Abstract
Rice beer is traditionally prepared and consumed by various ethnic populations in the Southeast Asian countries. To understand the probable effects of rice beer on human health, present research was aimed to study biochemical parameters, microbial diversity and metabolites of major rice beer varieties of Assam, namely Apong (Poro and Nogin), Xaaj and Joubishi. Alcoholic content of rice beer varieties varied from 9.41 to 19.33% (v/v). Free radical scavenging activity against DPPH· and ABTS+ were 1.94–4.14 and 1.69–3.91 mg of ascorbic acid/ml of rice beer, respectively. In relation to antioxidant activities, phenolic content varied from 2.07 to 5.40 mg gallic acid/ml of rice beer. Next-generation sequencing of 16S rDNA showed that 18 genera of bacteria were present irrespective of rice beer varieties in which lactic acid bacteria were the dominant group (90% abundance). Functional predictions based on the bacterial profiles indicated pathways, such as metabolisms of carbohydrate, amino acid, vitamins and cofactors, and xenobiotic biodegradation, to be active in the rice beer varieties. Out of 18 core bacterial genera, 7 had correlations with the predicted functions. Gas chromatography and mass spectroscopy-based metabolite analysis revealed that the metabolite profiles of the rice beer varieties consisted of 18 saccharides, 18 organic acids, 11 sugar alcohols, 8 amino acids, 1 vitamin and nutraceutical compounds thiocoumarine, carotene, oxazolidine-2-one and acetyl tyrosine. Due to the presence of potent prebiotics, probiotics and nutraceuticals, rice beer may have health benefits which need to be studied further.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1702-z) contains supplementary material, which is available to authorized users.
Keywords: Biochemical properties, Metabolites, Microbial diversity, Nutraceuticals
Introduction
The knowledge of fermentation traces back to ancient civilizations of the world where various forms of foods and beverages had been integral to human diet and lifestyle. Microbial enzymes during fermentation act on the complex substrates and convert it to simpler forms, thereby enhancing its nutritional properties and shelf life (Aidoo et al. 2006; Ray et al. 2016). Fermented foods and drinks are enriched in lactic acid bacteria (LAB) including the genera Leuconostoc, Lactobacillus and Streptococcus. Such bacteria produce an array of compounds such as vitamins, bacteriocins and organic acids which confer health benefits to the consumers (Yadav et al. 2016; Yadav and Shukla 2017). In addition to LAB and probiotics, fermented products are also enriched with prebiotic molecules which can modulate the indigenous intestinal flora of the host. LAB in combination with certain prebiotic compounds are also used in therapeutics against certain metabolic syndromes (Dahiya et al. 2017). Among fermented products, beverages are of utmost importance which can be classified into fruit-based wines, beer and cereal-based alcoholic beverages (Das et al. 2014; Zapata et al. 2019). In cereal-based alcoholic beverages, Saccharomycescerevisiae plays a key role in producing a large number of volatile compounds that enhance flavor and nutritional properties (Dangi et al. 2017). A wide variety of cereal-based alcoholic beverages are prepared and consumed by different ethnic communities as an integral part of their socio-cultural activities (Ghosh et al. 2014, 2016). Assam, a northeastern state of India is home to diverse ethnic communities, mostly of Mongoloid origin who are known for their skill in the preparation of rice beer. The methods of rice beer fermentation are similar among the ethnic groups with some variations in the starter cakes which contain various herbs and rice flour (Das et al. 2012). The rice beer varieties consumed in Assam have similarities with some cereal-based alcoholic beverages of the world, such as Shaosingiju and Laochao of China, Sake of Japan, Chongju and Tkaju of Korea, Brem-Bali and Tape-Ketan of Indonesia and Khaoumak of Thailand (Aidoo et al. 2006). The major rice beer varieties of Assam are Apong (Poro and Nogin), Xaaj and Joubishi prepared by the ethnic communities, namely Mishing, Ahom and Bodo, respectively (Das et al. 2016). It is believed that consumption of rice beer reduces stress and prevents gastrointestinal infections (Ray et al. 2016). Few studies had reported the microbial composition of the starter cake and the biochemical composition of the rice beer (Bhuyan et al. 2014; Das et al. 2014; Ghosh et al. 2015; Sha et al. 2017). However, a detailed study on the microbial diversity, their functionalities and the metabolites of rice beer varieties is not yet available to understand their probable effects on health. Therefore, this research aimed to study the microbial diversity and metabolites of major rice beer varieties of Assam, namely Apong (Poro and Nogin), Xaaj and Joubishi using culture-independent technique coupled with GC–MS analysis.
Materials and methods
Rice beer samples and reagents
A total of 27 rice beer samples of Apong (Poro and Nogin), Xaaj and Joubishi varieties were collected from nine villages in Assam, India, prepared by Mishing,Ahom and Bodo ethnic groups, respectively (Supplementary Table S1). The samples were stored at 4 °C during transit to the laboratory and stored at − 20 °C for further analysis. Triplicates samples from each location were pooled in equal quantities to form nine composite samples representing nine locations for the three ethnic groups. Details about the substrates (rice variety) and the herbs used in the starter cakes are presented in supplementary Table S2. All the reagents and enzymes were procured from Sigma-Aldrich (St. Louis, Missouri, USA) unless otherwise mentioned.
Biochemical analyses of rice beer
Estimation of alcohol
Alcohol contents of the rice beer samples were estimated following potassium dichromate method with some modifications (Caputi et al. 1968). Briefly, a 1000 µl rice beer sample was centrifuged at 8000 rpm for 5 min and a 100 µl of the supernatant was mixed with 125 µl of 1 M potassium dichromate and incubated at 62.5 °C for 15 min. A standard curve with ethanol (Merck Millipore, India) was prepared and the absorbance of both the standards and samples was recorded at 600 nm. Alcohol content was expressed as % (v/v).
Assay of antioxidant activity
Antioxidant activities of the rice beer samples were determined following 2,2-dipheny l-1-picrylhydrazyl (DPPH) method with some modifications (Ghosh et al. 2015). Briefly, 50 µl of rice beer was added to 450 µl of 0.1 mM of DPPH in methanol. After 30 min of incubation at 37 °C, free radical scavenging capacity was evaluated by measuring the decrease of absorbance at 517 nm. Antioxidant activity was also further supplemented by 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) or ABTS assay with some modification (Adak et al. 2013). Briefly, 20 µl of rice beer was added to 1000 µl 7 mM ABTS in 2.45 mM ammonium persulfate solution. After 1 min of incubation at room temperature, free radical scavenging capacity was evaluated by measuring the decrease of absorbance at 734 nm. A standard curve was prepared with ascorbic acid (Sigma-Aldrich, USA) and the antioxidant activity was expressed as mg of ascorbic acid equivalent to 1 ml of rice beer.
Scavenging activity (%) = (1 – Asample/Acontrol) × 100, where Asample was the absorbance in the presence of the sample or reference material and Acontrol was the absorbance of the control containing all the reaction reagents except the sample or reference material.
Estimation of total phenol
Total phenolic contents of the rice beer samples were determined following Folin–Ciocalteu (FC) method as earlier described by Singleton et al. (1999). In brief, 5 µl of rice beer supernatant was added with 150 µl of FC reagent (1:10) and incubated at 37 °C for 30 min. Thereafter, 100 µl of 0.1 M Na2CO3 was mixed with the reaction mixture and heated at 45 °C for 20 min. The reaction mixture was cooled to room temperature and the absorbance was measured at 765 nm. A standard curve of gallic acid (Sigma-Aldrich, USA) was prepared and phenolic content was expressed as mg of gallic acid/ml of rice beer sample.
Study on culture-independent bacterial diversity
Metagenomic DNA extraction
Metagenomic DNA from the rice beer samples were extracted following the method earlier described by Romi et al. (2015) with some modifications. Briefly, 5 mg of 0.1-mm Zirconia beads was added to 1 ml of rice beer and homogenized at 5000 rpm in a vortex (Genie® 2, MoBio Laboratories, Inc., USA) for 2 min. Thereafter, the samples were subjected to enzymatic lysis with 100 U/ml of mutanolysin, 0.1% of 10 mg/ml proteinase K and 20 mg/ml lysozyme for 15 min. Following enzymatic treatment, equal quantity of phenol:chloroform (1:1) was added and centrifuged at 12,000 rpm for 20 min. Following centrifugation, the phase-separated upper layer was collected in a fresh tube. DNA was precipitated by adding 3.5 M sodium acetate, washed with 70% ethanol and finally dissolved in 20 µl TE (Tris–EDTA) buffer.
Next-generation sequencing (NGS) analysis
NGS analysis was carried with the sequencing service provider Macrogen Inc. (Seoul, Republic of Korea). Pure double-stranded DNA was subjected to PCR amplification using 341F–805R primer pairs spanning the V3–V4 region of 16S rDNA gene. The NGS library was prepared using the Nextera XT library preparation kit according to the Illumina MiSeq protocol (Illumina Inc., 2017). Sequencing was carried out in an Illumina MiSeq machine (MiSEq 2500) following 2 × 300 bp paired end chemistry with the multiplexed pooled samples. All the sequences were deposited in the MG-RAST server under study ID mgp 85043 (Supplementary Table S3).
NGS data analysis
The NGS data were analyzed using QIIME2 (v 2018.6) pipeline (Caporaso et al. 2010) using various built-in plugins. DADA2 plugin (Callahan et al. 2016) was used for denoising, filtering, merging and chimera removal from the raw Illumina reads. Sequences having a minimum Phred score of 25 were considered for analysis and hence truncated at 270 bp for proper merging of forward and reverse sequences. For phylogenetic classification, the sequences were aligned with MAFFT (Katoh and Standley 2013) prior to phylogenetic tree construction using FastTree2 (Price et al. 2010). Taxonomic assignment was performed by training a Naïve Bayes classifier specific to V3–V4 region using the most recent SILVA 132 database (Quast et al. 2013). Taxonomic classification was collapsed and assigned at the genus level. For functional prediction, closed reference OTU picking was performed using VSEARCH (Rognes et al. 2016) on 97% similarity clustered OTUs using the Greengenes 13_5 database. The QIIME2 artifacts were converted to BIOM format and subjected to PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) analysis to predict the functional features of the bacterial profiles of the rice beer varieties (Langille et al. 2013). OTUs were normalized by copy number and predicted functional categories were created using the Kyoto encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2010).
Gas chromatography and mass spectrometry (GC–MS) analysis
The rice beer samples were extracted with a mixture of methanol and water (9:1) for 16 h. The mixture was centrifuged at 8000 rpm for 8 min and a 200 µl of the supernatant was further concentrated under vacuum at 45 °C for 60 min. To the concentrated sample, a 50 µl of methoxyamine hydrochloride containing 15 mg/ml pyridine was added and incubated at 70 °C for 45 min. After cooling, 50 µl of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) was added and incubated at 70 °C for 1 h (Das et al. 2014). The MSTFA-derivatized samples were subjected to GC–MS analysis in a Shimadzu GC 2010 Plus-triple Quadrupole (TP-8030) GC–MS/MS system fitted with EB-5MS column (length: 30 m, thickness: 0.25 µm and ID: 0.25 mm). The temperature of both the injector and detector was maintained at 200 °C. The oven temperature was maintained at 100 °C for 2 min and then allowed to rise up to 300 °C at a ramping rate of 10 °C/min and finally for 10 min at 300 °C. The mass spectrometer was operated in the electron ionization (EI) mode at 70 eV with a source temperature of 200 °C and a continuous scan from 45 to 600 m/z. The peaks were identified by matching the National Institute of Standards and Technology (NIST) library, USA. Noisy peaks and columns bleeds were removed prior to data analysis.
Statistical analysis
Statistical analyses were conducted within the R package v 3.5.1. All the biochemical tests were performed in triplicates and one-way ANOVA was performed to determine the differences in biochemical properties among the rice beer varieties. To analyze the differences in microbial diversities among the rice beer varieties, Kruskal–Wallis H test was performed. To understand the interactions among the microbes in the rice beer varieties, Spearman’s rank correlation analysis was carried out using the rcorr package (Harrell and Harrell 2018) and correlation matrix was visualized through corrplot package (Wei et al. 2017). Heatmap was constructed using gplots package (Warnes et al. 2009). All other graphs and illustrations were visualized using the ggplot2 package (Wickham 2016). Network analysis between core bacterial genera and predicted functions was visualized by Cytoscape 3.6.1 software (Shannon et al. 2003) using the Metscape app (Karnovsky et al. 2011). Statistical analysis on the metabolites detected in the GC–MS analysis was performed with MetaboAnalyst V 4.0 (Chong and Xia 2018). The differences in metabolites among the rice beer varieties were calculated based on partial least square discriminant analysis scores. Statistical significance was considered at p < 0.05 for all the analysess.
Results
Biochemical properties of the rice beer varieties
Alcohol contents of the rice beer varieties were within the range of 9.41–19.33% (v/v) (Table 1). The Joubishi sample RB09 had the highest content of alcohol (19.33%) whereas the NoginApong sample RB02 had the least alcohol content (9.41%) (p < 0.05). Free radical scavenging activity against DPPH· indicating antioxidant activities of the rice beer varieties was expressed as mg of ascorbic acid/ml of rice beer. The antioxidant activities varied within a range of 1.94–4.14 mg/ml of ascorbic acid. The Xaaj sample RB05 exhibited highest antioxidant activity (4.14 mg/ml), the Xaaj sample RB06 had least antioxidant activity 1.94 ± 0.37 mg/ml (p < 0.05). Similar trend was observed in case of free radical scavenging activity against ABTS+. The antioxidant activities varied within a range of 1.69–3.91 mg/ml of ascorbic acid. The Xaaj sample RB05 exhibited highest antioxidant activity (3.91 mg/ml) and the Xaaj sample RB06 had least antioxidant activity (1.69 mg/ml) (p < 0.05) (Table 1). Total phenolic content was expressed as mg of gallic acid/ml of rice beer. The phenolic contents were within a range of 2.07–5.40 mg/ml of rice beer. However, there was no significant difference in the phenolic contents of the rice beer samples.
Table 1.
Biochemical properties of rice beer varieties collected from different ethnic groups
| Sample information | Biochemical properties | |||||
|---|---|---|---|---|---|---|
| Community | Sample ID | Variety | Alcohol content (% v/v) | Antioxidant activity (ascorbic acid equivalent mg/ml) | Phenolic (gallic acid equivalent mg/ml) | |
| DPPH· | ABTS+ | |||||
| Mishing | RB01 | Poro Apong | 10.67 ± 1.02e | 3.01 ± 0.18c | 2.04 ± 0.06f | 2.99 ± 1.05a |
| RB02 | Nogin Apong | 9.99 ± 0.40c | 3.47 ± 0.06a | 3.23 ± 0.03c | 2.80 ± 0.21a | |
| RB03 | Nogin Apong | 10.37 ± 0.57d | 3.17 ± 0.08a | 1.80 ± 0.03g | 2.07 ± 0.25a | |
| Ahom | RB04 | Xaaj | 14.82 ± 0.25hf | 3.19 ± 0.14a | 2.85 ± 0.06d | 4.14 ± 0.38a |
| RB05 | Xaaj | 14.04 ± 0.81i | 4.14 ± 0.17a | 3.91 ± 0.04a | 3.93 ± 0.78a | |
| RB06 | Xaaj | 9.41 ± 0.66b | 1.94 ± 0.37b | 1.69 ± 0.34g | 2.64 ± 0.52a | |
| Bodo | RB07 | Joubishi | 12.26 ± 0.74bf | 3.60 ± 0.02ac | 3.48 ± 0.07b | 3.43 ± 0.48a |
| RB08 | Joubishi | 12.94 ± 0.19cg | 3.01 ± 0.31d | 2.58 ± 0.09e | 3.62 ± 0.89a | |
| RB09 | Joubishi | 19.33 ± 0.37a | 3.44 ± 0.09a | 3.90 ± 0.07a | 5.40 ± 1.35a | |
The values are expressed as mean ± SEM. Values within a column with the same letters (a, b, c, d, e, f, g, h, and i) are not significantly different (p < 0.05)
Culture-independent bacterial diversity
To understand the total bacterial diversity of the rice beer varieties, NGS analysis of 16S rDNA amplicons was performed and analyzed with the QIIME2 pipeline. An average of 30,000 high-quality reads per sample was considered for examining the bacterial diversity. Alpha diversity, a measure of species richness and evenness within a sample, was determined by considering the observed OTU, Faith’s phylogenetic diversity (FPD), Shannon’s diversity index and Pielou’s evenness (PE) (Table 2). Observed OTU, FPD, Shannon diversity and PE measure the unique number of features, community richness incorporating phylogenetic relationship between features, community richness and community evenness, respectively. Observed OTU varied across all the samples like FPD and Shannon diversity indices. The number of observed OTUs varied from 67 in the Joubishi sample RB07 to 753 in the NoginApong sample RB02. The NoginApong sample RB02 also had highest FPD and Shannon diversity indices (62.23 and 7.02, respectively) and the Joubishi sample RB07 had lowest (11.86 and 3.16, respectively). PE index ranges from 0 to 1, in which the higher value indicates more evenness in the species of a community. The PE value was highest in the Xaaj sample RB05 (0.31) and lowest in the NoginApong sample RB020 (0.73).
Table 2.
Microbial diversity indices of the rice beer varieties
| Sample ID | Faith PD | Observed OTUS | Pielou's evenness | Shannon index |
|---|---|---|---|---|
| RB01 | 21.27 | 298.00 | 0.61 | 4.99 |
| RB02 | 62.23 | 753.00 | 0.74 | 7.03 |
| RB03 | 29.44 | 339.00 | 0.63 | 5.34 |
| RB04 | 26.53 | 220.00 | 0.55 | 4.30 |
| RB05 | 29.38 | 201.00 | 0.31 | 2.39 |
| RB06 | 35.13 | 425.00 | 0.68 | 5.96 |
| RB07 | 11.90 | 67.00 | 0.52 | 3.16 |
| RB08 | 27.56 | 309.00 | 0.61 | 5.04 |
| RB09 | 44.27 | 175.00 | 0.59 | 4.42 |
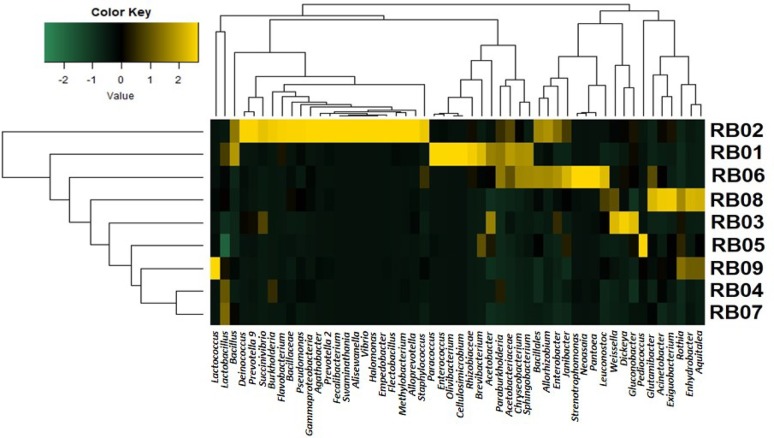
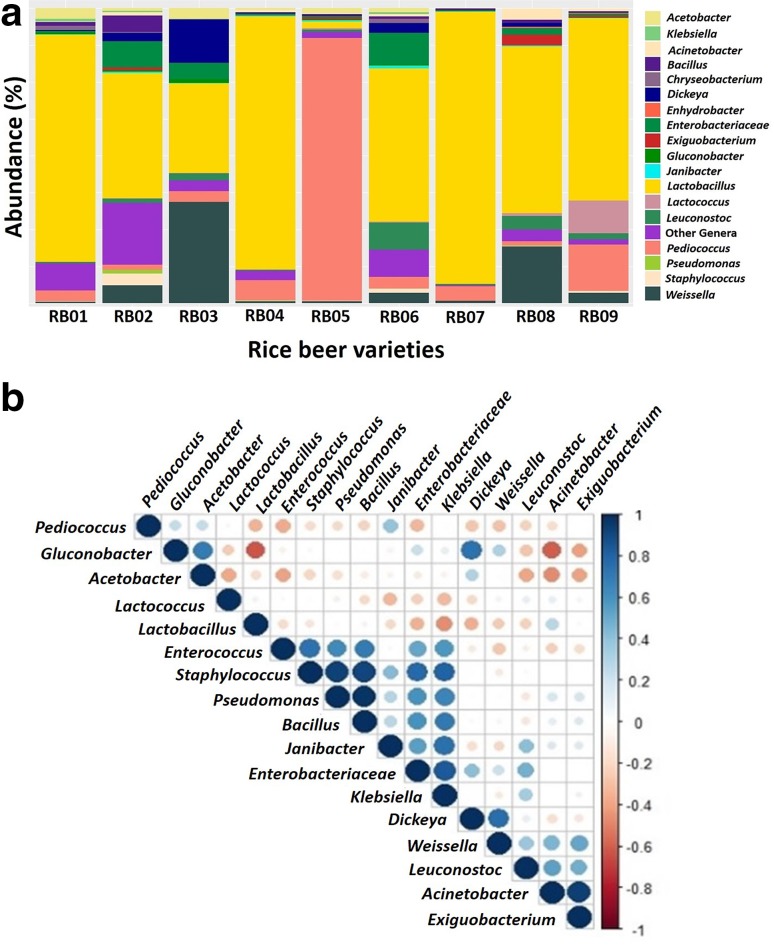
Taxonomic assignment was performed for the obtained features by training a naïve classifier against SILVA 132 database and a total of 257 genera were obtained. The differential abundance of the dominant genera (≥ 1% abundance) spanning all the samples are represented in a heatmap (Fig. 1). The NoginApong sample RB02 had a wider variety of bacteria dominated by LAB. In addition to LAB, a group of acetic acid-producing bacteria, environmental contaminants such as Vibrio and Pseudomonas were also present in the NoginApong samples. The PoroApong sample RB01 and the Xaaj sample RB06 were enriched with the bacterial genera Paracoccus, Enterococcus,Olivibacter and Cellulosimicrobium. The bacterial genera Stenotrophomonas, Neoasia, Pantoea,Dickeya and Rhizobiaceae were found in all the samples. Core bacterial diversity was computed by considering a minimum of 1% of abundance with occurrence in at least 80% of the samples. A total of 18 genera of bacteria were found as core bacteria across the samples in which LAB was the dominant group (Fig. 2a). Lactobacillus (42.36–92.25% abundance) was the predominant genus in all the rice beer samples except the NoginApong sample RB03 and the Xaaj sample RB05, in which Weissella and Pediococcus were the dominant genera (34.36% and 89.09% abundance, respectively). LAB, such as Leuconostoc and Lactococcus were also present in all the samples. Leuconostoc was dominant in the Xaaj sample RB06 (9.21% abundance), whereas Lactococcus was dominant in the Joubishi sample RB09 (11.21% abundance). Apart from LAB, Acetobacter, Acinetobacter, Bacillus, Dickeya, Enterococcus, Enterobacter, Exiguobacterium, Gluconobacter, Janibacteria, Rothia, Klebsiella, Pseudomonas and Staphylococcus were the members of the core bacteria in the rice beer varieties. Correlations among the core bacterial genera were determined based on Spearman’s rank correlation (Fig. 2b). A general negative correlation was observed between Lactobacillus with other LAB genera (r ≤ − 0.36, p < 0.05). Among the LAB, only Weissella and Leuconostoc were positively correlated to each other (r = 0.61, p < 0.05). Interestingly, positive correlations were observed between Acinetobacter and LAB (r ≥ 0.27, p < 0.05), with the exception of Pediococcus (r = − 0.47, p < 0.05).
Fig. 1.
Heatmap showing the differential abundance of bacterial genera in various rice beer samples. Bacterial abundance (≥ 1%) of the microbes were scaled in a range of − 2 to 2 and clustered according to Euclidian distance matrix. Sample codes Poro Apong (RB01), Nogin Apong (RB02 and RB03), Xaaj (RB04, RB05 and RB06) and Joubishi (RB07, RB08 and RB09)
Fig. 2.
a Relative abundance (%) of core bacterial genera in the rice beer samples. Sample codes Poro Apong (RB01), Nogin Apong (RB02 and RB03), Xaaj (RB04, RB05 and RB06) and Joubishi (RB07, RB08 and RB09). b Correlation matrix among the 18 core bacterial genera. The Spearman’s rank correlation coefficients ranged from 1.0 to − 1.0, corresponding to a strongly positive to a strongly negative correlations and has been represented in hierarchal order in the correlation plot
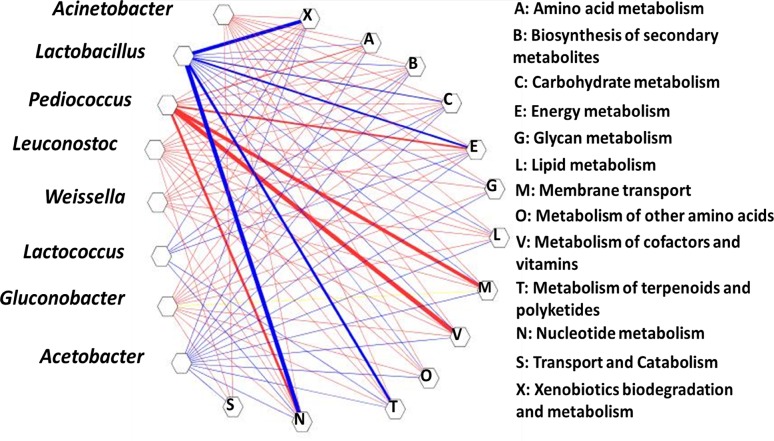
The functional features were predicted using PICRUSt in context of KEGG pathways. A total of 24 pathways were enriched in the rice beer samples depicting active microbial metabolisms. Spearman’s ranked correlations between the core bacteria and the predicted KEGG pathways were analyzed to understand the probable roles of the core bacteria. Lactobacillus, Lactococcus,Acinetobacter, Pediococcus, Leuconostoc, Weissella and Gluconobacter had correlations with 13 predicted pathways (p > 0.05) (Fig. 3). Lactobacillus was positively correlated with all the 13 pathways (r ≥ 0.45, p < 0.05), followed by Acetobacter (r ≥ 0.27, p < 0.05) and Lactococcus (r ≥ 0.086, p < 0.05). However, negative correlations with the predicted pathways were observed with Pediococcus (r ≤ − 0.48, p < 0.05), Leuconostoc (r ≤ − 0.095, p < 0.05) and Weissella (r ≤ − 0.126, p < 0.05).
Fig. 3.
Network analysis showing the linkages of core bacterial genera to predicted functional pathways. Blue and red branches connecting the nodes indicate positive and negative correlations, respectively. The width of the branch depicts the strength of correlation
Metabolite profiling by GC–MS analysis
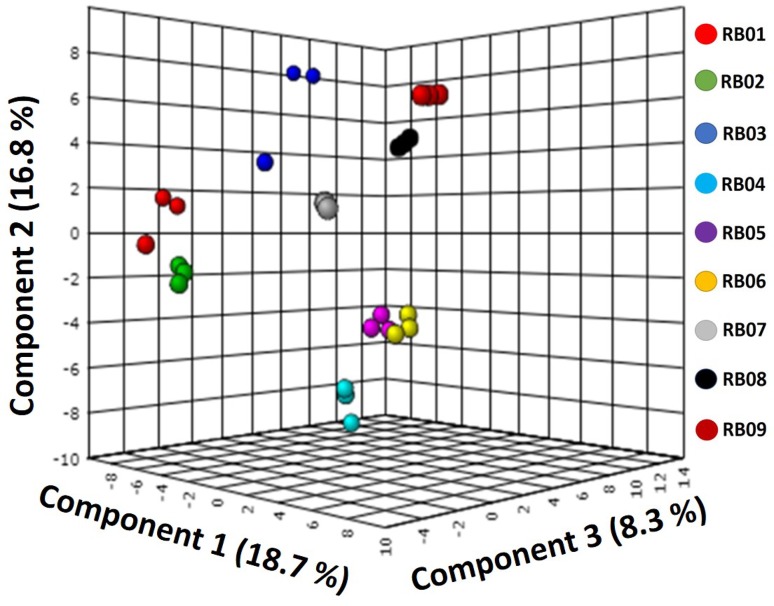
A total of 230 compounds were detected in all the samples. However, only 66 compounds having importance in metabolisms were taken into consideration for analysis. The profiles of these compounds included 18 saccharides, 18 organic acids, 11 sugar alcohols, 8 amino acids, 1 vitamin along with the compounds thiocoumarine, carotene, oxazolidine-2-one (pharmaceuticals), butane, acetyltyrosine, mono-ethylmalonate and mevalonic lactone (Table 3). The peak area (%) of metabolites detected by GC–MS analysis has been provided in the Supplementary file. Multivariate statistical analysis was performed with the data on metabolites using partial least squares discriminant analysis (PLS-DA) test among the rice beer samples (Fig. 4). In the PLS-DA plot, the rice beer samples clustered separately according to their origins. However, the Xaaj samples RB05 and RB06 clustered together depicting similarity in their metabolite profiles.
Table 3.
List of metabolites of the rice beer varieties as detected by GC–MS analysis
| Saccharides | Organic acids | Amino acids | Others |
|---|---|---|---|
| d-Mannopyranoside | Pentanoic acid (valeric acid) | Glycine | Butane |
| d-Glucopyranoside | Propanoic acid | l-Proline | Thiocoumarine |
| dl-Glyceraldehyde | Hexadecanoic acid (palmitic acid) | Serine | l-( + )-Ascorbic acid |
| Glyceryl-glycoside | Pentanedioic acid | Alanine–Thr | N-Acetyltyrosine |
| d-(−)-Ribofuranose | Octadecanoic acid (stearic acid) | Alanine | psi-psi-Carotene |
| d-( + )-Talose | Eicosanoic acid (arachidic acid) | l-Valine | Quinoxaline-2-carboxamide |
| d-Glucose | Tetradecanoic acid (myristic acid) | l-Leucine | Physovenine |
| d-Galactofuranose | Butanoic acid | l-Glutamic acid | Oxazolidin-2-one |
| Lactulose | Heptadecanoic acid | ||
| d-Lactose | Propanoic acid | Sugar alcohols | Sugar alcohols |
| 3-Mannobiose | d-Gluconic acid | Glycerol | Dulcitol |
| 2-Mannobiose | Galactonic acid | Pentitol | l-Threitol |
| d-Fructose | Butyric acid | Xylitol | 4-Hydroxyphenylethanol |
| d-( + )-Trehalose | Acetic acid | Glucitol | l-(−)-Arabitol |
| Maltose | Hydroxyisobutyric acid | l-Fucitol | Adonitol/ribitol |
| d-( + )-Cellobiose | d-Galactopyranosiduronic acid | Myo-inositol | |
| Sedoheptulose | Phthalic acid | ||
| Lyxopyranoside | Lactic acid |
Fig. 4.
Partial least square discriminant analysis (PLS-DA) plot based on the metabolite profiles of the rice beer samples. Sample codes Poro Apong (RB01), Nogin Apong (RB02 and RB03), Xaaj (RB04, RB05 and RB06) and Joubishi (RB07, RB08 and RB09)
Discussion
In Asia and Africa, the cereals rice, corn, wheat, millet, sorghum, etc. constitute more than 80% of the diet. Digestibility and nutritional value of a complex form of cereal depend on its processing as feed. Fermentation of cereals reduces antinutritional factors and provides more macro- and micronutrients (Shewry and Hey 2015; Sokrab et al. 2014). Rice beer is prepared using a starter cake made of rice flour and herbs which is mixed with cooked rice and fermented in a solid state for 7–10 days. After fermentation, the ferment is diluted and consumed as a refreshing drink. In cereal-based fermentation, the saccharolytic activity of microbes convert the complex starch into simple sugars which in turn are converted into organic acids and alcohol. The content of both acids and alcohols in a beverage depends on the microbial consortia in the starter cake and their succession during fermentation (Ghosh et al. 2015). Moreover, starch content of a rice variety used, the process of saccharification and the plant materials used in the starter cake determine the quality of the beverage (Huang et al. 2018).
In this study, the major rice beer varieties of Assam were characterized to understand the composition of microbes and their functions in the rice beer. The alcohol content of the Xaaj samples RB05 and RB06 and Joubishi sample RB09 was found to be higher which could be due to the use of glutinous variety of rice (Khan et al. 2017). The microbial enzymes deconjugate the phenolics from the fibres (Sokrab et al. 2014) and render them soluble in the rice beer. The free polyphenols either from the herbs or rice may have contributed to the antioxidant activities of the rice beer varieties (Deka et al. 2018). The antioxidant activity was highest in the Xaaj sample RB05, while phenolic content equivalent was higher in the Joubishi sample RB09. The results suggested that apart from phenolic compounds, other metabolites such as ascorbic acid and organic acids (such as acetic, butyric and propanoic acids) may also have a role in the antioxidant activity (Chen et al. 2018). Microbes have a very complex role in cereal fermentation and involves mixed cultures of yeasts, bacteria and fungi (Das et al. 2014). A study on Marcha and Thiat, a starter cake used for the preparation of alcoholic beverage in Sikkim and Meghalaya states in India showed that proteobacteria was a dominant phylum followed by Firmicutes and Actinobacteria (Sha et al. 2017). A study showed that succession of moulds and yeasts followed by LAB played an important role in fermentation of rice to produce alcohol and lactic acid (Ghosh et al. 2015). In the present study, metagenomic approach detected a wider diversity of bacteria in the rice beer varieties. Alpha diversity indices were used to determine the diversity within a sample and were expressed as observed OTU, FPD, Shannon and PE indices. The NoginApong sample RB02 had highest microbial diversity and species richness. On the contrary, the Xaaj sample RB06 had low bacterial diversity, probably with conserved functionalities. PE signifies the community structure in terms of equality in number of species. It was observed that PE index for the Xaaj sample RB05 was lowest suggesting dominance of a single genus or a group of highly conserved genera. Culture-independent study showed the presence of 18 core bacteria in which the LAB Lactobacillus, Leuconostoc, Pediococcus, Lactococcus and Weissella had higher abundances in all the rice beer varieties. Previous studies have also reported the dominance of Weissella, Pediococcus and Lactobacillus in the starter culture used for rice beer fermentation (Cai et al. 2018). Therefore, such LAB might have migrated from the starter cakes and played an important role in fermentation of rice beer. The group of LAB can convert hexoses into organic acids by homolactic or heterolactic fermentation. The homolactic fermenters Pediococcus, Lactococcus, Streptococcus and Lactobacillus were present across the samples and can produce lactic acid from hexose sugars. Moreover, heterolactic fermenters which include Leuconostoc and Weissella are known to convert those hexoses simultaneously into lactate, carbon dioxide and ethanol (Steinkraus 1983). Apart from their role in fermentation, a number of studies have reported LAB as probiotics those confer health benefits (Amara and Shibl 2015). In a more recent study, Lactobacillusplantarum isolated from fermented drink Raabadi exhibited a good hypocholesterolemic effect (Yadav et al. 2016). Presence of such bacteria in rice beer might be beneficial to the consumers. Moreover, the presence of Acetobacter and Gluconobacter and their positive correlations with Pediococcus indicated probable conversion of sugar or ethanol to acetic acid (Doran-Peterson et al. 2008). Production of organic acids adds flavor and increases the shelf life by decreasing the pH of rice beer (Vandenbergh 1993). Interestingly, the concordance of functional pathways with the core bacteria showed that Pediococcus had negative correlations with membrane transport and metabolisms of cofactors and vitamins. Besides, the positive correlations of Lactobacillus and Weissella with organic acid-producing bacteria indicated accumulation of organic acids that can prevent the growth of other bacteria by interfering with membrane potential, active transport and metabolism of cofactors (Davidson et al. 2013). The Pediococcus-dominant Xaaj sample RB05 had less dominance of other bacteria which indicated that it acted as an antagonist to other bacteria. Apart from Pediococcus, other LAB Lactobacillus and Lactococcus also had negative correlations with Klebsiella, Pseudomonas, Staphylococcus and Enterobacteria. The presence of Klebsiella, Rothia and Staphylococcus as core bacteria across the samples could be linked to the unhygienic practices during rice beer processing. In addition, Dickeya might have originated from plant parts used in the starter cake preparation (Yamazaki et al. 2011). Gluconobacter, Lactobacillus and Lactococcus are also involved in the production of aromatic compounds which may contribute towards the enhancement of flavor in rice beer (Zou et al. 2018). The genera such as Acinetobacter, Enterococcus, Exiguobacterium, Enterobacter, Gluconobacter,Janibacteria and Pseudomonas are common environmental bacteria. Due to their widespread occurrence in soil and water, these bacteria might have mixed with the starter cakes or rice beer during preparation. A similar functional core microbiota in a Chinese rice wine was reported by Huang et al. (2018).
In the GC–MS analysis, 18 organic acids were detected in all the rice beer samples of which acetic, butyric, propanoic and lactic acids are known to have potent antimicrobial and gut-health-promoting effects (Tanaka et al. 2016). Additionally, the oligosaccharides such as melibiose, cellobiose, mannobiose have been detected in rice beer which can be utilized by gut commensals to produce SCFAs. SCFAs are involved in maintaining the health of gut (Gibson et al. 2017). Lactulose, sedoheptulose and trehalose are used in the treatment of constipation and hepatic encephalopathy (Shukla et al. 2011). Glycerol is the major alcohol in beer that contributes to the sensory property and taste (Langstaff and Lewis 1993). Other sugar alcohols detected in the rice beer samples were pentitol, xylitol, glucitol, fucitol, dulcitol and threitol. Among these, xylitol has been used as dietary supplement and as laxative (Mäkinen 2016). In addition, physovenine, quinoxaline-2-carboxamide, oxazolidin-2-one, acetyl tyrosine and thiocoumarine were detected in the rice beer samples. Physovenine has been reported as acetyl cholinesterase (AChE) inhibitor and is used in the treatment of Alzheimer’s disease (Hostettmann et al. 2006). Quinoxaline-2-carboxamide and oxazolidin-2-one are used in the treatment of hypertensions (Wookey et al. 2004).
Moreover, there was concordance among the GC–MS-derived metabolites and microbiome-derived pathway predictions. The metabolites detected are either end products or intermediates produced in due course of microbial metabolisms as per the predicted pathways. The saccharides, amino acids, vitamins and other secondary metabolites detected in the GC–MS analysis can be linked to the predicted pathways of carbohydrate metabolism, amino acid metabolism and metabolism of vitamins and other cofactors, respectively. However, a robust linking of metabolites to the predicted pathways was not possible as many of the intermediate compounds involved in a pathway were not detected in the GC–MS analysis.
This study for the first time showed the microbial composition and their relation with metabolic pathways of the major rice beer varieties of Assam. The presence of LAB, Acetobacter as well as yeast and moulds plays an important role in the final characteristics of rice beer. The synergistic actions of the microbial consortia converted the starchy materials present in rice into potentially beneficial compounds which included mannobiose, sugar alcohol, organic acids and amino acids. The presence of LAB and nutraceuticals in the rice beer may provide health benefits to the consumers which needs to be studied further.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Institutional level Biotech Hub (DBT, Govt. of India) and Central instrumentation facility of IASST for providing the facilities. This research was funded by Department of Biotechnology (DBT) under the Unit of Excellence Project (BT/550/NE/U-Excel/2014). The authors would like to thank Mr. Anupam Bhattacharya, Research Associate, BIF centre IASST for assistance in computational analysis. The authors are also thankful to volunteers for providing samples.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
References
- Adak A, Maity C, Ghosh K, Halder SK, Mohapatra PKD, Mondal KC. Potentialities of newly isolated Bacillussubtilis and Lactobacillus sp. for curd preparation and a comparative study of its physico-chemical parameters with other marketed curds. Indian J Exp Biol. 2013;51:910–918. [PubMed] [Google Scholar]
- Aidoo KE, Nout MJ, Sarkar PK. Occurrence and function of yeasts in Asian indigenous fermented foods. FEMS Yeast Res. 2006;6:30–39. doi: 10.1111/j.1567-1364.2005.00015.x. [DOI] [PubMed] [Google Scholar]
- Amara AA, Shibl A. Role of probiotics in health improvement, infection control and disease treatment and management. Saudi Pharm J. 2015;23:107–114. doi: 10.1016/j.jsps.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuyan DJ, Barooah MS, Bora SS, Singaravadivel K. Biochemical and nutritional analysis of rice beer of North East India. Indian J Tradit Knowl. 2014;13:142–148. [Google Scholar]
- Cai H, Zhang T, Zhang Q, Luo J, Cai C, Mao J. Microbial diversity and chemical analysis of the starters used in traditional Chinese sweet rice wine. Food Microbiol. 2018;73:319–326. doi: 10.1016/j.fm.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi A, Ueda M, Brown T. Spectrophotometric determination of ethanol in wine. Am J Enol Vitic. 1968;19:160–165. [Google Scholar]
- Chen RH, Chen WX, Chen HM, Zhang GF, Chen WJ. Comparative evaluation of the antioxidant capacities, organic acids, and volatiles of papaya juices fermented by Lactobacillus acidophilus and Lactobacillusplantarum. J Food Qual. 2018 doi: 10.1155/2018/9490435. [DOI] [Google Scholar]
- Chong J, Xia J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics. 2018;27:4313–4314. doi: 10.1093/bioinformatics/bty528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya DK, Puniya M, Shandilya UK, et al. Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: a review. Front Microbiol. 2017;8:563. doi: 10.3389/fmicb.2017.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi AK, Dubey KK, Shukla P. Strategies to improve Saccharomycescerevisiae: technological advancements and evolutionary engineering. Indian J Microbiol. 2017;57:378–386. doi: 10.1007/s12088-017-0679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Deka S, Miyaji T. Methodology of rice beer preparation and various plant materials used in starter culture preparation by some tribal communities of North-East India: a survey. Int Food Res J. 2012;19:101. [Google Scholar]
- Das AJ, Khawas P, Miyaji T, Deka SC. HPLC and GC-MS analyses of organic acids, carbohydrates, amino acids and volatile aromatic compounds in some varieties of rice beer from northeast India. J Inst Brew. 2014;120:244–252. doi: 10.1002/jib.134. [DOI] [Google Scholar]
- Das G, Patra JK, Singdevsachan SK, Gouda S, Shin H-S. Diversity of traditional and fermented foods of the seven sister states of India and their nutritional and nutraceutical potential: a review. Front Life Sci. 2016;9:292–312. doi: 10.1080/21553769.2016.1249032. [DOI] [Google Scholar]
- Davidson PM, Taylor TM, Schmidt SE. Chemical preservatives and natural antimicrobial compounds. In: Doyle MP, Buchanan RL, editors. Food microbiology. Washington, DC: American Society of Microbiology; 2013. pp. 765–801. [Google Scholar]
- Deka AK, Handique P, Deka DC. Antioxidant-activity and physicochemical indices of the rice beer used by the Bodo community in North-East India. J Am Soc Brew Chem. 2018;76:112–116. [Google Scholar]
- Doran-Peterson J, Cook DM, Brandon SK. Microbial conversion of sugars from plant biomass to lactic acid or ethanol. Plant J. 2008;54(4):582–592. doi: 10.1111/j.1365-313X.2008.03480.x. [DOI] [PubMed] [Google Scholar]
- Ghosh K, Maity C, Adak A, et al. Ethnic preparation of haria, a rice-based fermented beverage, in the province of lateritic West Bengal, India. Ethnobot Res Appl. 2014;12:039–049. [Google Scholar]
- Ghosh K, Ray M, Adak A, et al. Microbial, saccharifying and antioxidant properties of an Indian rice based fermented beverage. Food Chem. 2015;168:196–202. doi: 10.1016/j.foodchem.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Rahaman L, Kaipeng DL, Deb D, Nath N, Tribedi P, Sharma BK. Community-wise evaluation of rice beer prepared by some ethnic tribes of Tripura. J Ethn Foods. 2016;3:251–256. doi: 10.1016/j.jef.2016.12.001. [DOI] [Google Scholar]
- Gibson GR, et al. Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- Harrell FE Jr, Harrell MFE Jr (2018) Package ‘Hmisc’ R foundation for statistical computing. http://biostat.mc.vanderbilt.edu/Hmisc
- Hostettmann K, Borloz A, Urbain A, Marston A. Natural product inhibitors of acetylcholinesterase. Curr Org Chem. 2006;10:825–847. doi: 10.2174/138527206776894410. [DOI] [Google Scholar]
- Huang ZR, Guo WL, Zhou WB, et al. Microbial communities and volatile metabolites in different traditional fermentation starters used for Hong Qu glutinous rice wine. Food Res Int. 2018 doi: 10.1016/j.foodres.2018.12.024. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2011;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MR, Bhaskar B, Adak A, Talukdar NC (2017) Rice based beverage with high alcohol content and method therefor (Application No. 201731006470, Publication Date: 17/03/2017)
- Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langstaff SA, Lewis MJ. The mouthfeel of beer—a review. J Inst Brewing. 1993;99:31–37. doi: 10.1002/j.2050-0416.1993.tb01143.x. [DOI] [Google Scholar]
- Mäkinen KK. Gastrointestinal disturbances associated with the consumption of sugar alcohols with special consideration of Xylitol: scientific review and instructions for dentists and other health-care professionals. Int J Dent Res. 2016;2016:1–16. doi: 10.1155/2016/5967907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M, Ghosh K, Singh S, Mondal KC. Folk to functional: an explorative overview of rice-based fermented foods and beverages in India. J Ethn Foods. 2016;3:5–18. doi: 10.1016/j.jef.2016.02.002. [DOI] [Google Scholar]
- Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romi W, Ahmed G, Jeyaram K. Three-phase succession of autochthonous lactic acid bacteria to reach a stable ecosystem within 7 days of natural bamboo shoot fermentation as revealed by different molecular approaches. Mol Ecol. 2015;24:3372–3389. doi: 10.1111/mec.13237. [DOI] [PubMed] [Google Scholar]
- Sha SP, Jani K, Sharma A, et al. Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci Rep. 2017;7:10967. doi: 10.1038/s41598-017-11609-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Hey SJ. The contribution of wheat to human diet and health. Food Energy Secur. 2015;4:178–202. doi: 10.1002/fes3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Shukla A, Mehboob S, et al. Meta-analysis: the effects of gut flora modulation using prebiotics, probiotics and synbiotics on minimal hepatic encephalopathy. Aliment Pharmacol Ther. 2011;33:662–671. doi: 10.1111/j.1365-2036.2010.04574.x. [DOI] [PubMed] [Google Scholar]
- Singleton VL, Orthofer R, Lamuela-Raventós RM. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. In: Packer L, editor. Methods in enzymology. Cambridge: Academic Press; 1999. pp. 152–178. [Google Scholar]
- Sokrab AM, Mohamed Ahmed IA, Babiker EE. Effect of fermentation on antinutrients, and total and extractable minerals of high and low phytate corn genotypes. J Food Sci Technol. 2014;51:2608–2615. doi: 10.1007/s13197-012-0787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KH. Effect of pre and post fermentation treatments on quality of red wine preparation from punjab purple grapes. Antonie van Leeuwenhoek. 1983;49:12. doi: 10.1007/BF00399508. [DOI] [Google Scholar]
- Tanaka S, Yamamoto K, Yamada K, et al. Relationship of enhanced butyrate production by colonic butyrate-producing bacteria to immunomodulatory effects in normal mice fed an insoluble fraction of Brassica rapa L. Appl Environ Microbiol. 2016;82:2693–2699. doi: 10.1128/AEM.03343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbergh PA. Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol Rev. 1993;12:221–237. doi: 10.1111/j.1574-6976.1993.tb00020.x. [DOI] [Google Scholar]
- Warnes GR et al (2009) gplots: Various R programming tools for plotting data R package version 2:1
- Wei T, Simko V, Levy M, et al. Package ‘corrplot’. Statistician. 2017;56:316–324. [Google Scholar]
- Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer
- Wookey A, Turner PJ, Greenhalgh JM, Eastwood M, Clarke J, Sefton C. AZD2563, a novel oxazolidinone: definition of antibacterial spectrum, assessment of bactericidal potential and the impact of miscellaneous factors on activity in vitro. Clin Microbiol Infect. 2004;10:247–254. doi: 10.1111/j.1198-743X.2004.00770.x. [DOI] [PubMed] [Google Scholar]
- Yadav R, Shukla P. Probiotics for human health: current progress and applications. In: Shukla P, editor. Recent advances in applied microbiology. Singapore: Springer; 2017. pp. 133–147. [Google Scholar]
- Yadav R, Puniya AK, Shukla P. Probiotic properties of Lactobacillus plantarum RYPR1 from an indigenous fermented beverage Raabadi. Front Microbiol. 2016;7:1683. doi: 10.3389/fmicb.2016.01683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki A, Li J, Hutchins WC, et al. Commensal effect of pectate lyases secreted from Dickeya dadantii on proliferation of Escherichia coli O157:H7 EDL933 on lettuce leaves. Appl Environ Microbiol. 2011;77:156–162. doi: 10.1128/AEM.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata PJ, Martínez-Esplá A, Gironés-Vilaplana A, et al. Phenolic, volatile, and sensory profiles of beer enriched by macerating quince fruits. LWT. 2019;103:139–146. doi: 10.1016/j.lwt.2019.01.002. [DOI] [Google Scholar]
- Zou W, Zhao C, Luo H. Diversity and function of microbial community in Chinese strong-flavor Baijiu ecosystem: a Review. Front Microbiol. 2018;9:671. doi: 10.3389/fmicb.2018.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.