Abstract
This study assesses the potential for the lipid production by the oleaginous yeast Cystobasidium oligophagum JRC1 using dairy industry waste cheese whey as a substrate. Cheese whey was used either untreated (UCW) or deproteinized (DCW) at different concentrations (25–100%) to serve as the carbon and energy source. Both UCW and DCW supported high biomass and lipid productivities. The biomass productivity of 0.076 ± 0.0004 and 0.124 ± 0.0021 g/L h, lipid productivity of 0.0335 ± 0.0004 and 0.0272 ± 0.0008 g/L h, and the lipid content of 44.12 ± 0.84 and 21.79 ± 1.00% were achieved for 100% DCW and UCW, respectively. The soluble chemical oxygen demand (sCOD) removal rate was 8.049 ± 0.198 and 10.61 ± 0.0165 g/L day (84.91 ± 0.155 and 86.82 ± 0.067% removal) for 100% DCW and UCW, respectively. Fatty acid methyl ester (FAME) composition obtained using GC-FID studies revealed the presence of C16 and C18 fatty acid in the lipid extract and the biodiesel properties were found to be in accordance with ASTM and EN standards. The study presents a method for the valorization of cheese whey waste into a feasible feedstock for biodiesel.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1701-0) contains supplementary material, which is available to authorized users.
Keywords: Cheese whey, Deproteinized cheese whey, Biodiesel, Oleaginous yeast, Rhodotorula, Cystobasidium oligophagum
Introduction
Research on renewable energy resources is gaining momentum considering the limited stocks of fossil reserves (Kumar 2018). Third-generation biofuels have emerged as promising alternatives to the existing transport fuel, but the process and the production cost are still higher when compared to conventional fossil fuels (Leong et al. 2018). Oil production from the oleaginous yeasts is one of the alternatives which circumvent some of the drawbacks associated with third generation biofuels (Shields-Menard et al. 2018). The second-generation biofuels depend on the lignocellulosic wastes (Leiva-Candia et al. 2014; Patel et al. 2017b; Wang et al. 2017). However, these wastes need elaborated pretreatment before they serve as a feedstock. Waste streams originating from food industries offer readily utilizable substrates for lipid accumulation. Cheese whey is one such waste which can be used as a suitable feedstock for lipid accumulation in oleaginous microbes.
Cheese whey is a greenish–yellow liquid residue resulting from the cheese making process of dairy industries. The chemical composition of the cheese whey varies with the type of cheese. It is typically composed of 92–95% water. The solid content is 5–8% of which lactose accounts for 60–80%, proteins account for 10% and trace elements, vitamins, fat and other important elements account for the remaining 10%. It contains 50–102 g/L chemical oxygen demand (COD) and 27–60 g/L biological oxygen demand (BOD) (Carvalho et al. 2013). The global production of the cheese whey in the year 2000 was in the order of 140 million tons (FAO Food and Agricultural Organization of the United Nations 2002). Cheese whey is nutrient rich and has high utilization potential, yet nearly 50% of the waste is discarded untreated. Disposing of untreated cheese whey leads to environmental problems like eutrophication, loss of agricultural land, etc. (Seo et al. 2014; Taskin et al. 2015). In order to address this, a number of processes have been developed by researchers to utilize the lactose and protein part of the cheese whey and convert them into important chemicals and biomolecules like ethanol, pigments, enzymes, lipids, single cell proteins, etc., (Guimarães et al. 2010).
Cheese whey has also been used as a substrate for lipid accumulation in oleaginous microbes. Some oleaginous yeasts were found to grow and accumulate lipid on the pretreated cheese whey substrates (Seo et al. 2014; Taskin et al. 2015; Arous et al. 2016, 2017). Since lactose is a major component of cheese whey, the oleaginous organisms should have lactose utilization pathway. Lactose assimilation ability is not so common amongst yeasts (Domingues et al. 2010). Cheese whey, therefore, needs pretreatment before its utilization as feedstock. The pretreatment includes deproteinization, alkali treatment, hydrodynamic cavitation (HC) treatment, etc., (Seo et al. 2014; Arous et al. 2016). Oleaginous yeast which can accumulate lipids directly on untreated cheese whey is desirable.
In this study, the oleaginous yeast C. oligophagum JRC1 was cultivated on cheese whey. The yeast was found to utilize lactose in the previous study (Vyas and Chhabra 2017). The yeast was grown on different concentrations of both deproteinized and untreated cheese whey and the lipid accumulation potential and COD removal was assessed. The oil composition was determined to check its suitability for biodiesel production.
Materials and methods
Materials
All the medium supplements and general chemicals were purchased from Hi-Media laboratories, Merck, SRL chemicals and Fischer scientific. The FAME standard (for GC analysis) and other chemicals such as methanolic-BF3 complex solution, etc., were purchased from Sigma-Aldrich.
Cheese whey waste characterization
The cheese whey from the cheddar cheese whey plant was collected from the nearby dairy industry and stored at − 20 °C in the sterile container till further use. The effluent was characterized for some basic parameters such as pH, chemical oxygen demand (COD), total reducing sugars and total Kjeldahl nitrogen (TKN).
Yeast strain and media conditions
Oleaginous yeast C. oligophagum JRC1 originally isolated from the site rich in cellulosic waste was used for the study (Vyas and Chhabra 2017). The yeast was cultivated on two different media. In one of the medium, the cheese whey was deproteinized using a heat treatment method. This involved heating at 121 °C for 15 min. The proteins which precipitated were separated using centrifugation followed by filtration using whatman filter paper. The filtrate so obtained was called deproteinized cheese whey (DCW) (Arous et al. 2016). The other medium involved direct utilization of the cheese whey without any pretreatment. This was called untreated cheese whey (UCW). The DCW and UCW both were utilized at 25, 50, 75 and 100% concentration. The dilutions were done with distilled water. The shake flasks experiments were conducted in 250 mL Erlenmeyer flasks containing 100 mL media supplemented with minerals at the concentration of (g/L): KH2PO4, 7; NaH2PO4, 3; MgSO4, 3. Other trace elements were added at the concentration of (g/L): CuSO4·5H2O, 0.0001; MnSO4·5H2O, 0.0001; ZnSO4·7H2O, 0.001; CO (NO3)3·3H2O, 0.0001. C. oligophagum JRC1 was grown on 100 mL YPD medium (yeast extract, 10; Peptone, 20; and glucose, 10 g/L) at 28 °C and 150 rpm and 5 mL of the culture during log phase of growth (approximately 1.74 × 108 cells/mL) was aseptically centrifuged at 8000 rpm for 10 min separating the cells and supernatant. The supernatant was discarded and the cells were re-suspended in sterile saline (0.85% NaCl) and centrifuged again. This step was repeated twice. The washed cells were finally re-suspended in 5 mL sterile saline and introduced into the autoclaved cheese whey media aseptically. All the flasks were incubated at 28 °C and 150 rpm.
Analytical methods
Biomass, residual sugar and soluble COD estimation
Biomass or cell dry weight (CDW) was obtained gravimetrically. Briefly, aliquots of 8 mL were withdrawn every 24 h and centrifuged at 8000 rpm for 10 min. Cell pellets were washed thrice with distilled water (Arous et al. 2016) and dried in a hot air oven (80 °C) until the constant weight was achieved. Total reducing sugar was measured using the DNS method (Miller 1959). Total Kjeldahl Nitrogen (TKN) was measured by standard methods (APHA 2005). Total soluble chemical oxygen demand (sCOD) was determined using Merck spectroquant and standard COD estimation kits as per manufacturer instructions (APHA 2005).
Total lipid extraction
The total lipids were extracted using a modified Bligh and Dyer method (Bligh and Dyer 1959). Briefly, the dry cell pellet was treated with 10 mL of 2 M HCl (for approximately 300 mg cell pellet) at 90 °C for 1 h in a water bath. The acid–hydrolyzed cells were then disrupted by sonication (20 kHz, 10 min) followed by stirring in 20 ml of 2:1 chloroform and methanol for 3 h. The mixture was centrifuged at 8000 rpm for 15 min to separate the aqueous and organic phases. The lower organic phase was aspirated, filtered and dried to obtain the lipid extract in the pre-weighed tube. The total lipid content (%) and lipid output (g/L) were determined gravimetrically.
FTIR, TLC and proton (1H) NMR analysis
The extracted lipids were dissolved in chloroform and were analyzed by Fourier Transform Infrared spectroscopy (FTIR) (Bruker-70 V, USA) in the range of 4000–400/cm. Triolein (glycerol trioleate) was used as a standard for the tri-acylglycerol (TAG). Thin layer chromatography (TLC) was performed as previously described Fei et al. (2009). Briefly, the triolein standard and the samples were resolved using a solvent comprising of hexane: diethyl ether: acetic acid (85:15:1, v/v/v). The TLC plate was treated with methanolic-MnCl2 (0.63 g MnCl2·4H2O in 60 mL water + 60 mL methanol + 4 mL of concentrated H2SO4) followed by drying at 110 °C for 20 min in a hot air oven for the quantification of the TAG. For proton (1H) nuclear magnetic resonance studies (NMR), 5 mg of lipid extract was dissolved in 0.7 mL deuterated chloroform (CDCL3) and spectra were recorded according to the methods described previously (Sarpal et al. 2014).
Transesterification of lipids and GC analysis
The lipid extract was subjected for transesterification using boron trifluoride (BF3) methanol as described previously (Morrison and Smith 1964). The FAME samples were analyzed using GC-FID chromatography (Scion-436 GC) (Column specification: Capillary, Rt-2560, 100 m × 0.25 mm × 0.2 µm thickness; Restek Corp, USA). The samples were resolved using helium as a carrier gas. The column temperature was programmed as follows: Ramping temperature from 100 to 240 °C at the rate of 3 °C/min and held for 15 min. The injector and detector temperature was set at 225 °C and 250 °C, respectively. Fatty acid identification and relative composition were carried out according to the standard method (AOAC Method Ce 1-62 2005) using commercially available FAME mixture (Supelco 37 component FAME mix). The mixture comprised of 37 FAME with known concentrations.
Estimation of biodiesel properties
The different biodiesel properties were calculated as described previously (Patel et al. 2017a; Deeba et al. 2018). The biodiesel characteristics were estimated using the data obtained by GC-FID. The formulas used were as shown below:
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
| 8 |
| 9 |
| 10 |
where M = molecular mass, F = percentage of each fatty acid, N = double bond number, PUFA = weight percentage of polyunsaturated fatty acids, MUFA = weight percentage of monounsaturated fatty acid.
Statistical analysis
All experiments were performed in triplicate. The results reported are the average values with the standard deviation ranging from 5 to 10%.
Results and discussion
Cheese whey waste characterization
The physical and chemical composition of the cheese whey from dairy industries may vary with the variation in the process, final product and operation methods. In general, the sCOD of the cheese whey falls in the range of 50–102 g/L. The primary constituents are lactose and fats (Carvalho et al. 2013). Both DCW and UCW were rich in sCOD with the values in the order of 66.35 ± 1.515 and 85.53 ± 1.323 g/L for DCW and UCW, respectively. The total reducing sugar obtained was 39.63 ± 0.67 and 56.47 ± 1.75 g/L for DCW and UCW, respectively. The pH values were 6.6 and 6.8 for DCW and UCW, respectively. This is consistent with the pH values observed with other types of cheese whey substrates (Carvalho et al. 2013). TKN concentrations were 0.711 ± 0.024 and 1.54 ± 0.098 g/L for DCW and UCW, respectively, which was within the general range of 0.01–1.7 g/L for cheese whey (Prazeres et al. 2012). The COD/N ratio was 93 and 55 for DCW and UCW, respectively.
Lipid production in deproteinized cheese whey in a shake flask experiment
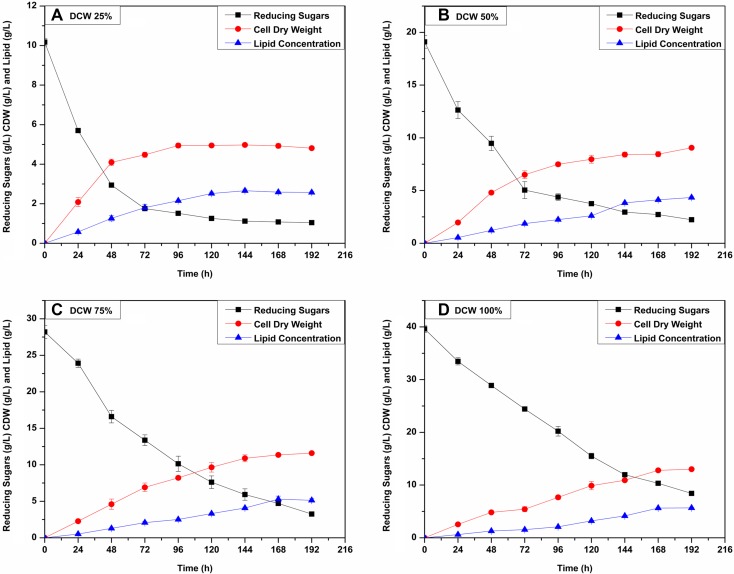
Very few yeasts possess lactose permease and β-galactosidase required for lactose metabolism (de la Fuente and Sols 1962; Domingues et al. 2010). Due to this reason, the growth of oleaginous yeast on cheese whey medium is limited. The yeast C. oligophagum JRC1 accumulated lipids in the medium containing lactose as the only carbon source (Vyas and Chhabra 2017). This trait helped us extrapolate its utility for the valorization of industrial waste such as cheese whey which is primarily composed of lactose. Cheese whey was deproteinized to increase C/N ratio and untreated cheese whey was also used to assess the growth and lipid accumulation by the yeast. The reducing sugar, CDW and lipid production for all four concentrations (25, 50, 75 and 100% DCW) were recorded for every 24 h as shown in Fig. 1. Different concentrations were tried to know the optimum concentration for yeast growth and lipid production. The approximate initial C/N ratio was calculated using the COD/N ratio as previously reported (Saenge et al. 2011). The initial TKN concentrations were 0.183 ± 0.002, 0.420 ± 0.004, 0.549 ± 0.014 and 0.711 ± 0.024 g/L (Initial C/N ratio: 99, 98, 98, 93) for 25, 50, 75 and 100% DCW media, respectively. As shown in Fig. 1, the initial reducing sugar concentrations were 10.17 ± 0.19, 19.10 ± 0.59, 28.20 ± 0.91, and 39.63 ± 0.67 g/L for 25, 50, 75 and 100% DCW media, respectively. The levels of reducing sugars dropped faster during the first few hours and stabilized at 96 h. This was followed by the lipid accumulation phase. Sugar consumption rate increased with the increase in the concentration of cheese whey from 25 to 100%. The CDW achieved also followed a similar trend with the highest concentration achieved at 100% DCW. The values were 4.92 ± 0.11, 8.45 ± 0.24, 11.35 ± 0.21, 12.79 ± 0.078 g/L, respectively. The lipid production were 2.58 ± 0.037, 4.11 ± 0.050, 5.29 ± 0.41 and 5.64 ± 0.07 g/L (52.48 ± 1.53, 48.74 ± 1.13, 46.63 ± 3.74 and 44.12 ± 0.84% of CDW) after 168 h for 25, 50, 75 and 100% DCW media, respectively. Cheese whey contains minimum levels of certain vitamins (Carvalho et al. 2013) which might have been useful for yeast growth. The C/N ratio is an important parameter for lipid accumulation. The nitrogen-limiting condition (higher C/N ratio) triggers the tri-acylglycerol (TAG) accumulation in the oleaginous yeasts (Sitepu et al. 2013) by channeling the excessive carbon into lipid bodies in the form of TAG (Ageitos et al. 2011). However, the biochemical response is not solely influenced by C/N ratio but also influenced by factors such as the type of carbon sources, nitrogen sources, growth factors and culture conditions (pH and temperature) (Papanikolaou and Aggelis 2011). The lipid accumulation in oleaginous yeast is induced at a C/N ratio > 20 (Papanikolaou and Aggelis 2011). In both UCW and DCW the C/N was > 20. However, the difference between C/N of 100% UCW and DCW was close to 38 and this led to a significant increase in oil accumulation in DCW. The lipid content increased from 21.79 ± 1.00 to 44.12 ± 0.84% of CDW. Similar results have been documented in earlier reports (Annamalai et al. 2018). Also, when comparing different concentrations of DCW and UCW, the difference in the C/N ratio was not significant as it was just diluted in buffer. Therefore, the differences in the cellular yield and lipid productivity can be attributed to the concentration of carbon source and other nutrients available for yeast growth. Since the concentration in the 100% cheese whey was the highest, it supported the highest lipid productivity in both DCW as well as UCW. All the results obtained in the present study were compared with the literature as shown in Table 1. The lipid productivity increased from 0.0153 ± 0.0002 for 25% DCW to 0.0335 ± 0.0004 g/L h for 100% DCW. Yeasts such as Debaryomyces etchellsii, Wickerhamomyces anomalus, Yarrowia lipolytica and Cryptococcus curvatus, etc., have been reported to accumulate lipids on DCW medium (Seo et al. 2014; Taskin et al. 2015; Arous et al. 2016, 2017). However, D. etchellsii could not assimilate lactose and utilized the DCW medium only as a nitrogen source and accumulated only 0.4 ± 0.05 g/L of lipid (15.9 ± 0.93% of CDW) (Arous et al. 2016). Also, W. anomalus accumulated 0.65 ± 0.01 g/L (24 ± 0.24% of CDW) after 96 h of incubation (Arous et al. 2017), while C. oligophagum JRC1 in the present study accumulated much higher lipid content with a value of 2.09 ± 0.02 g/L (27.30 ± 1.52% of CDW) at 96 h of incubation in 100% DCW medium. The growth of C. curvatus was supported after the pretreatment of cheese whey which involved alkaline treatment combined with hydrodynamic cavitations (HC) (Seo et al. 2014). In another study, C. curvatus NRRL Y-1511 yeast supported higher growth on ricotta cheese whey obtained after acid, salt and deproteinization pretreatment (Carota et al. 2017). Oleaginous yeast Y. lipolytica B9 strains grown in the non-sterile DCW medium and under optimized conditions achieved a lipid content of 2.73 ± 0.13 g/L (39% of CDW). External supplementation of lactose and ammonium sulfate increased the lipid content to 4.29 g/L (58% of CDW) (Taskin et al. 2015). In the present study, the yeast could accumulate lipids higher than the previous reports and up to 5.64 ± 0.075 g/L (44.12 ± 0.84% of CDW) lipid after 168 h of the incubation. The lipid productivities realized in this study are higher than most of the yeasts except for C. curvatus (Table 1). Longer incubation period leads to slightly low lipid productivity. However, the productivity can be enhanced by optimizing the bioprocess for various parameters such as feeding strategy, C/N etc. The lipid production was achieved without any external addition of C or N sources which accentuates that the given yeast is the better candidate for cheese whey utilization.
Fig. 1.
Lipid production with time on Deproteinized cheese whey (DCW), showing 1A (25% DCW), 1B (50% DCW), 1C (75% DCW) and 1D (100% DCW) as the time course of lipid production
Table 1.
Comparison of lipid production by oleaginous yeasts cultivated on different cheese whey sources
| Organism | Types of the media | Culture mode | Incubation period (h) |
Cell dry weight X (g/L) |
Lipid production Y (g/L) |
Lipid content (%) | Lipid productivity g/L h |
References |
|---|---|---|---|---|---|---|---|---|
|
Debaryomyces etchellsii |
Deproteinized cheese whey | Shake flask | 96 | 2.8 ± 0.11 | 0.4 ± 0.05 | 15.9 ± 0.93 | 0.0041 | (Arous et al. 2016) |
|
Wickerhamomyces anomalus |
Deproteinized Cheese whey | Shake flask | 96 | 2.61 ± 0.03 | 0.65 ± 0.01 | 24.00 ± 0.24 | 0.0067 | (Arous et al. 2017) |
|
Yarrowia Lipolytica B9 |
Deproteinized cheese whey | Shake flask; non-sterile | 120 | 7.0 ± 0.18 | 2.73 ± 0.13 | 39 | 0.022 | (Taskin et al. 2015) |
| Cryptococcus curvatus | Pretreated cheese whey (HC treatment) | Shake flask | 24 | 7.2 | 4.68 | 65 | 0.195 | (Seo et al. 2014) |
| Cryptococcus curvatus NRRL Y-1511 | Ricotta cheese whey | 3L stirred tank bioreactor | 72 | 10.77 ± 0.21 | 6.83 ± 0.14 | 63.41 | 0.094 | (Carota et al. 2017) |
|
Yarrowia lipolytica NCIM 3589 |
Cheese whey | Shake flask | 72 | 2.6 | 0.33 | 13 | 0.004 | (Katre et al. 2012) |
|
Cystobasidium oligophagum JRC1 |
Deproteinized cheese whey DCW 25% DCW 50% DCW 75% DCW 100% |
Shake flask | 168 |
4.92 ± 0.11 8.45 ± 0.24 11.35 ± 0.21 12.79 ± 0.078 |
2.58 ± 0.037 4.11 ± 0.050 5.29 ± 0.41 5.64 ± 0.07 |
52.48 ± 1.53 48.74 ± 1.13 46.63 ± 3.74 44.12 ± 0.84 |
0.0153 ± 0.0002 0.0245 ± 0.0002 0.0315 ± 0.0002 0.0335 ± 0.0004 |
(This study) |
|
Cystobasidium oligophagum JRC1 |
Untreated cheese whey UCW 25% UCW 50% UCW 75% UCW 100% |
Shake flask | 168 |
6.38 ± 0.10 12.55 ± 0.40 17.87 ± 0.90 20.98 ± 0.36 |
2.75 ± 0.029 3.59 ± 0.12 4.20 ± 0.057 4.57 ± 0.13 |
43.13 ± 0.27 28.63 ± 0.35 23.53 ± 1.05 21.79 ± 1.00 |
0.0163 ± 0.0001 0.0214 ± 0.0007 0.025 ± 0.0003 0.0272 ± 0.0008 |
(This study) |
Lipid production in untreated cheese whey in shake flask experiments
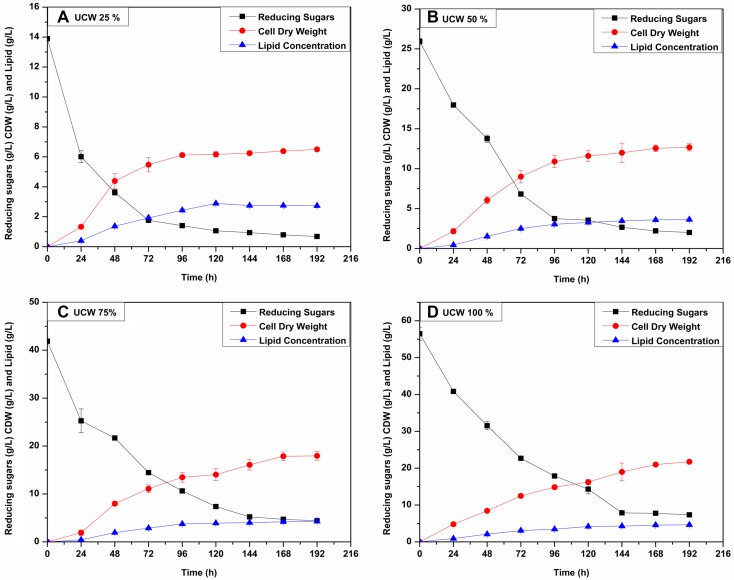
In another set of experiments, the cheese whey was utilized without any pretreatment. In this medium also no external carbon or nitrogen sources were added. The C/N ratio was lower in this case as expected. Initial C/N ratio was in the order of 59, 58, 56 and 55 for 25, 50, 75 and 100% UCW media, respectively. The reducing sugar, CDW and lipid accumulations were monitored every 24 h for all four concentrations and the profiles can be seen in Fig. 2. The cellular growth was higher in UCW as compared to DCW. Also, the CDW increased from 25 to 100% UCW. The initial reducing sugar concentrations were 13.88 ± 0.12, 25.94 ± 0.34, 41.88 ± 0.22 and 56.47 ± 1.75 g/L for 25, 50, 75 and 100% UCW media, respectively. The dry weight achieved was 6.38 ± 0.10, 12.55 ± 0.40, 17.87 ± 0.90 and 20.98 ± 0.36 g/L and lipid content was 2.75 ± 0.029, 3.59 ± 0.12, 4.20 ± 0.057, and 4.57 ± 0.13 (43.13 ± 0.27, 28.63 ± 0.35, 23.53 ± 1.05 and 21.79 ± 1.00% of CDW) at 168 h interval for 25, 50, 75 and 100% UCW, respectively. The lipid productivity observed was 0.0163 ± 0.0001, 0.0214 ± 0.0007, 0.025 ± 0.0003 and 0.0272 ± 0.0008 g/L h for 25, 50, 75 and 100% UCW, respectively. The lipid production in the present study was higher than Y. lipolytica NCIM 3589 (Katre et al. 2012). The lipid content was lower in UCW than DCW because of the low C/N ratio in UCW. The ability of C. oligophagum JRC1 to grow on untreated cheese whey and without any nutrient addition indicated its utility for cheese whey valorization in a sustainable way. Also, to date, no other oleaginous yeast has been explicitly reported to grow and accumulate lipids on untreated cheese whey. The cultivation process can be further optimized for various biochemical parameters to further increase lipid productivity.
Fig. 2.
Lipid production with time on Untreated Cheese Whey (UCW), showing 2A (25% UCW), 2B (50% UCW), 2C (75% UCW) and 2D (100% UCW) as the time course of lipid production
Soluble COD removal and lipid generation
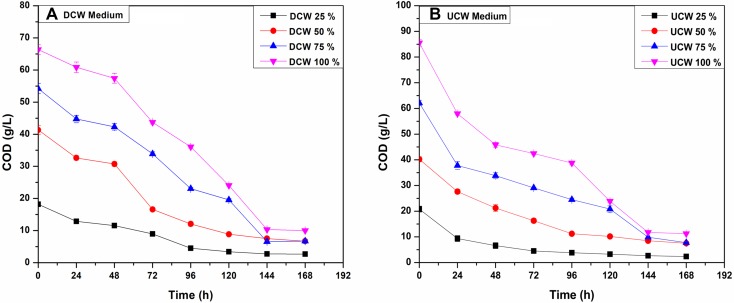
Cheese whey is characterized by high COD which is due to the presence of oxidizable organic matters present in it (Carvalho et al. 2013). The growth of yeast on cheese whey converts the soluble COD to volatile suspended solids (VSS) thereby treating it and rendering it suitable for discharge. The sCOD was monitored every 24 h and the profile is shown in Fig. 3. The sCOD decreased from 66.35 ± 1.515 g/L to 10.00 ± 0.132 g/L after 7 days in 100% DCW at the rate of 8.049 ± 0.198 g/L day. Similarly, sCOD reduced down from 85.53 ± 1.323 to 11.26 ± 0.175 g/L after 7 days of incubation in 100% UCW at the rate of 10.61 ± 0.165 g/L day. The total sCOD removal was observed at 84.91 ± 0.155 and 86.82 ± 0.067% for 100% DCW and UCW, respectively. The initial sCOD removal rate was higher for UCW media than that of DCW media. The remainder of the sCOD might be due to the presence of inert molecules which resist degradation by yeasts. The sCOD removal rate and total removal achieved was higher for this yeast as compared to other yeasts reported in the literature. Different values for the COD removal are compared with the literature in Table 2. D. etchellsii showed 58.3% of COD removal at the rate of 14.05 g/L day in an expired soft drink medium mixed with cheese whey (1:1) (Arous et al. 2016). In another study, 94.22% COD removal at the rate of 0.106 g/L day was achieved for paper and pulp industry wastewater by Rhodosporidium kratochvilovae HIMPA1 (Patel et al. 2017b). In yet another study, 68% COD removal at the rate of 6.025 g/L day was achieved for acetone–butanol–ethanol fermentation wastewater using Trichosporon cutaneum CH002 (Xiong et al. 2015). Similarly, T. cutaneum ACCC20271 treated cellulosic ethanol fermentation wastewater and removed 55.05% COD at the rate of 13.054 g/L day (Wang et al. 2017). In the present study, the initial sCOD was considerably higher and the total sCOD removal was also higher than most of the studies involving yeasts.
Fig. 3.
Change in sCOD with time for DCW and UCW, showing soluble COD values for the time course of all 3A (DCW) and 3B (UCW)
Table 2.
Comparison of COD removal from different types of wastewaters using oleaginous yeasts
| Organism name | Type of the wastewater | Cell dry weight X (g/L) | Lipid content (%) |
Initial COD (g/L) |
Final COD (g/L) |
COD removal (%) | References |
|---|---|---|---|---|---|---|---|
| Debaryomyces etchellsii |
50% Expired soft drink + 50% Cheese whey 62.4% expired soft drinks + 37.6% olive mill wastewater |
7.90 4.6 |
14.90 28.90 |
96.36 113.26 |
40.13 66.48 |
58.35 41.30 |
(Arous et al. 2016) |
| Rhodosporidium kratochvilovae HIMPA1 | Paper and pulp Industry wastewater | 13.87 | 61.71 | 0.675 | 0.039 | 94.22 | (Patel et al. 2017b) |
|
Trichosporon cutaneum CH002 |
Acetone-Butanol-ethanol (ABE) Fermentation wastewater | 4.9 | 14.7 | 18.050 | 6.00 | 68 | (Xiong et al. 2015) |
|
Trichosporon cutaneum ACCC20271 |
Cellulosic ethanol fermentation wastewater | 16.20 | 13.33 | 118.58 | 53.31 | 55.05 | (Wang et al. 2017) |
|
Cystobasidium oligophagum JRC1 |
Deproteinized cheese whey (100%) Untreated cheese whey (100%) |
12.79 ± 0.078 20.98 ± 0.36 |
44.12 ± 0.84 21.79 ± 1.00 |
66.35 ± 1.515 85.53 ± 1.323 |
10.00 ± 0.132 11.26 ± 0.175 |
84.91 ± 0.155 86.82 ± 0.067 |
(This study) (This study) |
FTIR, TLC and 1H NMR analysis
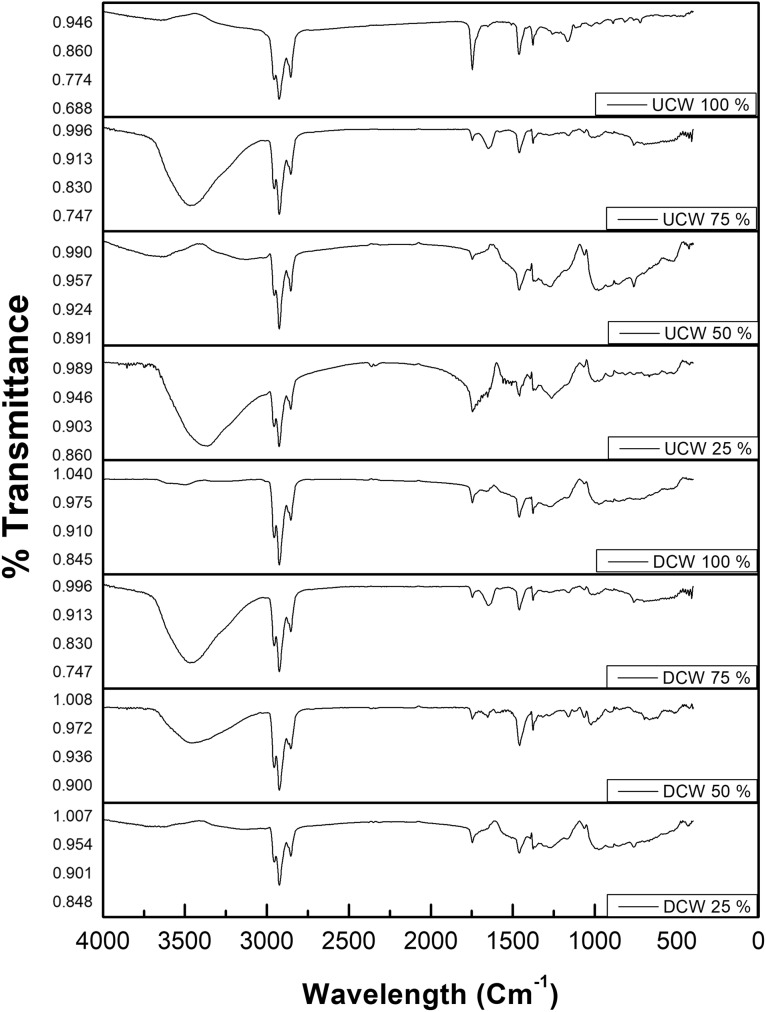
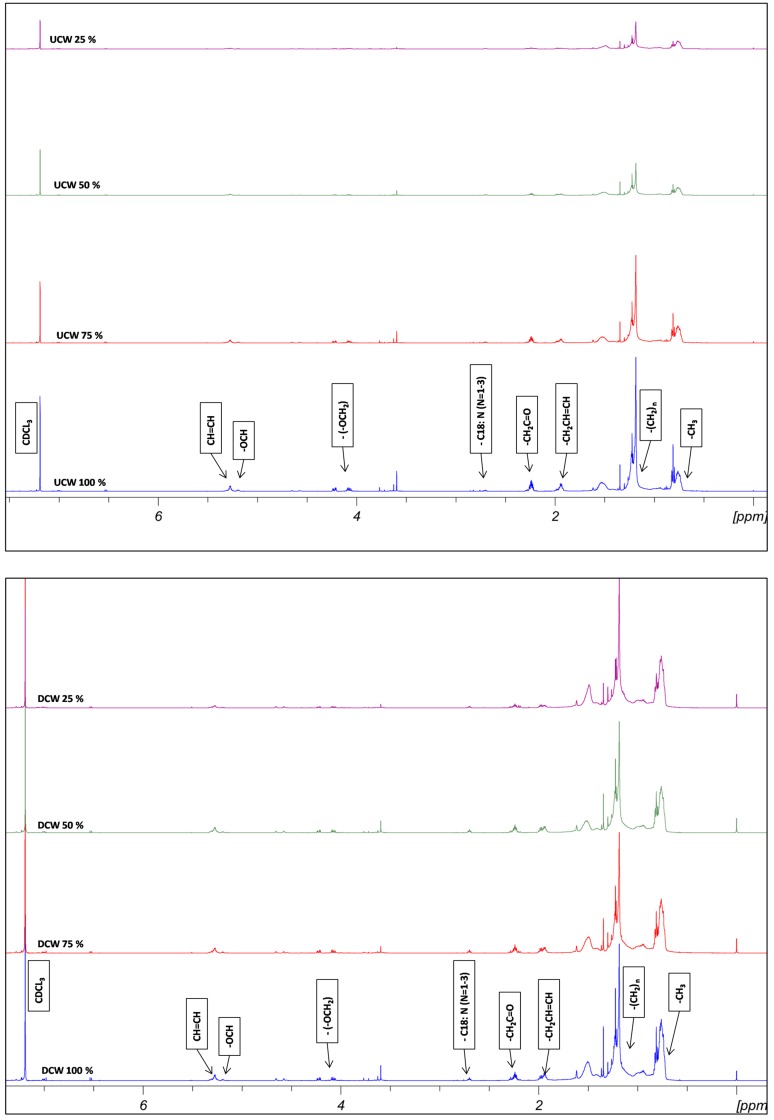
The FTIR analysis was done for all the lipid extracts. The peak assignment was done according to previous studies (Tariq et al. 2011; Vyas and Chhabra 2017). As shown in Fig. 4, the broader peak at 3400/cm indicates the stretching vibration of –O–H group due to the presence of water molecule. The lipid extracts from DCW and UCW showed three characteristic peaks in the range of 2800–3000/cm. These peaks appear due to the presence of symmetric vibrations of –CH3, –CH2 and –CH. The peaks in the range of 1450–1370, 1300–1100 and peaks nearby 720/cm show the asymmetric vibrations for –CH3, –CH2 and –CH, respectively. A sharp peak obtained at nearly 1750/cm in all samples confirms the presence of the carbonyl (–C=O) functional group. TLC was performed on a silica gel and lipid extracts from all the different experiments were analyzed. Triolein standard was used as a standard for the TLC analysis. After incubation at higher temperature different spots were identified on the TLC plates (Online Resource 1). The concentration of the TAG was the highest followed by DAG and MAG.1H NMR spectra of the lipid extract were recorded and the characteristic peaks for triglycerides were assigned according to the previous studies (Tariq et al. 2011; Sarpal et al. 2014). As shown in Fig. 5, all the samples showed peaks at 4.0–4.35 ppm indicating the presence of (–OCH2) functional group. Other peak values such as 2.30, 5.20, 5.05–5.65, 2, 1.26 and 0.85 ppm were observed which indicated the presence of –CH2C=O, –OCH, –CH=CH, –CH2CH=CH, –(CH2)n and –CH3 groups, respectively. The sharp peaks in between 3 and 4 ppm are due to glycol/phospholipids. The triplet at 2.3 ppm represents free fatty acids. The peaks at 2.7–2.8 ppm show the presence of C18: N (N = 1–3), which were comparable with the GC results.
Fig. 4.
FTIR spectra recorded for the lipid extracts obtained from yeast cultivated in DCW and UCW
Fig. 5.
Proton NMR spectra recorded for the lipid extracts obtained from yeast cultivated in DCW and UCW
Lipid profile and GC analysis of the samples
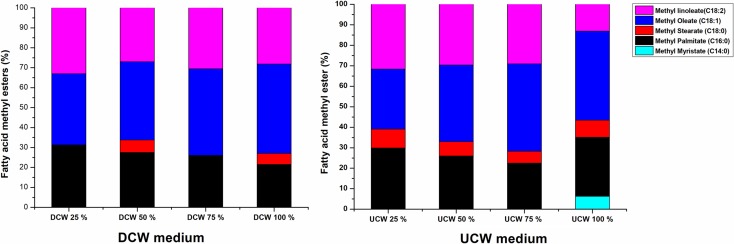
The FAME profile of C. oligophagum JRC1 cultivated on different concentrations of DCW and UCW measured at 168 h of the fermentation are shown in Fig. 6. Of the total FAME, 93.7–99.9% were a mixture of C16 and C18 saturated fatty acids (SFA) as well as unsaturated fatty acids. The FAME profiles for DCW and UCW were having slight differences in the concentrations of SFA. The FAME profiles for both DCW as well as UCW were comparable to the FAME composition obtained on glucose medium as previously reported (Vyas and Chhabra 2017). The lipid samples contained 68–72 and 56–71% of unsaturated fatty acid methyl ester for DCW and UCW, respectively. The linoleic acid methyl esters (C18:2) were in the range of 28–32 and 13–31, while oleic acid methyl esters (C18: 1) were in the range of 35–44 and 29–43% for the DCW and UCW, respectively. SFA such as palmitic acid methyl ester (C16:0) was in the range of 21–31 and 22–29, while stearic acid methyl ester (C18:0) was in the range of 5–6 and 5–9% for DCW and UCW media, respectively. The values were comparable with other oleaginous yeasts. W. anomalus grown on DCW medium showed similar composition containing palmitic acid (C16:0) and stearic acid (C18:0) methyl ester, while oleic acid (C18:1) methyl esters and linoleic acid methyl ester (C18:2) were lower than in the present studies (Arous et al. 2017). The higher amounts of oleic acid (C18:1) methyl esters indicated better biodiesel properties (Sitepu et al. 2014; Arous et al. 2017). When compared with other oleaginous yeast cultivated on cheese whey, the lipid profiles were nearly the same (Arous et al. 2016). The differences in the lipid composition are inevitable considering the heterogeneity of substrates used and the yeast species employed (Sitepu et al. 2014). In general, longer incubation period supports a better fatty acid profile for biodiesel generation (Sitepu et al. 2013). Yet, the overall FAME composition was of high quality and comparable with different plant and vegetable oils (Ramos et al. 2009).
Fig. 6.
FAME compositions obtained by GC-FID, showing comparison of FAME compositions for the lipid extracts from 6A (DCW) and 6B (UCW)
Biodiesel potential of FAME
Based on the data available from the GC experiments, some predictions on biodiesel properties can be made using the formulas shown in the appendix A section (Patel et al. 2017a; Deeba et al. 2018). The calculated values were compared with ASTM D6751 (US biodiesel specification) and EN14214 (European biodiesel specification) standards as shown in Table 3. These properties are used to assess the ability of feedstock to be used as automotive fuel and the values must be within the prescribed ranges. The higher cetane number (CN), longer oxidative stability and low cold filter plug point are the criteria for good engine performance (Deeba et al. 2018). The CN is a measurement of fuel’s autoignition quality (Knothe 2014). As shown in Table 3, the CN was in the range of the standard values of 49–60. The CN value above 50 reduces the formation of white smoke (Balat and Balat 2010). Oxidative stability, which determines the shelf-life of the fuel (Patel et al. 2017a), was higher for UCW lipids (11.62 h) as compared to DCW media (6 h). However, both of the values fall in the required range. Kinematic viscosity (KV) determines the ability of the fuel to flow. The values were in the standard range of 1.9–6.0 mm2/s. Iodine value (IV) is a measurement of unsaturation with all the samples exhibiting moderate unsaturation. Cold filter plug point (CFPP) determines the freezing point of the fuel and as per the standard ASTM or EN, the CFPP values were low and better than that of vegetable oils and jatropha oils (Ramos et al. 2009).
Table 3.
Biodiesel properties predicted for DCW and UCW from the GC data
| Quality parameters | DCW 25% |
DCW 50% |
DCW 75% | DCW 100% |
UCW 25% |
UCW 50% |
UCW 75% |
UCW 100% | ASTM standard | EU standard |
|---|---|---|---|---|---|---|---|---|---|---|
| Saponification value (SV) (mg KOH) | 205.02 | 203.33 | 203.95 | 202.90 | 204.57 | 203.81 | 203.08 | 203.63 | ND | ND |
| Iodine value (IV) (g I2/100 g) | 91.84 | 84.12 | 94.29 | 91.29 | 83.56 | 87.26 | 90.95 | 50.93 | ND | 120 (max.) |
| Cetane number (CN) (min) | 49.51 | 51.69 | 49.02 | 49.92 | 51.68 | 51.25 | 49.97 | 60.11 | 47 | 51 |
| Degree of Unsaturation (DU) (%wt) | 101.67 | 93.16 | 104.42 | 101.12 | 92.48 | 96.61 | 100.72 | 69.60 | ND | ND |
| Long chain saturation factor (LCSF) (%wt) | 3.132 | 5.887 | 2.608 | 4.912 | 7.621 | 6.095 | 5.145 | 7.052 | ND | < 5/< − 20 |
| Cold filter plug points (CFPP) (°C) | − 5.77 | 3.63 | − 7.566 | 0.314 | 9.56 | 4.34 | 1.1 | 7.619 | ND | ND |
| High Heating value (HHV) (MJ/Kg) | 39.640 | 39.833 | 39.655 | 39.740 | 39.796 | 39.764 | 39.739 | 40.317 | ND | ND |
| Kinematic viscosity (KV) (mm2/s) | 3.7073 | 4.4827 | 3.7073 | 4.4827 | 4.4827 | 4.4827 | 4.4827 | 4.9726 | 1.9–6.0 | 3.5–5.0 |
| Density (D) (g/cm3) | 0.887 | 0.886 | 0.887 | 0.886 | 0.886 | 0.886 | 0.886 | 0.885 | ND | 0.86–0.90 |
| Oxidative stability (OS) (h) | 6.16 | 6.96 | 6.45 | 6.78 | 6.32 | 6.57 | 6.653 | 11.62 | ND | ≥ 6 |
ND no limit designated by standards for biodiesel ASTM D6751 and EU14214
Conclusions
The oleaginous yeast C. oligophagum JRC1 was successfully grown on cheese whey. The deproteinized cheese whey supported higher lipid content while untreated cheese whey supported high cellular biomass. Higher sCOD removal was obtained in both of the conditions. The lipid productivities were considerably higher. The lipid profile was assessed and the composition was suitable for biodiesel feedstock. The predicted biodiesel properties were also in accordance with ASTM and EN standards. The study shows the potential of the given yeast for the utilization of cheese whey. Further, process optimization can increase cellular yield and lipid yield.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Author MC thanks IIT Jodhpur for providing various research facilities. Author SV thanks the Ministry of Human Resource Development (MHRD) India, for his senior research fellowship (SRF). Authors thank Dr. Mahesh Balwant Khot (SERB, National Postdoctoral Fellow, Biofuels division, CSIR-Indian Institute of Petroleum, Dehradun, India) for GC-FID analysis.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Contributor Information
Sachin Vyas, Email: vyas.1@iitj.ac.in.
Meenu Chhabra, Phone: +91 291 2801205, Email: meenuchhabra@iitj.ac.in.
References
- Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG. Oily yeasts as oleaginous cell factories. Appl Microbiol Biotechnol. 2011;90:1219–1227. doi: 10.1007/s00253-011-3200-z. [DOI] [PubMed] [Google Scholar]
- Annamalai N, Sivakumar N, Oleskowicz-Popiel P. Enhanced production of microbial lipids from waste office paper by the oleaginous yeast Cryptococcus curvatus. Fuel. 2018;217:420–426. doi: 10.1016/j.fuel.2017.12.108. [DOI] [Google Scholar]
- AOAC Method Ce 1-62 (2005) Fatty acid composition by gas chromatography. In: Firestone D (ed) Official methods of analysis of AOAC, 18th edn, AOAC International, Gaithersburg, Maryland.
- APHA . Standard methods for the examination of water and wastewater. 21. Washington: American Public Health Association; 2005. [Google Scholar]
- Arous F, Frikha F, Triantaphyllidou IE, et al. Potential utilization of agro-industrial wastewaters for lipid production by the oleaginous yeast Debaryomyces etchellsii. J Clean Prod. 2016;133:899–909. doi: 10.1016/j.jclepro.2016.06.040. [DOI] [Google Scholar]
- Arous F, Atitallah IB, Nasri M, Mechichi T. A sustainable use of low-cost raw substrates for biodiesel production by the oleaginous yeast Wickerhamomyces anomalus. 3 Biotech. 2017;7:268. doi: 10.1007/s13205-017-0903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balat M, Balat H. Progress in biodiesel processing. Appl Energy. 2010;87:1815–1835. doi: 10.1016/j.apenergy.2010.01.012. [DOI] [Google Scholar]
- Bligh EG, Dyer WJA. Rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- Carota E, Crognale S, D’Annibale A, et al. A sustainable use of Ricotta Cheese Whey for microbial biodiesel production. Sci Total Environ. 2017;584–585:554–560. doi: 10.1016/j.scitotenv.2017.01.068. [DOI] [PubMed] [Google Scholar]
- Carvalho F, Prazeres AR, Rivas J. Cheese whey wastewater: characterization and treatment. Sci Total Environ. 2013;445–446:385–396. doi: 10.1016/j.scitotenv.2012.12.038. [DOI] [PubMed] [Google Scholar]
- de la Fuente G, Sols A. Transport of sugars in yeasts II. Mechanisms of utilization of disaccharides and related glycosides. Biochim Biophys Acta. 1962;56:49–62. doi: 10.1016/0006-3002(62)90526-7. [DOI] [PubMed] [Google Scholar]
- Deeba F, Patel A, Arora N, et al. Amaranth seeds (Amaranthus palmeri L.) as novel feedstock for biodiesel production by oleaginous yeast. Environ Sci Pollut Res. 2018;25:353–362. doi: 10.1007/s11356-017-0444-x. [DOI] [PubMed] [Google Scholar]
- Domingues L, Guimarães PMR, Oliveira C. Metabolic engineering of Saccharomyces cerevisiae for lactose/whey fermentation. Bioeng Bugs. 2010;1:164–171. doi: 10.4161/bbug.1.3.10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Wang H, Fu X, et al. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem J. 2009;424:61–67. doi: 10.1042/BJ20090785. [DOI] [PubMed] [Google Scholar]
- Food and Agricultural Organization of the United Nations . FAO Yearbook Production, 2002. Rome: Food and Agricultural Organization of the United Nations; 2002. [Google Scholar]
- Guimarães PMR, Teixeira JA, Domingues L. Fermentation of lactose to bio-ethanol by yeasts as part of integrated solutions for the valorisation of cheese whey. Biotechnol Adv. 2010;28:375–384. doi: 10.1016/j.biotechadv.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Katre G, Joshi C, Khot M, et al. Evaluation of single cell oil (SCO) from a tropical marine yeast Yarrowia lipolytica NCIM 3589 as a potential feedstock for biodiesel. AMB Express. 2012;2:36. doi: 10.1186/2191-0855-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knothe G. A comprehensive evaluation of the cetane numbers of fatty acid methyl esters. Fuel. 2014;119:6–13. doi: 10.1016/j.fuel.2013.11.020. [DOI] [Google Scholar]
- Kumar A. Global warming, climate change and greenhouse gas mitigation. In: Kumar A, Ogita S, Yau Y-Y, editors. Biofuels: greenhouse gas mitigation and global warming: next generation biofuels and role of biotechnology. New Delhi: Springer; 2018. pp. 1–16. [Google Scholar]
- Leiva-Candia DE, Pinzi S, Redel-Macias MD, et al. The potential for agro-industrial waste utilization using oleaginous yeast for the production of biodiesel. Fuel. 2014;123:33–42. doi: 10.1016/j.fuel.2014.01.054. [DOI] [Google Scholar]
- Leong WH, Lim JW, Lam MK, et al. Third generation biofuels: a nutritional perspective in enhancing microbial lipid production. Renew Sustain Energy Rev. 2018;91:950–961. doi: 10.1016/j.rser.2018.04.066. [DOI] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Morrison WR, Smith LM. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride–methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part II: technology and potential applications. Eur J Lipid Sci Technol. 2011;113:1052–1073. doi: 10.1002/ejlt.201100015. [DOI] [Google Scholar]
- Patel A, Arora N, Mehtani J, et al. Assessment of fuel properties on the basis of fatty acid profiles of oleaginous yeast for potential biodiesel production. Renew Sustain Energy Rev. 2017;77:604–616. doi: 10.1016/j.rser.2017.04.016. [DOI] [Google Scholar]
- Patel A, Arora N, Pruthi V, Pruthi PA. Biological treatment of pulp and paper industry effluent by oleaginous yeast integrated with production of biodiesel as sustainable transportation fuel. J Clean Prod. 2017;142:2858–2864. doi: 10.1016/j.jclepro.2016.10.184. [DOI] [Google Scholar]
- Prazeres AR, Carvalho F, Rivas J. Cheese whey management: a review. J Environ Manag. 2012;110:48–68. doi: 10.1016/j.jenvman.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Ramos MJ, Fernández CM, Casas A, et al. Influence of fatty acid composition of raw materials on biodiesel properties. Bioresour Technol. 2009;100:261–268. doi: 10.1016/j.biortech.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Saenge C, Cheirsilp B, Suksaroge TT, Bourtoom T. Efficient concomitant production of lipids and carotenoids by oleaginous red yeast Rhodotorula glutinis cultured in palm oil mill effluent and application of lipids for biodiesel production. Biotechnol Bioprocess Eng. 2011;16:23–33. doi: 10.1007/s12257-010-0083-2. [DOI] [Google Scholar]
- Sarpal AS, Silva PRM, Martins JL, et al. Biodiesel potential of oleaginous yeast biomass by NMR spectroscopic techniques. Energy Fuels. 2014;28:3766–3777. doi: 10.1021/ef402516x. [DOI] [Google Scholar]
- Seo YH, Lee I, Jeon SH, Han JI. Efficient conversion from cheese whey to lipid using Cryptococcus curvatus. Biochem Eng J. 2014;90:149–153. doi: 10.1016/j.bej.2014.05.018. [DOI] [Google Scholar]
- Shields-Menard SA, Amirsadeghi M, French WT, Boopathy R. A review on microbial lipids as a potential biofuel. Bioresour Technol. 2018;259:451–460. doi: 10.1016/j.biortech.2018.03.080. [DOI] [PubMed] [Google Scholar]
- Sitepu IR, Sestric R, Ignatia L, et al. Manipulation of culture conditions alters lipid content and fatty acid profiles of a wide variety of known and new oleaginous yeast species. Bioresour Technol. 2013;144:360–369. doi: 10.1016/j.biortech.2013.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitepu IR, Garay LA, Sestric R, et al. Oleaginous yeasts for biodiesel: current and future trends in biology and production. Biotechnol Adv. 2014;32:1336–1360. doi: 10.1016/j.biotechadv.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Tariq M, Ali S, Ahmad F, et al. Identification, FT-IR, NMR (1H and 13C) and GC/MS studies of fatty acid methyl esters in biodiesel from rocket seed oil. Fuel Process Technol. 2011;92:336–341. doi: 10.1016/J.FUPROC.2010.09.025. [DOI] [Google Scholar]
- Taskin M, Saghafian A, Aydogan MN, Arslan NP. Microbial lipid production by cold-adapted oleaginous yeast Yarrowia lipolytica B9 in non-sterile whey medium. Biofuels Bioprod Biorefining. 2015;9:595–605. doi: 10.1002/bbb.1560. [DOI] [Google Scholar]
- Vyas S, Chhabra M. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: a cellulase and lipase producing oleaginous yeast. Bioresour Technol. 2017;223:250–258. doi: 10.1016/j.biortech.2016.10.039. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Zhang H, Bao J. Converting chemical oxygen demand (COD) of cellulosic ethanol fermentation wastewater into microbial lipid by oleaginous yeast Trichosporon cutaneum. Appl Biochem Biotechnol. 2017;182:1121–1130. doi: 10.1007/s12010-016-2386-z. [DOI] [PubMed] [Google Scholar]
- Xiong L, Huang C, Li XM, et al. Acetone–butanol–ethanol (ABE) fermentation wastewater treatment by oleaginous yeast Trichosporon cutaneum. Appl Biochem Biotechnol. 2015;176:563–571. doi: 10.1007/s12010-015-1595-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.