Abstract
The treatment of bacterial infections is becoming increasingly ineffective due to rapid mutation which leads to antibiotic resistant and resistant bacteria become more prevalent. As a result the existing antibiotics are gradually obsolete and again new drugs are needed to be designed for the same threat. However, the prediction of evolutionary processes/antibiotic resistance is uncertain. Still, the understanding of mode of evolution of resistance in bacteria is a determining step in the preclinical development of new antibiotics, because drug developers assess the risk of resistance arising against a drug during preclinical development. Multidrug efflux pump systems play an important role for making multidrug resistance to a range of clinically important antibiotics in gram-negative bacteria like Pseudomonas aeruginosa, which lower the intracellular drug concentration by exporting incoming antibiotics across the membranes. We tried to show that the wild type susceptible bacteria P. aeruginosa modified its genetic makeup at mutational hotspots under stress. This strain may either become multidrug resistant or remain susceptible depending on position of amino acid changes in regulatory proteins of efflux pump. Multidrug resistant strain made significant changes at the amino acid positions, 103rd (G → A) and 126th (E → V) through the mutation on the nucleotide position of 308th (G → C); both 377th (A → T) and 378th (G → T), respectively in mexR, a repressor of mexAB-oprM efflux pump. This mutant protein showed low affinity with their operator. But the alteration at 103th position (G → A) in mexR may provide almost similar structural and functional stability as wild type. It was found that mutation was seemed to be well regulated within the limit and position specific under stress which might be back to its original form by supplying counter stress unless addition or deletion takes place.
Electronic supplementary material
The online version of this article (10.1007/s12088-019-00781-6) contains supplementary material, which is available to authorized users.
Keywords: Antibiotics, Antibiotic resistant, Adaptation, Stress, Mutational hotspots, Nucleotide
Antibiotic resistance is a growing global public health issue. The evolution of antibiotic resistant genes is dominated by the relative rates of two processes: horizontal gene transfer (i.e. genetic material is acquired from a distinct bacterial strain) and spontaneous mutation. But antibiotic resistance can evolve through the sequential accumulation of mutations [1]. We categorised emerging problem of resistance and susceptibility of P. aeruginosa into five case studies from the published paper of Yen and Papin in Plos Biology, where they grown separately the wild-type susceptible strain in increasing concentrations, log2 (minimum inhibitory concentration) (μg/ml) of three different antibiotics [piperacillin (PIP), tobramycin (TOB), and ciprofloxacin (CIP)] for 20 days followed by 20 more days to each of the other drugs or incubation to lysogeny broth (LB) media without a drug. It has been suggested that particular order of drugs is useful in antibiotic therapy based on past adaptation in bacteria to a single antibiotic that impact on the evolution of antibiotic resistant genes during subsequent adaptation to a different antibiotic [2, 3].
A set of various regulatory protein, mexR, mexS, nfxB and mexZ of efflux pump, mexAB-orpM, mexEF-orpN, mexCD-oprJ and mexXY-oprM, respectively of P. aeruginosa were retrieved from NCBI for this study. The peptides were analysed through blast of mexR, mexS, nfxB and mexZ gene sequence of strain PA14 of P. aeruginosa with its various multidrug resistant strains and showed base substitution at a specific position. Amino acid sequences of mexR in wild- susceptible strain, multidrug resistant strain (having both 103G → 103A and 126E → 126 V substitution of mexR of the strain PA14) and two mutant strain (one having 103G → 103A and another 126E → 126 V substitution of mexR) of PA14 were used as target sequences which showed 98.64%, 100% and 99.32%, respectively identity with template for the modelling. The total energy in an additive force field was calculated by using software Swiss-PdbViewer 4.1.0. E values for wild and mutated protein of mexR were − 17042.246 (wild), − 17151.564 (mutated, 103G → 103A substitution), − 16681.887 (mutated, 126E → 126 V substitution) and − 16832.822 (mutated, 103G → 103A and 126E → 126 V substitution). B-form 3DNA of known nucleotide sequence (5′-AAATGTGGTTGATCCAGTCAACTATTTTGCTTATTTTAGTTGACCTTATCAACCTTGTTT-3′) was made by using w3DNA software (http://w3dna.rutgers.edu/protein/option) and DNA-interaction sites were predicted from a list adjustment residue, DISPLAR (http://pipe.scs.fsu.edu/displar/) for active site prediction on interacting protein with DNA [4, 5]. Prepared/modelled proteins and DNA structures were further analysed by DNA–Protein docking (http://hdock.phys.hust.edu.cn/) to see the interaction between repressor protein mexR (active site) and its operator at 5′-GTTGA-3′ inverted repeat during stress [6, 7]. We used wild and all the reported mutant protein of mexR of the strain PA14 for docking with known operator sequence.
On the basis of available published data and in silico study we analysed the expression profile of P. aeruginosa in response to 3 clinically relevant drugs-PIP, TOB, and CIP at different concentrations with times and tried to show how reverse/back mutation, SOS and ROS responses made fully susceptible again or increased resistance to first antibiotic depending on series of antibiotic treatment, where P. aeruginosa developed resistance from susceptibility in first antibiotic for 20 days incubation followed by subsequent 20 days adaptation in a new antibiotic. Some of the important molecular mechanisms of gene action in bacteria which lead to evolution under antibiotic stress are discussed herein.
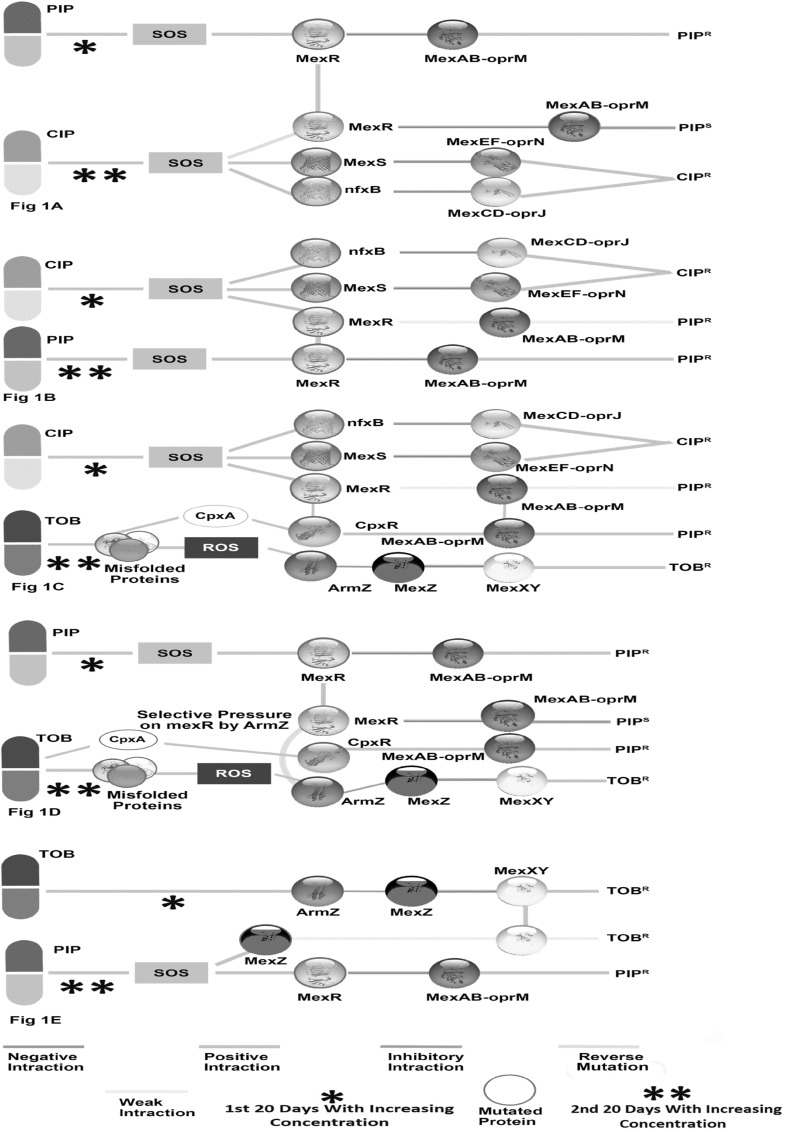
Case one, 20 days adaptation in PIP followed by subsequent 20 days adaptation in CIP led to significantly less minimum inhibition concentration for PIP (MICPIP) than that of 20 days PIP (The MICPIP of Day 40 PIP CIP) by causing mutation in the genes- mexA, mexS, nfxB, transcription regulator, two component sensor (histidine sensor kinase/phosphatase CpxA and its cognate response regulator CpxR) etc., and no mutation in mexR gene [2].
PIP adaptation which caused mutation in mexR by SOS response led to resistant in PIP. But subsequent adaptation to CIP, mexR got its original function by reverse mutation repressing mexAB-oprM which made PIP susceptible lineage. There was also mutation in mexS (repressor of mexEF-oprN efflux pump) and nfxB (repressor of mexCD-oprJ efflux pump) which made the bacteria resistant in CIP. The SOS response which was induced in the treatments with these two antibiotics made to use partially overlapping transcription profiles (Fig. 1a) [8]. Mutation in CpxA caused by Day 40 PIP CIP adaptation lineage, did not have significant effect on the activation of CpxR. It is a regulator of the cell envelope stress response which happens to activate mexAB-oprM pump in presence or absence of mexR [9, 10].
Fig. 1.
Schematic representation of regulation of mexAB-oprM, mexCD-oprJ, mexEF-oprN, mexXY-oprM efflux pump. amexR which is mutated in adaptation to PIP expressing mex-AB-oprM is turned back to normal repressing mexAB-oprM efflux pump by reverse mutation during subsequent adaptation to CIP; b normal expression of mexCD-oprM, mexEF-oprN efflux pump is turned on by mutation in nfxB and mexS respectively and also weak expression of mexAB-oprM by mutation in mexR in CIP adaptation which is became partially resensitive by selection on specific changed base position in subsequent PIP adaptation. In subsequent adaptation also tunes to express mexAB-oprM by mutation in mexR; c CIP adaptation in first treatment effect in the same way as earlier Fg1B, but mexAB-oprM is activated by CpxR which is got kinase activity by CpxA in responses of misfolded proteins in subsequent adaptation to TOB. mexXY-oprM is also activated by an antirepressor, ArmZ of repressor mexZ; d PIP adaptation activates mexAB-oprM by mutation in mexR. During subsequent TOB adaptation, mexAB–oprmM continues to do so by CpxR which is activated by mutation in phosphatase region of CpxA, because no mutation in any efflux pump regulatory genes is thought to be operated by selection of AmrZ on mexR; e mexXY-oprM efflux pump which is activated in TOB adaptation by activation of AmrZ is also partially opened by mutation in mexZ in absence of ArmZ in subsequent adaptation to PIP
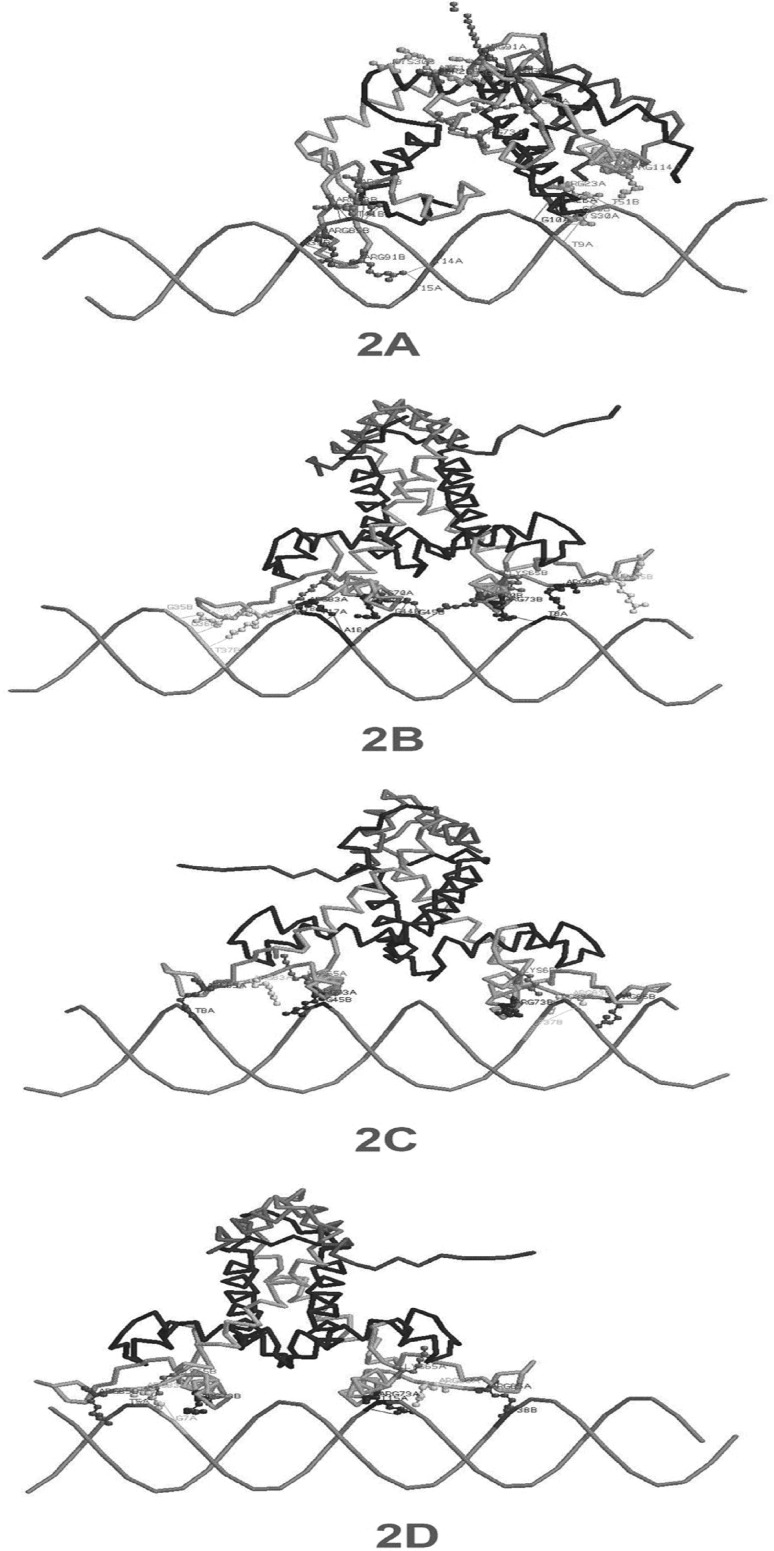
Mutation in mexR during PIP adaptation may cause the changes at some specific base positions (96th G to A, 201st G to A, 308th G to C, 377th A to T, 378th G to T, 384th G to A, 411th G to A) with high frequency which was supported by NCBI blast data. It was tabulated by comparing various strains of Pseudomonas aeruginosa with its PA14 strain available in NCBI (Table S1). However, these particular nucleotide positions might have flexibility to undergo base substitution in either way depending on forces acted on it. Therefore, those positions may call as mutational hotspot vicinity within which these changed bases caused during adaptation to one antibiotic stress turn to its original form of before stress in subsequent adaption to another antibiotic. It was found that mutation at nucleotide positions, 308th (G → C); both 377th and 378th (A → T and G → T) of mexR corresponded to the changes in amino acid sequences, G103 → A103 and E126 → V126, respectively. These changes may reduce the structural and functional stability of mexR protein (RMSD 117.04 and E value − 16832.822) which made the bacteria resistant to antibiotics. If the mutation restricted only to amino acid position 103 i.e., G103 → A103 may not lead to resistance but to susceptible by repressing mexAB-oprM, which was supported by RMSD 82.44 and E value − 17151.564 as wild type (RMSD 69.11 and E value − 17042.246) (Fig. 2a, b, respectively) [2]. Both mutant (G103 → A103) and wild susceptible strain may show similar structural and functional activity as because E value and RMSD of both of them are almost same.
Fig. 2.
DNA-protein docking to see the interaction between repressor protein mexR (active site) and its operator at 5′-GTTGA-3′ inverted repeat during stress: a thirteen different hydrogen and electrostatic bonds are contributed by wild mexR protein-DNA (A:ARG23-B:T51, A:ARG23-A:T9, B:ARG83-B:T41, B:ARG83-B:T41, B:ARG85-B:G39, B:ARG91-A:T14, B:ARG91-A:T15, A:ARG114-B:T51, B:ARG73-B:T41, A:CYS30-A:T9, SER26-A:G10, A:ARG23-B:C50, B:ARG73-B:T41) having RMSD 69.11; b change of amino acid at 103rd position (G → A) in mexR protein (RMSD 82.44) also shows thirteen different hydrogen and electrostatic bonds (A:LYS65-A:C18, A:ARG70-B:G44, A:ARG70-B:G45, A:ARG83-A:T17, A:ARG85-B:T37, A:ARG91-B:G36, A:ARG91-B:G35, B:ARG70-B:G45, B:ARG73-A:T8, B:ARG83-A:T8, B:ARG85:NH1-B:A45:O1P, A:ARG70-B:G44, A:ARG83-A:A16); c six different bonds (A:ARG73-B:G45, A:ARG85-A:T8, B:LYS65-B:G35, B:ARG83-B:T37, A:ARG85-A:T8, A:ARG73-B:G45) are found in docking of multidrug resistant mexR protein (G103 → A103) and E126 → V126) having RMSD 117.04; d mutation of E126 → 126 V in mexR (RMSD 103.31) shows five different types of bonds (A:ARG73-A:T15, A:ARG85-B:T38, B:LYS65-A:T5, B:ARG83-A:G7, A:ARG85-B:T38)
Case two, 20 days adaptation in ciprofloxacin followed by adaptation in piperacillin for subsequent 20 days led to a higher final MICPIP than the reverse order by significant mutation in mexR and a partial resensitization to ciprofloxacin (The MICPIP of Day 40 CIP PIP). No such pronounced mutation is found in mexS and nfxB [2].
CIP adaptation induced SOS response which caused mutation in mexS and nfxB expressing mexEF-oprN, mexCD-oprJ efflux pump, respectively and also weak expression of mexAB-oprM efflux pump which may be explained by mutation in coding sequence of mexR at E126 → 126 V (RMSD 103.31 and E value − 16681.887) (Fig. 2d). mexAB-oprM expression was increased in subsequent adaptation to PIP by SOS response which exerted mutation force at G103 → 103A and E126 → 126 V on mexR (RMSD 117.04 and E value − 16832.822) and finally completely inactivated it (Fig. 2c). A partial resensitization to CIP was also appeared spontaneously by selection in devoid of CIP stress (Fig. 1b), because mutations in mexS and nfxB which was caused by CIP stress induced such changes in bases i.e., within a mutational hotspot vicinity (321st G to A, 435th T to C, 477th A to G, 990th C to T for mexS and 104th A to G, 168th C to G, 267th G to C, 336th A to G, 444th C to T, 465th G to C for nfxB) and that was partially turned back again in absence of CIP stress during subsequent adaptation to PIP (Table S2 and S3).
Case three, MICCIP decrease i.e., partial resensitization was occurred in a series of treatment- Day 40 CIP TOB or LB. CIP adaptations first initially made collateral sensitivity to PIP, but became resistant again to PIP in subsequent adaptation to TOB having mutation in efflux pump related genes [2].
MICCIP decreased in either treatment i.e., TOB or LB in subsequent adaptation as because of large mutational hotspot vicinity in mexS and nfxB compared to mexR (Table S2 and S3). CpxR, ArmZ which were activated in subsequent TOB adaptation induced to express mexAB-oprM making resistant to PIP and mexXY-oprM efflux pump, respectively [9, 10]. Misfolded proteins or aberrant proteins were formed during subsequent TOB adaptation, which became e prone to oxidation leading to ROS. This ROS response activates ArmZ, an antirepressor of mexZ and mexZ is the repressor on mexXY operon [10, 11]. Misfolded envelop proteins also transduces signal to CpxR from CpxA by conserved phosphorylation (aspartate residue on CpxR) [10, 12]. Finally, this bacterial strain became multidrug resistant (Fig. 1c).
Case four, MICPIP is increased having no mutations in efflux pump-related genes, but carried significant mutation in two component sensor protein in series treatment of Day 40 PIPR TOBR [2].
The results showed no mutation in efflux pump related genes in the Day 40 PIPR TOBR lineages which led to less MICTOB than that of days 20 TOB, because ROS induced activation of AmrZ. The activated AmrZ, which was masked by sub inhibitory concentration of TOB of days 20 TOB led to less resistance in TOB. Adaptation in PIP induced to express mexAB-oprM through mexR mutation by SOS response. During subsequent adaptaion to TOB, ArmZ which imposed selective pressure on mexR and made normal in function by reverse mutation modulates to express mexXY-oprM. Mutation in CpxA induced to express CpxR which happens to activate the expression of mexAB-oprM in presence or absence of mexR. CpxA has both kinase and phosphatase activity, therefore probably mutation which abolishes phosphatase activity, not kinase activity always make operative the CpxR [9, 10, 13] (Fig. 1d).
Case five, Day 20 TOB was evolved to CIP, PIP and LB, which caused partial re-sensitization to TOB, but subsequent adaptation to PIP led to a partial re-sensitization to TOB which was not as much as CIP and LB did in subsequent adaptation. There were small numbers of mutation in transcription regulation [2].
TOB adaptation activated ArmZ which helped in mexXY expression making TOB resistant. SOS response which was induced during subsequent adaptation to PIP expressed the mexAB-oprM efflux pump by mutation in mexR gene. SOS also managed to operate a little expression of mexXY operon by mutation in mexZ in absence of proper activation of ArmZ and made resistance to TOB [13, 14]. Mutational hotspot vicinity of mexZ (78th T to C, 123rd T to C, 150th C to T, 195th A to G, 268th C to T, 314th G to T, 315th A to C, 399th G to C, 439th T to C, 458th G to A, 492th T to C, 516th C to G) made less susceptible compared to CIP and LB stress in a subsequent adaptations (Table S4) (Fig. 1e).
Conclusions and Future Perspectives
It is obvious to describe that the bacterial evolution is meant for overcoming the instability and developed diversified characters against stress. Besides, it is speculated that verily susceptible bacteria would be multidrug resistant based on available empirical data. This sign has created fear among all of us to combat against infectious bacteria. Antibiotic stress is the main weapon to cure bacterial infection, but it is also one of the main factors of developing multidrug resistant bacteria. After analyzing the whole operating pathway related to antibiotic stress, it was observed that still there was hope and clear cut indication of recovery from multidrug resistance of bacteria. Some specific forces in form of reverse/back mutation can counteract the evolutionary forces within mutational hotspot vicinity generated during stress. Therefore, looking at the sequence of treatment with antibiotics, it is the time to research on antibiotic treatment to find out the counteract forces to turn back collateral susceptibility from multidrug resistant strain. Evolutionary conserved sequences and position specific mutation which is developed in response to particular drug may help to choose different restriction enzymes. This restriction digestion may guide to select specific drug for fighting against this resistant strain. Moreover, the alternative option for the treatment of pathogenic MDR strain would be operated by understanding quorum sensing (QS) and Quorum sensing inhibitors (QSIs). Synergism between antibiotics and QSIs are found to be effective in enhancing the action of antibiotics [15–17].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Raju Biswas is thankful to CSIR for Junior Research Fellowship (File No: 09/025(0216)/2015-EMR-I) and Principal, Symsundar College, Shymsundar, Burdwan for conducting the research. Authors are thankful to UGC-Center of Advanced Study and DST-FIST, Department of Botany, The University of Burdwan for pursuing research activities.
Author’s Contributions
RB 1 and 2 adopted the idea. RB 1 and ASP conducted some in silico work and interpreted the data. RB 1, 2 and ASP wrote and all authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors report no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Toprak E, Veres A, Michel BJ, Chait R, Hart DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2011;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen P, Papin JA. History of antibiotic adaptation influences microbial evolutionary dynamics during subsequent treatment. PLoS Biol. 2017;15:e2001586. doi: 10.1371/journal.pbio.2001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.York A. Historical influences on antibiotic resistance. Nat Rev Microbiol. 2017 doi: 10.1038/nrmicro.2017.111. [DOI] [PubMed] [Google Scholar]
- 4.Kelly E, Lateef A, Keith P. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucl Acids Res. 2007;35:1465–1477. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucl Acids Res. 2017;1:1. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel L, Keith Natali N CJ. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biochem Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 8.Blázquez J, Gómez-Gómez JM, Oliver A, Juan C, Kapur V, Martín S. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol Microbiol. 2006;62:84–99. doi: 10.1111/j.1365-2958.2006.05366.x. [DOI] [PubMed] [Google Scholar]
- 9.Mahoney TF, Silhavy TJ. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol. 2013;195:1869–1874. doi: 10.1128/JB.02197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian Z, Yi X, Cho A, O’Gara F, Wang Y. CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathogen. 2016;12:e1005932. doi: 10.1371/journal.ppat.1005932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermann T. Drugs targeting the ribosome. Curr Opin Struct Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- 13.Fraud S, Poole K. Oxidative stress induction of the MexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011;55:1068–1074. doi: 10.1128/AAC.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay T, Fraud S, Lau CH, Gilmour C, Poole K. Antibiotic Inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS ONE. 2013;8:e56858. doi: 10.1371/journal.pone.0056858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalia VC. Microbes, antimicrobials and resistance: the battle goes on. Indian J Microbiol. 2014;54:1–2. doi: 10.1007/s12088-013-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Kalia VC, Patel SKS, Kang YC, Lee JK. Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol Adv. 2019;37:68–90. doi: 10.1016/j.biotechadv.2018.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.