Abstract
Histological chorioamnionitis (HCA) is an infection of fetal membranes and complicates 5.2% to 28.5% of all live births. HCA is associated with increased mortality and morbidity in both premature and term neonates. Exposure to HCA may have long-term consequences, including an increased risk for allergic disorders and asthma later in childhood, the mechanism(s) of which are still not yet well understood. The objective of this study was to determine the mRNA transcriptome of cord blood mononuclear leukocytes from term neonates to identify key genes and pathways involved in HCA. We found 366 differentially expressed probe IDs with exposure to HCA (198 upregulated, 168 downregulated). These transcriptomes included novel genes and pathways associated with exposure to HCA. The differential gene expression included key genes regulating inflammatory, immune, respiratory and neurological pathways, which may contribute to disorders in those pathways in neonates exposed to HCA. Our data may lead to understanding of the role of key genes and pathways identified on the long-term sequelae related to exposure to HCA, as well as to identifying potential markers and therapies to prevent HCA-associated complications.
Introduction
Chorioamnionitis (CA) is an infection of the fetal membranes and placenta that complicates 5.2% of all live births1. The prevalence of histological CA (HCA) in term neonates with spontaneous labor is 23.6–28.7%2,3. Approximately 500,000 to one million infants in the United States are born each year to mothers diagnosed with HCA. Exposure to HCA may have long-term consequences, including an increased risk for allergic disorders and asthma later in childhood4–6. Additionally, studies have demonstrated that exposure to HCA can also lead to developmental delay and cerebral palsy in preterm and term infants7–9. Asthma and other allergic chronic diseases during childhood affect greater than 7 million children in the United States10. Although HCA is associated with the development of asthma, allergic disorders, and neurodevelopmental impairment, the exact mechanism for these complications is unknown. Exposure to infection during late fetal life is likely to incite epigenetic changes, which may modulate the immune and neurological systems and increase the risk for development of neurological and allergic disorders later in life.
Gene expression studies of cells and tissues have become a major tool for discovery in the pathogenesis of various diseases. Global gene expression by transcriptomic analysis can uncover gene signatures and help delineate molecular pathways involved in the development of asthma, allergy, and neurodevelopmental impairment in neonates born to mothers with HCA. Previous studies have reported differential gene expression patterns associated with preterm labor and lipopolysaccharide (LPS) stimulation of cord blood leukocytes11,12. More recently, differential gene expression in whole blood from preterm neonates exposed to HCA has been published13. However, fetal inflammatory and immune response to microbial infection differ between preterm (born before 37 weeks gestation) and term infants14–16. The effects of HCA on gene expression profile on cord blood mononuclear leukocytes in human term neonates have not been characterized. Our objective was to determine the mRNA transcriptome of cord blood mononuclear leukocytes from term neonates and identify key genes and pathways involved in HCA.
Results
Ten term infants were enrolled in the study. Five infants had histological chorioamnionitis (HCA group), and five infants with no evidence of histological chorioamnionitis on placental histopathology served as the control group.
Differential Gene Expression
Comparison of the HCA group array data with the control group using transcriptome array console software revealed that 366 probe IDs were differentially expressed with a fold change ≥ 1.5 (p < 0.05). Of these, 198 probe IDs were significantly up-regulated (Supplementary Table 1), 60 with annotated genes and 138 non-annotated genes. One hundred and sixty-eight probe IDs were significantly down-regulated (Supplementary Table 2), of which 105 had annotation with gene symbols and 63 were not annotated. The top 10 up- and down-regulated genes based on the fold change are reported in Table 1. The top 10 up-regulated genes included chemokine (C-C motif) receptor-2 (CCR2), a pro-inflammatory chemokine important in inflammatory diseases including asthma17,18. The top down-regulated genes included two important antimicrobial proteins, lactoferrin (LTF) and cathelicidin antimicrobial peptide (CAMP).
Table 1.
Top 10 differentially up- or down-regulated genes after exposure to HCA.
| ProbeID | Gene Symbol | HCA Group Average Expression | Control Group Average Expression | Fold Change | Up/Down | P-value |
|---|---|---|---|---|---|---|
| TC17001604.hg.1 | LRRC37A4P | 1389.16 | 344.89 | 4.01 | Up | 0.0245 |
| TC04002938.hg.1 | LOC285505 | 364.56 | 136.24 | 2.68 | 0.012 | |
| TC07000288.hg.1 | LINC01061 | 1652 | 643.59 | 2.58 | 0.0117 | |
| TC03000255.hg.1 | CCR2 | 765.36 | 304.44 | 2.51 | 0.0417 | |
| TC04001682.hg.1 | FAM198B | 643.59 | 256 | 2.5 | 0.0167 | |
| TC17000604.hg.1 | LRRC37A2 | 13216.02 | 5556.65 | 2.37 | 0.0312 | |
| TC07001299.hg.1 | TRGV4 | 140.07 | 60.13 | 2.32 | 0.0002 | |
| TC07001300.hg.1 | TRGV3 | 179.77 | 79.89 | 2.24 | 4.50xE-05 | |
| TC04002939.hg.1 | TMEM144 | 240.52 | 112.99 | 2.12 | 0.0173 | |
| TC17001603.hg.1 | LRRC37A4P; LRRC37A2 | 20594.91 | 9946.68 | 2.08 | 0.0478 | |
| TC01003638.hg.1 | PTGS2 | 252.48 | 9089.59 | −35.91 | Down | 0.0205 |
| TC04002953.hg.1 | AREG | 313 | 1884.54 | −6.03 | 0.001 | |
| TC01001749.hg.1 | G0S2 | 2352.53 | 13034.07 | −5.54 | 0.008 | |
| TC05001854.hg.1 | HBEGF | 385.34 | 2005.85 | −5.18 | 0.0022 | |
| TC19001593.hg.1 | PLAUR | 1448.15 | 7082.29 | −4.89 | 0.0138 | |
| TC21001069.hg.1 | SAMSN1 | 3956.48 | 17438.64 | −4.42 | 0.0257 | |
| TC12001216.hg.1 | OLR1 | 178.53 | 765.36 | −4.27 | 0.0365 | |
| TC03000276.hg.1 | CAMP | 657.11 | 2683.69 | −4.09 | 0.0011 | |
| TC09000508.hg.1 | NR4A3 | 143.01 | 580.04 | −4.07 | 0.0128 | |
| TC03001357.hg.1 | LTF | 2683.69 | 9345.14 | −3.49 | 0.0208 |
Differential Gene Expressions Related to Immune Response
Thirty genes related to immune response and important for regulation of inflammatory pathways were identified (Table 2). Twelve genes were up-regulated and 18 genes were down-regulated. The up-regulated genes have been previously associated with asthma; hyper-reactivity and allergy (CCR2, HRH2, PTEN); lung development and apoptosis (CHRNA7, RTKN2); and immune function (GIMAP4, GIMAP7, KLRC3 and CD99) (Table 3). Similarly, the down-regulated genes have roles in asthma and airway modeling (PTGS2, AREG, HBEGF, PLAUR, CAMP, LTF, FCAR, PDE4B, ZNF331 and IRAK2) and immune function (NR4A3, NFIL3, ADGRE3, TARM1 and ZC3H12A). The down-regulated gene CXCL1 has a role in lung development and BPD (Table 4).
Table 2.
Differential expression of genes related to immune response after exposure to HCA.
| Gene Symbol | Up/Down | Fold Change | P-value | HCA Group Average Expression | Control Group Average Expression |
|---|---|---|---|---|---|
| CCR2 | Up-regulated Genes | 2.51 | 0.0417 | 765.36 | 304.44 |
| TRGV4 | 2.32 | 0.0002 | 140.07 | 60.13 | |
| GIMAP4 | 2.29 | 0.0118 | 52498.92 | 23010.42 | |
| TRGV3 | 2.24 | 0.000045 | 179.77 | 79.89 | |
| KLRC3 | 1.8 | 0.0433 | 1458.23 | 809.00 | |
| HRH2 | 1.74 | 0.0233 | 1734.13 | 1002.93 | |
| CHRNA7 | 1.61 | 0.0201 | 33.36 | 20.68 | |
| GIMAP7 | 1.61 | 0.0086 | 4482.23 | 2778.33 | |
| RTKN2 | 1.59 | 0.0294 | 153.28 | 96.34 | |
| CD99 | 1.56 | 0.0121 | 2646.74 | 1698.45 | |
| PTEN | 1.55 | 0.0275 | 694.58 | 448.82 | |
| GIMAP1-GIMAP5; GIMAP5; GIMAP1 | 1.53 | 0.0155 | 989.12 | 648.07 | |
| PTGS2 | Down-regulated Genes | −35.91 | 0.0205 | 252.48 | 9089.59 |
| AREG | −6.03 | 0.001 | 313.00 | 1884.54 | |
| HBEGF | −5.18 | 0.0022 | 385.34 | 2005.85 | |
| PLAUR | −4.89 | 0.0138 | 1448.15 | 7082.29 | |
| CAMP | −4.09 | 0.0011 | 657.11 | 2683.69 | |
| NR4A3 | −4.07 | 0.0128 | 143.01 | 580.04 | |
| LTF | −3.49 | 0.0208 | 2683.69 | 9345.14 | |
| NFIL3 | −2.77 | 0.0262 | 1192.69 | 3304.00 | |
| FCAR | −2.35 | 0.0023 | 2256.70 | 5330.30 | |
| PDE4B | −2.33 | 0.0125 | 280.14 | 652.58 | |
| ADGRE3 | −2.3 | 0.042 | 680.29 | 1573.76 | |
| CXCL1 | −2.05 | 0.0404 | 112.21 | 229.13 | |
| TARM1 | −2.02 | 0.0059 | 116.16 | 235.57 | |
| SNAI1 | −1.93 | 0.0083 | 47.18 | 91.14 | |
| ZNF331 | −1.77 | 0.0308 | 288.01 | 512.00 | |
| ZC3H12A | −1.76 | 0.0422 | 256.00 | 451.94 | |
| IRAK2 | −1.74 | 0.048 | 164.28 | 284.05 | |
| NFKBIE | −1.59 | 0.0351 | 413.00 | 657.11 |
Table 3.
Up-regulated genes after exposure to HCA and their functions.
| Gene Symbol | Gene name | Function |
|---|---|---|
| CCR2 | C-C Motif Chemokine Receptor 2, Receptor for monocyte chemoattractant protein-1 | Role in asthma, inflammation |
| GIMAP4 | GTPase, Immunity-Associated Protein family member 4 | T- and B-cell development and survival, T-cell apoptosis |
| KLRC3 | Killer Cell Lectin-Like Receptor Subfamily C, Member 3 | Natural Killer receptor gene, role in cell proliferation |
| TRGV3 | T Cell Receptor Gamma Variable 3 | Peptide antigen binding, immune response |
| HRH2 | Histamine Receptor H2 | Histamine receptor gene, role in hypersensitiviy |
| CHRNA7 | Cholinergic Receptor Nicotinic Alpha 7 Subunit | Alter airway morphometry and lung function |
| GIMAP7 | GTPase, Immune-Associated Nucleotide-Binding Protein 7 | Regulators of lymphocyte survival and homeostasis (Schwefel D 2013) |
| RTKN2 | Rhotekin 2 | Role in apoptosis |
| CD99 | Cluster of differentiation 99, Single-chain type-1 glycoprotein | Role in cell adhesion, migration, death, differentiation and diapedesis, and it influences processes associated with inflammation, immune responses (pasello M 2018) |
| VNN2 | Vanin 2 | Hematopoietic cell trafficking, Role in oxidative-stress |
| PTEN | Phosphatase And Tensin Homolog | Role in asthma, hyper-reactivity |
Table 4.
Down-regulated genes after exposure to HCA and their functions.
| Gene symbol | Gene name | Function |
|---|---|---|
| PTGS2 | Prostaglandin-Endoperoxide Synthase 2 | Role in asthma and lung inflammation, hyperoxia induced lung injury |
| GOS2 | G0/G1 Switch 2 | Regulation of lipid metabolism |
| AREG | Amphiregulin, Colorectum Cell-Derived Growth Factor | Role in asthma, airway modelling, BPD |
| HBEGF | Heparin-Binding Epidermal Growth Factor | Role in asthma, airway modelling |
| PLAUR | Plasminogen Activator, Urokinase Receptor | Role in asthma, COPD |
| CAMP | Cathelicidin Antimicrobial Peptide | Role in asthma, lung infection |
| NR4A3 | Nuclear Receptor Subfamily 4 Group A Member 3 | Regulates neutrolphils number and survival, Treg cell development through activation of Foxp3 |
| LTF | Lactotransferrin | Role in asthma, BPA VAP |
| FCAR | Fc Fragment Of Immunoglobulin Alpha Receptor | SNP associated with allergic asthma (Jasek M 2004) |
| NFIL3 | Nuclear Factor, Interleukin 3 Regulated | Critical regulator for IgE production and airway hyper-responsiveness. |
| PDE4B | Phosphodiesterase 4B | Role in asthma, inhibitors use as treatment |
| ADGRE3 | Adhesion G Protein-Coupled Receptor E3 | Modulator of immune cell funtion |
| CXCL1 | C-X-C Motif Chemokine Ligand 1 | Role in BPD, lung alveolization |
| TARM1 | T Cell-Interacting, Activating Receptor On Myeloid Cells 1 | Upregulated by LPS, recruitment to site of inflammation |
| SNAI1 | Snail Family Transcriptional Repressor 1 | Β-catenin target gene, lung fibrosis |
| ZNF331 | Zinc Finger Protein 331 | Pathogenesis of asthma |
| ZC3H12A | Zinc Finger CCCH-Type Containing 12A | Regulates the development and function of IL-5-producing TH2 cells through the Notch/Gata3 pathway |
| IRAK2 | Interleukin 1 Receptor Associated Kinase 2 | IRAK2 Attenuate the Proinflammatory Effects of IL-33 in Asthma-like Mouse Models. |
Ingenuity Pathway Analysis
Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) by loading 366 probe sets that were differentially expressed with HCA19. Seventy-three functions were modified by differential gene expression with exposure to HCA. Important functions modified with the exposure to HCA are listed in Table 5. The modification of functions with HCA included genes related to immune system, inflammatory response, connective tissue disorders, neurodevelopmental disorders, hematological development and disorders, and respiratory development and disorders. Ingenuity Canonical Pathway Analysis identified 207 pathways that were modified after exposure to HCA, of which 19 pathways are known to be important in immune regulation and inflammatory responses (Table 6).
Table 5.
Functions modified with exposure to HCA.
| Modified Functions | Number of Genes Involved | Range of p-value for genes involved |
|---|---|---|
| Cellular Movement | 46 | 1.26E-13-1.52E-03 |
| Immune Cell Trafficking | 38 | 1.26E-13-1.03E-03 |
| Inflammatory Disease | 46 | 7.43E-13-1.25E-03 |
| Cell Death and Survival | 60 | 1.5E-12-1.46E-03 |
| Immunological Disease | 47 | 2.74E-12-1.25E-03 |
| Connective Tissue Disorders | 38 | 3.3E-12-1.47E-03 |
| Inflammatory Response | 55 | 3.55E-11-1.38E-03 |
| Neurological Disease | 32 | 1.29E-10-1.38E-03 |
| Cell-To-Cell Signaling and Interaction | 40 | 3.45E-10-1.51E-03 |
| Hematological System Development and Function | 44 | 3.45E-10-1.51E-03 |
| Cellular Growth and Proliferation | 49 | 2.3E-07-1.46E-03 |
| Hematopoiesis | 27 | 4.16E-07-9.74E-04 |
| Lymphoid Tissue Structure and Development | 33 | 4.16E-07-9.74E-04 |
| Cellular Function and Maintenance | 42 | 4.95E-07-1.52E-03 |
| Humoral Immune Response | 15 | 5.54E-07-1.4E-03 |
| Infectious Diseases | 28 | 6.5E-07-1.39E-03 |
| Respiratory Disease | 19 | 6.5E-07-1.23E-03 |
| Cell Cycle | 26 | 6.84E-07-1.49E-03 |
| Gene Expression | 37 | 6.84E-07-1.5E-03 |
| Free Radical Scavenging | 14 | 7.69E-06-9.02E-06 |
| Cell-mediated Immune Response | 15 | 2.3E-05-5.63E-04 |
| DNA Replication, Recombination, and Repair | 11 | 1.06E-04-4.46E-04 |
| Antimicrobial Response | 9 | 2.39E-04-2.39E-04 |
| Nervous System Development and Function | 15 | 3.11E-04-1.19E-03 |
| Post-Translational Modification | 14 | 3.43E-04-3.43E-04 |
| Hypersensitivity Response | 5 | 3.8E-04-3.8E-04 |
| Hematological Disease | 39 | 4.13E-04-1.09E-03 |
| Cellular Assembly and Organization | 6 | 8.04E-04-8.04E-04 |
| Psychological Disorders | 3 | 9.59E-04-9.59E-04 |
| Respiratory System Development and Function | 2 | 9.59E-04-9.59E-04 |
| Developmental Disorder | 12 | 1.24E-03-1.24E-03 |
| Behavior | 17 | 1.47E-03-1.47E-03 |
Table 6.
Canonical pathways in ingenuity pathway analysis associated with differentially expressed genes between control and HCA groups.
| Ingenuity Canonical Pathways | −log10(p-value) | Molecules involved in pathways |
|---|---|---|
| Protein Kinase A Signaling | 4.46 | PTEN,PTGS2,PTPRE,DUSP6,MYH10, NFKBIE,PDE4B,ADD2,DUSP2,NFKBID |
| PI3K/AKT Signaling | 3.32 | PTEN,PTGS2,CDKN1A,NFKBIE,NFKBID |
| IL-17A Signaling in Airway Cells | 3.12 | PTEN,CXCL1,NFKBIE,NFKBID |
| MIF-mediated Glucocorticoid Regulation | 3.09 | PTGS2,NFKBIE,NFKBID |
| MIF Regulation of Innate Immunity | 2.83 | PTGS2,NFKBIE,NFKBID |
| iCOS-iCOSL Signaling in T Helper Cells | 2.46 | PTEN,HLA-DQB1,NFKBIE,NFKBID |
| TNFR2 Signaling | 2 | NFKBIE,NFKBID |
| IL-17 Signaling | 1.99 | PTGS2,CXCL1,TIMP1 |
| 4-1BB Signaling in T Lymphocytes | 1.91 | NFKBIE,NFKBID |
| IL-1 Signaling | 1.9 | NFKBIE,IRAK2,NFKBID |
| PPAR Signaling | 1.9 | PTGS2,NFKBIE,NFKBID |
| IL-17A Signaling in Fibroblasts | 1.84 | NFKBIE,NFKBID |
| B Cell Activating Factor Signaling | 1.71 | NFKBIE,NFKBID |
| IL-8 Signaling | 1.7 | HBEGF,PTGS2,CXCL1,IRAK2 |
| p53 Signaling | 1.68 | PTEN,CDKN1A,TNFRSF10B |
| PI3K Signaling in B Lymphocytes | 1.54 | PTEN,NFKBIE,NFKBID |
| Glucocorticoid Receptor Signaling | 1.49 | CCNH,PTGS2,CDKN1A,NFKBIE,NFKBID |
| IL-10 Signaling | 1.29 | NFKBIE,NFKBID |
| T Helper Cell Differentiation | 1.29 | HLA-DQB1,IL23R |
IPA Networks
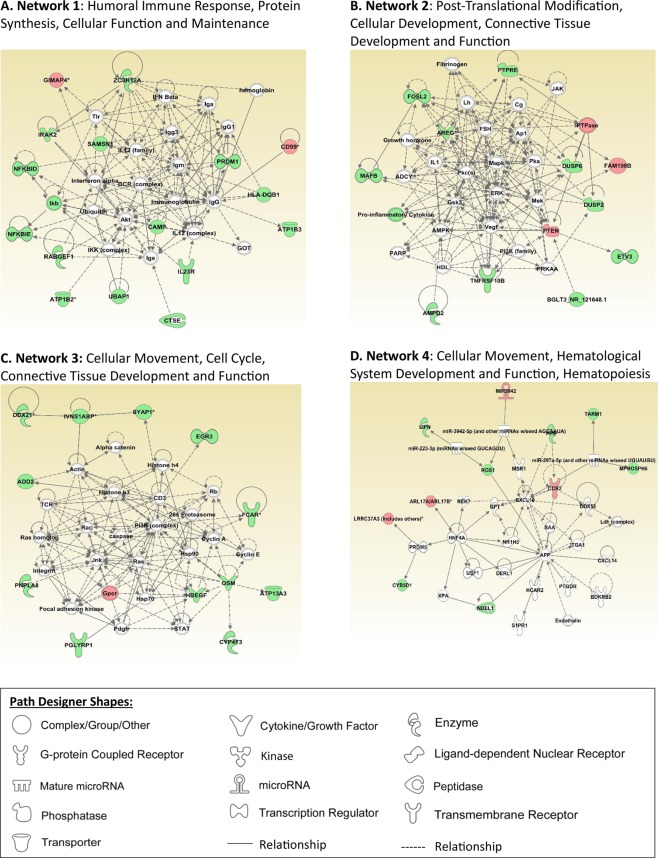
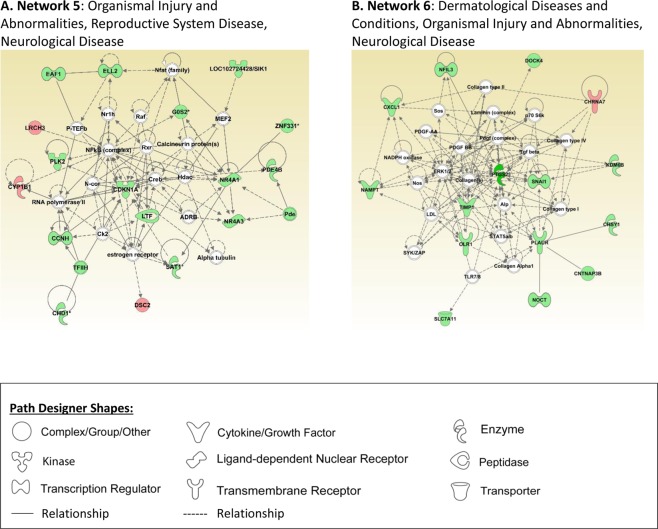
IPA picked up important networks related to immune, inflammatory, and neurological pathways. Four were closely related to immune modulation and inflammatory response and two were related to neurologic disease. Networks 1 through 4 are shown in Fig. 1. Network 1 is related to humoral immune response, protein synthesis, cellular function and maintenance. Network 2 is related to post-translational modification, cellular development, connective tissue development and function. Network 3 is related to cellular movement, cell cycle, connective tissue development, and function. Network 4 is related to cellular movement, hematological system development and function, and hematopoiesis. Networks 5 and 6 are closely related to neurological disease (Fig. 2). Network 5 involves organismal injury and abnormalities, reproductive system disease, and neurological disease. Network 6 involves dermatological diseases and conditions, organismal injury and abnormalities, as well as neurological disease.
Figure 1.
Ingenuity pathway analysis networks related to immune modulation and inflammatory response are presented in this figure (A–D). Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) by loading 366 probe sets that were differentially expressed with HCA19.
Figure 2.
Ingenuity pathway analysis networks related to neurological disease are presented in this figure (A,B). Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) by loading 366 probe sets that were differentially expressed with HCA19.
Quantitative Real Time PCR Verification
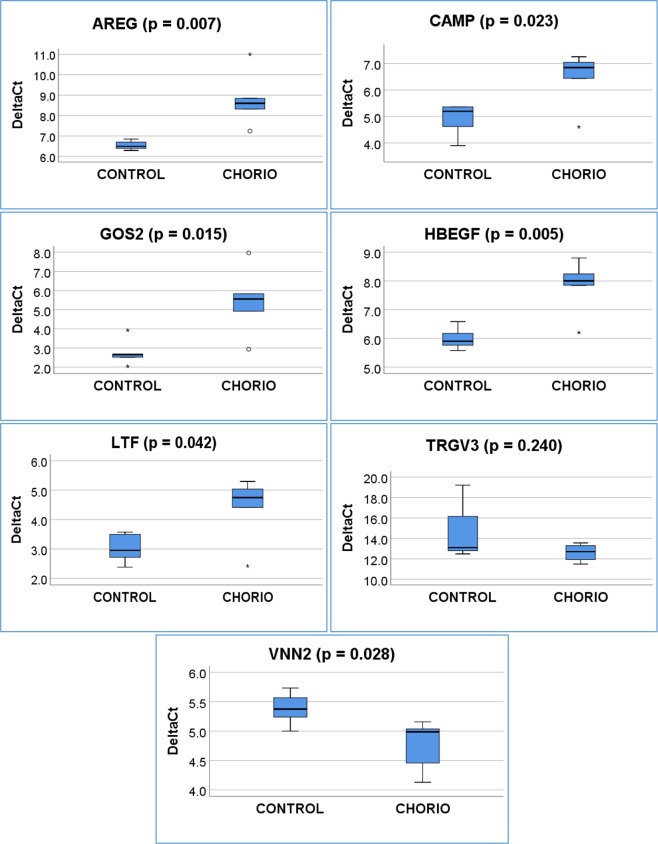
To confirm gene expression results, we selected seven genes from the differentially expressed gene list that were closely related to inflammatory response and immune modulation. A quantitative Real-Time PCR was performed using total RNA to validate the microarray data from the same neonates studied by microarray analysis (Fig. 3). Of the seven selected differentially expressed genes, five were down-regulated (GOS2, AREG, HBEGF, CAMP and LTF) and two were up-regulated (VNN2 and TRGV3). Fold-changes obtained by RT-PCR were consistent with microarray results. All fold changes in RT-PCR results reached significance (p ≤ 0.05) except for TRGV3 (p = 0.24). A comparison of the microarray data with RT-PCR results is shown in Table 7.
Figure 3.
Box plot for real-time PCR data on selected genes in the two groups (Control and HCA) are presented in this figure.
Table 7.
Validation of microarray data with real-time PCR.
| Gene Symbol | Microarray | Real-Time PCR | ||
|---|---|---|---|---|
| Fold Change | p-value | Fold Change | p-value | |
| GOS2 | −3.18 | 0.011 | −6.42 | 0.015 |
| AREG | −2.54 | 0.006 | −4.78 | 0.007 |
| HBEGF | −2.15 | 0.008 | −3.53 | 0.005 |
| CAMP | −1.83 | 0.005 | −2.93 | 0.023 |
| LTF | −1.73 | 0.044 | −2.56 | 0.042 |
| VNN2 | 1.56 | 0.021 | 1.55 | 0.028 |
| TRGV3 | 1.78 | 9.18E-04 | 3.67 | 0.240* |
Discussion
Histological chorioamnionitis is associated with significant long-term morbidities including asthma and allergic and neurological disorders in preterm and term infants4–9. The exact mechanism of increased risk for asthma and allergic disorders in neonates born to mothers with HCA is not well understood. Inflammatory mediators released with HCA may reprogram the developing immune, respiratory, and neurological systems, leading to allergy, asthma, and immunological and neurological disorders20,21. To our knowledge, this is the first study reporting the differential gene expression profile in cord blood mononuclear leukocytes in term infants exposed to HCA. Our data indicate that the key genes regulating inflammatory, immune, respiratory, and neurological pathways are differentially expressed in cord blood leukocytes from term infants born to mothers with HCA.
Based on fold change, the top up-regulated genes included chemokine receptor CCR2, T-cell receptor gamma variable-3 (TRGV3), and TRGV4. CCR2 is an important chemokine receptor that induces macrophage and monocytes recruitment to sites of inflammation22. CCR2 is up-regulated on T-helper cells in an animal model of allergic asthma and blocking of CCR2 suppressed airway inflammation18.The top down-regulated genes included HBEGF, LTF, CAMP, and NR4A3. HBEGF is a potent vasodilator of intestinal microvasculature, and supplementation of HBEGF reduced intestinal injury in animal models of necrotizing enterocolitis (NEC)23–25.
In utero exposure to HCA leads to fetal inflammatory response syndrome. Understanding the effects of the fetal inflammatory response on the programming of immune, respiratory, and neurological systems in neonates could lead to new ways of modulating these responses and improving long-term consequences. We have identified several key genes involved in immune function, inflammation, and lung development that are expressed differentially after exposure to HCA. These candidate genes are likely to play a critical role in long-term consequences including bronchopulmonary dysplasia (BPD), asthma, allergy, and immunological disorders in neonates exposed to HCA. This warrants further functional investigation.
We report that several important genes involved in the immune system are up-regulated in cord blood mononuclear cells after exposure to HCA. RTKN2 has anti-apoptotic properties, is highly expressed in T-cells, and plays a role as a key signaling switch for regulation of genes involved in T-cell survival26. RTKN2 may prolong survival of inflammatory cells and contribute to an exaggerated inflammatory response in neonates exposed to HCA. CD99 is a cell surface protein involved in cell adhesion, migration, differentiation, and apoptosis, and influences processes associated with inflammation and immune response27. GIMAP family members play a role in T-helper cell differentiation28. GIMAP4 and GIMAP5 have a role in T-cell survival. GIMAP7 is also a regulator of lymphocyte survival and homeostasis29. NR4A3, a gene involved in protein coding that regulates Treg cell development through activation of FoxP3, and which may have potential for therapeutic target for immune disorders, was down-regulated30. NR4A3 also positively regulates neutrophil survival and homeostasis31.
Our data indicate that several key pro-inflammatory mediators are down-regulated in cord blood mononuclear leukocytes after exposure to HCA. The down-regulation of a few pro-inflammatory mediators likely is a modulating mechanism for the exaggerated inflammatory response induced by HCA. The inflammatory mediators downregulated by HCA include PDE4B, CXCL1, TARM1, and IRAK2. PDE4B degrades cyclic AMP, a second messenger involved in inflammatory process. Selective inhibition of PDE4B is a therapeutic target for inflammatory and allergic disorders32. CXCL1 is a potent neutrophil chemoattractant that plays a role in the development of BPD33. IRAK2 forms a Myddosome complex with Myd88 and IRAK4, activating downstream NF-kB and MAPKs P38 and JNK, leading to inflammatory response34. The roles of these pro-inflammatory mediators downregulated by exposure to HCA need further investigation.
We also report that several key genes involved in lung development, BPD, and asthma are expressed differentially in cord blood mononuclear cells after exposure to HCA. Exposure to HCA upregulates CHRNA7, a key mediator of the biological effects of nicotine. CHRNA7 plays a critical role in lung development as well in the pathogenesis of asthma35,36, and inhibition of CHRNA7 is an important therapeutic target for asthma36. Amphiregulin (AREG) is a member of the epidermal growth factor family and contributes to the regulation of cell proliferation. Recombinant AREG suppresses epithelial cell apoptosis in LPS-induced lung injury in mice37. Down-regulation of AREG by HCA may contribute to the development of BPD by increasing epithelial cell apoptosis. CAMP plays a critical role in innate immunity against invasive bacterial infections. CAMP is an important host defense against respiratory pathogens. Suppression of CAMP by HCA may increase the risk for recurrent respiratory infections in neonates exposed to HCA. Ramos-Martinez et al. showed that treatment with vitamin D reduced respiratory infections in patients with asthma, and this effect was mediated by increase in CAMP38. LTF is an important mucosal antimicrobial protein that is also downregulated in neonates exposed to HCA. Revenis showed that the level of LTF was lower in tracheal aspirate samples from premature infants who developed BPD39. Additionally, LTF can down-regulate allergic airway inflammation in asthma40. In an animal model of allergic rhinitis, LTF administration in the nostril alleviated allergic rhinitis and its mechanisms41. NFIL3 is the most strongly induced transcription factor by IL-4 stimulation and is a critical regulator of IgE production and airway hyper-responsiveness42,43. The target genes (SNAI1, ZNF331, Zc3h12a) for the canonical Wnt/B-catenin pathway are down-regulated after exposure to HCA. These target genes of Wnt/B-catenin pathways play a role in the pathogenesis of asthma44–46. Peng et al. demonstrated that Zc3h12a-knockout mice have severe spontaneous lung inflammation with an increase in IL-5- and IL-13-producing cells in the lung45. ZNF331 was expressed differentially in bronchial alveolar lavage cells from patients with asthma46. PTEN regulates airway smooth muscle contraction in allergic asthma47. Epithelial-mesenchymal transition (EMT) accounts for accumulation of subepithelial mesenchymal cells and contributes to airway hyper-responsiveness and remodeling48. Up-regulating expression of PTEN inhibits EMT and may be protective on airway modeling in asthma and BPD. PTGS2/COX2 increases lung inflammation and impairs lung development33,49. HBEGF overexpression is associated with airway remodeling and asthma50. Interestingly, we found that expression of PTGS2 and HBEGF was down-regulated in cord blood mononuclear cells after exposure to HCA. HBEGF was also downregulated in lung tissue after intra-amniotic LPS administration in an animal model of HCA51. The role of PTGS2 and HBEGF on lung development and lung inflammation needs further investigation.
Infants and children born to mothers with HCA are at not only an increased risk for development of inflammatory, immune, and allergic diseases, but they are also predisposed to abnormal neurodevelopmental disorders7–9. Our pathway analysis indicates that exposure to HCA affects genes involved in nervous system development, neurological disease, behavioral, developmental, and psychological disorders. Allard et al. showed that female rats exposed to in utero placental inflammation showed hyperactive behavior after puberty52. IPA analysis of our data indicates that HCA differentially expressed genes in the IL-17 signaling pathway, IL-17A signaling in airway cells, and IL-17A signaling in fibroblasts. Recent findings suggests that IL-17A in the fetal and maternal inflammatory response leads to fetal brain injury and neurological sequelae including cerebral palsy and potentially autism, schizophrenia, and multiple sclerosis later in life53.
Our study has several strengths. This is the first study reporting differential gene expression in cord blood from term neonates exposed to HCA. A similar study reported differential gene expression after exposure to HCA in preterm infants using whole blood, which does not allow distinction of the relative contribution of leukocytes. Our study is unique as global gene expression was performed in cord blood mononuclear leukocytes, not in whole blood. Further, our global gene expression data were validated by Real-Time PCR for the identified target genes. Six out of seven genes validated with Real-Time PCR showed significant change in the same direction as microarray data, with the seventh gene displaying trends in the same direction. Neonates are vulnerable subjects, and drawing a large amount of blood from them is ethically unacceptable. By using umbilical cord blood, we were able to collect a large amount of blood non-invasively without additional risks to the neonates.
Our study also has several limitations. Our sample size of 10 neonates is small, but similar sample sizes have been used commonly in studies investigating differential gene expression using microarray12,54. There is an inherent chance of finding differences in the gene expression due to multiple comparison; however, our results were validated by using Real-Time PCR.
In conclusion, HCA induces differential gene expression in cord blood mononuclear leukocytes from term neonates. We found novel genes and pathways associated with exposure to HCA, including key genes regulating inflammatory, immune, respiratory and neurological pathways. This differential gene expression may contribute to inflammatory, immunological, and neurological disorders in neonates exposed to HCA. Future studies can further validate differential expression of target genes in a larger cohort of neonates exposed to HCA. Our data may lead to understanding the role of key genes and pathways identified in this study on the long-term sequelae related to exposure to HCA. Functional studies on the identified genes and pathways could lead to the development of potential markers for the diseases caused by exposure to HCA and possible therapy to prevent those complications.
Methods
Ethical approval: human study protocol and institutional biosafety approvals
All human protocols and procedures described in this study were approved by the Institutional Review Board of Thomas Jefferson University Hospital. All experiments performed in this study were approved and in accordance with the Nemours Institutional Biosafety Committee. The Institutional Review Board waived informed consent as this study was performed on discarded blood and placental tissue samples.
Study Design
This is a prospective observational study that examines differential gene expression in mononuclear leukocytes isolated from cord blood of term infants born to mothers with HCA. Cord blood and fetal membranes were collected from term neonates (37–40 weeks of gestation). Exclusion criteria were maternal infections other than HCA and complications of pregnancy including diabetes, hypertension, major congenital/chromosomal anomalies, and intrauterine growth restriction.
Cord blood collection and isolation of leukocytes
At the time of delivery, the umbilical cord was disinfected and cut at the placental side of the clamp. Cord blood was collected in sterile EDTA tubes, mixed thoroughly, and checked for blood clots. Mononuclear leukocytes were isolated by Ficoll-Paque Plus density gradient (GE Healthcare Biosciences, Pittsburgh, PA), following the manufacturer’s protocol and the method of Normann et al.55. In brief, collected blood was diluted with an equal amount of phosphate buffered saline (PBS). 1 ml of PBS was mixed thoroughly with 16 ml of Ficoll-Paque plus and used as density gradient. 25 to 30 ml of diluted blood was layered on top of the Ficoll mixture and centrifuged at 400xg for 30 minutes. The upper plasma layer was collected and centrifuged to remove platelets. The cellular interface was then collected in a separate tube and washed with PBS containing 1 mM EDTA. Cells were washed with autologous plasma to remove platelets. Finally, the mononuclear leukocytes were washed with PBS, and 2 million packed cells per vial were saved at −80 °C for RNA isolation and microarray analysis.
Fetal membrane collection, processing, staining, and diagnosis of HCA
Small pieces of fetal membranes tissue were washed with cold PBS and fixed in 10% neutral buffered formalin for 24–48 hours. Membrane pieces were processed and paraffin embedded in Histoplast LP (Thermo Fisher Scientific, Fremont, CA). Samples were sectioned at 5 µm on a Leica RM2255 microtome (Leica, Buffalo Grove, IL) and floated onto Superfrost® Plus slides (Thermo Fisher Scientific, Fremont, CA). The sections were heat immobilized for 60 minutes at 60 °C and were subsequently equilibrated to room temperature prior to staining. The slides were placed on Tissue-Tek Prizma stainer (Sakura Finetek USA, Torrance, CA) and were deparaffinized with xylene, hydrated through a graded series of alcohols, and hydrated to water. Finally, the slides were stained with Harris hematoxylin (BBC Biomedical, Mount Vernon, WA) and eosin (Acros Organics, Cole-Parmer, Vernon Hills, IL) and were dehydrated, cleared, and mounted in Permount® (Thermo Fisher Scientific, Fremont, CA). Tissue samples were processed by the Nemours Histochemistry and Tissue Processing Core using standard workflows and operating procedures. The stained slides were examined by a pathologist (JC) and classified either as HCA (placental membranes score ≥ stage 1) or no HCA (no histological inflammatory changes in fetal membranes)56.
RNA Isolation and Gene Expression Study
Total RNA was isolated using Qiagen miRNeasy Mini Kit (Qiagen, Germantown, MD). RNA was quantified on a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and quality was assessed by an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA). 100 ng of RNA was used from each sample to prepare fragmented biotin-labeled cDNA by GeneChip WT PLUS reagent kit (Affymetrix, Santa Clara, CA). The Affymetrix gene chips and Human Transcriptome Array 2.0 were hybridized with 5 µg of fragmented biotin-labeled cDNA in 220 µl hybridization cocktail, followed by target denaturation at 99 °C for 5 min and then 45 °C for 5 min. Hybridization was performed for 16 hours at 45 °C with a rotation of 60 rpm. An Affymetrix GeneChip hybridization wash and stain kit was used to wash and stain the arrays in GeneChip Fluidic Station 450. Chips were scanned on an Affymetrix GeneChip Scanner 3000 using Command Console Software (Thermo Fisher Scientific, Waltham, MA). Expression Console Software v1.4.1 was used to perform quality control.
Real-Time PCR
Selected genes from microarray data were validated by Quantitative Real Time PCR with TaqMan Gene Expression Assay Mix from Applied Biosystems (Applied Biosystems, Foster City, CA) following the manufacturer’s protocol. In brief, 200 ng of total RNA was reverse transcribed with a High-Capacity cDNA RT kit (Applied Biosystems, Foster City, CA) in a total reaction volume of 20 µl. Real-Time PCR was performed in duplicate wells with 0.5 µl of cDNA preparation using TaqMan Universal PCR Master Mix and TaqMan Gene Expression Assay mixes specific for different genes. Actin B was used as a housekeeping gene, and data were collected by Quant Studio 12K Flex Thermal Cycler (Applied Biosystems, Foster City, CA).
Statistical Analysis
Expression Console Software was used to generate a Chp file from Affymetrix after sst-rma normalization. The HCA Group was compared with the control group using Transcriptome array console software v 1.4.1 (Thermo Fisher Scientific, Waltham, MA). Genes with fold change ≥1.5 and p < 0.05 were identified as differentially expressed. Student t-test was performed for comparison of the two groups. Gene expression data are available at the Gene Expression Omnibus (GEO) database of the NIH under accession number GSE120855. Data were analyzed through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)19. Real-Time data were compared using Student t-test or Wilcoxon signed rank test between the HCA and control group.
Supplementary information
Acknowledgements
This work was supported in part by the Institutional Development Award (IDeA) (Aghai) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number U54-GM104941 (PI: Binder-Macleod) and NIH COBRE P30GM114736 (PI: Thomas H Shaffer). Md Jobayer Hossain, PhD, Manager Biostatistics Core, Nemours Biomedical Research, Wilmington, provided statistical assistance for the manuscript.
Author Contributions
S.G.B., S.M. contributed to concept and design, sample collection and processing, acquisition and assembly of data, data analysis and interpretation and manuscript writing. M.F., G.F., contributed to sample collection, acquisition and analysis of data and manuscript writing. J.S.C., S.A. contributed to data analysis and interpretation. V.B., I.R., T.S.and J.G. contributed to concept and design, data analysis and interpretation. Z.A. contributed to concept and design, data analysis and interpretation and manuscript writing. All authors have approved the version of the submitted manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42205-x.
References
- 1.Kim CJ, et al. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol. 2015;213:S29–52. doi: 10.1016/j.ajog.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torricelli M, et al. Histologic chorioamnionitis at term: implications for the progress of labor and neonatal wellbeing. J Matern Fetal Neonatal Med. 2013;26:188–192. doi: 10.3109/14767058.2012.722724. [DOI] [PubMed] [Google Scholar]
- 3.Mi Lee S, et al. The frequency and risk factors of funisitis and histologic chorioamnionitis in pregnant women at term who delivered after the spontaneous onset of labor. J Matern Fetal Neonatal Med. 2011;24:37–42. doi: 10.3109/14767058.2010.482622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Getahun D, et al. Effect of chorioamnionitis on early childhood asthma. Arch Pediatr Adolesc Med. 2010;164:187–192. doi: 10.1001/archpediatrics.2009.238. [DOI] [PubMed] [Google Scholar]
- 5.De Luca G, et al. Fetal and early postnatal life roots of asthma. J Matern Fetal Neonatal Med. 2010;23(Suppl 3):80–83. doi: 10.3109/14767058.2010.509931. [DOI] [PubMed] [Google Scholar]
- 6.McDowell KM, et al. Pulmonary Morbidity in Infancy after Exposure to Chorioamnionitis in Late Preterm Infants. Ann Am Thorac Soc. 2016;13:867–876. doi: 10.1513/AnnalsATS.201507-411OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shevell A, Wintermark P, Benini R, Shevell M, Oskoui M. Chorioamnionitis and cerebral palsy: lessons from a patient registry. Eur J Paediatr Neurol. 2014;18:301–307. doi: 10.1016/j.ejpn.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Pappas A, et al. Chorioamnionitis and early childhood outcomes among extremely low-gestational-age neonates. JAMA Pediatr. 2014;168:137–147. doi: 10.1001/jamapediatrics.2013.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shatrov JG, et al. Chorioamnionitis and cerebral palsy: a meta-analysis. Obstet Gynecol. 2010;116:387–392. doi: 10.1097/AOG.0b013e3181e90046. [DOI] [PubMed] [Google Scholar]
- 10.Akinbami, L. J., Simon, A. E. & Rossen, L. M. Changing Trends in Asthma Prevalence Among Children. Pediatrics137, 10.1542/peds.2015-2354 (2016). [DOI] [PMC free article] [PubMed]
- 11.Erez O, et al. Differential expression pattern of genes encoding for anti-microbial peptides in the fetal membranes of patients with spontaneous preterm labor and intact membranes and those with preterm prelabor rupture of the membranes. J Matern Fetal Neonatal Med. 2009;22:1103–1115. doi: 10.3109/14767050902994796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson D, et al. Gene expression profile of endotoxin-stimulated leukocytes of the term new born: control of cytokine gene expression by interleukin-10. PLoS One. 2013;8:e53641. doi: 10.1371/journal.pone.0053641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weitkamp JH, et al. Histological chorioamnionitis shapes the neonatal transcriptomic immune response. Early Hum Dev. 2016;98:1–6. doi: 10.1016/j.earlhumdev.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peoples JD, et al. Neonatal cord blood subsets and cytokine response to bacterial antigens. Am J Perinatol. 2009;26:647–657. doi: 10.1055/s-0029-1220788. [DOI] [PubMed] [Google Scholar]
- 15.Yoon BH, et al. Differences in the fetal interleukin-6 response to microbial invasion of the amniotic cavity between term and preterm gestation. J Matern Fetal Neonatal Med. 2003;13:32–38. doi: 10.1080/jmf.13.1.32.38. [DOI] [PubMed] [Google Scholar]
- 16.Tatad AM, et al. Cytokine expression in response to bacterial antigens in preterm and term infant cord blood monocytes. Neonatology. 2008;94:8–15. doi: 10.1159/000112541. [DOI] [PubMed] [Google Scholar]
- 17.Singh SR, et al. CCL2 release by airway smooth muscle is increased in asthma and promotes fibrocyte migration. Allergy. 2014;69:1189–1197. doi: 10.1111/all.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang A, et al. CCL2/CCR2-dependent recruitment of Th17 cells but not Tc17 cells to the lung in a murine asthma model. Int Arch Allergy Immunol. 2015;166:52–62. doi: 10.1159/000371764. [DOI] [PubMed] [Google Scholar]
- 19.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leviton A, et al. Persistence after birth of systemic inflammation associated with umbilical cord inflammation. J Reprod Immunol. 2011;90:235–243. doi: 10.1016/j.jri.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Kallapur SG, et al. Intra-amniotic IL-1beta induces fetal inflammation in rhesus monkeys and alters the regulatory T cell/IL-17 balance. J Immunol. 2013;191:1102–1109. doi: 10.4049/jimmunol.1300270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakos E, et al. CCR2 Regulates the Immune Response by Modulating the Interconversion and Function of Effector and Regulatory T Cells. J Immunol. 2017;198:4659–4671. doi: 10.4049/jimmunol.1601458. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Brigstock D, Besner GE. Heparin-binding EGF-like growth factor is a potent dilator of terminal mesenteric arterioles. Microvasc Res. 2009;78:78–85. doi: 10.1016/j.mvr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, Radulescu A, Zorko N, Besner GE. Heparin-binding EGF-like growth factor increases intestinal microvascular blood flow in necrotizing enterocolitis. Gastroenterology. 2009;137:221–230. doi: 10.1053/j.gastro.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng J, El-Assal ON, Besner GE. Heparin-binding EGF-like growth factor (HB-EGF) and necrotizing enterocolitis. Semin Pediatr Surg. 2005;14:167–174. doi: 10.1053/j.sempedsurg.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Collier FM, et al. RTKN2 Induces NF-KappaB Dependent Resistance to Intrinsic Apoptosis in HEK Cells and Regulates BCL-2 Genes in Human CD4(+) Lymphocytes. J Cell Death. 2009;2:9–23. doi: 10.4137/JCD.S2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasello M, Manara MC, Scotlandi K. CD99 at the crossroads of physiology and pathology. J Cell Commun Signal. 2018;12:55–68. doi: 10.1007/s12079-017-0445-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinonen MT, et al. GIMAP GTPase family genes: potential modifiers in autoimmune diabetes, asthma, and allergy. J Immunol. 2015;194:5885–5894. doi: 10.4049/jimmunol.1500016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwefel D, et al. Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure. 2013;21:550–559. doi: 10.1016/j.str.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Won HY, Hwang ES. Transcriptional modulation of regulatory T cell development by novel regulators NR4As. Arch Pharm Res. 2016;39:1530–1536. doi: 10.1007/s12272-016-0803-z. [DOI] [PubMed] [Google Scholar]
- 31.Prince LR, et al. NR4A orphan nuclear receptor family members, NR4A2 and NR4A3, regulate neutrophil number and survival. Blood. 2017;130:1014–1025. doi: 10.1182/blood-2017-03-770164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contreras S, Milara J, Morcillo E, Cortijo J. Selective Inhibition of Phosphodiesterases 4A, B, C and D Isoforms in Chronic Respiratory Diseases: Current and Future Evidences. Curr Pharm Des. 2017;23:2073–2083. doi: 10.2174/1381612823666170214105651. [DOI] [PubMed] [Google Scholar]
- 33.Londhe VA, et al. CXCR2/CXCR2 ligand biological axis impairs alveologenesis during dsRNA-induced lung inflammation in mice. Pediatr Res. 2005;58:919–926. doi: 10.1203/01.PDR.0000181377.78061.3E. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Small Molecule Mimetics of alpha-Helical Domain of IRAK2 Attenuate the Proinflammatory Effects of IL-33 in Asthma-like Mouse Models. J Immunol. 2018;200:4036–4043. doi: 10.4049/jimmunol.1700693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wongtrakool C, Wang N, Hyde DM, Roman J, Spindel ER. Prenatal nicotine exposure alters lung function and airway geometry through alpha7 nicotinic receptors. Am J Respir Cell Mol Biol. 2012;46:695–702. doi: 10.1165/rcmb.2011-0028OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bencherif M, Lippiello PM, Lucas R, Marrero MB. Alpha7 nicotinic receptors as novel therapeutic targets for inflammation-based diseases. Cell Mol Life Sci. 2011;68:931–949. doi: 10.1007/s00018-010-0525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogata-Suetsugu S, et al. Amphiregulin suppresses epithelial cell apoptosis in lipopolysaccharide-induced lung injury in mice. Biochem Biophys Res Commun. 2017;484:422–428. doi: 10.1016/j.bbrc.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 38.Ramos-Martinez E, et al. Reduction of respiratory infections in asthma patients supplemented with vitamin D is related to increased serum IL-10 and IFNgamma levels and cathelicidin expression. Cytokine. 2018;108:239–246. doi: 10.1016/j.cyto.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Revenis ME, Kaliner MA. Lactoferrin and lysozyme deficiency in airway secretions: association with the development of bronchopulmonary dysplasia. J Pediatr. 1992;121:262–270. doi: 10.1016/S0022-3476(05)81201-6. [DOI] [PubMed] [Google Scholar]
- 40.Kruzel ML, Bacsi A, Choudhury B, Sur S, Boldogh I. Lactoferrin decreases pollen antigen-induced allergic airway inflammation in a murine model of asthma. Immunology. 2006;119:159–166. doi: 10.1111/j.1365-2567.2006.02417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SB, et al. Lactoferrin administration into the nostril alleviates murine allergic rhinitis and its mechanisms. Scand J Immunol. 2013;78:507–515. doi: 10.1111/sji.12118. [DOI] [PubMed] [Google Scholar]
- 42.Rothman, P. B. The transcriptional regulator NFIL3 controls IgE production. Trans Am Clin Climatol Assoc121, 156–171, discussion 171 (2010). [PMC free article] [PubMed]
- 43.Kashiwada M, et al. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc Natl Acad Sci USA. 2010;107:821–826. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh M. Multilayered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/betacatenin signaling activation (Review) Int J Mol Med. 2018;42:713–725. doi: 10.3892/ijmm.2018.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng H, et al. Monocyte chemotactic protein-induced protein 1 controls allergic airway inflammation by suppressing IL-5-producing TH2 cells through the Notch/Gata3 pathway. J Allergy Clin Immunol. 2018;142:582–594 e510. doi: 10.1016/j.jaci.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan Y, et al. Uncovering potential key genes associated with the pathogenesis of asthma: A microarray analysis of asthma-relevant tissues. Allergol Immunopathol (Madr) 2017;45:152–159. doi: 10.1016/j.aller.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Khalifeh-Soltani, A. et al. The Mfge8-alpha8beta1-PTEN pathway regulates airway smooth muscle contraction in allergic inflammation. FASEB J, fj201800109R, 10.1096/fj.201800109R (2018). [DOI] [PubMed]
- 48.Yang N, Zhang H, Cai X, Shang Y. Epigallocatechin-3-gallate inhibits inflammation and epithelialmesenchymal transition through the PI3K/AKT pathway via upregulation of PTEN in asthma. Int J Mol Med. 2018;41:818–828. doi: 10.3892/ijmm.2017.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Britt RD, Jr., Velten M, Tipple TE, Nelin LD, Rogers LK. Cyclooxygenase-2 in newborn hyperoxic lung injury. Free Radic Biol Med. 2013;61:502–511. doi: 10.1016/j.freeradbiomed.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Li H, Yao Y, Xia D, Zhou J. The overexpression of heparin-binding epidermal growth factor is responsible for Th17-induced airway remodeling in an experimental asthma model. J Immunol. 2010;185:834–841. doi: 10.4049/jimmunol.0901490. [DOI] [PubMed] [Google Scholar]
- 51.Hillman NH, Gisslen T, Polglase GR, Kallapur SG, Jobe AH. Ventilation-induced increases in EGFR ligand mRNA are not altered by intra-amniotic LPS or ureaplasma in preterm lambs. PLoS One. 2014;9:e96087. doi: 10.1371/journal.pone.0096087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allard MJ, Brochu ME, Bergeron JD, Sebire G. Hyperactive behavior in female rats in utero-exposed to group B Streptococcus-induced inflammation. Int J Dev Neurosci. 2018;69:17–22. doi: 10.1016/j.ijdevneu.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Lawrence SM, Ruoss JL, Wynn JL. IL-17 in neonatal health and disease. Am J Reprod Immunol. 2018;79:e12800. doi: 10.1111/aji.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathias B, et al. LPS Stimulation of Cord Blood Reveals a Newborn-Specific Neutrophil Transcriptomic Response and Cytokine Production. Shock. 2017;47:606–614. doi: 10.1097/SHK.0000000000000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Normann E, Lacaze-Masmonteil T, Winkler-Lowen B, Guilbert LJ. Isolation of non-activated monocytes from human umbilical cord blood. Am J Reprod Immunol. 2010;63:66–72. doi: 10.1111/j.1600-0897.2009.00767.x. [DOI] [PubMed] [Google Scholar]
- 56.Redline RW, et al. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2003;6:435–448. doi: 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.