical filaments, producing dome-like end cap structures. On the basis of these results, we propose a model where CHMP2A-CHMP3 filaments assemble in a bud neck (Fig. 5A) and VPS4 ATP catalysis drives constriction from a diameter of ~50 nm to a few nanometers. Because CHMP2A-CHMP3 filament formation is likely directional, the positioning of the VPS4 hexamer within the tubular structure is important, thereby producing either clockwise (Fig. 5B) or anticlockwise rotational movements (Fig. 5, B and C) to thread substrate via its central pore (34). Correct positioning of VPS4B is further corroborated by the inactivity of a VPS4B I124E linker mutant that retains some ATPase activity but fails to constrict, cleave, and disassemble CHMP2A-CHMP3 tubes. The importance of positioning of VPS4B on ESCRT-III filaments is consistent with the observation that constriction starts asymmetrically and progresses to symmetric constriction (Fig. 5, B and C). In this scenario, the successive decrease of the CHMP2A-CHMP3 filament diameter is likely achieved by removal of individual CHMP molecules, thereby curling up the filament, cleaving it, and generating a dome-like end cap structure (Fig. 5B). These structures have been proposed to be able to constrict a membrane tube to 6 nm in silico (44), thereby setting the stage for membrane fission (Fig. 5B). Generation of one dome-like end cap on the cytoplasmic side of a budding virus could suffice to induce membrane constriction (Fig. 5B). Alternatively, two VPS4 complexes are recruited to the fission site, one acting on the cytoplasmic side and one “inside” the virion (Fig. 5C), consistent with the detection of small amounts of VPS4 and ESCRT-III proteins inside mature HIV-1 virions by proteomics analyses (49). Furthermore, super-resolution imaging proposed that ESCRT-III complexes might act from within a virus bud (50). Because cellular imaging predicts the presence of two or more VPS4 complexes at budding sites (37, 39), VPS4 dodecamer formation, which is dispensable for its enzymatic activity (51), might serve to position two side-by-side acting VPS4 complexes constricting the membrane from two starting points. We propose that this might occur via one VPS4 hexamer rotating and shrinking clockwise and another one in an anticlockwise direction during constriction (Fig. 5C).
Fig. 5. Model of membrane constriction induced by remodeling of CHMP2A-CHMP3 filaments.
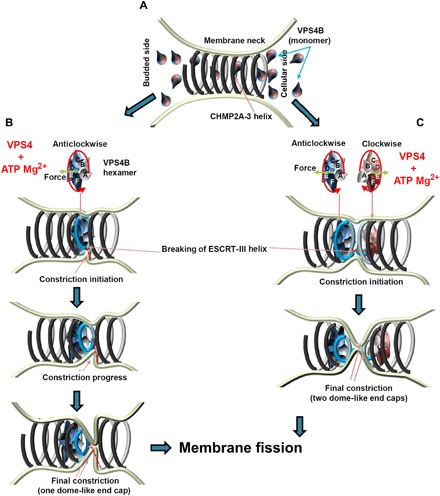
(A) CHMP2A-CHMP3 helical filaments assemble within a membrane neck structure such as a vesicle or virus bud or at the midbody. Currently, we do not know how many turns assemble in vivo. (B) VPS4 forms an asymmetric hexamer structure in the presence of ATP and Mg2+. This structure needs to assemble on functional ESCRT-III filaments, and ATP-driven rotation threads the substrate via its central pore. Because CHMP2A-CHMP3 polymer assembly is directional, the assembly of VPS4 acting clockwise or anticlockwise is likely to be important. The assembly of one VPS4 complex might be sufficient to induce CHMP2A-CHMP3 constriction, which often starts asymmetrically (middle). This can lead to membrane constriction and cleavage of the CHMP2A-CHMP3 filament. (C) Alternatively, two adjacent VPS4B complexes remodel CHMP2A-CHMP3 filaments acting clockwise and anticlockwise, thereby leading to the generation of two dome-like end caps and cleavage of the CHMP2A-CHMP3 filament. In both scenarios, constriction of CHMP2A-CHMP3 could prime the site for fission, and cleavage of the filament might play an important role in tension release as proposed (53).
Our in vitro system does not directly yield kinetic parameters on constriction, as we cannot determine the exact VPS4B concentration at the site of constriction. This is due to the fact that VPS4 needs to diffuse into the lumen of the CHMP2A-CHMP3 tubular structure, thereby encountering a multitude of low-affinity MIM-binding sites (32, 33). Nevertheless, we find that the kinetics of constriction and cleavage depends on the VPS4 concentration. Our recorded time scales for single-constriction events observed between 100 and 200 s are consistent with the short residence time of VPS4 at HIV-1 budding sites (38, 41). The fast constriction kinetics observed at 10 μM VPS4 indicates that a high local concentration of VPS4 favors disassembly, probably by efficient recycling of VPS4 protomers during the conveyor belt–like action of hexameric VPS4 during ESCRT-III protein translocation via the central pore (34). Following constriction and filament cleavage, CHMP2A-CHMP3 filaments are completely disassembled, consistent with recycling (31) and their disassembly at HIV-1 budding sites (41).
Although VPS4 remodeling of ESCRT-III filaments assembled on flat membranes has been reported previously, our results provide two important observations that distinguish them from the results obtained by Mierzwa et al. (29). First, remodeling on flat ESCRT-III polymers starts only from the outer circles, the end of the filament (29), while in case of the CHMP2A-CHMP3 tubular structures, filament constriction can start from within a filament. Second, flat filaments are not cleaved by VPS4 (29), while the CHMP2A-CHMP3 filaments are efficiently cleaved by VPS4B during the constriction process. Cleavage of ESCRT-III has been suggested to induce tension release required for constriction (52, 53). It is thus possible that VPS4 constriction of CHMP2A-CHMP3 filaments generates tension as well, which is ultimately released by cleavage, thereby contributing to the fission process.
Important open questions concern the nature of the ESCRT-III filaments assembled in vivo. First, while CHMP2A-CHMP3 or CHMP1B-IST1 tubular helical structures are formed by thin filaments (24, 43), ESCRT-III spirals imaged at the cytokinetic midbody are much wider and likely composed of multiple filaments (29, 30). However, these spirals have not been detected at vesicle or enveloped virus budding sites and may thus be specific for cytokinesis, which requires constriction from larger diameters on. Second, it is not clear how many ESCRT-III filament turns are present at vesicle or enveloped virus budding sites. Recent imaging suggested up to 50 Vps24 (CHMP3) molecules at endosomal budding sites (37), which could form one and a half turn of a CHMP2A-CHMP3 helical filament (43). Last, our system to image CHMP2A-CHMP3 filament constriction and cleavage in vitro can now be extended to wrap membrane around the tubular structure to determine whether the system suffices to cleave membrane tubes.
MATERIALS AND METHODS
Protein expression, purification, and CHMP2A-CHMP3 tube formation
Full-length CHMP3, CHMP2AΔC containing residues 9 to 161 fused to the maltose-binding protein (MBP) VPS4B, and VPS4B E235Q and I124E mutants were expressed and purified as described (12, 22). A final purification step includes size exclusion chromatography in HBS buffer A [20 mM Hepes (pH 7.6) and 150 mM NaCl] for all proteins. For tube formation, 10 μM CHMP2AΔC and 15 μM CHMP3 were mixed and incubated overnight. Tubes were concentrated by centrifugation at 15,000 rpm in a Sorvall centrifuge.
Surface preparation for HS-AFM
To reduce the surface interaction to ESCRT tubes, we have used a lipid bilayer on top of freshly cleaved mica, as modified from literature (21, 54, 55). The lipid bilayer was formed by absorption of large unilamellar vesicles (LUVs) onto freshly cleaved mica. To prepare LUVs, the lipids, DOPC, and DOPS from Avanti, polar lipids were used. Sixty percent DOPC and 40% DOPS (mol:mol) containing 1 mg/ml of total lipids were mixed in 200 μl of chloroform in a small glass vial. Next, chloroform was evaporated using argon gas while slowly rotating the vial to produce a lipid film on the glass wall. Last, the film was completely dried in a vacuum desiccator for 30 to 45 min. Rehydration of the lipid film was performed with 200 μl of buffer B [10 mM Hepes (pH 7.4), 100 mM NaCl, and 50 mM sucrose]. The mixture was immediately vortexed for 30 s, followed by three cycles of freeze-thaw using liquid nitrogen. Small aliquots of LUVs were stored at −20°C and used within 1 month after preparation. LUVs were thawed at room temperature and diluted to a concentration of 0.2 mg/ml in the recording buffer C [10 mM Hepes, 10 mM tris-HCl (pH 7.5), and 150 mM NaCl] before use. Two microliters of diluted LUVs was incubated on a freshly cleaved, 1-mm-diameter mica disc attached to a small glass rod (HS-AFM sample stage) (56). After incubation for 5 to 10 min, the surface was cleaned three to five times with cleaning buffer D [10 mM tris-HCl (pH 7.5), 100 mM NaCl, and 10 mM MgCl2].
HS-AFM experiments
Before immobilization, the ESCRT tubes were treated for 30 min with 1 mM tobacco etch virus protease to remove the MBP tag from CHMP2AΔC. After removal of MBP, 2 μl of ESCRT tubes was incubated on the previously prepared lipid bilayer for 15 min. After the incubation time, the surface was cleaned gently with recording buffer. The glass rod was then attached to the piezo head of the HS-AFM with a small amount of wax. All HS-AFM data were taken in amplitude modulation mode using a sample scanning HS-AFM [Research Institute of Biomolecule Metrology (RIBM), Japan] (47, 56). Short cantilevers (USC-F1.2-k0.15, NanoWorld, Switzerland) with spring constant of 0.15 N/m, resonance frequency around 0.6 MHz, and a quality factor of ∼2 in buffer were used. The cantilever-free amplitude is 3 nm, and the set-point amplitude for the cantilever oscillation was set around 2.7 nm. Images were taken at 0.2 to 1 frame/s depending on the size of the image. Unless mentioned, all the HS-AFM recordings were performed in recording buffer. For all experiments with VPS4B and/or ATP Mg2+, the indicated concentrations of VPS4B ranging from 1 to 10 μM (dissolved in recording buffer) were applied directly to the sample chamber during continuous imaging. The ESCRT-III CHMP2A-CHMP3 tubes were first imaged in recording buffer, and then, VPS4B was added at indicated concentrations and incubated for 3 to 4 min (to provide sufficient time for diffusion of VPS4B inside the tubes) before the addition of ATP Mg2+. Because of the perturbation by the added liquid, sometimes tip-sample interactions were briefly affected, but never longer than 30 s. The time mentioned in the figure panels represents time starting at the first image, that is, within 30 s from the moment of VPS4B/ATP addition.
Data analysis and image processing
All the HS-AFM images were processed primarily with Igor Pro with a built-in script from RIBM (Japan) and ImageJ software with additional home-written plugins. The HS-AFM images/movies were only processed minimally through tilt correction, drift correction, and brightness correction. The kymographs were obtained from the cross section at a fixed position (marked for each image) over the entire movie. It represents the height distribution along the line cross section as a function of time. For each experimental condition, at least four tubes were investigated. For the control experiments with VPS4B E235Q and VPS4B I124E, the number of local deformation sites was measured for at least nine tubes. For the VPS4B I124E, we found several filaments without any local deformation; therefore, the calculation of the filament length per deformation for this particular case is underestimated, and the real value should be higher. Therefore, it does not affect our final conclusion.
Electron microscopy
For negative-staining EM, samples were applied to the clean side of carbon on mica (carbon/mica interface) and negatively stained with 2% sodium silicotungstate (pH 7.0). A grid was placed on top of the carbon film covered by the sample, which was subsequently air-dried. Micrographs were taken with a T12 FEI microscope at 120 kV and a magnification of ×30,000. For cryo-EM, vitrified samples were prepared according to the method of Dubochet and co-workers (57). Sample (3.5 μl; CHMP2A-CHMP3 tubes) at 1 mg/ml was applied to glow-discharged R2/2 Quantifoil grids (Quantifoil Micro Tools GmbH, Germany). The excess liquid was blotted with filter paper for 2 s and rapidly plunged into liquid ethane at −175°C using a Vitrobot (20°C, 100% humidity, and force 1). Specimens were observed with a Polara electron microscope at 300 kV. Movies (40 frames of 0.1 s and a dose of 1 electron/Å2 per frame) were recorded in an automatic mode on a K2 summit direct electron detector using Latitude S (Gatan) at a nominal magnification of ×30,000 (1.2 Å per pixel at the image level) with defocus values between 1.2 and 3.5 μm.
Supplementary Material
Acknowledgments
We thank M. Punter (Rijksuniversiteit Groningen) for support in implementing the ImageJ script. Funding: This work was supported by the ANR (ANR-14-CE09-0003-01) and the Institut Universitaire de France (IUF) to W.W., an MSCA Individual fellowship (INTERACT 751404) to S.M., and an NWO Vidi grant to W.H.R. We further acknowledge the platforms of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) supported by the French Infrastructure for Integrated Structural Biology Initiative (FRISBI; ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB) and the FRM and GIS IBiSA for support of the EM platform. Author contributions: W.W. initiated the study. W.W., S.M., and W.H.R. designed and implemented the study. S.M. performed AFM experiments and data analysis. C.C. and N.M. prepared ESCRT-III polymers. G.Su., G.E., and G.Sc. performed the EM analysis. W.W. and W.H.R. wrote the manuscript. All authors discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaau7198/DC1
Fig. S1. Stability of VPS4B-treated CHMP2A-CHMP3 tubular structure over an extended period of time.
Fig. S2. Constriction time for 5 μM VPS4B-treated CHMP2A-CHMP3 tube in the presence of ATP Mg2+.
Fig. S3. Effect of VPS4B E235Q and I124E mutants on CHMP2A-CHMP3 tube remodeling.
Fig. S4. Snapshots of constricted tubes.
Fig. S5. Negative staining EM images of CHMP2A-CHMP3 tubes.
Fig. S6. Cryo-EM images of CHMP2A-CHMP3 tubes incubated with 50 μM VPS4B in the presence of 50 μM adenylyl-imidodiphosphate (AMP-PNP) Mg2+.
Fig. S7. Negative staining images of CHMP2A-CHMP3 tubes incubated with 5 μM VPS4B and 200 μM ATP Mg2+, indicating asymmetric and symmetric constriction sites, along with dome-like end cap formation at the constriction sites.
Movie S1. CHMP2A-CHMP3 tube immobilized on a membrane bilayer without VPS4B.
Movie S2. CHMP2A-CHMP3 tube immobilized on a membrane bilayer with 10 μM VPS4B.
Movie S3. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B.
Movie S4. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 5 μM VPS4B.
Movie S5. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B E235Q mutant and 200 μM ATP Mg2+.
Movie S6. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B I124E and 200 μM ATP Mg2+.
Movie S7. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B.
Movie S8. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 5 μM VPS4B.
Movie S9. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 3 μM VPS4B.
Movie S10. CHMP2A-CHMP3 tubes immobilized on membrane bilayer, preincubated with 1 μM VPS4B.
REFERENCES AND NOTES
- 1.Henne W. M., Stenmark H., Emr S. D., Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 5, a016766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCullough J., Colf L. A., Sundquist W. I., Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scourfield E. J., Martin-Serrano J., Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem. Soc. Trans. 45, 613–634 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Juan T., Fürthauer M., Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 74, 66–77 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Stoten C. L., Carlton J. G., ESCRT-dependent control of membrane remodelling during cell division. Semin. Cell Dev. Biol. 74, 50–65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadoul R., Laporte M. H., Chassefeyre R., Chi K. I., Goldberg Y., Chatellard C., Hemming F. J., Fraboulet S., The role of ESCRT during development and functioning of the nervous system. Semin. Cell Dev. Biol. 74, 40–49 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Weissenhorn W., Poudevigne E., Effantin G., Bassereau P., How to get out: ssRNA enveloped viruses and membrane fission. Curr. Opin. Virol. 3, 159–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurley J. H., ESCRTs are everywhere. EMBO J. 34, 2398–2407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teis D., Saksena S., Emr S. D., Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Carlson L.-A., Hurley J. H., In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc. Natl. Acad. Sci. U.S.A. 109, 16928–16933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morita E., Sandrin V., McCullough J., Katsuyama A., Baci Hamilton I., Sundquist W. I., ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muzioł T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H., Weissenhorn W., Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821–830 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W.-M. B., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I., Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16, 754–762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao J., Chen X.-W., Davies B.-A., Saltiel A. R., Katzmann D. J., Xu Z., Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 20, 3514–3524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y., Kimpler L. A., Naismith T. V., Lauer J. M., Hanson P. I., Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 280, 12799–12809 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Lata S., Roessle M., Solomons J., Jamin M., Gőttlinger H. G., Svergun D. I., Weissenhorn W., Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 378, 818–827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pires R., Hartlieb B., Signor L., Schoehn G., Lata S., Roessle M., Moriscot C., Popov S., Hinz A., Jamin M., Boyer V., Sadoul R., Forest E., Svergun D. I., Göttlinger H. G., Weissenhorn W., A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17, 843–856 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobro M. J., Samson R. Y., Yu Z., McCullough J., Ding H. J., Chong P. L.-G., Bell S. D., Jensen G. J., Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol. Biol. Cell 24, 2319–2327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Q.-T., Schuh A. L., Zheng Y., Quinney K., Wang L., Hanna M., Mitchell J. C., Otegui M. S., Ahlquist P., Cui Q., Audhya A., Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol. 206, 763–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang S., Henne W. M., Borbat P. P., Buchkovich N. J., Freed J. H., Mao Y., Fromme J. C., Emr S. D., Structural basis for activation, assembly, and membrane binding of ESCRT-III Snf7 filaments. eLife 4, e12548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiaruttini N., Redondo-Morata L., Colom A., Humbert F., Lenz M., Scheuring S., Roux A., Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866–879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gőttlinger H. G., Weissenhorn W., Helical structures of ESCRT-III are disassembled by VPS4. Science 321, 1354–1357 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henne W. M., Buchkovich N. J., Zhao Y., Emr S. D., The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151, 356–371 (2012). [DOI] [PubMed] [Google Scholar]
- 24.McCullough J., Clippinger A. K., Talledge N., Skowyra M. L., Saunders M. G., Naismith T. V., Colf L. A., Afonine P., Arthur C., Sundquist W. I., Hanson P. I., Frost A., Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–1551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson P. I., Roth R., Lin Y., Heuser J. E., Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389–402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodon G., Chassefeyre R., Pernet-Gallay K., Martinelli N., Effantin G., Hulsik D. L., Belly A., Goldberg Y., Chatellard-Causse C., Blot B., Schoehn G., Weissenhorn W., Sadoul R., Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J. Biol. Chem. 286, 40276–40286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cashikar A. G., Shim S., Roth R., Maldazys M. R., Heuser J. E., Hanson P. I., Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife 3, e02184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., Gerlich D. W., Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Mierzwa B. E., Chiaruttini N., Redondo-Morata L., von Filseck J. M., König J., Larios J., Poser I., Müller-Reichert T., Scheuring S., Roux A., Gerlich D. W., Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19, 787–798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goliand I., Adar-Levor S., Segal I., Nachmias D., Dadosh T., Kozlov M. M., Elia N., Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM. Cell Rep. 24, 1756–1764 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Babst M., Wendland B., Estepa E. J., Emr S. D., The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I., ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L., Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Han H., Monroe N., Sundquist W. I., Shen P. S., Hill C. P., The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. eLife 6, e31324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghazi-Tabatabai S., Saksena S., Short J. M., Pobbati A. V., Veprintsev D. B., Crowther R. A., Emr S. D., Egelman E. H., Williams R. L., Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16, 1345–1356 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Caillat C., Macheboeuf P., Wu Y., McCarthy A. A., Boeri-Erba E., Effantin G., Göttlinger H. G., Weissenhorn W., Renesto P., Asymmetric ring structure of Vps4 required for ESCRT-III disassembly. Nat. Commun. 6, 8781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adell M. A. Y., Migliano S. M., Upadhyayula S., Bykov Y. S., Sprenger S., Pakdel M., Vogel G. F., Jih G., Skillern W., Behrouzi R., Babst M., Schmidt O., Hess M. W., Briggs J. A. G., Kirchhausen T., Teis D., Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 6, e31652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jouvenet N., Zhadina M., Bieniasz P. D., Simon S. M., Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13, 394–401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baumgärtel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P. W., Kräusslich H.-G., Bräuchle C., Müller B., Lamb D. C., Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 13, 469–474 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Prescher J., Baumgärtel V., Ivanchenko S., Torrano A. A., Bräuchle C., Müller B., Lamb D. C., Super-resolution imaging of ESCRT-proteins at HIV-1 assembly sites. PLOS Pathog. 11, e1004677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson D. S., Bleck M., Simon S. M., Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. eLife 7, e36221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D., Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Effantin G., Dordor A., Sandrin V., Martinelli N., Sundquist W. I., Schoehn G., Weissenhorn W., ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell. Microbiol. 15, 213–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabrikant G., Lata S., Riches J. D., Briggs J. A. G., Weissenhorn W., Kozlov M. M., Computational model of membrane fission catalyzed by ESCRT-III. PLOS Comput. Biol. 5, e1000575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lata S., Schoehn G., Solomons J., Pires R., Göttlinger H. G., Weissenhorn W., Structure and function of ESCRT-III. Biochem. Soc. Trans. 37, 156–160 (2009). [DOI] [PubMed] [Google Scholar]
- 46.McCullough J., Frost A., Sundquist W. I., Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 34, 85–109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ando T., Kodera N., Naito Y., Kinoshita T., Furuta K., Toyoshima Y. Y., A high-speed atomic force microscope for studying biological macromolecules in action. Chemphyschem 4, 1196–1202 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M., Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Schwedler U. K., Stuchell M., Müller B., Ward D. M., Chung H.-Y., Morita E., Wang H. E., Davis T., He G.-P., Cimbora D. M., Scott A., Kräusslich H.-G., Kaplan J., Morham S. G., Sundquist W. I., The protein network of HIV budding. Cell 114, 701–713 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Van Engelenburg S. B., Shtengel G., Sengupta P., Waki K., Jarnik M., Ablan S. D., Freed E. O., Hess H. F., Lippincott-Schwartz J., Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science 343, 653–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monroe N., Han H., Gonciarz M. D., Eckert D. M., Karren M. A., Whitby F. G., Sundquist W. I., Hill C. P., The oligomeric state of the active Vps4 AAA ATPase. J. Mol. Biol. 426, 510–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elia N., Sougrat R., Spurlin T. A., Hurley J. H., Lippincott-Schwartz J., Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U.S.A. 108, 4846–4851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elia N., Fabrikant G., Kozlov M. M., Lippincott-Schwartz J., Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys. J. 102, 2309–2320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyagi A., Chipot C., Rangl M., Scheuring S., High-speed atomic force microscopy shows that annexin V stabilizes membranes on the second timescale. Nat. Nanotechnol. 11, 783–790 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Takeda T., Kozai T., Yang H., Ishikuro D., Seyama K., Kumagai Y., Abe T., Yamada H., Uchihashi T., Ando T., Takei K., Dynamic clustering of dynamin-amphiphysin helices regulates membrane constriction and fission coupled with GTP hydrolysis. eLife 7, e30246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchihashi T., Kodera N., Ando T., Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Lepault J., Booy F. P., Dubochet J., Electron microscopy of frozen biological suspensions. J. Microsc. 129, 89–102 (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaau7198/DC1
Fig. S1. Stability of VPS4B-treated CHMP2A-CHMP3 tubular structure over an extended period of time.
Fig. S2. Constriction time for 5 μM VPS4B-treated CHMP2A-CHMP3 tube in the presence of ATP Mg2+.
Fig. S3. Effect of VPS4B E235Q and I124E mutants on CHMP2A-CHMP3 tube remodeling.
Fig. S4. Snapshots of constricted tubes.
Fig. S5. Negative staining EM images of CHMP2A-CHMP3 tubes.
Fig. S6. Cryo-EM images of CHMP2A-CHMP3 tubes incubated with 50 μM VPS4B in the presence of 50 μM adenylyl-imidodiphosphate (AMP-PNP) Mg2+.
Fig. S7. Negative staining images of CHMP2A-CHMP3 tubes incubated with 5 μM VPS4B and 200 μM ATP Mg2+, indicating asymmetric and symmetric constriction sites, along with dome-like end cap formation at the constriction sites.
Movie S1. CHMP2A-CHMP3 tube immobilized on a membrane bilayer without VPS4B.
Movie S2. CHMP2A-CHMP3 tube immobilized on a membrane bilayer with 10 μM VPS4B.
Movie S3. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B.
Movie S4. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 5 μM VPS4B.
Movie S5. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B E235Q mutant and 200 μM ATP Mg2+.
Movie S6. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B I124E and 200 μM ATP Mg2+.
Movie S7. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 10 μM VPS4B.
Movie S8. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 5 μM VPS4B.
Movie S9. CHMP2A-CHMP3 tubes immobilized on a membrane bilayer, preincubated with 3 μM VPS4B.
Movie S10. CHMP2A-CHMP3 tubes immobilized on membrane bilayer, preincubated with 1 μM VPS4B.


