Abstract
Animal models support a role for the gut microbiota in the development of hypertension. There has been a lack of epidemiologic cohort studies to confirm these findings in human populations. We examined cross-sectional associations between measures of gut microbial diversity and taxonomic composition and blood pressure in 529 participants of the biracial (African- and European-American) Coronary Artery Risk Development in Young Adults (CARDIA) study. We sequenced V3-V4 regions of the 16S ribosomal RNA marker gene using DNA extracted from stool samples collected at CARDIA’s Year 30 follow-up examination (2015-16; aged 48-60 years). We quantified associations between blood pressure [hypertension (defined as systolic blood pressure (SBP) ≥ 140 mmHg and/or diastolic blood pressure ≥ 90 mmHg and/or antihypertension medication use) and SBP] and with- and between-person diversity measures. We conducted genera-specific multivariable-adjusted regression analysis, accounting for multiple comparisons using the false discovery rate. Hypertension and SBP were inversely associated with measures of alpha-diversity, including richness and the Shannon Diversity Index, and were distinguished with respect to principal coordinates based on a similarity matrix of genera abundance. Several specific genera were significantly associated with hypertension and SBP, though results were attenuated with adjustment for body mass index. Our findings support associations between within-person and between-person gut microbial community diversity and taxonomic composition and blood pressure in a diverse population-based cohort of middle-aged adults. Future work is needed to define functional pathways that underlie observed associations and identify specific microbial targets for intervention.
Keywords: Blood pressure, hypertension, gastrointestinal microbiome, epidemiology, population
Introduction
There is growing evidence that the gut microbiota may influence cardiovascular disease (CVD)1–5. Proposed mechanisms include gut microbial effects on systemic inflammation6, 7 and the production of CVD-related metabolites, such as trimethylamine N-oxide (TMAO)1, 2 and short-chain fatty acids. Findings from animal and human studies are consistent with a role for the gut microbiota in obesity3, 8, type 2 diabetes9–12, dyslipidemia3, metabolic syndrome5, 13, and lifetime CVD risk4. Animal models have demonstrated gut microbial effects on blood pressure14–17}. Decreased microbial diversity has been observed in both animal models of hypertension and human samples16. Population-based human studies have revealed significant associations between microbial metabolites and blood pressure18, 19}. However, there has been a lack of data on the gut microbiome and blood pressure in population-based and sociodemographically-diverse samples.
In the current study, we examined cross-sectional associations between gut microbial diversity and taxonomic composition and blood pressure in 529 middle-aged adults recruited from 4 U.S. urban field centers in the Coronary Artery Risk Development in Young Adults (CARDIA) study. CARDIA is a population-based and sociodemographically-diverse sample of African- and European-American participants, with clinic-based measurement of blood pressure and extensive data on relevant covariates, including diet, antihypertensive medication use, and body mass index (BMI). At CARDIA’s Year 30 follow-up examination in 2015–16, we collected stool samples and sequenced the 16S ribosomal RNA (rRNA) prokaryotic marker gene. We hypothesized that: 1) the gut microbial community differs significantly according to blood pressure, 2) within-person diversity of the gut microbiota is inversely associated with hypertension and systolic blood pressure (SBP), and 3) hypertension and SBP are associated with specific taxa, such as those involved in the production of the short-chain fatty acid butyrate.
Methods
Data and code availability
All data used in the present analysis are available from the CARDIA Study Data Coordinating Center at the University of Alabama at Birmingham. The process for obtaining data through CARDIA is outlined at: https://www.cardia.dopm.uab.edu/publications-2/publications-documents. Computer code/scripts used in the generation of data and statistical analysis are available from the authors upon request.
Study participants
The CARDIA study is a prospective multicenter cohort study designed to study the evolution of CVD over adulthood. CARDIA began in 1985–86 and enrolled 5,115 participants aged 18–30 years from 4 U.S. urban centers (Birmingham, AL; Chicago, IL; Minneapolis, MN; Oakland, CA)20. Since baseline, there have been 8 follow-up examination (years 2, 5, 7, 10, 15, 20, 25, and 30) with retention among survivors of 91%, 86%, 81%, 79%, 74%, 72%, 72%, and 71%, respectively.
A microbiome sub-study was initiated at the Year 30 follow-up examination (2015–2016). Participants were recruited sequentially until the target sample size of 600 was reached (n=300 from the Chicago, IL, field center and n=100 from each of the other 3 field centers). Exclusions were based on a screening questionnaire administered at the time of CARDIA contact, including if participants: were pregnant at the time of the clinic exam; used antibiotics in the past month; had ever been diagnosed with inflammatory bowel disease; reported having had a gastrointestinal illness, vomiting, diarrhea, or atypical constipation in the past week. The present analysis includes data from CARDIA participants with complete sequencing data (n=538). Participants were excluded from analysis if they were missing data on blood pressure medication (n=2), diet (n=2), or physical activity (n=5), for an analytic sample of n=529 participants. CARDIA was approved by institutional review boards of each field center; each study participant provided informed written consent for both the CARDIA core examination and the microbiome sub-study.
Measurement of sociodemographic, behavioral, and clinical characteristics
Standard questionnaires were used to obtain demographic and health behavioral data at the CARDIA field centers during the core examination. The interviewer-administered CARDIA Physical Activity Questionnaire queried past-year engagement in activities, from which a total activity score was calculated21. Participants reported their use of medication for hypertension, lipid-lowering, and diabetes; and their current and historic use of tobacco products. A brief 23-item qualitative diet assessment was completed by participants in the microbiome sub-study22. A summary measure of diet quality (a priori diet quality score) was derived as previously in CARDIA23 and other studies24. Based on hypothesized impact on health, foods were classified into beneficial, adverse or neutral quality and quartiles were created: beneficial foods were scored positively (0 to 3 for the 1st to 4th quartile, respectively), adverse foods were scored inversely (3 to 0 for the 1st to 4th quartile, respectively), and neutral foods were not scored. Scores were summed over all foods; higher scores reflect higher diet quality.
Standardized protocols were used by trained staff for all clinic measures. Resting SBP and diastolic blood pressure (DBP) measures were taken in the seated position with elbow and forearm resting on the chair armrest. Blood pressure values were calculated as the mean of the second and third of three measurements taken with oscillometer (OmROn HEM907XL automated/oscillometric blood pressure monitor) calibrated to a random-zero sphygmomanometer. Arm cuff bladder size was based on arm circumference as follows (9 cm cuff for 17.0–22.5 cm arm; 12.0 cm for 22.6–32.5 cm arm; 15.0 cm for 32.6–42.5 cm arm; and 17.5 cm cuff for 42.6–50.0 cm arm). For individuals with an arm circumference >50 cm, a thigh cuff was used with an OmROn 108ML aneroid/manual blood pressure monitor. Hypertension was defined as current use of anti-hypertensive medication, a SBP ≥ 140 mmHg, and/or a DBP ≥ 90 mmHg. Height and weight were measured to the nearest 0.5 cm and 0.2 kg, respectively for body mass index (BMI, kg/m2). Fasting serum glucose was measured using hexokinase coupled to glucose-6-phosphate dehydrogenase. Diabetes was defined as having fasting glucose ≥126 mg/dL (7 mmol/L), 2-hour OGTT ≥200 mg/dL (11.1 mmol/L), HbA1c ≥6.5% (48 mmol/mol), or the use of hypoglycemic medications.
Microbiome data collection, sequencing, and data processing
We followed standard protocols for stool collection and processing25, 26. Briefly, stool samples were collected by participants in their home, shipped overnight to the study lab at UNC-Chapel Hill, where they were stored at −80 °C until processing. Collection tubes were pre-filled with RNAlater to stabilize microbial DNA during transport to the lab. At the time of their stool collection, participants completed a short survey, including the time and date of their collection, past year antibiotic use, use of fiber supplements or probiotics, and their average weekly stool frequency.
DNA was extracted from 0.20 grams of stool using the MoBio PowerSoil kit. The V3-V4 hypervariable regions were amplified (primers: 341F/785R) and sequenced using the Illumina MiSeq platform (2×300). Processing of sequence data was completed with BioLockJ, a Java-based pipeline for metagenomics analysis27. Paired-end sequences were merged with Paired-End reAd mergeR (PEAR, v 0.9.8)28 using default arguments; sequences for which primers did not match or for which 10 base pairs did not overlap were excluded. Taxonomic assignment was with the Ribosomal Database Project (RDP) Classifier v2.12 (confidence threshold=80%)29.
The R package ‘vegan’ was used to generate measures of microbial diversity30. Within-person diversity (alpha diversity) measures included the Shannon Diversity Index and richness, both derived at the genus level31. Richness was calculated as the number of distinct genera per participant, with total per-participant abundance rarified through random sampling to the minimum genera count across all participants. We used Principal Coordinates Analysis (PCoA) to assess between-participant diversity (beta-diversity) in microbial community composition. PCoA was used to generate orthogonal summary measures of microbial composition based on a distance matrix of microbial abundance (Bray-Curtis)32. We report on PCoA axes that explain at least 5% of the variability in the taxonomic similarity measure. To assess sensitivity of findings to distance matrix, we additionally generated factors with principal components analysis (PCA) based on a Euclidean distance matrix.
Statistical analysis
We compared the analytic sample (n=529) to CARDIA participants who attended the Year 30 follow-up examination, but did not participate in the Microbiome Study (n=2,752). We assessed differences in categorical variables with chi-square and continuous variables with non-parametric test for comparing means or medians if the variable was not normally distributed.
Primary outcomes were hypertension and SBP. We controlled for antihypertensive medication use in analysis of SBP, as medications may alter the gut microbiota33, 34. We conducted multivariable-adjusted regression models for alpha- and beta-diversity measures of microbial community composition, as well as for analysis of individual taxa. We tested for differences in beta-diversity, represented using PCoA, with permutational multivariate analysis of variance using distance matrices (PERMANOVA), through which pseudo-F ratios are generated and p-values obtained through permutations35(here, 1000 permutations were used). In an effort to evaluate the connection between specific taxa and PCoA ordination, we overlaid biplots with vectors for the 10 most abundant genera that significantly differentiate the PCoA axes. Vectors point in the direction of taxa-specific associations with respect to the PCoA axes, with vector length proportional to the correlation between the specific taxa and PCoA axes.
We conducted separate regression models for each taxonomic group with respect to: 1) hypertension and 2) systolic blood pressure. We focused our primary analysis on genera, the lowest level of classification from our 16S rRNA sequences. In addition, we conducted secondary analysis at the family level, which allows us to test associations between blood pressure and families that have been shown to carry genes for butyrate production pathways36. To account for spurious findings due to rare taxa, we restricted analysis to taxa that were present in at least 25% of participants; after this restriction, 149 genera and 42 families remained from among 379 genera and 100 families originally identified in the data37. Raw taxonomic counts were transformed for analysis as log10[(RC/n)(x/N)+1], where RC is the total raw taxon count for a participant and n is the total count across all taxa for a participant, x is the total across all OTUs and participants and N is the total number of participants38. We controlled taxonomic analysis for multiple comparisons using the Benjamini-Hochberg method for false discovery rate (FDR)39.
We conducted several multivariable-adjusted regression models. A minimal model (Model 1) adjusted for sequencing run. Model 2 included additional covariate adjustment for field center (4 categories), sex (male/female), race (African-/European-American), age (continuous), and educational attainment (continuous). In a more fully adjusted model (Model 3), we adjusted for physical activity (continuous), smoking status (current, former, never), and diet quality score (continuous). We additionally adjusted for antihypertensive medication use in Model 3 analysis of SBP. Finally, in Model 4, we adjusted for BMI (continuous), a potential intermediate of microbiome effects on blood pressure. Data analysis was conducted in R 3.4.2 (http://www.r-project.org) and SAS version 9.4 (SAS Institute Inc., Cary, NC).
Results
Study participants differed slightly from the full CARDIA cohort examined at the Year 30 follow-up examination. Microbiome Study participants were generally similar to non-participants (except for study center, by design of the Microbiome Study), though participants had statistically significantly lower mean BMI and waist circumference, and a smaller proportion had hypertension (Table 1). The relative abundance (percentage) of the 149 genera included in analysis (after removing rare taxa) ranged from a mean of 0.00044% for Parascardovia to 31.38% for Bacteroides (S1).
Table 1.
| Participant characteristic | Participants | Non-Participants | p-value3 |
|---|---|---|---|
| N | 529 | 2,752 | |
| Age, yr | 55.3 (3.4) | 55 (3.6) | 0.068 |
| Female, % | 53.9 | 57.9 | 0.09 |
| African-American, % | 44.4 | 48.4 | 0.09 |
| Field center, % | <0.0001 | ||
| Birmingham | 13.4 | 24.6 | |
| Chicago | 53.4 | 14.9 | |
| Minneapolis | 19.5 | 27.4 | |
| Oakland | 13.8 | 33.1 | |
| Educational attainment, yr | 15.9 (2.6) | 15.8 (3.1) | 0.17 |
| Smoking status, % | 0.30 | ||
| Current | 12.3 | 14.5 | |
| Former | 22.2 | 23.3 | |
| Physical activity, intensity units, med(IQR) | 277 (127, 510) | 252 (116, 456) | 0.14 |
| BMI, kg/m2 | 29.4 (6.3) | 30.8 (7.4) | 0.0003 |
| Waist circumference, cm | 94.4 (15.6) | 96.6 (16.6) | 0.024 |
| Diabetes, % | 12.4 | 14.8 | 0.15 |
| Systolic blood pressure, mmHg | 119.4 (15.8) | 119.8 (16.1) | 0.92 |
| Diastolic blood pressure, mmHg | 72.9 | 73.2 | 0.91 |
| Antihypertensive medication use, % | 29.2 | 34.1 | 0.09 |
| Hypertension, % | 35.1 | 40.3 | 0.03 |
Mean (SD) unless noted.
Non-participants who attended the c examination.
Differences in categorical variables assessed with chi-square and continuous variables with a non-parametric test for comparing means (or medians for physical activity).
Several measures of gut microbial composition varied significantly with respect to sociodemographic and anthropometric variables in univariate analysis (S2-S7). There were significant differences (PERMANOVA p-value=0.001) in between-person diversity (beta-diversity) for all variables: age, gender, race, BMI, and waist circumference. In Within-person diversity (alpha-diversity) was inversely associated with BMI and waist circumference, but was not associated with age, gender, or race (S2). Specific genera were significantly associated with sociodemographic and anthropometric variables in FDR-adjusted univariate analysis of 149 genera. At an FDR-adjusted p-value threshold of 0.05, 5 genera were associated with age, 44 were associated with gender, 89 were associated with race, 63 were associated with BMI, and 58 were associated with waist circumference (S3-S7, respectively).
Regression results were consistent with an inverse association between blood pressure measures and microbial diversity, in particular microbial richness (Table 2). In multivariable-adjusted regression models, hypertension was statistically significantly inversely associated with genera richness (Model 3; OR=0.75 (95% CI: 0.60, 0.94 for a standard deviation (SD)-unit increase in richness), but was not significantly associated with the Shannon diversity index. An SD-unit increase in richness was associated with −1.52 (−2.92, −0.12) mmHg lower SBP, and an SD-unit increase in Shannon diversity index was associated with −1.44 (−2.72, −0.16) mmHg lower SBP. Inverse relations were attenuated after further adjustment for BMI, a potential intermediate (Model 4).
Table 2.
Multivariable-adjusted1 associations between gut microbial alpha diversity2 and blood pressure measures3 in CARDIA4.
| Hypertension | Systolic blood pressure | |||
|---|---|---|---|---|
| Model specification | Odds ratio (95% CI) | p-value | Beta coefficient (95% CI) | p-value |
| Shannon index | ||||
| Model 1 | 0.83 (0.69, 1.00) | 0.053 | −1.69 (−2.99, −0.28) | 0.018 |
| Model 2 | 0.82 (0.67, 1.00) | 0.052 | −1.50 (−2.78, −0.22) | 0.022 |
| Model 3 | 0.86 (0.70, 1.06) | 0.10 | −1.44 (−2.72, −0.16) | 0.028 |
| Model 4 | 0.88 (0.71, 1.10) | 0.26 | −1.33 (−2.60, −0.052) | 0.042 |
| Model 5 | 0.90 (0.72, 1.11) | 0.32 | −1.29 (−2.57, −0.0087) | 0.049 |
| Richness | ||||
| Model 1 | 0.72 (0.59, 0.88) | 0.0014 | −1.75 (−3.19, −0.31) | 0.018 |
| Model 2 | 0.70 (0.56, 0.87) | 0.0013 | −1.71 (−3.09, −0.33) | 0.016 |
| Model 3 | 0.75 (0.60, 0.94) | 0.012 | −1.52 (−2.92, −0.12) | 0.033 |
| Model 4 | 0.78 (0.61, 0.99) | 0.037 | −1.37 (−2.76, 0.031) | 0.056 |
| Model 5 | 0.79 (0.62, 1.00) | 0.048 | −1.32 (−2.73, 0.083) | 0.065 |
Multivariable-adjusted regression models from –glm– command in R: family=binomial for odds ratios; family=Gaussian for beta coefficients. Model 1 adjusted for sequencing run. Model 2 additionally adjusted for age, gender, race, clinical field center, and education. Model 3 additionally adjusted for diet quality score, physical activity, and smoking status. Model 3 was also adjusted for antihypertensive medication use in models for systolic blood pressure. Model 4 included Model 3 covariates, additionally adjusted for BMI. Model 5 included Model 3 covariates, additionally adjusted for waist circumference.
Associations are per standard deviation unit of genus-level diversity measures: Shannon index mean (SD)=2.46 (0.35); richness (SD)=90.2 (11.3).
Hypertensive was defined as taking an antihypertensive medication, having systolic blood pressure ≥ 140, or having diastolic blood pressure ≥ 90.
n=529 participants at the CARDIA Year 30 exam (2015-2016).
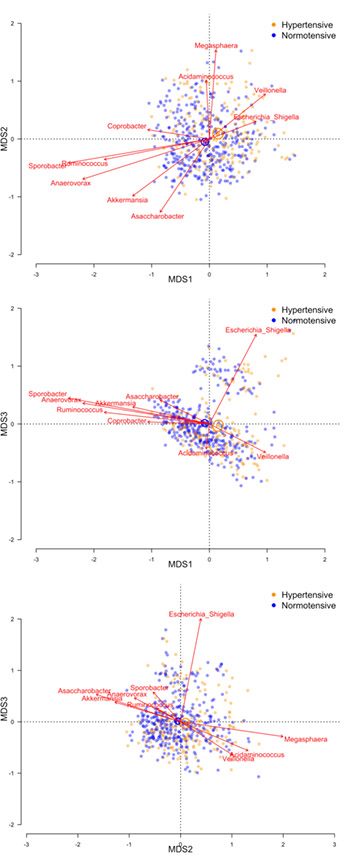
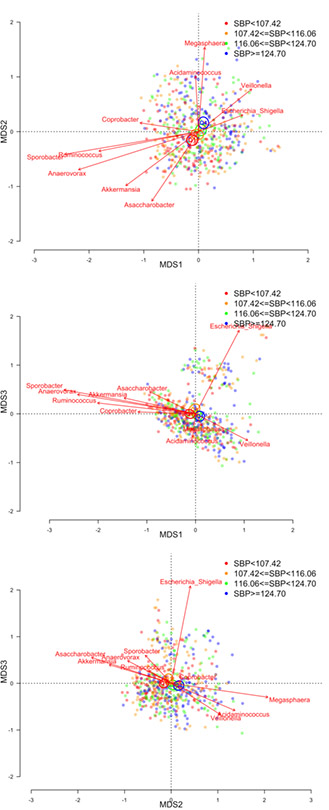
Beta-diversity was significantly associated with both hypertension and systolic blood pressure in all multivariable-adjusted models (all PERMANOVA p-values = 0.001). Figures 1 and 2 presents biplots for each pairwise comparison of the first 3 PCoA axes with respect to hypertension and systolic blood pressure, respectively. Each of the first 3 PCoA axes explained at least 5% of the variability in the microbial similarity measure (11.2%, 8.6%, and 5.4% for the first, second, and third PCoA axes, respectively). Vectors indicate the 10 most abundant genera that were significantly differentiated between PCoA axes. We note that significant vectors do not necessarily indicate that specific taxa are significantly associated with blood pressure. Still, the vectors are generally consistent with what we observed in taxa-specific analysis. Vectors Akkermansia, Ruminococcus, Anaerovorax, Sporobacter, and Asaccharobacter tended to align in direction with individuals who were normotensive (Figure 1) or had lower systolic blood pressure (Figure 2), while Veillonella aligned with individuals who were hypertensive (Figure 1) or had higher systolic blood pressure (Figure 2). Furthermore, spearman correlation coefficients for genera and PCoA axes were consistent with the displays of generat vectors and PCoA axes (S1). For example, Akkermansia was negatively correlated with PCoA axes 1 and 3, which were positively associated with hypertension and SBP; while Akkermansia was positively associated with PCoA 2, which was negatively associated with blood pressure.
Figure 1.
Microbial similarity biplots (joint PCoA axes) for study participants with hypertension (orange) or normal blood pressure (blue). Biplots shown for PCoA axes that explain at least 5% of variability in microbial similarity. Centroids illustrate the 95% confidence interval for the mean location of each population group. The ten most abundant genera are shown with respect to their directional association along PCoA axes, with vector length indicating the strength of association. PERMANOVA p-values were 0.001 for hypertension in each of the five multivariable-adjusted models. Model 1 adjusted for sequencing run; Model 2 additionally adjusted for age, race, gender, study center, and educational attainment; Model 3 additionally adjusted for smoking, physical activity, and diet quality score; Model 4 additionally adjusted for BMI; Model 5: adjusted for Model 3 covariates, with the addition of waist circumference.
Figure 2.
Microbial similarity biplots (joint PCoA axes) for study participants with respect to quartiles (Q1-Q4) of systolic blood pressure (Q1: red; Q2: brown; Q3: green; Q4: blue). Biplots shown for PCoA axes that explain at least 5% of variability in microbial similarity. Centroids illustrate the 95% confidence interval for the mean location of each population group. The ten most abundant genera are shown with respect to their directional association along PCoA axes, with vector length indicating the strength of association. PERMANOVA p-values were 0.001 for systolic blood pressure in each of the five multivariable-adjusted models. Model 1 adjusted for sequencing run; Model 2 additionally adjusted for age, race, gender, study center, and educational attainment; Model 3 additionally adjusted for smoking, physical activity, diet quality score, and antihypertensive medication; Model 4 additionally adjusted for BMI; Model 5: adjusted for Model 3 covariates, with the addition of waist circumference.
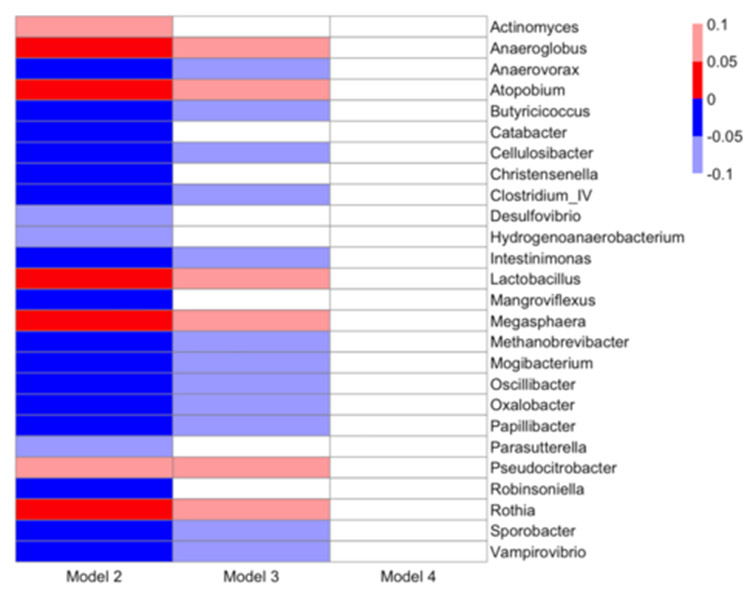
We conducted multivariable-adjusted regression analysis of individual genera with respect to blood pressure (Figures 3 and 4; S8 and S9). In unadjusted analysis, a large number of genera were associated with hypertension (65 genera with FDR-adjusted p-value <0.05 and 74 with FDR-adjusted p-value <0.10) and SBP (34 genera with FDR-adjusted p-value <0.05, and 51 with FDR-adjusted p-value <0.10) (Figure 3 and S8). Findings attenuated with multivariable adjustment. Upon adjustment for sociodemographic variables age, race, sex, field center, and educational attainment (Model 2), hypertension remained significantly associated with 21 genera at FDR-adjusted p-value <0.05 and with 5 genera at FDR-adjusted p-value <0.10 (Figure 2). Several genera were significantly associated with hypertension in directions consistent with PCoA findings (S1), such as positive associations between hypertension and genera Anaerovorax, Clostridium IV, Oscillibacter, and Sporobacter. With further adjustment for health behaviors (diet, physical activity, and smoking), 18 genera remained associated with hypertension with FDR-adjusted p-values <0.10, including positive associations with Anaerovorax, Clostridium IV, Oscillibacter, and Sporobacter. Following control for BMI, a potential mediator, no genus was associated with hypertension with an FDR-adjusted p-value <0.25. Results were not meaningfully different with adjustment for waist circumference instead of BMI (S8).
Figure 3.
Heatmap of associations between genera and hypertension from multivariable-adjusted models. Direction of association is indicated by color (blue: negative, red: positive) and FDR-adjusted p-values (q-values) are indicated by shading (bold: q-value<0.05, light: 0.05≤q-value<1.0). Multivariable-adjusted regression models adjusted for: Model 2: sequencing run, age, race, gender, study center, educational attainment; Model 3: additionally adjusted for smoking, physical activity, and diet quality score; Model 4: additionally adjusted for BMI. Results are not shown for Model 5, which adjusted for Model 3 covariates plus waist circumference, as Model 5 results were not meaningfully different from Model 4 results.
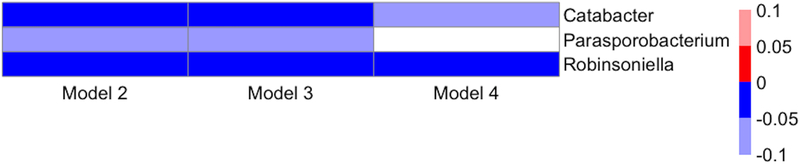
Figure 4.
Heatmap of associations between genera and systolic blood pressure from multivariable-adjusted models. Direction of association is indicated by color (blue: negative, red: positive) and FDR-adjusted p-values (q-values) are indicated by shading (bold: q-value<0.05, light: 0.05≤q-value<1.0). Multivariable-adjusted regression models adjusted for: Model 2: sequencing run, age, race, gender, study center, educational attainment; Model 3: additionally adjusted for smoking, physical activity, diet quality score, and antihypertensive medication use; Model 4: additionally adjusted for BMI. Results are not shown for Model 5, which adjusted for Model 3 covariates plus waist circumference, as Model 5 results were not meaningfully different from Model 4 results.
Findings between specific genera and SBP were appreciably weaker (Figure 4 and S9). SBP was positively associated with Catabacter and Robinsoneilla at FDR-adjusted p-value <0.05 and with Parasporobacterium at FDR-adjusted p-value <0.10 in Models 2 and 3; SBP remained positively associated with Catabacter (FDR-adjusted p-value <0.10) and Robinsoneilla (FDR-adjusted p-value <0.05) with additional adjustment for BMI (Model 4). Similarly, both Catabacter and Robinsoleilla remained significant at FDR-adjusted p-value<0.05 when the model adjusted for waist circumference instead of BMI (S9). Catabacter and Robinsoleilla were positively associated with hypertension in Model 2 (FDR-adjusted p-value <0.05), but these findings were attenuated with additional adjustment (FDR-adjusted p-values 0.11 and 0.40 in Models 3 and 4, respectively, for Catabacter, and FDR-adjusted p-values 0.11 and 0.40 in Models 3 and 4, respectively, for Robinsoleilla).. We noted the possibility that results of hypertension may reflect use of antihypertensive medication, and considered stratified analysis to distinguish hypertension and medication use. However, there was insufficient sample size to examine subgroups robustly. Among the 183 participants with hypertension in the analysis, 153 reported taking antihypertensive medication, of whom 128 were controlled (SBP <140 mmHg and DBP <90 mmHg). Therefore, analysis of medication use in participants with normal hypertension would have been limited to 128 individuals, and analysis of hypertension in participants not taking antihypertensive medication would have been limited to the 30 individuals we were able to examine
Given observed differences in the microbial community by race and sex (S4 and S5)40, we assessed the possibility of effect modification by race and sex by including a cross-product term for race or sex in regression models. These tests were not statistically significant, and we therefore present results adjusted for race and sex.
Secondary analysis of family level data did not in general support a hypothesis of inverse associations between blood pressure and families that we considered relevant for butyrate production, based on published literature36. We focused on six families that include numerous species shown to carry genes for enzymes in the acetyl-CoA butyrate-production pathway: Lachnospiraceae, Peptostreptococcaceae, Clostridiales incertae sedix XI, Clostridiaceae, Ruminococcaceae, and Veillonellaceae.36. Two families were significantly associated with hypertension in univariate and semi-adjusted models: Ruminococcaceae was inversely associated through Model 2 (sociodemographics-adjusted); Veillonellacaeae was positively through Model 3 (sociodemographics and health behaviors) (S10). No family was significantly associated with systolic blood pressure in univariate or multivariable-adjusted models (S11).
Discussion
In the CARDIA Microbiome Study, several measures of the gut microbiota were significantly associated with blood pressure in cross-sectional analysis. Measures of within-person microbial diversity were inversely associated with hypertension and SBP, and there was significant separation of blood pressure according to microbial dissimilarity in PCoA. These differences remained significant after adjustment for a range of demographic and health behavior covariates. Several specific taxonomic groups appeared associated with hypertension and SBP, though these findings were sensitive to covariate adjustment, in particular BMI, and generally were not statistically significant after adjustment for multiple comparisons.
A growing body of literature supports a role of the gut microbiota in CVD risk, but there have been relatively few population-based studies of gut microbiota and CVD risk factors and we know of no study that has focused on blood pressure. In a case-control analysis of 112 participants from the Bogalusa Heart Study, Kelly et al. found that microbial richness and several distinct taxonomic groups were associated with a measure of lifetime CVD risk score comprising fasting glucose, LDL-C, and SBP4. Similar to Kelly, et al.4, and other studies of CVD risk factors10, our findings for microbial community richness were more robust than findings for diversity measures that incorporate both richness and evenness such as the Shannon Diversity Index.
There has been a lack of analysis on blood pressure in population-based adult cohorts. Hypertension has been induced in normotensive rats through transplantation of cecal contents from hypertensive rats15, 41. Data support microbiota-dependent production of short-chain fatty acids (SCFAs), such as butyrate, acetate, and propionate, as one mechanism through which gut microbiota may influence blood pressure 42–45. In a study of 205 overweight and obese pregnant women, blood pressure was associated with gut microbiota composition and, specifically, inversely associated with butyrate-producing bacteria in the gut microbiota45. Microbiota-generated SCFAs have been shown to influence blood pressure through olfactory receptors expressed in the vasculature and kidneys42, 43. Additional microbial metabolites of dietary components may also play a role. In a population-based metabolomics analysis, urinary hippurate was inversely associated with blood pressure in the INTERMAP study18; serum hydroxy-hippurate was positively associated with incident hypertension in ARIC19.
Results were more robust for microbial diversity (alpha- and beta-diversity) measures, as compared to analysis of specific taxa. These findings are consistent with previous support for community-level measures3–5. Analyses of microbial diversity will be more powerful than multiple-comparisons-adjusted analyses of individual taxa. In addition, these results may reflect aspects of the gut microbial community not captured in analysis of specific genera. Diversity measures remained significant after adjustment for potential confounders, but many individual genera lost statistical significance after adjustment for demographic, health behavior, and clinical covariates. In particular, BMI, a potential intermediate, appeared to account for much observed attenuation, especially in analysis of hypertension. At this stage of microbiome research, the relevance of various covariates has not been firmly established, and studies have varied with respect to adjustments included in analysis. For example, adjustment for diet had a modest impact in our analysis, but was an important covariate in Bogalusa4; diet was not included as a covariate in analyses of Lifelines-DEEP or METSIM3, 5. Future work and a growing number of studies with microbiome data will contribute to our understanding46, 47.
The larger number of significant associations of genera with hypertension, as compared to SBP, may reflect the greater severity of the phenotype. We also considered the possibility that the use of antihypertensive treatment may influence the gut microbiome, but were unable to disentangle associations between hypertension and medication use in our analysis, given the very small number of individuals who were discordant on hypertension status and antihypertensive medication use. Data from animal models are supportive of a causal effect of the gut microbiome on blood pressure15, 41–43, but longitudinal data and larger samples are needed to confirm the relevance of the gut microbiome in human populations and robustly identify specific bacteria that may serve as targets for intervention.
Our analysis provides two approaches to assessing specific taxa with respect to blood pressure. Several of the most abundant genera that differentiated PCoA axes have been previously associated with pathways that may influence blood pressure. Notably, Akkermansia—aligned with axes associated with normotension in analysis—may signal improved gut epithelial integrity, and has been associated with obesity, diabetes, and inflammation48–50. Sporobacter and Ruminococcus—both aligned with axes associated with normotension—are members of the Ruminococcaceae family within the phylum Firmicutes. Several genera were associated with blood pressure in both PCoA and taxa-specific analysis; for example, Sporobacter and Anaerovorax were inversely associated with hypertension in both analyses. Taxa-specific analyses revealed additional genera-blood pressure associations, with Robinsoniella and Catabacter positively associated with both hypertension and SBP.
Based on our 16S rRNA sequence data, results did not support our hypothesis that taxa, at the family level, related to the production of short-chain fatty acids, particularly butyrate, are inversely associated with blood pressure. For this analysis, we focused on taxa at the family level, given the availability of published data for genes in butyrate-producing pathways at family and species, but not genus, levels36. One challenge with this approach is that our data may not be sufficiently precise, as we would expect appreciable variability in function within taxonomic levels available from 16S rRNA data. Analysis at the gene level, using whole-metagenomic sequencing, would allow improved pathway assignment based on the presence of relevant genes. Furthermore, short-chain fatty acids are a diverse class of molecules, with reported inverse associations between blood pressure and acetate51, propionate44, and formate18. Vital, et al. provide a catalog of families and species relevant for butyrate production36, but there is a paucity of literature defining the full set of pathways for comprehensive analysis of short-chain fatty acids.
Our paper addresses a lack of population-based studies of the gut microbiota with respect to blood pressure. The CARDIA cohort allowed analysis of a socio-demographically diverse group of adults at a critical life period for increasing CVD risk. Blood pressure was measured by trained field center clinic staff using a standardized protocol. We used validated protocols for the collection and processing of samples and 16S rRNA sequencing. CARDIA collects extensive covariate information using standardized and validated instruments and we were able to control for major potential confounders, including health behaviors, anti-hypertensive medication use, and clinic-based assessment of anthropometry.
Our study also has limitations. This was the first collection of microbiome data in CARDIA, which prevents the establishment of temporality in these cross-sectional analyses. In particular, we cannot distinguish the role of the gut microbiota in the development or progression of hypertension from the possibility that blood pressure itself, or associated covariates, may alter the microbiota. The gut microbiome has been associated with several CVD risk factors that correlate with blood pressure3–5; aside from BMI and health behaviors, we did not include other risk factors in regression modeling and it is possible that some of the microbial variability associated with blood pressure reflects other clinical measures. Our sample size was along the lines of other epidemiologic studies of gut microbiota, but it is possible that we lacked power in our multiple comparisons analysis of individual genera. In addition, there are few population-based studies with microbiome data, particularly with representation similar to CARDIA, and we lacked data for replication of results. Future analysis of blood pressure and microbiome in independent samples is needed to confirm our findings. Our analysis was limited to 16S rRNA sequence data, which yields compositional measures and taxonomic analysis to the genus level.
Perspectives
Our findings support an association between the gut microbiota and blood pressure in a biracial middle-aged population-based cohort. Microbial diversity was inversely associated with hypertension and SBP. Several specific genera were significantly associated with blood pressure after adjustment for potential confounders and for multiple comparisons, but findings were attenuated upon adjustment for BMI. Further studies are needed to quantify prospective associations in larger samples and with functional measures and refined compositional information to assess sub-genus taxonomies and functional differences that underlie observed associations.
Supplementary Material
Novelty and Significance: 1) What is new, 2) What is relevant?
What Is New?
To our knowledge, this is the first population-based cohort study focused on investigating associations between gut microbial community composition and blood pressure.
What Is Relevant?
Animal models have demonstrated mechanistic pathways through which the gut microbiota may influence blood pressure.
Our results demonstrate significant associations between gut microbial composition and blood pressure.
Summary
In this population-based cohort of middle-aged U.S. Caucasians and African-Americans, there were significant differences in the composition of the gut microbiota with respect to blood pressure. Gut microbial diversity was inversely associated with both hypertension and systolic blood pressure. These results support additional research to understand the role of the gut microbiome in blood pressure.
Acknowledgements
We thank all CARDIA investigators, staff, and participants for their valuable contributions. This manuscript has been reviewed by CARDIA for scientific content. Additional information about CARDIA can be found at https://www.cardia.dopm.uab.edu.
Sources of Funding
The Coronary Artery Risk Development in Young Adults Study is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of Alabama at Birmingham (HHSN268201800005I & HHSN268201800007I), Northwestern University (HHSN268201800003I), University of Minnesota (HHSN268201800006I), and Kaiser Foundation Research Institute (HHSN268201800004I). The Microbiome Study was funded by K01-HL127159, the Intramural Research Program of the National Institute on Aging, the UNC Nutrition Research Institute, the UNC Office of the Vice Chancellor for Research, P30-DK056350, and UL1TR002489.
Footnotes
Conflicts of Interest/Disclosures
None
References
- 1.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu J, Bonder MJ, Cenit MC, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ Res. 2015;117:817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF, Correa A, He J. Gut Microbiome Associates With Lifetime Cardiovascular Disease Risk Profile Among Bogalusa Heart Study Participants. Circ Res. 2016;119:956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, Soininen P, Wang Z, Ala-Korpela M, Hazen SL, Laakso M, Lusis AJ. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–81. [DOI] [PubMed] [Google Scholar]
- 7.Cani PD and Delzenne NM. Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr Opin Pharmacol. 2009;9:737–43. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. [DOI] [PubMed] [Google Scholar]
- 10.Lambeth SM, Carson T, Lowe J, Ramaraj T, Leff JW, Luo L, Bell CJ, Shah VO. Composition, Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. J Diabetes Obes. 2015;2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- 12.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pluznick JL, Protzko RJ, Gevorgyan H, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adnan S, Nelson JW, Ajami NJ, Venna VR, Petrosino JF, Bryan RM Jr, Durgan DJ Alterations in the gut microbiota can elicit hypertension in rats. Physiol Genomics. 2017;49:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65:1331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics. 2015;47:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Yu B, Alexander D, Mosley TH, Heiss G, Nettleton JA, Boerwinkle E. Metabolomics and incident hypertension among blacks: the atherosclerosis risk in communities study. Hypertension. 2013;62:398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr., Liu K, Savage PJ CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 1988;41:1105–16. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs DR Jr., Hahn LP, Haskell WL, Pirie P, Sidney S Validity and reliability of short physical activity history: CARDIA and Minnesota Heart Health Program. J Cardiopulm Rehabil. 1989;9:448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rifas-Shiman SL, Willett WC, Lobb R, Kotch J, Dart C, Gillman MW. PrimeScreen, a brief dietary screening tool: reproducibility and comparability with both a longer food frequency questionnaire and biomarkers. Public Health Nutr. 2001;4:249–54. [DOI] [PubMed] [Google Scholar]
- 23.Sijtsma FP, Meyer KA, Steffen LM, Shikany JM, Van Horn L, Harnack L, Kromhout D, Jacobs DR Jr. Longitudinal trends in diet and effects of sex, race, and education on dietary quality score change: the Coronary Artery Risk Development in Young Adults study. Am J Clin Nutr. 2012;95:580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mursu J, Steffen LM, Meyer KA, Duprez D, Jacobs DR Jr. Diet quality indexes and mortality in postmenopausal women: the Iowa Women’s Health Study. Am J Clin Nutr. 2013;98:444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J, Garrett WS, Chan AT, Huttenhower C. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A. 2014;111:E2329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Hullar MA, Beresford SA, Lampe JW. Variation of glucoraphanin metabolism in vivo and ex vivo by human gut bacteria. Br J Nutr. 2011;106:408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sioda M, Fodor AA. BioLockJ [computer program]. 2018. https://github.com/msioda/BioLockJ [Google Scholar]
- 28.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. Community Ecology Package. CRAN: 2017. [Google Scholar]
- 31.Peet RK. The measurement of species diversity. Annual Review of Ecology and Systematics. 1974;5:285–307. [Google Scholar]
- 32.Faith DP, Minchin PR, Belbin L. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio. 1987;69:57–68. [Google Scholar]
- 33.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:23–46. [Google Scholar]
- 36.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winglee K, Howard AG, Sha W, Gharaibeh RZ, Liu J, Jin D, Fodor AA, Gordon-Larsen P. Recent urbanization in China is correlated with a Westernized microbiome encoding increased virulence and antibiotic resistance genes. Microbiome. 2017;5:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodology). 1995;57:289–300. [Google Scholar]
- 40.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durgan DJ, Ganesh BP, Cope JL, Ajami NJ, Phillips SC, Petrosino JF, Hollister EB, Bryan RM Jr. Role of the Gut Microbiome in Obstructive Sleep Apnea-Induced Hypertension. Hypertension. 2016;67:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pluznick J A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2014;5:202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pluznick JL. Renal and cardiovascular sensory receptors and blood pressure regulation. Am J Physiol Renal Physiol. 2013;305:F439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartolomaeus H, Balogh A, Yakoub M, et al. The short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. December 4, 2018. doi: 10.1161/CIRCULATIONAHA.118.036652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M, Group ST. Increased Systolic and Diastolic Blood Pressure Is Associated With Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension. 2016;68:974–81. [DOI] [PubMed] [Google Scholar]
- 46.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4. [DOI] [PubMed] [Google Scholar]
- 47.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–36. [DOI] [PubMed] [Google Scholar]
- 50.Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. [DOI] [PubMed] [Google Scholar]
- 51.Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El-Osta A, Mackay CR, Kaye DM. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation. 2017;135:964–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in the present analysis are available from the CARDIA Study Data Coordinating Center at the University of Alabama at Birmingham. The process for obtaining data through CARDIA is outlined at: https://www.cardia.dopm.uab.edu/publications-2/publications-documents. Computer code/scripts used in the generation of data and statistical analysis are available from the authors upon request.