A nitrogen-vacancy center in diamond is used for magnetic imaging of intracellular proteins in a single cell at 10-nm scale.
Abstract
The in situ measurement of the distribution of biomolecules inside a cell is one of the important goals in life science. Among various imaging techniques, magnetic imaging (MI) based on the nitrogen-vacancy (NV) center in diamond provides a powerful tool for the biomolecular research, while the nanometer-scale MI of intracellular proteins remains a challenge. Here, we use ferritin as a demonstration to realize the MI of endogenous proteins in a single cell using the NV center as the sensor. With the scanning, intracellular ferritins are imaged with a spatial resolution of ca. 10 nm, and ferritin-containing organelles are colocalized by correlative MI and electron microscopy. The approach paves the way for nanoscale MI of intracellular proteins.
INTRODUCTION
Improving the spatial resolution of the imaging technique in biology and medicine has been essential in reaching continuing demand. Among various kinds of techniques, the magnetic imaging (MI) technique is widely used in biology and medicine. One of the MI techniques is magnetic resonance imaging (MRI) to measure the distribution of nuclear spins. Recent approaches based on conventional MRI reach ca. 1 μm in nuclear spin imaging (1), while the resolution is limited by their electrical detection sensitivity (2). To break the resolution barrier, a series of techniques, such as superconducting quantum inference device (3) and magnetic resonance force microscopy (4), have been developed to detect and image the electron and nuclear spins at nanoscale resolution. However, the reported schemes require a cryogenic environment and high vacuum, which severely makes the experiment complicated.
A recently developed quantum sensing method, based on the nitrogen-vacancy (NV) center in diamond, has radically pushed the boundary of MI techniques. NV detections of magnetic resonance spectrum at the nanoscale and single-molecule level have been reported for organic samples (5–7) and proteins (8, 9) in vitro. By combining quantum sensing with NV centers and scanning probe microscopy, nanoscale MRI has been demonstrated for single electron spin (10) and small nuclear spin ensemble (11, 12). The NV center has also been used as a biocompatible magnetometer to image the ferromagnetic particles inside cells in a noninvasive manner at the subcellular scale, i.e., 0.4 μm (13). Another important progress is that, as a wideband magnetometer, the depolarization of the NV center is used for detecting the magnetic fluctuating noise (14) from metal ions (15–17) and nuclear spins (18). However, nanoscale MI of proteins in single cells has not yet been reported, and the primary remaining barrier is the accessibility of the samples to a scanning NV center.
We report here two technical advancements to enable nanoscale MI of intracellular proteins in a cell. In the studies, the cell was fixed to solid state and then sectioned to a cube shape and placed at a tuning fork scanning probe of an atomic force microscope (AFM), with the flat cross section of the cell exposed to the air (Fig. 1A). This sample placement setup allows positioning of the NV sensor within 10 nm to the target proteins and the use of the AFM to suppress the thermal drift in sample positioning. In addition, by fabricating trapezoidal cylinder–shaped nanopillars at a bulk diamond surface and implanting NV centers at the top of the pillars (Fig. 1A), the acquisition time for an image was shortened by nearly one order (19, 20). In situ MI of the magnetic fluctuating noise of intracellular ferritin proteins has been successfully carried out by using this experimental setup.
Fig. 1. Schematic of the setup and experimental principle.
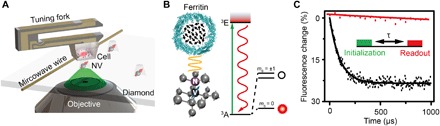
(A) Schematic view of the experimental setup. The cell embedded in resin is attached to a tuning fork and scans above the diamond nanopillar that contains a shallow NV center. A copper wire is used to deliver the microwave pulse to the NV center. A green laser (532 nm) from the CFM is used to address, initialize, and read out the NV center. (B) Left: Crystal lattice and energy level of the NV center. The NV center is a point defect that consists of a substitutional nitrogen atom and an adjacent vacancy in diamond. Right: Schematic view of a ferritin. The black arrows indicate the electron spins of Fe3+. (C) Experimental demonstration of the spin noise detection with and without ferritin in the form of polarization decay for the same NV center. The inset is the pulse sequence for detection and imaging of the ferritin. A 5-μs green laser is used to initialize the spin state to ms = 0, followed by a free evolution time τ to accumulate the magnetic noise, and finally the spin state is read out by detecting the fluorescence intensity. The pulse sequence is repeated about 105 times to acquire a good signal-to-noise ratio (SNR). The relaxation time is fitted to be 0.1 and 3.3 ms by exponential decay for the case with and without ferritin, respectively, indicating a spin noise of 0.01 mT2.
RESULTS
Ferritin is a globular protein complex with an outer diameter of 12 nm (Fig. 1B), and within its 8-nm-diameter cavity, the protein can house up to 4500 iron atoms (21). The magnetic noise of the ferric ions can be detected by their effects on the T1 relaxation time of an NV center. Previous work has shown that the presence of ferritin decreases T1 of the NV centers (16, 22), and we confirmed this observation by fluorescence measurements of the time-dependent decay of the population of the mS = 0 state of NV centers on a diamond surface coated with ferritins (Fig. 1C). Furthermore, it can be detected in a label-free manner using the NV center (16, 22) and transmission electron microscopy (TEM). This enabled the development of a correlated MI and TEM scheme to obtain and verify the first nanoscale MI of a protein in situ.
This work used hepatic carcinoma HepG2, a widely used cell line for the iron metabolism study. The cells were treated with ferric ammonium citrate (FAC) (Fig. 2A). As a result, the amount of ferritin increased significantly as verified by the results of confocal microscopy (CFM), Western blot, and TEM (fig. S3). Figure 2B and fig. S3E show that ferritins localized primarily in intracellular puncta around the nucleus among the cytoplasm. Bulk electron paramagnetic resonance (EPR) spectroscopy confirmed the paramagnetic properties of ferritins in FAC-treated HepG2 cells (Fig. 2C), and mass spectrometry measurements precluded interference due to other paramagnetic metal ions.
Fig. 2. The preparation and characterization of ferritin-rich HepG2 cell samples.
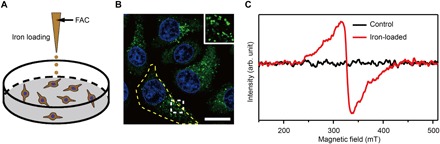
(A) Schematic view of the treatment to cultured cells. Following iron loading or no treatment, the HepG2 cells were examined for fluorescence images and EPR spectra, respectively. For the MI and TEM imaging, cell samples were treated through high-pressure freezing, freeze substitution, and sectioning. (B) Representative CFM image of ferritin structures (green) in iron-loaded HepG2 cells. The ferritin proteins were immunostained by anti-ferritin light chain antibody. The nuclei are indicated by 4′,6-diamidino-2-phenylindole (DAPI) in the blue channel. Inset displays magnified ferritin structures. The yellow dashed line outlines the contour of a cell. Scale bar, 20 μm. (C) EPR spectra of control and iron-loaded HepG2 cells at T = 300 K. HepG2 cells, harvested by trypsinization, were fixed with paraformaldehyde and immersed in phosphate-buffered saline (PBS).
All intracellular components of the Fe-loaded cells were immobilized by ultrafast high-pressure freezing. This treatment stabilized the intracellular structures and molecules and minimized Brownian motions that cause the proteins to randomly move on the scale up to a hundred nanometers (23). The frozen cells were embedded and polymerized in the LR White medium, with the concerned intracellular structures remaining in situ. The embedded cell sample was then glued on the AFM tuning fork and shaped to the frustum of a prism with a few cells on the tip. The tip surface was sectioned to nanometer flatness by a diamond knife on the ultramicrotome and examined under AFM. During this sectioning step, a cell was dissected with a certain amount of ferritins remaining just on or a few nanometers below the surface. The tuning fork was then placed on a sample stage, which contained a two-dimensional tilt that allows fine adjustments of the relative angle between the cell and diamond surface. After the adjustment, the nanopillar can parallel contact the surface of the cell cube.
MI images of ferritins were acquired by scanning the cell cube across the diamond nanopillar while simultaneously measuring the NV spin depolarization rate under the “leapfrog” scanning mode. Given the hardness of the diamond, the leapfrog mode maximally reduced the abrasion of the cell samples and allowed the reproduction of the results (12). Instead of measuring the whole depolarization curve, at each pixel, fluorescence decay was measured at a fixed free evolution time of τ = 50 μs to reveal the degree of NV sensor spin polarization, which is correlated with the amount of ferritin within the sensing volume. The total time for one whole scan to acquire an image was about 2 hours, during which the positioning thermal drift was only few tens of nanometers.
A correlated MI and TEM scheme was developed to speed up and verify the nanoscale MI images. We first cut off a cell section with a thickness of about 100 nm and prelocate the ferritin clusters by TEM (Fig. 3A). Then, with the MI setup, we relocated these clusters and scanned near their locations to search for the spin signal. Figure 3 shows an example of the MI and TEM correlated result of intracellular ferritins with a pixel size of 43 nm. The diameter of ferritin clusters was between 100 and 500 nm. Given the different axial detection ranges between TEM (here, ~100 nm) and MI [around 10 nm under the experimental signal-to-noise ratio (SNR); fig. S8], the respective images showed microdomains with different distributions of ferritins in a membrane-bound organelle. Some of the clusters appeared in both TEM and MI images, while others were not observed in MI (Fig. 3, B to D). Control measurements from CFM, EPR, TEM, and mass spectrometry confirmed that the spin noise causing the depolarization of the NV center was from intracellular ferritins.
Fig. 3. Correlative MI and TEM images.
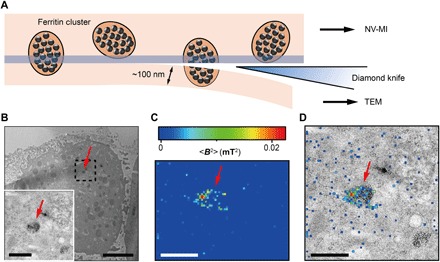
(A) Schematic view of sectioning for correlative MI and TEM imaging. The last section and the remaining cube were transferred for TEM imaging and MI scanning, respectively. The sectioning resulted in some split ferritin clusters that could be imaged under both microscopes. A transparent blue strip of ~10 nm indicates the imaging depth of the MI, while in the TEM, the imaging depth is ~100 nm. (B) Distribution of ferritins from the last ultrathin section under TEM. Inset: Magnified figure of the part in black dashed box. (C) MI result of the remaining cell cube. The pixel size is 43 nm. (D) The merged MI and TEM micrograph shows ferritins in a membrane-bound organelle. The red arrows in (B) to (D) indicate the same ferritin cluster. Scale bars, 5 μm (B) and 1 μm [B (inset), C, and D].
To obtain the detail of ferritins in clusters, we acquired a high-resolution MI image by minimizing the pixel size to 8.3 nm (Fig. 4). The image shows ferritins in a cluster of ca. 100 nm with an SNR (peak to peak) of >2. We observed several transitions from the background to the ferritin cluster with a minimal rising edge width of 8.3 nm, which was the same size as a single pixel (Fig. 4C). The spin noise from intracellular ferritins was measured at around 0.02 mT2. Using a model of 4000 Fe3+ uniformly distributed inside an 8-nm ferritin spherical core, the measured magnetic noise gives an estimation of approximately 9 nm for the vertical distance between the NV sensor and the center of the shallowest ferritin. We further calculated the point spread function of the measured signal and found that the spatial resolution determined by full width at half maximum was ~10 nm. This was close to the size of a single ferritin core and agrees well with the observed sharp transition at the edge of the pixel (Fig. 4C). Consistent with these analyses, other lines did not show this sudden transition (fig. S9). This is mostly because the ferritins on the cluster boundary were less shallow beneath the cell section surface, and the signal dwindled as the ferritin was farther from the NV sensor.
Fig. 4. High-resolution MI image of an intracellular ferritin cluster.
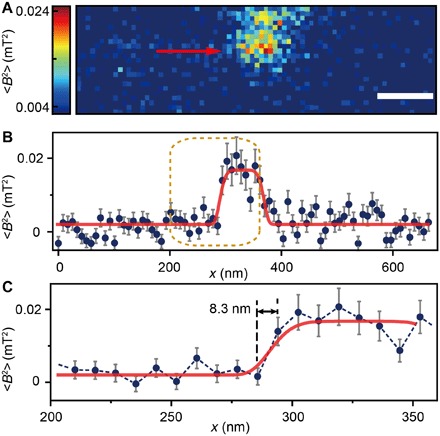
(A) Ferritin cluster imaged by the NV sensor with 80 × 24 pixels and a pixel size of 8.3 nm. Scale bar, 100 nm. (B) Trace data of the scanning line in (A) directed by the red arrow. The platform indicates the ferritin cluster. The red curve fitted by a plateau function serves as a guide to the eye. (C) Magnified figure of the gold dashed box in (B). The sharp transition indicated by the red arrow around x = 283 nm shows the scanning from the blank area to the area with ferritins.
DISCUSSION
The NV centers have been shown to be a sensitive and appropriate sensor for applications in biology. Here, we developed the methodologies based on such a sensor to obtain in situ the first 10-nm-resolution MI of a protein. With additional improvements in stability and sensitivity, we can speed up the scanning and image a larger region of interest to locate ferritins in a cell, such as around the nucleus or in peripheral, and associate them with more organelles. It will contribute to further studies on the iron storage and release in cells and the regulatory mechanism of iron metabolism disorders related to iron at nanoscale, such as hemochromatosis, anemia, liver cirrhosis, and Alzheimer’s disease (24, 25). For other cellular components with paramagnetic signal, such as magnetic molecules, metalloproteins, and special spin-labeled proteins, one may take a similar approach to image them and study their functions in situ. Further studies will explore additional suitable targets for correlated MI and TEM or optical microscopy detection, extend to protein nuclear spin MRI (5–7, 18), and perform three-dimensional cell tomography (26). The work reported here represents a significant step forward in pushing the boundary of MI on endogenous intracellular nanostructure and macromolecules.
MATERIALS AND METHODS
Experimental setup
The experiment was carried out on a homebuilt setup (fig. S1), which combined optically detected magnetic resonance microscopy (ODMR) with AFM. The ODMR was used for addressing, controlling, and measuring single NV centers in diamond, while the AFM controlled the distance between the cell sample and the diamond nanopillars. In the ODMR part, the CFM enabled the state initialization and readout of the electron spin state via a laser with 532-nm wavelength. The laser light passed twice through an acousto-optic modulator (ISOMET 1250C) and then was focused on an NV center by an objective [Olympus, LUCPLFLN 60×; numerical aperture (NA), 0.7]. The phonon sideband fluorescence with wavelengths of 650 to 775 nm was detected to determine the state of the electron spin. The fluorescence passed through the same objective and was collected by an Avalanche Photodiodes (Perkin Elmer SPCM-AQRH-14). A light-emitting diode (LED) illuminator with a 470-nm wavelength (Thorlabs M470L3-C1) and a charge-coupled device (CCD) camera were used for wide-field and interference fringe views.
The AFM based on a tuning fork was controlled by a commercial controller (Asylum Research MFP3D) and mounted with a homebuilt AFM probe head. A translation stage based on a piezo motor (Physik Instrumente Q-522) was used for the coarse approach. Each AFM probe head and the diamond base plate contained a three-axis tilt platform driven by a stepper motor actuator (Thorlabs ZFS25B), allowing accurate tilting adjustment.
The whole probe head was placed in a temperature-stabilized chamber to minimize the positioning thermal drift. A three-level temperature control system was set to control the temperature: (i) The ambient temperature of the laboratory was controlled by air-conditioning. (ii) A soundproof box was used to isolate noise and stabilize the temperature. (iii) The third level included a temperature controller (PTC10 from Stanford Research System) and a copper chamber with a heating resistor. As the core region of the MI system, the scanning part of the AFM could be stabilized at a temperature fluctuation of ±5 mK and a thermal drift of ±20 nm per day by using these multilevel temperature controls.
Fabrication of NV sensors
The NV centers were created by 10-keV 14N2+ implantation in [100] face bulk diamond and annealing for 2 hours at 800°C in high vacuum (27). The typical depth of the 14N2+ ions was estimated to be 5 to 11 nm below the diamond surface (28). We fabricated trapezoidal cylinder–shaped nanopillars on the diamond using electron beam lithography and reactive ion etching (RIE).
For fabrication, the negative electron beam–resisting hydrogen silsesquioxane (HSQ) was spun on the diamond with a thickness of 350 nm (fig. S2). Then, the HSQ layer was patterned by electron beam writing and followed by 5- min tetramethyl ammonium hydroxide (4%) developing. The nanopillars were formed from RIE with mixed CHF3 and O2, with an etching depth of about 400 nm. Last, the HF was used to remove the resist, and the diamond was thermally oxidized for 15 min at 580°C in atmosphere to improve the NV stability and depolarization time. After the etching and before changing the cell sample, the diamond was cleaned in boiling 1:1:1 mixture of H2SO4:HNO3:HClO4 for 6 hours. To remove the Fe3+ that remained on the diamond after scanning the cell sample, the diamond was additionally cleaned in the boiling deionized water for 8 hours.
We measured all the NV centers in the nanopillars and chose very shallow NV centers with short relaxation time as the sensors for MI. The fluorescence counts increased from ~20 to ~200 k/s under half-saturated laser power, which reduced the experiment time by nearly an order of magnitude under the same SNR.
Cell sample preparation
Cell culture
Hepatic carcinoma HepG2 cells were grown at 37°C in a 5% CO2 humidified environment in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Biological Industries), penicillin (100 U/ml), and streptomycin (100 μg/ml; HyClone). Cells were treated with FAC (200 μg/ml; F5879-100G, Sigma) for 3 days to induce iron-replete conditions. Before being collected, cells were washed with phosphate-buffered saline (PBS) five times.
Immunoblotting assay
HepG2 cells grown in six-well plates were treated with or without FAC. After washing with PBS, whole-cell lysates were prepared in lysis buffer [20 mM tris-HCl (pH 8.0), 137 mM NaCl, 2 mM EDTA, 1% NP-40, and 1× protease inhibitor cocktail], centrifuged to remove the precipitation, and mixed with SDS–polyacrylamide gel electrophoresis sample loading buffer. Immunoblots were stained sequentially with mouse anti-ferritin light chain (Santa Cruz Biotechnology, sc-74513, 1:800 dilution) and mouse anti–β-actin (Sigma, clone AC-74, 1:2000 dilution) monoclonal antibodies, and with horseradish peroxidase–labeled goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA).
Immunofluorescence microscopy
For immunostaining, cells were fixed in −20°C precooled 100% methanol for 5 min and washed with PBS five times at room temperature, each for 5 min. Subsequently, cell samples were blocked in blocking solution [2% bovine serum albumin (BSA), 1× PBS, 0.1% Triton X-100, and 0.05% Na azide] for 30 min at room temperature. Mouse anti-ferritin light chain monoclonal antibody (Santa Cruz Biotechnology, sc-74513) was used at a 1:200 dilution in incubation buffer (1% BSA, 1× PBS, 0.1% Triton X-100, and 0.05% Na azide) overnight at 4°C. Alexa Fluor 488 AffiniPure Goat Anti-Mouse IgG (H+L) (Jackson ImmunoResearch, 115-545-003) was used at a dilution of 1:600 and incubated for 2 hours at room temperature. After extensive washing and 4′,6-diamidino-2-phenylindole (DAPI) staining, the cells were used for confocal imaging. Multiwavelength fluorescence images were acquired using a FLUOVIEW FV1000 microscope (Olympus, Tokyo, Japan) equipped with a 100× NA 1.45 oil-immersion objective and captured using FV10-ASW version 3.1 software. Images were analyzed, and fluorescence intensities were quantified in ImageJ 1.45m (W. Rasband, National Institutes of Health).
Embedded cell sample preparation and TEM imaging
The resin-embedded cell samples were prepared by high-pressure freezing and freeze substitution. The treated cells were instantaneously frozen in a BAL-TEC HPM 010 high-pressure freezer (BAL-TEC). The freeze substitution was performed in an automatic freeze-substitution unit (Leica, AFS 2), and 1% osmium tetroxide (OsO4) was used to fix the cells and quench the autofluorescence in cells. As a result, the fluorescence photon count decreased from several million to several thousand per second.
In the following infiltration and polymerization, the LR White embedding medium (EMS) was chosen for its low fluorescence and EPR signals (fig. S4). The resin-embedded cells were polymerized for 48 hours at 60°C. For using other polymerization processes or some other resins, such as HM20, KM40, and GMA, either the polymerized resins showed strong autofluorescence at 532-nm laser illumination or the polymerization using 365-nm ultraviolet irradiation generated many unknown radicals in the sample, which appeared strong in the EPR spectroscopy. The ultrathin sections (70 to 100 nm) were examined in an HT-7700 TEM system equipped with an AMT (Advanced Microscopy Techniques) CCD camera operated at 80 kV.
For MI imaging, the polymerized cell sample was trimmed with blade and fixed at the end of the AFM-used tuning fork for sectioning. To minimally disturb the property of the tuning fork, the size of the quadrate sample was limited to less than 1 mm × 1 mm × 1 mm. On an ultramicrotome (Leica EM UC7), the sample tip was exquisitely trimmed to a trapezoidal frame by a glass knife, with a size of 50 μm × 50 μm at the bottom and 10 μm × 10 μm at the top and with a height of 50 μm for high-resolution MI in Fig. 4. Then, the surface of the 10 μm × 10 μm cross section was sectioned by a diamond knife (DiATOME) to form a smooth surface with a flatness of a few nanometers, examined by the AFM (fig. S5). Last, we verified that there were one or more cells in the exposed surfaces on the light microscope. For correlated imaging, the tips of the cell samples were trimmed to cubes with a size of 50 μm × 50 μm × 50 μm, whose size was big enough to ensure that we could be able to collect the sections. The last piece of ultrathin sections (100 nm) was collected for the TEM examination.
EPR and mass spectrometric assays
The ensemble EPR was carried out at room temperature on a JES-FA200 spectrometer. The EPR results of liquid samples and embedded samples are shown in Fig. 2 and fig. S4, respectively. Unlike the in vitro measurement with purified ferritin, there was a chance that some other paramagnetic species contributed the spin noise signal, such as manganese (Mn). We carried out mass spectrometric detection to preclude the influence of other possible paramagnetic metal ions. First, cells were washed for five times with PBS and collected. Then, 200 μl of concentrated HNO3 was added into the cells and heated for 2 hours at 90°C. The final cell lysates were diluted with ddH2O to 10 ml. The mass spectra were measured via inductively coupled plasma mass spectrometry. The mass spectra showed a significant increase of the iron concentration in iron-loaded cells compared with that in control cells (0.484 μg/ml versus 0.042 μg/ml). However, there was nearly no detectable Mn in both groups (0.005 and 0.003 μg/ml).
Experimental methods
Tilt between cell cube and diamond nanopillars
As shown in fig. S6, the adjustment of the angle θ between the lower surface of the cell cube and the upper surface of the diamond nanopillar was based on the interference fringes of the blue LED light guided through the objective lens. When the cell cube was several tens of microns above the diamond surface, the interference fringes of the blue LED light between the cell cube and the diamond were clear under the CCD integrated in the confocal microscope. The small angle can be written as θ = λ/(2δl), where λ = 470 nm is the center wavelength of the blue LED light and δl is the distance between two fringes. The fringe direction shows the perpendicular direction to the tilt. For the SNR shown in fig. S6, it was able to adjust the tilt when less than 1/4 fringe is viewed in the CCD, corresponding to δl ~ 200 μm and θ < 2 mrad in the angle. In the situation that the diameter of the nanopillars was 400 nm and under the assumption that the cell cube surface is perfectly flat, the maximal gap between the top surface of the nanopillars and the cell cube was controlled within 1 nm.
Supplementary Material
Acknowledgments
We thank C. Zou for help on diamond annealing, P. Z. Qin for help on the presentation of the work, L. Bai and X. Xu for their help on cell culture and Western blot, and W. Ji for helpful discussions. The fabrication of diamond nanopillars was performed at the University of Science and Technology of China (USTC) Center for Micro and Nanoscale Research and Fabrication. Funding: The authors in USTC are supported by the National Key R&D Program of China (grant nos. 2018YFA0306600 and 2016YFA0502400), the National Natural Science Foundation of China (grant nos. 81788101, 11722544, 11227901, 31470835, 91636217, 11761131011, and 31600685), the CAS (grant nos. GJJSTD20170001 and QYZDY-SSW-SLH004), the Anhui Initiative in Quantum Information Technologies (grant no. AHY050000), the Anhui Provincial Natural Science Foundation (grant nos. 1808085J09 and 1608085QC63), and the Fundamental Research Funds for the Central Universities. The authors in the Institute of Biophysics are supported by the Ministry of Science and Technology Program (grant no. 2016YFA0500203), the National Natural Science Foundation of China (grant nos. 31421002 and 31730054), and the Beijing Municipal Science & Technology Commission (grant no. Z181100004218002). Author contributions: J.D. and T.X. supervised the project and proposed the idea. J.D. and T.X. designed the proposal. P.W., M.G., S.P., and F.S. built the experimental setup and carried out the MI experiments. S.C., R.Z., and M.G. prepared and characterized the cell samples. S.C., M.C., J.S., and X.R. measured the EPR and mass spectra. M.W. fabricated and imaged the diamond sensors. P.W., M.G., and W.M. simulated the data. P.W., S.C., M.G., F.S., T.X., and J.D. wrote the manuscript. All authors participated in the discussion and analysis of the data and manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaau8038/DC1
Section S1. Principle of spin noise measurement based on the NV center
Section S2. Calculation of the spin noise intensity from the single-τ measurement
Section S3. Point spread function of a single ferritin
Fig. S1. Experimental setup.
Fig. S2. Images of the nanopillars on diamonds.
Fig. S3. The treatment of FAC significantly increased the ferritins in HepG2 cells.
Fig. S4. Electron spin resonance spectra of resin-embedded HepG2 cells.
Fig. S5. The AFM characterization of the surface of cell sample.
Fig. S6. Adjusting the distance between the NV center and the cell section.
Fig. S7. Calculated spin noise intensity as a function of the distance between NV center and ferritin.
Fig. S8. Simulation of noise intensity at different vertical distances between a single ferritin and NV center.
Fig. S9. Trace data of Fig. 4A.
Reference (29)
REFERENCES AND NOTES
- 1.Lee S.-C., Kim K., Kim J., Lee S., Han Yi J., Kim S. W., Ha K.-S., Cheong C., One micrometer resolution NMR microscopy. J. Magn. Reson. 150, 207–213 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Glover P., Mansfield S. P., Limits to magnetic resonance microscopy. Rep. Prog. Phys. 65, 1489 (2002). [Google Scholar]
- 3.Vasyukov D., Anahory Y., Embon L., Halbertal D., Cuppens J., Neeman L., Finkler A., Segev Y., Myasoedov Y., Rappaport M. L., Huber M. E., Zeldov E., A scanning superconducting quantum interference device with single electron spin sensitivity. Nat. Nanotechnol. 8, 639–644 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Rugar D., Budakian R., Mamin H. J., Chui B. W., Single spin detection by magnetic resonance force microscopy. Nature 430, 329–332 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Staudacher T., Shi F., Pezzagna S., Meijer J., Du J., Meriles C. A., Reinhard F., Wrachtrup J., Nuclear magnetic resonance spectroscopy on a (5-nanometer)3 sample volume. Science 339, 561–563 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Mamin H. J., Kim M., Sherwood M. H., Rettner C. T., Ohno K., Awschalom D. D., Rugar D., Nanoscale nuclear magnetic resonance with a nitrogen-vacancy spin sensor. Science 339, 557–560 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Aslam N., Pfender M., Neumann P., Reuter R., Zappe A., de Oliveira F. F., Denisenko A., Sumiya H., Onoda S., Isoya J., Wrachtrup J., Nanoscale nuclear magnetic resonance with chemical resolution. Science 357, 67–71 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Shi F., Zhang Q., Wang P., Sun H., Wang J., Rong X., Chen M., Ju C., Reinhard F., Chen H., Wrachtrup J., Wang J., Du J., Single-protein spin resonance spectroscopy under ambient conditions. Science 347, 1135–1138 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Lovchinsky I., Sushkov A. O., Urbach E., de Leon N. P., Choi S., De Greve K., Evans R., Gertner R., Bersin E., Müller C., McGuinness L., Jelezko F., Walsworth R. L., Park H., Lukin M. D., Nuclear magnetic resonance detection and spectroscopy of single proteins using quantum logic. Science 351, 836–841 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Grinolds M. S., Warner M., De Greve K., Dovzhenko Y., Thiel L., Walsworth R. L., Hong S., Maletinsky P., Yacoby A., Subnanometre resolution in three-dimensional magnetic resonance imaging of individual dark spins. Nat. Nanotechnol. 9, 279–284 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Häberle T., Schmid-Lorch D., Reinhard F., Wrachtrup J., Nanoscale nuclear magnetic imaging with chemical contrast. Nat. Nanotechnol. 10, 125–128 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Rugar D., Mamin H. J., Sherwood M. H., Kim M., Rettner C. T., Ohno K., Awschalom D. D., Proton magnetic resonance imaging using a nitrogen–vacancy spin sensor. Nat. Nanotechnol. 10, 120–124 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Le Sage D., Arai K., Glenn D. R., DeVience S. J., Pham L. M., Rahn-Lee L., Lukin M. D., Yacoby A., Komeili A., Walsworth R. L., Optical magnetic imaging of living cells. Nature 496, 486–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall L. T., Cole J. H., Hill C. D., Hollenberg L. C. L., Sensing of fluctuating nanoscale magnetic fields using nitrogen-vacancy centers in diamond. Phys. Rev. Lett. 103, 220802 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Steinert S., Ziem F., Hall L. T., Zappe A., Schweikert M., Götz N., Aird A., Balasubramanian G., Hollenberg L., Wrachtrup J., Magnetic spin imaging under ambient conditions with sub-cellular resolution. Nat. Commun. 4, 1607 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Ermakova A., Pramanik G., Cai J.-M., Algara-Siller G., Kaiser U., Weil T., Tzeng Y.-K., Chang H. C., McGuinness L. P., Plenio M. B., Naydenov B., Jelezko F., Detection of a few metallo-protein molecules using color centers in nanodiamonds. Nano. Lett. 13, 3305–3309 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Simpson D. A., Ryan R. G., Hall L. T., Panchenko E., Drew S. C., Petrou S., Donnelly P. S., Mulvaney P., Hollenberg L. C. L., Electron paramagnetic resonance microscopy using spins in diamond under ambient conditions. Nat. Commun. 8, 458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood J. D. A., Tetienne J.-P., Broadway D. A., Hall L. T., Simpson D. A., Stacey A., Hollenberg L. C. L., Microwave-free nuclear magnetic resonance at molecular scales. Nat. Commun. 8, 15950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momenzadeh S. A., Stöhr R. J., de Oliveira F. F., Brunner A., Denisenko A., Yang S., Reinhard F., Wrachtrup J., Nanoengineered diamond waveguide as a robust bright platform for nanomagnetometry using shallow nitrogen vacancy centers. Nano. Lett. 15, 165–169 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Rong X., Wang M., Geng J., Qin X., Guo M., Jiao M., Xie Y., Wang P., Huang P., Shi F., Cai Y.-F., Zou C., Du J., Searching for an exotic spin-dependent interaction with a single electron-spin quantum sensor. Nat. Commun. 9, 739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chasteen N. D., Harrison P. M., Mineralization in ferritin: An efficient means of iron storage. J. Struct. Biol. 126, 182–194 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Schäfer-Nolte E., Schlipf L., Ternes M., Reinhard F., Kern K., Wrachtrup J., Tracking temperature-dependent relaxation times of ferritin nanomagnets with a wideband quantum spectrometer. Phys. Rev. Lett. 113, 217204 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Di Rienzo C., Piazza V., Gratton E., Beltram F., Cardarelli F., Probing short-range protein Brownian motion in the cytoplasm of living cells. Nat. Commun. 5, 5891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentze M. W., Muckenthaler M. U., Galy B., Camaschella C., Two to tango: Regulation of Mammalian iron metabolism. Cell 142, 24–38 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Quintana C., Bellefqih S., Laval J. Y., Guerquin-Kern J. L., Wu T. D., Avila J., Ferrer I., Arranz R., Patiño C., Study of the localization of iron, ferritin, and hemosiderin in Alzheimer’s disease hippocampus by analytical microscopy at the subcellular level. J. Struct. Biol. 153, 42–54 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Sigal Y. M., Speer C. M., Babcock H. P., Zhuang X., Mapping synaptic input fields of neurons with super-resolution imaging. Cell 163, 493–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abrams D. S., Lloyd S., Quantum algorithm providing exponential speed increase for finding eigenvalues and eigenvectors. Phys. Rev. Lett. 83, 5162–5165 (1999). [Google Scholar]
- 28.J. F. Ziegler, J. P. Biersack, in Treatise on Heavy-Ion Science: Volume 6: Astrophysics, Chemistry, and Condensed Matter, D. A. Bromley, Ed. (Springer US, 1985), pp. 93–129. [Google Scholar]
- 29.Ziem F. C., Götz N. S., Zappe A., Steinert S., Wrachtrup J., Highly sensitive detection of physiological spins in a microfluidic device. Nano. Lett. 13, 4093–4098 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaau8038/DC1
Section S1. Principle of spin noise measurement based on the NV center
Section S2. Calculation of the spin noise intensity from the single-τ measurement
Section S3. Point spread function of a single ferritin
Fig. S1. Experimental setup.
Fig. S2. Images of the nanopillars on diamonds.
Fig. S3. The treatment of FAC significantly increased the ferritins in HepG2 cells.
Fig. S4. Electron spin resonance spectra of resin-embedded HepG2 cells.
Fig. S5. The AFM characterization of the surface of cell sample.
Fig. S6. Adjusting the distance between the NV center and the cell section.
Fig. S7. Calculated spin noise intensity as a function of the distance between NV center and ferritin.
Fig. S8. Simulation of noise intensity at different vertical distances between a single ferritin and NV center.
Fig. S9. Trace data of Fig. 4A.
Reference (29)


