Abstract
Toxigenic Vibrio cholerae of the O139 serogroup have been responsible for several large cholera epidemics in South Asia, and continue to be of clinical and historical significance today. This serogroup was initially feared to represent a new, emerging V. cholerae clone that would lead to an eighth cholera pandemic. However, these concerns were ultimately unfounded. The majority of clinically relevant V. cholerae O139 isolates are closely related to serogroup O1, biotype El Tor V. cholerae, and comprise a single sublineage of the seventh pandemic El Tor lineage. Although related, these V. cholerae serogroups differ in several fundamental ways, in terms of their O-antigen, capsulation phenotype, and the genomic islands found on their chromosomes. Here, we present four complete, high-quality genomes for V. cholerae O139, obtained using long-read sequencing. Three of these sequences are from toxigenic V. cholerae, and one is from a bacterium which, although classified serologically as V. cholerae O139, lacks the CTXφ bacteriophage and the ability to produce cholera toxin. We highlight fundamental genomic differences between these isolates, the V. cholerae O1 reference strain N16961, and the prototypical O139 strain MO10. These sequences are an important resource for the scientific community, and will improve greatly our ability to perform genomic analyses of non-O1 V. cholerae in the future. These genomes also offer new insights into the biology of a V. cholerae serogroup that, from a genomic perspective, is poorly understood.
Introduction
Vibrio cholerae is the aetiological agent of cholera, an acute, life-threatening diarrhoea which has spread worldwide in seven pandemics since the nineteenth century. V. cholerae is typically sub-classified into serogroups on the basis of its somatic O-antigen. Despite there being over 200 serogroups of V. cholerae1,2, only serogroup O1 has caused large scale epidemics historically3. Previous cholera pandemics have been caused by the classical biotype of V. cholerae O1, whereas the ongoing seventh pandemic, which began in the 1960s, is caused by the El Tor biotype of V. cholerae O13,4. Non-O1 serogroups of V. cholerae do not appear to cause pandemics, though they may cause outbreaks of disease. This is exemplified by an outbreak in Sudan in 1968, caused by V. cholerae O37, which was subsequently found to be genetically related to pandemic V. cholerae O15–8.
In 1992, a V. cholerae clone of serogroup O139 caused a large cholera epidemic which spread rapidly across Bangladesh and India9,10. Due to the geographic location of the epidemic, this clone was given the name Vibrio cholerae O139 Bengal10 (dubbed V. cholerae O139 hereafter). V. cholerae O139 caused substantial numbers of cholera cases in Southeast Asia in the early 1990s, and was anticipated to emerge as the aetiological agent of an eighth cholera pandemic11–13. However, rather than causing a pandemic, V. cholerae O139 was associated only with a low-level incidence of cholera cases after the initial 1992–93 epidemic, until a second large outbreak occurred in Bangladesh in the Spring of 200214. The re-emergence of this serogroup renewed fear that an eighth pandemic of cholera was beginning, driven by V. cholerae O13915. Once again, V. cholerae O139 did not proceed to cause a cholera pandemic, and although it no longer appears to be causing epidemic cholera, this serogroup has continued to be isolated since 2002. Recently, non-toxigenic V. cholerae O139 have been isolated in Thailand16. Toxigenic V. cholerae O139 have been isolated in China as recently as 201317, and continue to be isolated in Bangladesh. Six V. cholerae O139 strains were isolated in Bangladesh between 2013 and 201418 (four included in this analysis), and three more strains were isolated between 2015 and 2017.
Despite the low incidence of cholera caused by V. cholerae O139, this serogroup continues to be important to both the research and public health communities, not least because the disease caused by V. cholerae O139 is clinically indistinguishable from that caused by V. cholerae O110. Accordingly, V. cholerae O139 continues to be the subject of surveillance in Southeast Asia and is included in cholera vaccine formulations19–21. Early genetic and biochemical studies demonstrated that O139 strains were closely related to O1 seventh pandemic El Tor strains, and it was suggested that V. cholerae O139 had arisen from an O1 El Tor ancestor9,22–24. This was subsequently confirmed using whole-genome sequencing, which showed that toxigenic V. cholerae O139 formed a discrete sub-lineage within the seventh pandemic El Tor (7PET) lineage25,26.
Although the clinical diseases caused by V. cholerae O1 and O139 are indistinguishable, there are notable differences between V. cholerae O139 and V. cholerae O1 in addition to their serogroup. For instance, V. cholerae O139 expresses a polysaccharide capsule, which 7PET V. cholerae O1 isolates lack27. The capsule is encoded by genes not found in other 7PET V. cholerae O1 genomes, which are located adjacent to the locus encoding lipopolysaccharide (LPS) biosynthesis genes in V. cholerae O13915,28–32. It has also been reported that the complement of genomic islands found in the genome of MO10, a V. cholerae O139 strain, differs from that found in 7PET V. cholerae O125. The MO10 genome sequence is currently used to represent V. cholerae O139 in comparative genomic analyses, but its genome sequence is incomplete and comprises 84 contigs (assembly accession number GCA_000152425.1).
V. cholerae O139 caused 93% of laboratory-confirmed cholera cases in China in the early 2000s19. Such data demonstrate why V. cholerae O139 has twice been feared to be responsible for an eighth cholera pandemic. Despite the clinical importance of this serogroup which continues to be isolated today, a closed reference genome for V. cholerae O139 has not yet been published. Here, we report the first high-quality, closed reference genome sequences for this V. cholerae serogroup. We have used long-read sequencing to obtain the complete genome sequences of four recent V. cholerae O139 isolates from Bangladesh18. Three of these are toxigenic members of the 7PET lineage, two of which were isolated from asymptomatic patients from within a household where there had been a confirmed cholera case. The fourth isolate was acquired from a patient suffering from diarrhoea, vomiting and dehydration (see ref.18 for full details of the clinical history surrounding these four isolates), but is a non-toxigenic V. cholerae O139 variant that is not part of the 7PET lineage, and does not harbour the CTXφ bacteriophage18. This non-7PET genome offers an opportunity to study the genetic factors that enable non-toxigenic V. cholerae to cause diarrhoea in patients.
Results
Structure of CTXφ tandem arrays in V. cholerae O139
Using long-read sequencing read data, we generated single, contiguous genome sequences for the two chromosomes of each isolate sequenced in this study. Use of the corresponding short-read data for each isolate to correct these assemblies did not improve the assemblies. None of these isolates contained a third replicon, as has been reported elsewhere in other V. cholerae33. In order to estimate genetic distance, we mapped each sample’s corresponding short-read data18 to the 7PET reference genome N16961 and called single nucleotide variants (SNVs) between the mapped reads for these isolates and N16961. These SNV data are provided in Table 1, together with summary statistics for these closed assemblies. Since the three toxigenic samples were found to have near-identical genomes, varying in size by 4 bases at most, and differing by fewer than five SNVs between one another (SNVs were determined relative to N16961), 48853_H01 was selected as an exemplar sequence for further analyses.
Table 1.
Summary of the V. cholerae O139 genome assemblies generated in this study.
| Internal sequence ID (PacBio) | Sample Name | CTXφ present? | Accession (PacBio reads) | Accession (closed chromosomal assembly) | Accession (Illumina reads) | Genome Size (bp) | Average coverage of de novo assembly with long reads (X) | Coverage of N16961 (%) | Number of SNVs relative to N16961 |
|---|---|---|---|---|---|---|---|---|---|
| 48853_F01 | MP_070116 | No | ERR1716489 | LT992490-LT992491 | ERR568405 | 4123525 | 165.76 | 58.5 | 122865 |
| 48853_G01 | P_0684000 | Yes | ERR1716490 | LT992486-LT992487 | ERR568406 | 4092641 | 170.56 | 97 | 271 |
| 48853_H01 | ICVB_2236_02 | Yes | ERR1716491 | LT992488-LT992489 | ERR568407 | 4092645 | 147.29 | 97 | 270 |
| 48853_A02 | SMIC_67_01 | Yes | ERR1716492 | LT992492-LT992493 | ERR568408 | 4092644 | 165.65 | 97 | 274 |
The accession numbers for both the long-read sequences and assemblies generated in this study, and the original short reads used for assembly polishing, SNV calling, and phylogenetic analyses (see Methods) are reported. The SNV counts reported do not account for the removal of recombinogenic sequences, since the non-toxigenic isolate was not included in the recombination analysis. Average coverage values taken from de novo HGAP assemblies.
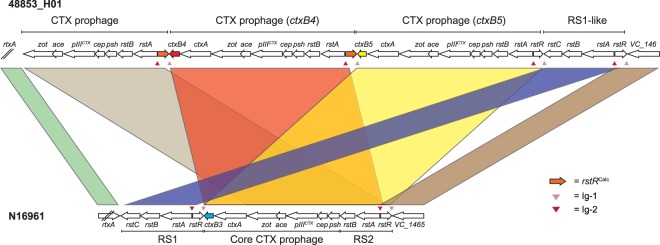
The CTXφ bacteriophage integrates into the V. cholerae chromosome in a XerCD-dependent manner, by recombination between the CTXφ attP site and bacterial attB site, which produces hybrid attL and attR sequences34,35. This occurs after the replicative, circular form of the viral genome forms after infection of the cell, and this integration usually involves tandem integrations of CTXφ into the genome36. CTXφ replication is achieved by the production of ssDNA from chromosomal tandem arrays of CTXφ in a manner dependent on the CTXφ-encoded RstA protein, where RstA nicks the CTXφ replication origin located in the intergenic region Ig-136,37. The exposed 3′ site permits synthesis of CTXφ DNA up until the second, tandem CTXφ replication origin is encountered, which is also a substrate for RstA cleavage, creating a free CTXφ genome36,37. We observed two additional CTXφ bacteriophage sequences in tandem, relative to that found in the N16961 reference genome, located between VC_1450 and VC_1467 in the larger chromosome in the three toxigenic isolates (Fig. 1). A partial third repeat of CTXφ was evident, comprising the genes between and including zot and rstA, and an rstR open reading frame corresponding to rstRCalc 38 (Fig. 1). We identified a complete attL sequence adjacent to the VC_1465 locus in 48853_H0135. The phage sequence in the attR site adjacent to rtxA is not identical to that reported by Huber and Waldor35, although the attR sequence does contain both the central recombination identity sequence and the residual bacterial attB sequence.
Figure 1.
Tandem copies of the CTXφ bacteriophage in V. cholerae O139. An illustration of the genomic organisation of three tandem copies of CTXφ in the toxigenic V. cholerae O139 samples in this study. Two copies of the ctxAB operon are present in these genomes, which harbour different ctxB alleles to one another and to that of N16961. Exemplar data from the assembly for 48853_H01 are presented. Loci are not to scale. Figure annotation based on ref.36.
Although tandem repeats of CTXφ genes in V. cholerae O139 have been reported previously15,38,39, difficulty in assembling these repetitive regions with short-read sequencing data meant that these repeats were not identified in our original sequencing of these isolates18. We mapped the short-read data to the long-read assemblies for each of these genomes, and to the N16961 reference, to confirm that short reads mapped to both the ctxB4 and ctxB5 variant CTXφ regions, and that when mapped to N16961, the coverage of the CTXφ region was approximately double that of the surrounding chromosome (Supplementary Fig. S1). Manual inspection of these mapping data showed that reads from both ctxB alleles mapped to the N16961 ctxB locus.
We noted that the two ctxB genes in these genomes are of different alleles (Fig. 1). The ctxB gene closest to rtxA in these assemblies was a ctxB4 allele, and the second ctxB was a ctxB5 allele. Both of these are ctxB alleles that have been found in V. cholerae O139 strains previously40,41. The presence of more than one ctxB allele in the same V. cholerae genome has not been reported previously to our knowledge, though it has been reported that V. cholerae O139 can harbour more than one type of CTXφ phage simultaneously15,38,42.
Genomic islands and antimicrobial resistance genes in V. cholerae O139
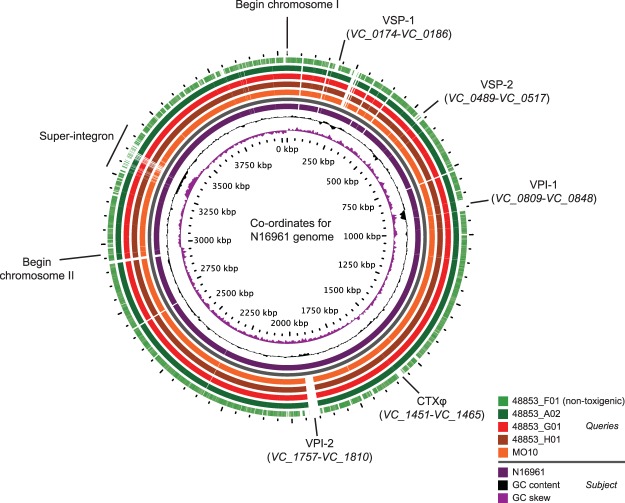
Having observed these unusual CTXφ configurations, we scanned the four assemblies for the presence and absence of the genomic islands that are associated with pandemic V. cholerae: VSP-1, VSP-2, VPI-1, VPI-2, and the drug resistance genetic element SXT25. We identified VPI-1 and VSP-2 islands in all three of the toxigenic V. cholerae O139 isolates, and we confirmed that VPI-2 is severely truncated to the point of absence, as described previously for MO1025,43 (Fig. 2). We identified a genomic island integrated into the VC_0659 locus (encoding peptide chain release factor 3) in each of the three toxigenic V. cholerae O139 assemblies, identical to SXT, which is also known as ICEVchInd444. SXT is integrated into the same locus as it is in MO10. We also identified an insertion into VC_0659 in the non-toxigenic genome assembly, which was 64% identical to ICEVchInd4 (Table 2). All of these observations agree with data from Chun et al.25 on the distribution of genomic islands in the V. cholerae O139 MO10 genome.
Figure 2.
BLAST atlas comparing the genomes used in this study and MO10 to the N16961 reference genome. The presence of VPI-1, VSP-1, and VSP-2 in the three toxigenic V. cholerae O139 assemblies, as well as the truncation of VPI-2, is indicated. The non-toxigenic strain did not harbour VSP-1, VSP-2, or VPI-2 (indicated), although a region homologous to part of VPI-1 (VC_0809 to VC_0816) was detected on the larger chromosome. The sequences of both V. cholerae chromosomes were concatenated to generate this figure; the boundary between the chromosomes is denoted.
Table 2.
Presence and absence of selected genomic islands in V. cholerae O139 genome assemblies.
| Sample Name | VSP-1 (VC_0174-VC_0186) | VSP-2 (VC_0489-VC_0517) | VPI-1 (VC_0809-VC_0848) | VPI-2 (VC_1757-VC_1810) | CTXφ (VC_1451-VC_1465) | SXT (VC_0659 insertion) |
|---|---|---|---|---|---|---|
| 48853_F01 | Absent | Absent | Partially present (VC_0809-VC_0816; deletion of VC_0817-VC_0848) | Absent | Absent | 64% match to ICEVchInd4 |
| 48853_G01 | Present, and duplication of VC_0175–0186 on chr2 | Present | Present | Deletion of VC_1761–1787 | Present, in more than one copy | 100% match to ICEVchInd4 |
| 48853_H01 | Present, and duplication of VC_0175–0186 on chr2 | Present | Present | Deletion of VC_1761–1787 | Present, in more than one copy | 100% match to ICEVchInd4 |
| 48853_A02 | Present, and duplication of VC_0175–0186 on chr2 | Present | Present | Deletion of VC_1761–1787 | Present, in more than one copy | 100% match to ICEVchInd4 |
Similarity percentages were obtained by comparing SXT element sequences to that of ICEVchInd4 using BLASTn. chr2 = chromosome 2.
In the three toxigenic V. cholerae O139 isolates, we detected the VSP-1 element integrated on the larger chromosome between genes VC_0173 and VC_0187, as found in N1696125 (Fig. 2). However, we also observed a sequence of DNA on the smaller chromosome of each of the toxigenic isolates, integrated between VC_A0695 and VC_A0696, that was 99% identical to VSP-1 (VC_0175 to VC_0186; Supplementary Fig. S2). This suggested that a second copy of the VSP-1 element was present on the second chromosome in each of these genomes. We mapped the previously-published Illumina reads for these genomes18 to the N16961 reference genome and plotted the read depth for VSP-1 relative to the surrounding genome (Supplementary Fig. S2), which further supported the conclusion that these genomes contain a second copy of VSP-1.
This VSP-1 duplication was detected in each of the three toxigenic V. cholerae O139 genome assemblies (summarised in Table 2). It is known that VSP-1 is capable of excising from the larger V. cholerae chromosome45, and it has been previously reported that the Matlab variant V. cholerae O1 strain MJ-1236 harbours a second copy of VSP-1 integrated between VC_A0695 and VC_A069646. Grim et al.46 used PCR to identify a single clinical isolate of V. cholerae O139 from Bangladesh that harboured an insertion between VC_A0695 and VC_A0696 resembling VSP-1, but this isolate was not described further. This phenomenon is likely to be that which we have now confirmed to be present in these three V. cholerae O139 genomes.
We also compared the genomic island complement of these four sequences with that of MO10. MO10 harbours a kappa prophage (GI-1125), which is absent from N16961 and also absent from the four sequences in this study (Supplementary Fig. S3). Likewise, MO10 harbours the Vibrio VSK prophage (GI-1625), which is absent from both N16961 and the O139 sequences in this study (Supplementary Fig. S3). MO10 does not appear to harbour the second VSP-1 copy on chromosome 2 which we identified in the three toxigenic isolates. The SXT variant harboured by MO10 is expanded relative to that found in these strains (Supplementary Fig. S3), and this expansion includes genes conferring resistance to the antimicrobials streptomycin (strAB), sulfamethoxazole (sul2), trimethoprim (dfr18), and chloramphenicol (floR). We scanned the assemblies for the four O139 genomes for antimicrobial resistance genes. The non-toxigenic 48853_F01 genome does not harbour any known antimicrobial resistance genes. The three toxigenic O139 genomes also do not contain any antimicrobial resistance genes, though they do harbour a catB9 gene that is known not to confer antibiotic resistance47. These data are concordant with the original antimicrobial sensitivity testing of these isolates, which found that they were resistant only to nalidixic acid18. We confirmed that these four isolates harbour an S83I mutation in GyrA, and that 48853_F01 also contains an A171S mutation in GyrA and a S85L mutation in ParC. All of these mutations are associated with nalidixic acid resistance in V. cholerae48. We also scanned the assembled genomes of the four isolates for the presence of V. cholerae accessory virulence genes, to determine whether candidate virulence genes were present in the genome of the otherwise non-toxigenic V. cholerae O139 isolate18 (Table 3). We did not identify any virulence determinants in the 48853_F01 genome assembly other than those typically found in V. cholerae3.
Table 3.
Presence and absence of accessory virulence genes in V. cholerae O139 genome assemblies.
| Accessory virulence gene (N16961 locus ID or accession number) | Present in 48853_F01 | Present in 48853_G01 | Present in 48853_H01 | Present in 48853_A02 |
|---|---|---|---|---|
| ToxR (VC_0984) | Yes | Yes | Yes | Yes |
| Zona occludens toxin, Zot (VC_1458) | No | Yes | Yes | Yes |
| Accessory cholera enterotoxin, Ace (VC_1459) | No | Yes | Yes | Yes |
| Haemolysin, hlyA (VC_A0219) | Yes | Yes | Yes | Yes |
| Mannose-sensitive haemagglutinin, MSHA (VC_0398..VC_0414) | Yes | Yes | Yes | Yes |
| MARTX toxin, rtxA (VC_1451) | Yes | Yes | Yes | Yes |
| MARTX toxin accessory gene, rtxC (VC_1450) | Yes | Yes | Yes | Yes |
| HA/protease, hapA (VC_A0865) | Yes | Yes | Yes | Yes |
| Heat-stable enterotoxin NAG-ST (Accession # M85198.1) | No | No | No | No |
| Type III secretion system from V. cholerae AM_19226 (typically present in lieu of VPI-2; accession # AATY01000000) | No | No | No | No |
Gene presence and absence was determined using ACT67 to visualise BLASTn synteny plots, and using tBLASTx to scan assemblies using the NAG-ST nucleotide sequence as a query.
Phylogenetic analysis
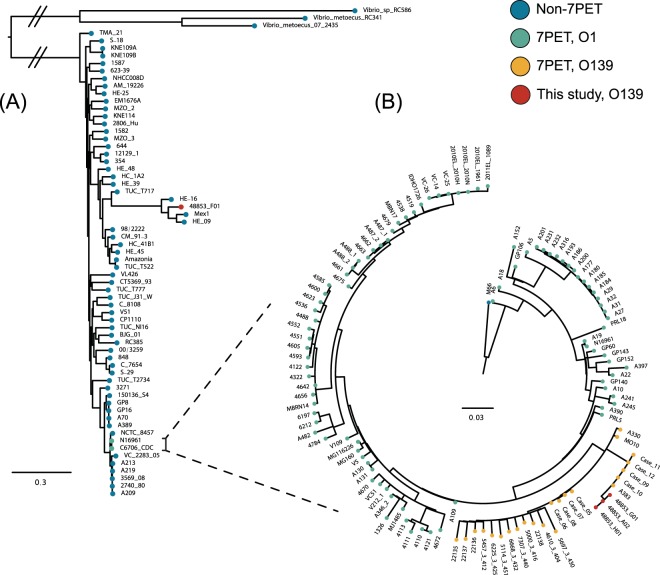
We constructed a maximum-likelihood phylogeny from an alignment of core genes from 65 diverse V. cholerae genomes, and confirmed that despite its serogroup, 48853_F01 is not a member of the 7PET O139 sublineage (Fig. 3A). We did find that the three toxigenic genomes were members of 7PET, and we used the previously-published short-read data for these genomes to place these isolates into phylogenetic context with 114 other V. cholerae, including 23 O139 genome sequences16,18,26 (117 genomes in total; Supplementary Table S1). We found that the three toxigenic isolates in this study clustered together with other 7PET V. cholerae O139 sequences from Bangladesh and India from 1992 to 2002 (Fig. 3B). The closest relatives of these three strains, which were isolated in 2013 and 2014, were isolated from Bangladesh in 2002 (A383, Case_09–12). All of these were also closely related to V. cholerae O139 samples from 1992–1995, including a recently-sequenced collection of V. cholerae O139 from Thailand16. These results recapitulate and reinforce previously-published data18, adding to the utility of these genomes as reference sequences for toxigenic V. cholerae O139. Moreover, there are no complete genome assemblies for non-toxigenic V. cholerae O139. Given that this isolate is clearly distinct from the 7PET O139 sublineage, we anticipate that this genome sequence will enable comparative genomic studies of V. cholerae other than 7PET isolates.
Figure 3.
Phylogenetic analysis. (A) A total of 168,476 variable sites from a core-gene alignment of 2,103 genes from 65 genomes were used to generate a maximum-likelihood phylogenetic tree of the V. cholerae species, rooted on Vibrio metoecus and Vibrio sp. RC586. Two genomes that are representative of the 7PET lineage were included. The non-toxigenic isolate 48853_F01 clustered together with other non-toxigenic, non-O1/O139 V. cholerae. (B) A maximum-likelihood phylogeny of the 7PET lineage constructed using 1,629 non-recombinant variable sites across 117 V. cholerae genomes, rooted on M66-2. The three toxigenic samples in this study clustered together with other toxigenic O139 genome sequences, all of which form a discrete sub-lineage within 7PET. Hatch marks denote branches that were shortened artificially for illustrative purposes; an unedited tree is presented in Supplementary Fig. S5. Trees were also computed using an ascertainment bias correction model (see Methods). These are presented in Supplementary Fig. S6.
The previous report of these genome sequences, obtained using short-read technology alone, examined the structure of the capsule and lipopolysaccharide (LPS) synthesis loci. These analyses were performed using incompletely-assembled genome sequences18. We used the closed sequences obtained in this project to compare these loci across the strains in this study to N16961 and MO10 (Supplementary Fig. S4). We confirmed that the three toxigenic strains contain O139 LPS operons that strongly resemble that found in MO10 (Supplementary Fig. S4). The equivalent region in the non-toxigenic isolate 48853_F01 is less similar, although this strain exhibits a strong O139-positive phenotype using the rapid dipstick assay and slide agglutination tests18. In our phylogenetic analyses, we noted that 48853_F01 clustered with two Haitian non-O1 V. cholerae isolates from 2010 and a Mexican isolate from 1991 for which there are no serotype data (Fig. 3A; Supplementary Table S1). We found that, although these three non-O1 V. cholerae share capsule biosynthesis genes with 48853_F01, they do not harbour the same LPS operon (Supplementary Fig. S4). These Haitian and Mexican isolates therefore are unlikely to be V. cholerae of serogroup O139.
Discussion
It is essential to have complete and accurate reference sequences to perform bacterial genomic analysis. Although several studies have provided closed V. cholerae sequences49–53, none to date have provided reference sequences for V. cholerae O139. The sequences in this study will serve as an important community resource in future studies of V. cholerae genomics and phylogenetics. For example, access to the closed sequences of the O139 LPS and capsule biosynthesis operons from these four strains means that it should be possible to serotype V. cholerae O139 sequences in silico. The fact that the LPS biosynthesis loci in these toxigenic and non-toxigenic strains are similar but not identical (Supplementary Fig. S4), and that the non-toxigenic strain is distantly related to the toxigenic V. cholerae O139 in this study (Fig. 3), suggests that there may be more than one genetic configuration that confers an O139 serogroup phenotype. In the absence of candidate virulence genes, putative or otherwise, we also cannot exclude the possibility that the non-toxigenic isolate was obtained from a patient who was co-infected with another toxigenic organism such as enterotoxigenic Escherichia coli54–56.
Many of the observations in this study could only be made because of the resolution offered by long-read sequencing. For example, the observation that several ctxB alleles can co-exist in a single genome is striking. The co-existence of several CTXφ sequences in tandem has been reported before, such as in the O395 classical reference sequence52,57 and in the PA1849 second-pandemic classical isolate4. However, in PA1849, the tandem bacteriophages are of the same ctxB allele, and in O395, the CTXφ array on the larger chromosome consists of one intact CTXφ and one partial prophage sequence57. Although these V. cholerae O139 had been sequenced previously, it had not been possible to assemble these genomes fully with the short-read Illumina technology used at the time. Consequently, in all three assemblies, CTXφ was not assembled into a single contig, and only one of the two ctxB genes was identifiable (our Illumina assemblies for 48853_G01 and 48853_H01 contain a ctxB4 allele in a small contig, and 48853_A02 contains ctxB5 in a larger contig). In future studies of V. cholerae O139, mapping sequencing reads against these reference sequences will address this problem, which will not be resolved if data are exclusively mapped to N16961 or related sequences.
Furthermore, we note that the oligonucleotides used for PCR based ctxB typing40 are 100% homologous to regions upstream and downstream of both ctxB loci in these three genomes. It would therefore not be possible to discriminate between ctxB types based on Sanger sequencing of these amplicons. This suggests that caution should be used in the interpretation of PCR-based ctxB typing data in the epidemiological study of cholera outbreaks, particularly if this CTXφ configuration is present in other V. cholerae lineages.
The functions of VSP-1 and VSP-2 are not fully understood, although it is well-accepted that these two genomic islands are found in V. cholerae which are members of the seventh pandemic25. It is known that DncV, encoded by VSP-1, represses V. cholerae chemotaxis58 and that repression of chemotaxis has been linked to improved intestinal colonisation by V. cholerae59. DncV is also known to be upregulated under conditions of gastrointestinal infection, in response to conditions that activate the ToxT transcription factor via the TarB small RNA which prevents the production of VPI-1-encoded VspR. It is interesting to speculate that the duplication of VSP-1 might further attenuate V. cholerae chemotaxis under colonisation conditions via a gene dosage effect, thereby modulating the ability of these V. cholerae O139 strains to colonise the intestine.
Here, by sequencing V. cholerae O139 using long-read technology, we have highlighted genomic features that emphasise the genetic distinctions between V. cholerae O139 and V. cholerae O1. We have identified differences between these recent strains, N16961, and MO10, the V. cholerae O139 strain used for previous comparative analyses. We have also described unusual phenomena in V. cholerae O139 genome biology – namely, the co-existence of more than one ctxB allele, and the cross-chromosome duplication of VSP-1. There also appears to have been genetic changes within V. cholerae O139 that has occurred since its first identification in 1992, typified by the MO10 isolate. Given that V. cholerae O139 is a member of 7PET, has several characteristics of a sublineage with the potential to cause pandemic disease, and continues to be isolated in recent years, research into this serogroup should continue. These reference sequences enable such research, and as well as providing interesting insights into the genome structure of recent V. cholerae O139, these sequences are an important resource for future genomic studies of V. cholerae as a pathogen and as a species.
Methods
Isolates and sequences used in this study
Four previously-described V. cholerae O139 isolates18 were selected for re-sequencing on the PacBio RSII platform. A set of 178 genomes in addition to these were included in comparative genome analyses (182 genomes in total; Supplementary Table S1).
DNA isolation
Genomic DNA was prepared from 25 ml cultures of bacterial isolates grown overnight at 37 °C in LB media. Cells were harvested by centrifugation and resuspended in 2.0 ml of 25% w/v sucrose in TE buffer (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0). Nuclei Lysis Solution (Promega, #A7941, 6.0 ml) was added and samples were lysed by incubation at 80 °C for five minutes. Samples were mixed with proteinase K (250 μg/ml final concentration), RNase A Solution (Promega, #A797A, 15 μg/ml final concentration), EDTA pH 8.0 (25 mM final concentration) and aqueous SDS solution (0.3% final concentration). Mixtures were incubated on ice for two hours and then at 50 °C overnight. Following enzymatic treatment, TE buffer was added to each sample (12 ml final volume). DNA was then isolated by phenol-chloroform extraction. Nucleic acids were precipitated in absolute ethanol, washed in ethanol (70% v/v), and resuspended in approximately 350 μl Tris (10 mM; pH 8.0). EDTA was omitted from the resuspension solution, to avoid interference with PacBio sequencing chemistry.
Long-read sequencing
SMRTbell libraries were created from approximately 10 μg DNA according to the manufacturer’s protocol (15 kb library size, no size selection). Long reads were generated by sequencing on the PacBio RSII platform using polymerase version P6 and C4 sequencing chemistry. Sequence reads were assembled using HGAP v360 of the SMRT analysis software v2.3.0. The fold coverage to target when picking the minimum fragment length for assembly was set to 30 and the approximate genome size was set to 3 Mbp. The HGAP assembler assembled the reads from sample 48853_G01 into three contigs. However, assembling this sample with Canu v1.161 produced an assembly of two contigs, one per chromosome, and this was used for subsequent analysis. Assemblies were circularised using Circlator v1.1.362 and the pre-assembled reads (also known as corrected reads). The circularised assemblies were polished using the PacBio RS_Resequencing protocol and Quiver v1 of the SMRT analysis software v2.3.0. Automated annotation was performed using Prokka v1.1163 and genus specific databases from RefSeq64. Pilon v1.1965 did not identify any SNVs in any of the PacBio assemblies using the corresponding short-read data – accordingly, no short-read corrections were made to these assemblies. Raw sequencing reads and the genome assemblies described in this study have been deposited into the European Nucleotide Archive (Table 1; Supplementary Table S1).
Comparative genomics and BLAST atlas construction
The four annotated genome assemblies were compared to one another, and to the N16961 and MO10 reference genomes (see Supplementary Table S1 for accession numbers), using BLASTn66. These comparisons were visualised using ACT67 and by BLAST atlas comparison using the GView web server (https://server.gview.ca/).
Read alignment, SNV identification, and core gene alignment
Paired-end Illumina reads from 116 7PET V. cholerae O1 and O139 samples, together with the M66-2 pre-pandemic strain (117 genomes in total; Supplementary Table S1), were mapped to the V. cholerae O1 El Tor N16961 reference genome (see Supplementary Table S1 for accession numbers) using SMALT v.0.7.4. Variable sites were identified using samtools mpileup v0.1.19, with parameters “-d 1000 –DsugBf”, and bcftools v0.1.1968. High quality SNVs were determined as previously described69, and putative recombinant regions were detected and filtered from the alignment using Gubbins70, to produce a final alignment of 1,629 SNVs.
Prokka-annotated assemblies for 63 non-7PET and two 7PET genomes63,71 were used to generate a species-level pan-genome using Roary72 with the following arguments: “-e–mafft -s -cd 97”. Poorly-aligned and gap-rich sites were removed from an alignment of 2,103 core gene sequences using trimAl v1.4.rev573 with the “-automated1” argument. A total of 168,476 variable sites were identified in the resultant alignment using SNP-sites v2.3.274.
Phylogenetic analysis
Maximum likelihood phylogenetic trees were constructed using RAxML v8.2.875 under the GTR model with the gamma distribution to model site heterogeneity (GTRGAMMA), using 500 bootstrap replicates. An alignment composed of 1,629 non-recombinant variable sites was used to generate a reference-based 7PET phylogeny. An alignment of 168,476 variable sites from 2,103 core genes was used to generate a core-gene V. cholerae species phylogeny. Trees were also computed using the GTR + ASC model in IQ-Tree v1.5.576, optimised for an input containing no invariant nucleotides, and were supported by 5,000 ultrafast bootstrap approximations and approximate likelihood ratio tests77–79. Phylogenetic trees were visualised using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and the interactive tree of life (iTOL) v380.
Identification of antimicrobial resistance genes
Genome assemblies were scanned for the presence of antimicrobial resistance genes using the ResFinder web server v2.1 (https://cge.cbs.dtu.dk/services/ResFinder/)81.
Supplementary information
Supplementary material for High quality reference genomes for toxigenic and non-toxigenic <i>Vibrio cholerae</i> serogroup O139
Acknowledgements
We acknowledge the support of the dedicated field and laboratory workers at the icddr,b involved in this study, and thank Derek Pickard for technical help with DNA extractions. We also thank the sequencing and Pathogen Informatics teams at the Wellcome Sanger Institute for help with processing samples and depositing sequencing data. This work was supported by Wellcome (grants 098051, 206194) and by the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b). This study was supported by grants from the National Institutes of Health, including grants from the National Institute of Allergy and Infectious Diseases (AI106878 [F.Q.], AI058935 [F.Q.], and AI103055 [F.Q.]). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. icddr,b is grateful to the Governments of Bangladesh, Canada, Sweden and UK for providing core/unrestricted support. M.J.D. is supported by a Wellcome Sanger Institute PhD Studentship.
Author Contributions
N.R.T. and F.Q. designed and supervised the study. S.S. and Y.A.B. co-ordinated and performed experimental work. M.J.D., D.D. and M.I.U. analysed the data. M.J.D., D.D. and M.I.U. wrote the manuscript, with major contributions from M.H.A., F.Q. and N.R.T. All authors contributed to the editing of the manuscript.
Data Availability
Sequencing reads generated during this project have been deposited into the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under study accession number PRJEB14661. Assembled genome sequences have been deposited under accession numbers LT992486- LT992493. Sequence alignments, phylogenetic tree data, and other supporting information are available in Figshare (10.6084/m9.figshare.6480266).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Matthew J. Dorman, Daryl Domman and Muhammad Ikhtear Uddin contributed equally.
Contributor Information
Firdausi Qadri, Email: fqadri@icddrb.org.
Nicholas R. Thomson, Email: nrt@sanger.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41883-x.
References
- 1.Shimada T, et al. Extended serotyping scheme for Vibrio cholerae. Curr. Microbiol. 1994;28:175–178. doi: 10.1007/BF01571061. [DOI] [Google Scholar]
- 2.Chapman C, et al. Scanning the landscape of genome architecture of non-O1 and non-O139 Vibrio cholerae by whole genome mapping reveals extensive population genetic diversity. PLoS ONE. 2015;10:e0120311. doi: 10.1371/journal.pone.0120311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaper JB, Morris JG, Levine MM. Cholera. Clin. Microbiol. Rev. 1995;8:48–86. doi: 10.1128/CMR.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devault AM, et al. Second-pandemic strain of Vibrio cholerae from the Philadelphia cholera outbreak of 1849. N. Engl. J. Med. 2014;370:334–340. doi: 10.1056/NEJMoa1308663. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea YA, Reen FJ, Quirke AM, Boyd EF. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 2004;42:4657–4671. doi: 10.1128/JCM.42.10.4657-4671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinnaka Y, Carpenter CC. An enterotoxin produced by noncholera vibrios. Johns Hopkins Med. J. 1972;131:403–411. [PubMed] [Google Scholar]
- 7.Bik EM, Gouw RD, Mooi FR. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltrán P, et al. Genetic diversity and population structure of Vibrio cholerae. J. Clin. Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albert MJ. Vibrio cholerae O139 Bengal. J. Clin. Microbiol. 1994;32:2345–2349. doi: 10.1128/jcm.32.10.2345-2349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cholera Working Group, icddr,b, et al. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae 0139 synonym Bengal. The Lancet342, 387–390 (1993). [PubMed]
- 11.Finkelstein, R. A. Cholera, Vibrio cholerae O1 and O139, and other pathogenic Vibrios. Medical Microbiology [Baron, S. (ed.)] (University of Texas Medical Branch at Galveston 1996). [PubMed]
- 12.WHO. WHO | 1998 - Cholera - Vibrio cholerae O139 strain. WHO Available at: http://www.who.int/csr/don/1998_09_22/en/ (Accessed: 3rd January 2018)
- 13.Faruque AS, Fuchs GJ, Albert MJ. Changing epidemiology of cholera due to Vibrio cholerae O1 and O139 Bengal in Dhaka, Bangladesh. Epidemiol. Infect. 1996;116:275–278. doi: 10.1017/S0950268800052572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque SM, et al. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg. Infect. Dis. 2003;9:1116–1122. doi: 10.3201/eid0909.020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faruque SM, et al. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. USA. 2003;100:1304–1309. doi: 10.1073/pnas.0337468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siriphap A, et al. Characterization and genetic variation of Vibrio cholerae isolated from clinical and environmental sources in Thailand. PLoS ONE. 2017;12:e0169324. doi: 10.1371/journal.pone.0169324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi Y, et al. Genome sequence and comparative analysis of a Vibrio cholerae O139 strain E306 isolated from a cholera case in China. Gut Pathog. 2014;6:3. doi: 10.1186/1757-4749-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury F, et al. Vibrio cholerae serogroup O139: Isolation from cholera patients and asymptomatic household family members in Bangladesh between 2013 and 2014. PLoS Negl. Trop. Dis. 2015;9:e0004183. doi: 10.1371/journal.pntd.0004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. WHO | Weekly Epidemiological Record, 30 July 2004, vol. 79, 31 (pp 281–288). (2004). Available at: http://www.who.int/wer/2004/wer7931/en/. (Accessed: 5th July 2017).
- 20.WHO | Weekly Epidemiological Record, 25 August 2017, vol. 92, 34 (pp. 477–500). WHO Available at: http://www.who.int/wer/2017/wer9234/en/. (Accessed: 28th January 2018)
- 21.Saha A, et al. Safety and immunogenicity study of a killed bivalent (O1 and O139) whole-cell oral cholera vaccine Shanchol, in Bangladeshi adults and children as young as 1 year of age. Vaccine. 2011;29:8285–8292. doi: 10.1016/j.vaccine.2011.08.108. [DOI] [PubMed] [Google Scholar]
- 22.Berche P, et al. The novel epidemic strain O139 is closely related to the pandemic strain O1 of Vibrio cholerae. J. Infect. Dis. 1994;170:701–704. doi: 10.1093/infdis/170.3.701. [DOI] [PubMed] [Google Scholar]
- 23.Calia KE, Waldor MK, Calderwood SB. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect. Immun. 1998;66:849–852. doi: 10.1128/iai.66.2.849-852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall RH, Khambaty FM, Kothary MH, Keasler SP, Tall BD. Vibrio cholerae non-O1 serogroup associated with cholera gravis genetically and physiologically resembles O1 E1 Tor cholera strains. Infect. Immun. 1994;62:3859–3863. doi: 10.1128/iai.62.9.3859-3863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun J, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weintraub A, et al. Vibrio cholerae O139 Bengal possesses a capsular polysaccharide which may confer increased virulence. Microb. Pathog. 1994;16:235–241. doi: 10.1006/mpat.1994.1024. [DOI] [PubMed] [Google Scholar]
- 28.Bik EM, Bunschoten AE, Gouw RD, Mooi FR. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sozhamannan S, et al. Cloning and sequencing of the genes downstream of the wbf gene cluster of Vibrio cholerae serogroup O139 and analysis of the junction genes in other serogroups. Infect. Immun. 1999;67:5033–5040. doi: 10.1128/iai.67.10.5033-5040.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stroeher UH, Parasivam G, Dredge BK, Manning PA. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J. Bacteriol. 1997;179:2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamasaki S, Garg S, Nair GB, Takeda Y. Distribution of Vibrio cholerae O1 antigen biosynthesis genes among O139 and other non-O1 serogroups of Vibrio cholerae. FEMS Microbiol. Lett. 1999;179:115–121. doi: 10.1111/j.1574-6968.1999.tb08716.x. [DOI] [PubMed] [Google Scholar]
- 32.Waldor MK, Colwell R, Mekalanos JJ. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc. Natl. Acad. Sci. USA. 1994;91:11388–11392. doi: 10.1073/pnas.91.24.11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada K, et al. Characterization of 3 megabase-sized circular replicons from Vibrio cholerae. Emerg. Infect. Dis. J. 2015;21:1262. doi: 10.3201/eid2107.141055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLeod SM, Waldor MK. Characterization of XerC- and XerD-dependent CTX phage integration in Vibrio cholerae. Mol. Microbiol. 2004;54:935–947. doi: 10.1111/j.1365-2958.2004.04309.x. [DOI] [PubMed] [Google Scholar]
- 35.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 36.McLeod SM, Kimsey HH, Davis BM, Waldor MK. CTXϕ and Vibrio cholerae: exploring a newly recognized type of phage–host cell relationship. Mol. Microbiol. 2005;57:347–356. doi: 10.1111/j.1365-2958.2005.04676.x. [DOI] [PubMed] [Google Scholar]
- 37.Moyer KE, Kimsey HH, Waldor MK. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXϕ. Mol. Microbiol. 2001;41:311–323. doi: 10.1046/j.1365-2958.2001.02517.x. [DOI] [PubMed] [Google Scholar]
- 38.Davis BM, Kimsey HH, Chang W, Waldor MK. The Vibrio cholerae O139 Calcutta bacteriophage CTXϕ is infectious and encodes a novel repressor. J. Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das B, Kumar Ghosh R, Sharma C, Vasin N, Ghosh A. Tandem repeats of cholera toxin gene in Vibrio cholerae 0139. The Lancet. 1993;342:1173–1174. doi: 10.1016/0140-6736(93)92157-O. [DOI] [PubMed] [Google Scholar]
- 40.Bhuiyan NA, et al. Changing genotypes of cholera toxin (CT) of Vibrio cholerae O139 in Bangladesh and description of three new CT genotypes. FEMS Immunol. Med. Microbiol. 2009;57:136–141. doi: 10.1111/j.1574-695X.2009.00590.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim EJ, Lee CH, Nair GB, Kim DW. Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends Microbiol. 2015;23:479–489. doi: 10.1016/j.tim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Kimsey HH, Nair GB, Ghosh A, Waldor MK. Diverse CTXΦs and evolution of new pathogenic Vibrio cholerae. The Lancet. 1998;352:457–458. doi: 10.1016/S0140-6736(05)79193-5. [DOI] [PubMed] [Google Scholar]
- 43.Klinzing DC, et al. Hybrid Vibrio cholerae El Tor lacking SXT identified as the cause of a cholera outbreak in the Philippines. mBio. 2015;6:e00047–15. doi: 10.1128/mBio.00047-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldor MK, Tschäpe H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J. Bacteriol. 2008;190:636–647. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grim CJ, et al. Occurrence of the Vibrio cholerae seventh pandemic VSP-I island and a new variant. OMICS J. Integr. Biol. 2010;14:1–7. doi: 10.1089/omi.2009.0087. [DOI] [PubMed] [Google Scholar]
- 47.Rowe-Magnus DA, Guerout A-M, Mazel D. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 2002;43:1657–1669. doi: 10.1046/j.1365-2958.2002.02861.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y, et al. Accumulation of mutations in DNA gyrase and topoisomerase IV genes contributes to fluoroquinolone resistance in Vibrio cholerae O139 strains. Int. J. Antimicrob. Agents. 2013;42:72–75. doi: 10.1016/j.ijantimicag.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu D, et al. Origins of the current seventh cholera pandemic. Proc. Natl. Acad. Sci. 2016;113:E7730–E7739. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin C-S, et al. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng L, et al. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS ONE. 2009;3:e4053. doi: 10.1371/journal.pone.0004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez Chaparro PJ, et al. Whole genome sequencing of environmental Vibrio cholerae O1 from 10 nanograms of DNA using short reads. J. Microbiol. Methods. 2011;87:208–212. doi: 10.1016/j.mimet.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Chowdhury F, et al. Concomitant enterotoxigenic Escherichia coli infection induces increased immune responses to Vibrio cholerae O1 antigens in patients with cholera in Bangladesh. Infect. Immun. 2010;78:2117–2124. doi: 10.1128/IAI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qadri F, Svennerholm A-M, Faruque ASG, Sack RB. Enterotoxigenic Escherichia coli in developing countries: Epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 2005;18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faruque A, Mahalanabis D, Islam A, Hoque S. Severity of cholera during concurrent infections with other enteric pathogens. J. Diarrhoeal Dis. Res. 1994;12:214–218. [PubMed] [Google Scholar]
- 57.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype Vibrio cholerae: Functional phage genes but dysfunctional phage genomes. J. Bacteriol. 2000;182:6992–6998. doi: 10.1128/JB.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butler SM, et al. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 2006;60:417–426. doi: 10.1111/j.1365-2958.2006.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 61.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736, 10.1101/gr.215087.116 (2017). [DOI] [PMC free article] [PubMed]
- 62.Hunt M, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 64.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker BJ, et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Carver TJ, et al. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 68.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page AJ, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb. Genomics. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Page AJ, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genomics. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- 78.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 79.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for High quality reference genomes for toxigenic and non-toxigenic <i>Vibrio cholerae</i> serogroup O139
Data Availability Statement
Sequencing reads generated during this project have been deposited into the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under study accession number PRJEB14661. Assembled genome sequences have been deposited under accession numbers LT992486- LT992493. Sequence alignments, phylogenetic tree data, and other supporting information are available in Figshare (10.6084/m9.figshare.6480266).