Abstract
Background
Although embryo accounts for only 2–3% of the total weight of a rice grain, it is a good source of various nutrients for human health. Because enlarged embryo size causes increase of the amount of nutrients and bioactive compounds stored within rice grain, giant embryo mutants of rice (Oryza sativa L.) are excellent genetic resources for improving the nutritional value of rice grains.
Results
Three giant embryo mutants, including large embryo (le), giant embryo (ge) and super-giant embryo (ges), with variable embryo size were used in this study. We investigated whether genes controlling embryo size in these mutants (le, ge and ges) were allelic to each other. Although ge and ges was allelic to GIANT EMBRY (GE), le was not allelic to ge and ges in allelism test. The GE gene carried a unique nucleotide substitution in each of the two mutants (ge and ges), resulting in non-synonymous mutations in exon 2 of GE in both mutants. However, the GE gene of the le mutant did not carry any mutation, suggesting that the enlarged embryo phenotype of le was governed by another gene. Using map-based cloning, we mapped the LE gene to the short arm of chromosome 3. The le mutant showed mild enlargement in embryo size, which resulted from an increase in the size of scutellar parenchyma cells. The LE encodes a C3HC4-type RING finger protein and was expressed to relatively high levels in seeds at a late developmental stage. Knockdown of LE expression using RNA interference increased the embryo size of rice grains, confirming the role of LE in determining the embryo size.
Conclusion
Overall, we identified a new gene controlling embryo size in rice. Phenotypic and molecular characterization results suggest that the le mutant will serve as a valuable resource for developing new rice cultivars with large embryos and nutrient-dense grains.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0277-y) contains supplementary material, which is available to authorized users.
Keywords: Rice, Large embryo, Giant embryo, C3HC4-type RING finger protein, Nutritional value
Background
With an ever-increasing demand for high quality rice (Oryza sativa L.), several attempts have been made to improve the quality of rice (Fitzgerald et al. 2009). A mature rice embryo typically accounts for only 2–3% of the total kernel weight, but has enriched various nutrients, such as proteins, lipids, vitamins, minerals, and phytochemicals (Gopala Krishna et al. 1984; Tanaka et al. 1977). The high nutritional content of embryo makes it popular to use brown rice, polished embryonic rice, and byproduct (bran) generated after polishing.
Breeding efforts to improve the nutritional quality of rice have focused on the induction and isolation of mutants. Various mutants with distinct embryo and endosperm phenotypes have been reported in rice, including endospermless, embryoless, reduced embryo, and giant embryo mutants (Satoh and Omura 1981; Hong et al. 1995). Among these, the giant embryo mutants have often been used as breeding materials to improve the nutritional value of rice. To date, several giant embryo rice cultivars have been reported (Maeda et al. 2001; Takahashi et al. 2009; Hong et al. 2012; Han et al. 2012). Brown rice of these rice mutants contain higher amounts of proteins, lipids, essential amino acids, vitamins (B1, B2, and E), minerals (Ca, Fe, Mg, K, and P), and bioactive compounds [γ-aminobutyric acid (GABA) and γ-oryzanol] than those of wild-type rice cultivars with normal embryo size (Koh et al. 1993; Zhang et al. 2005; Jeng et al. 2012; Kim et al. 2013).
Until now, only two genes related to the enlargement of embryo size have been identified in rice. GIANT EMBRYO (GE) was mapped on chromosome 7 (Satoh and Iwata 1990; Koh et al. 1996), and Nagasawa et al. (2013) revealed that GE gene encodes CYP78A13, a cytochrome P450 protein, and functions in determining embryo/endosperm size. The loss-of-function GE exhibits an enlargement of embryo and shrinkage of endosperm. Additionally, GE plays critical role in shoot apical meristem (SAM) maintenance, and GE overexpression promotes cell proliferation, plant growth and grain yield (Yang et al. 2013). In maize, the insertion of a transposable element in ZmGE2, a homolog of OsGE, has resulted in a reduction in its expression and an increase in the embryo to endosperm ratio (EER) (Zhang et al. 2012). ZmGE2 encodes a cytochrome P450 protein of the CYP78A family, and overexpression of OsGE in maize results in a reduction in EER. These results suggest that cytochrome P450 proteins play an important role in regulating embryo size, and the mechanism of EER determination is evolutionarily conserved in rice and maize (Zhang et al. 2012). GOLIATH (GO), another gene related to embryo size, has been mapped to the long arm of chromosome 3 (Taramino et al. 2003). Independently identified mutant, plastochron3 (PLA3) is allelic to go mutant. The loss of function PLA3, which encodes glutamate carboxypeptidase, exhibits pleiotropic phenotypes including enlarged embryo (caused by an increase in the number of cells), seed vivipary, defects in SAM maintenance, aberrant leaf morphology (Kawakatsu et al. 2009). Recently, new giant embryo breeding materials have been reported; these display an enlarged embryo and higher content of triacylglycerol compared with the wild type. The gene responsible for these phenotypes is not allelic to GE and maps to the short arm of chromosome 3 (Sakata et al. 2016).
In this study, we performed allelism tests to determine if genes responsible for the enlarged embryo phenotypes of the embryo mutants, ge, ges, and le, are allelic to each other and identified a new gene controlling embryo size. Our results will contribute to understanding genetic regulation of embryo size. Furthermore, the series of mutants may facilitate the breeding of embryonic cultivar with high nutritional value.
Results
Characterization of Three Giant Embryo Rice Mutants
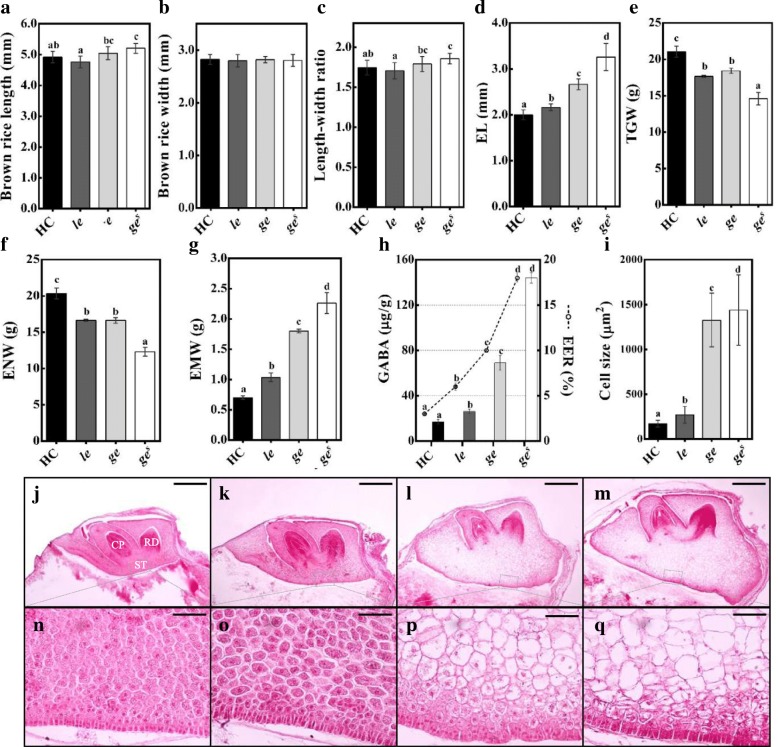
All three mutants exhibited enlarged embryo phenotype compared with Hwacheong (HC). Depending on the embryo size, the three giant embryo mutants were designated as le, ge, and ges; the embryo size was relatively smaller in le, larger in ges, and intermediate in ge mutant (Figs. 1 and 2d). We evaluated the relative embryo size of these mutants using the EER, which is defined as the weight of the embryo relative to that of the endosperm. The EER gradually increased in the order of le, ge, and ges mutants, and the GABA content of brown rice followed the same trend (Fig. 2h). The enlarged embryo phenotype of all mutants resulted from a conspicuous enlargement of scutellar parenchyma cells (Fig. 2i and n-q). In addition to the embryo size, other phenotypes were also investigated. Although none of the giant embryo mutants showed any major defects in shoot and radicle differentiation, the shoot size of ge and ges mutants was slightly smaller than that of le mutant and wild-type HC (Fig. 2j-m). Among grain characteristics, the width of mutant rice grains was comparable to that of HC; however, the length and length to width ratio (LWR) of the ges grains were significantly higher than those of HC (Fig. 2a-c). The giant embryo mutants showed significantly lower thousand-grain weight (TGW) and endosperm weight (ENW) but higher embryo weight (EMW) than HC (Fig. 2e-g). Compared with HC, the ge and ges mutants showed a dramatic increase in EMW of 157% and 223%, respectively, whereas the le mutant exhibited only a 48% increase in EMW (Fig. 2g). Among other agronomic traits, culm length (CL), grain width (GW), grain length (GL), and LWR of the le mutant were significantly different from those of HC (Additional file 3: Figure S2).
Fig. 1.

Grain and embryo phenotypes of HC and giant embryo mutants. Images from left to right are of HC, le, ge, and ges in this order. Scale bars = 2 mm
Fig. 2.
Characterization of phenotypic traits of rice grains and embryos of HC and giant embryo mutants. a-i Evalucation of various seed characters. Brown rice length (a), width (b), and LWR (c); Embryo length (EL) (d); Thousand-grain weight (TGW) (e); Endosperm weight (ENW, 1000 seeds) (f); Embryo weight (EMW, 1000 seeds) (g); GABA content of EER (h); Size of scutellar parenchyma cells (i). j-q Longitudinal sections of embryo at 20 days after pollination (DAP): wild type (j and n), le (k and o), ge (l and p), and ges (m and q). CP, coleoptiles; RD, radicle; ST, scutellum (n-q). The means followed by lowercase letters indicate statistically significant difference (p < 0.05, ANOVA) according to Duncan test. Error bars indicate ± s.d. of replicates. Scale bars = 0.5 mm (j-m), Scale bar = 50 μm (n-q)
Genetic Analysis of Giant Embryo Mutants
In order to confirm allelism and heredity among genes responsible for enlarged embryo phenotype in three mutants, we investigated the segregation ratio of F2 seeds derived from reciprocal crosses among HC, le, ge, and ges (Table 1). Data showed that the embryo phenotype of F2 seeds derived from reciprocal crosses between each mutant and HC segregated in a 3: 1 ratio (normal: enlarged), indicating that the enlarged embryo phenotype of each mutant was controlled by a single recessive gene. Because we previously showed that ges is allelic to ge (Kim et al. 1992), we inferred that the enlarged embryo phenotype of ges mutant was induced by a mutation in GIANT EMBRYO (GE) gene. In reciprocal crosses between ge and ges, the EL of F1 seeds was increased, and embryos of all F2 seeds showed the enlarged phenotype, suggesting that the ge locus is identical to that of ges (Table 1). On the other hand, F2 seeds derived from reciprocal crosses between le and each of the other two giant embryo mutants, ge and ges, showed three different embryo phenotypes with 9:3:4 segregation ratio (normal type: le type: ge/ges type) (Additional file 4: Figure S3), suggesting that LE is not allelic to GE.
Table 1.
Segregation of F2 seeds derived from the reciprocal crosses among HC and three giant embryo mutants
| Cross combination | EL of F1 seed (mm) | Number of different embryo type | Expected ratio | x 2 | p-value | |||
|---|---|---|---|---|---|---|---|---|
| nor | le | ge | ge s | |||||
| HC/le | 2.10a | 299 | 99 | – | – | 3:1 | 0 | 1 |
| le/HC | 260 | 100 | – | – | 3:1 | 1.34 | 0.25 | |
| HC/ge | 2.16a | 348 | – | 134 | – | 3:1 | 1.87 | 0.17 |
| ge/HC | 269 | – | 83 | – | 3:1 | 0.31 | 0.58 | |
| HC/ges | 2.12a | 246 | – | – | 76 | 3:1 | 0.27 | 0.61 |
| ges/HC | 265 | – | – | 84 | 3:1 | 0.12 | 0.73 | |
| le/ge | 2.15a | 177 | 64 | 71 | – | 9:3:4 | 1.16 | 0.56 |
| ge/le | 145 | 48 | 63 | – | 9:3:4 | 0.02 | 0.99 | |
| le/ges | 2.21a | 178 | 60 | – | 78 | 9:3:4 | 0.02 | 0.99 |
| ges/le | 160 | 51 | – | 63 | 9:3:4 | 0.67 | 0.72 | |
| ges/ge | 3.12b | – | – | 296 | 117 | 3:1 | 2.27 | 0.13 |
| ge/ges | – | – | 106 | 45 | 3:1 | 1.61 | 0.2 | |
The means followed by lowercase letters are significantly different (p < 0.05, ANOVA) according to Duncan test. EL embryo length, nor normal type
Sequencing Analysis of GE Locus in the Three Giant Embryo Mutants
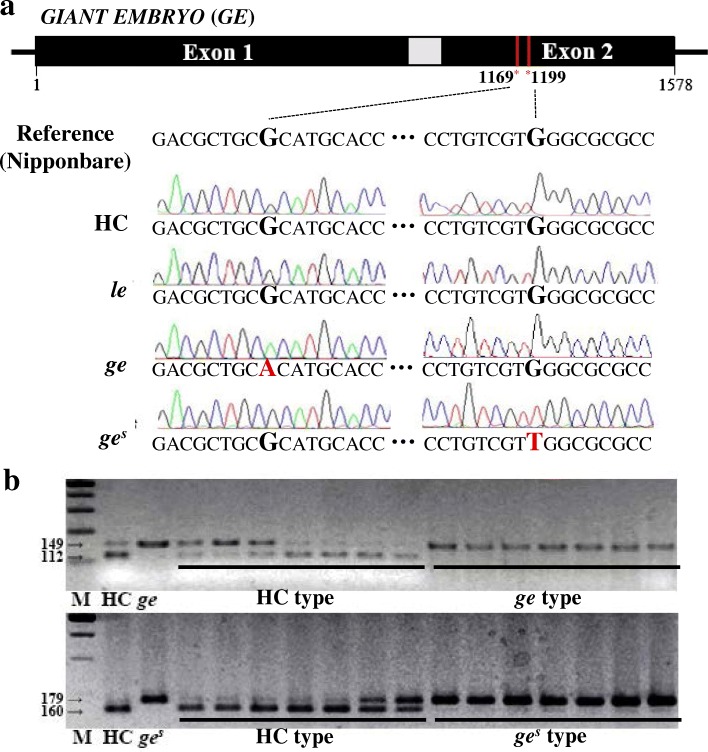
We sequenced the GE locus (Os07g0603700) of all three giant embryo mutants. Data revealed G to A single base change at nucleotide position 1169 in the GE locus of ge mutant, resulting in arginine (Arg) to histidine (His) amino acid substitution at position 390, and G to T single base change at nucleotide position 1199 in the GE locus of ges mutant, resulting in tryptophan (Trp) to leucine (Leu) amino acid substitution at position 400 (Fig. 3a). These mutations co-segregated with the enlarged embryo phenotype in F2 seeds derived from crosses between HC and each of these two mutants (ge and ges) (Fig. 3b). However, no nucleotide substitutions were detected in the GE locus of le mutant. Additionally, no genetic polymorphism was present in the PLA3/GO locus of le mutant (data not shown). These data suggest that, while mutations in GE were responsible for the enlarged embryo phenotypes of ge and ges mutants, another gene was responsible for the enlarged embryo phenotype of le mutant. These results were consistent with those of our allelism tests described above.
Fig. 3.
Sequence analysis of the GE locus. a Identification of mutations in GE locus. b Co-segregation test of mutation point and enlarged embryo phenotype in F2 seeds derived from HC/ge and HC/ges crosses
Map-Based Cloning of LE Gene
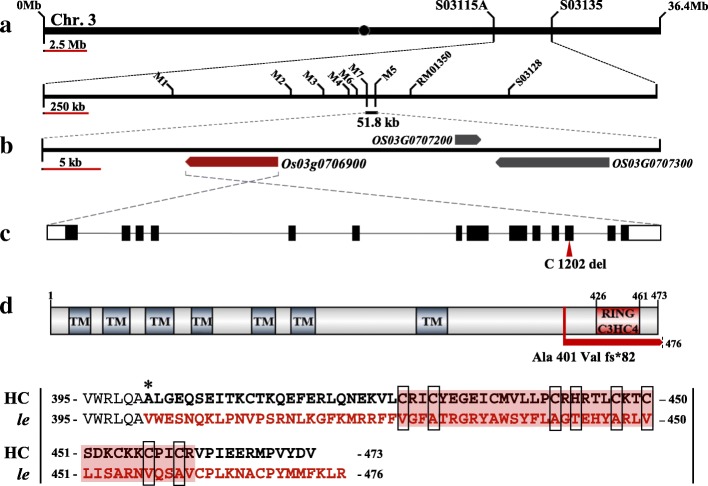
To isolate gene responsible for the enlarged embryo phenotype of le mutant, we performed map-based cloning using F2 and F3 populations derived from a cross between le mutant and rice cv. Hangangchal1 (Tongil-type). Bulked segregant analysis (BSA) showed that the LE gene was linked with the InDel marker S03128 on the short arm of chromosome 3. To determine the exact position of LE locus, we developed a new set of InDel markers on the basis of genetic polymorphisms between sequences of indica and japonica rice (http://www.ncbi.nlm.nih.gov/). Using these markers, LE was mapped to an approximately 52-kb region between M7 and M5 markers (Fig. 4a). Three predicted open reading frames (ORFs) were located within this candidate region (Fig. 4b), all of which were sequenced in the le mutant and HC. Sequence analysis revealed the deletion of a single nucleotide ‘C’ at position 1202 in exon 12 of the locus Os03g0706900 in le mutant (Fig. 4c). This locus encodes a C3HC4-type RING finger protein. The deletion of ‘C’ at position 1202 was predicted to cause frameshift mutation and termination of the ORF after 82 aberrant amino acids (Fig. 4d and Additional file 5).
Fig. 4.
Map based cloning of the LE gene. a Schematic diagram of mapping of LE locus. b Candidate ORFs in delimited candidate region. c LE gene structure and splicing pattern. Black lines, white solid boxes, and black solid boxes indicate intron, UTRs, and exons, respectively. The mutation position is marked with red arrow head. d LE protein structure and comparison of amino acid sequence of the frameshift region between the HC and le mutant. Asterisk represents the location of amino acid substitution caused by one base deletion in le mutant. Amino acids in the region of frameshift (fs) are indicated in red. Red background and black open boxes indicate amino acid sequences of RING C3HC4 domain and conserved residues in the domain
To perform co-segregation analysis, we developed a derived cleaved amplified polymorphic sequences (dCAPS) marker. The dCAPS genotype of F2 individuals co-segregated with the enlarged embryo phenotype of the F3 seeds type. These data showed that the deletion of C base at nucleotide position 1202 of the locus Os03g0706900 was responsible for the enlarged embryo phenotype of le mutant (Additional file 6). To determine whether this mutant variation is present as a natural variation in the rice germplasm, we genotyped the Os03g0706900 locus in 12 O. sativa cultivars (six japonica, two tongil-type, and four indica) and one O. nivara accession using the dCAPS marker. However, none of these rice genotypes harbored the mutation present in the Os03g0706900 locus of le mutant (Additional file 6). Therefore, we focused on Os03g0706900 as a candidate for LE.
Effect of LE Knockdown on Embryo Size
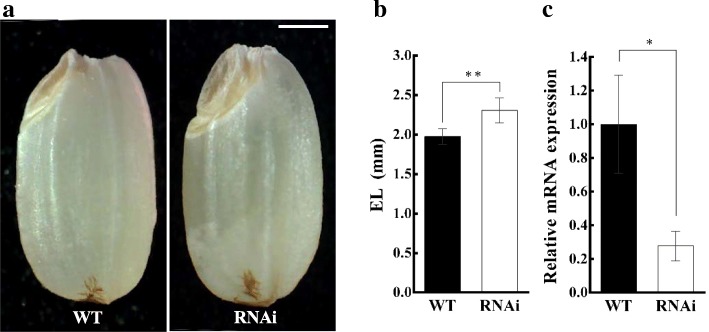
To confirm whether the LE gene regulates embryo size in rice, we used dsRNA-mediated interference (RNAi) to knockdown the expression of LE in the japonica cv. Dongjin. The embryo size of RNAi T1-plant grains was significantly bigger than that of wild-type grains (Fig. 5a and c). Quantitative real time PCR (qRT PCR) analysis using leaf samples revealed that the expression of LE gene in RNAi T1 plants was significantly lower than that in wild-type (Fig. 5c), indicating that LE controls embryo size in rice.
Fig. 5.
Comparison of embryo phenotype and LE expression between RNAi transgenic plant and wild type (Dongjin). a Seed phenotype of WT and RNAi plants. b Comparison of embryo length between WT and RNAi plants. c Relative expression of LE (normalized to UBIQUITIN) in WT and RNAi plants. Error bars indicate ± s.d. of three replicates. Asterisks represent significant difference, as determined by a two-tailed Student’s t-test (*, p < 0.05; **, p < 0.01). Scale bar = 1 mm
Expression Pattern of LE
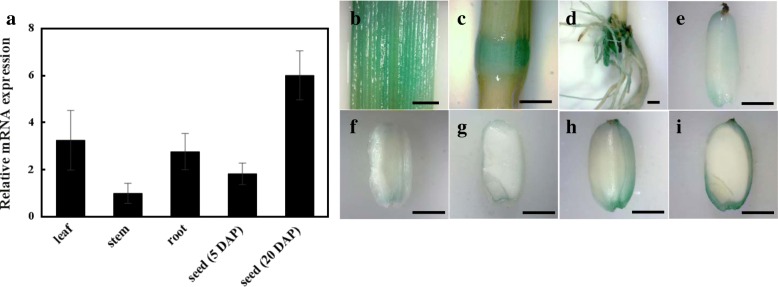
We conducted qRT PCR analysis to monitor the expression of LE in various organs, including leaves, stems, roots, and seeds (Fig. 6). LE was expressed in all organs in HC (Fig. 6a). The expression of LE in stems was lower than that in leaves and roots. In seeds, the expression level of LE was much lower at 5 DAP than at 20 DAP, indicating that the expression of LE increases as seeds mature (Fig. 6a). We also performed β-glucuronidase (GUS) staining experiments using transgenic rice plants expressing GUS reporter under the control of LE gene promoter (proLE::GUS). GUS staining data were consistent with the results of qRT PCR analysis (Fig. 6b-i). GUS expression was observed in all organs, including leaves, stems, roots, and seeds (Fig. 6b-i), and the expression of GUS in seeds increased over time (Fig. 6f-i). Interestingly, GUS staining data showed that LE is expressed in the outer part of rice seed (Fig. 6g and i).
Fig. 6.
Tissue-specific RNA expression pattern of LE. a qRT-PCR analysis of LE gene in leaf, stem, root and seed (5 and 20 DAP). b-i GUS expression pattern of various tissues in ProLE:GUS transgenic line [leaf (b); stem (c); root (d); seed at 5 DAP (e); seed at 10 DAP (f and g); seed at 20 DAP (h and i)]. Error bars indicate ± s.d. of replicates. Scale bars = 2 mm
Discussion
In this study, we performed map-based cloning to identify the LE gene responsible for the enlarged embryo phenotype of the rice le mutant. We showed that LE is not allelic to GE. Although LE was mapped to the same chromosome with GO (Kawakatsu et al. 2009) and new giant embryo locus (Sakata et al. 2016), the candidate region of LE locus was far from the position of GO and the other locus. RNAi transgenic approach confirmed that LE, Os03g0706900, is the gene responsible for the large embryo in the le mutant.
The LE protein consists of 473 amino acid residues with a C3HC4-type RING finger domain at the C-terminus and seven transmembrane motifs at the N-terminus predicted using TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Additional file 5). Multiple sequence alignment of LE orthologs revealed that the amino acid sequence of the predicted transmembrane motifs and the consensus sequence of RING finger domain (Cys-× 2-Cys-X11-Cys-X-His-X3-Cys-× 2-Cys-X6-Cys-X2-Cys) were highly conserved across monocots and dicots. However, the consensus sequence of LE in le mutant was highly dissimilar because of the frameshift generated by the single nucleotide deletion. (Fig. 4d and Additional file 5). These results indicate that the functional role of LE orthologs is conserved in plants and is dependent on the C3HC4-type RING finger domain. The sequence of LE is identical to that of OsRHC1 cloned and characterized previously (Cheung et al. 2007). OsRHC1 is differentially expressed in bacterial blight resistant near-isogenic lines (NILs) containing Xa14 or Xa23 as well as in NILs susceptible to bacterial blight (Cheung et al. 2007). Additionally, transgenic Arabidopsis plants overexpressing OsRHC1 are more resistant to bacterial pathogens than wild-type Arabidopsis (Ma et al. 2009). In rice, the function of C3HC4-type RING finger proteins is not well understood. To date, only a few C3HC4-type RING finger proteins have been reported in rice, including OsCOP1, OsCOIN1, and OsXB3; these are involved in photomorphogenesis, abiotic stress tolerance, and disease resistance, respectively (Tsuge et al. 2001; Liu et al. 2007; Wang et al. 2006). However, there is no report that C3HC4-type RING finger protein is involved in embryo morphology and development in rice. By contrast, the function of C3HC4-type RING finger proteins in Arabidopsis is well characterized, and several genes encoding C3HC4-type RING finger proteins have been identified, including AtCOP1 and AtCIP8 (photomorphogenesis) (von Arnim and Deng 1993; Hardtke et al. 2002), AtTED3 (light signaling) (Pepper and Chory 1997), AtPEX10 and AtPEX12 (peroxisome biogenesis) (Schumann et al. 2003; Fan et al. 2005), AtXBAT32 (root development), and AtSDIR1 (stress tolerance) (Zhang et al. 2007). Among these, AtPEX10 and AtPEX12 contain two transmembrane helices and a C3HC4-type RING finger domain, which are similar in sequence to those of LE. Both AtPEX10 and AtPEX12 are required for peroxisome biogenesis, and their dysfunction leads to lethality at the heart stage of embryogenesis.
Both transmembrane motifs at the N-terminus and a C3HC4-type RING domain at the C-terminus predicted were considered essential for the function and localization of LE protein. Amino acid sequences of LE orthologs were highly conserved among monocots and dicots. ZmZF13, a maize ortholog of OsLE, has been previously identified from a cDNA library obtained from developing maize seeds (Wang et al. 2010). There is, however, no published information on the functions of these LE orthologs, except for pathogen defense of OsRHC1. Therefore, further studies are needed to understand the mechanism of action of LE, and its regulation of embryo size in rice grains.
With the increase in the embryo size of giant embryo mutants (le < ge < ges), the GABA content of brown rice also increased by 150%, 400%, and 850% compared with wild type, respectively. In addition to GABA content, other nutrients and micronutrients such as protein, lipid, vitamin B1 and vitamin E were also higher in ges and le mutant compared to normal rice (Koh et al. 1993; Kim and Kim 2013; Chung et al. 2014a). Giant embryo mutants with high nutritional content are very useful breeding materials to improve the nutritional value of rice. Therefore, the creation and discovery of new genetic variation controlling embryo size is very important for breeding cultivars with high nutritional value. We used our three giant embryo mutants to breed high nutritional rice cultivar and had registered four cultivars to plant variety protection national list in Korea (soenong8 and seonong16 derived from ge; seonong14 derived from ges; seonong17 derived from le).
Enlarged embryo size and easy embryo detachment are desirable traits of rice cultivars used for bran or bran oil production. However, High EER giant embryo mutants such as ge and ges are can’t be used to increase the nutritional value of milled or polished rice, because most of the embryo are separated from the rice grain during the milling process. Therefore, most of giant embryo rice cultivars are supplied as unpolished rice (brown rice) to customers. On the other hand, seonong17, a cultivar derived from le mutant with relatively low EER, showed 88% of embryo residual rate with developed milling process (Kim and Kim 2013). This result implicates that the le mutant, with mildly enlarged embryo, is expected to serve as an appropriate breeding material for improving the nutritional quality of milled rice because large part of embryo remain attached to the endosperm even after milling.
Generally, giant embryo mutants exhibit problematic germination and seedling growth. For example, the go mutant shows pleiotropic phenotypes, including viviparous seed, shortened plastochron, and conversion of panicle primary branches to vegetative shoots, in addition to the enlarged embryo phenotype (Kawakatsu et al. 2009). Similarly, the ge mutant shows abnormal seedling growth characterized by the production of malformed leaves and dwarfism, which eventually result in death (Yang et al. 2013). By contrast, the le mutant showed normal germination rate and seedling emergence (data not shown). Besides, Seonong17 is effective in lowering body fat and has high hypolipidemic and antioxidant activities (Chung et al. 2014b, 2016). These observations suggest that the le mutant is a highly valuable genetic resource for improving the nutritional quality of rice.
Overall, we identified a new gene, LE, responsible for the enlarged embryo phenotype of the le mutant of rice, and characterized the potential of the le mutant as a breeding material. The embryo of le mutant was only mildly enlarged comparing to the embryos of ge and ges mutants. LE encodes a C3HC4-type RING finger protein, expecting new molecular mechanism to control embryo size. Although further investigations are needed to understand the function of LE, this study provides novel insights into the genetic mechanism of embryo development in rice and introduces new breeding material for high nutritional rice cultivars with large embryos.
Conclusion
The rice le mutant showed a mild enlargement of embryo and increased GABA content compared with wild type. Through allelism tests and sequence analysis, we showed that the enlarged embryo phenotype of le mutant was not controlled by previously reported genes regulating embryo size but by LE, a new gene controlling embryo size. The LE gene encodes a C3HC4-type RING finger protein. The disruption of the RING domain led to the enlargement of embryo size, possibly due to the increase in size of scutellar parenchyma cells. LE was highly expressed during the late stage of seed development. These results extend our understanding of embryo development in rice and facilitate the breeding of new giant embryo rice cultivars.
Methods
Plant Materials
Three giant embryo mutants were generated from a fertilized egg cell of japonica cv. HC treated with N-methyl-N-nitrosourea and designated as le, ge, and ges, in ascending order of embryo size (Kim et al. 1991, 1992; gem which is nomenclature of the mutant least altered embryo size was renamed as le to avoid confusion with alleles of GE gene). Wild-type japonica cv. HC was used as a control in phenotype analysis. The F2 seeds derived from a cross between le mutant and HC were used to calculate the segregation ratio. Map-based cloning was conducted using F2 and F3 populations derived from a cross between le mutant and cv. Hangangchal1 (Tongil-type). All plant materials were grown under normal cultivation conditions in the experimental paddy field of the Seoul National University, Suwon, Korea.
Phenotypic Analysis
Dimensions of rice grains were measured from images captured under a microscope with HD’MEASURE software (HANA Vision, Korea). To measure ENW, EMW, and EER, embryos and endosperms were dissected from husked seeds (14% water content) and weighed separately using an electronic balance (CAS, USA). Agronomic traits such as days to heading (DTH), culm length (CL), panicle length (PL), panicle number (PN), spikelet number per panicle (SPP), and spikelet fertility (SF) were also measured to compare morphological differences between HC and giant embryo mutants. All phenotypic data collected from HC and mutants were statistically analyzed using IBM SPSS Statistics software version 24 (IBM, USA).
Histological Analysis
Seeds of HC and giant embryo mutants harvested at 20 DAP were fixed in formalin-acetic acid-alcohol (FAA) fixative (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) for 24 h at 4 °C. Seeds were dehydrated by soaking them in a series of ethanol solutions of increased concentration for 2 h per each solution. After final dehydration using 100% ethanol, seeds were cleaned using a series of cleaning solution with progressively decreasing ethanol concentration and increasing concentration of histoclear, followed by soaking in 100% histoclear solution overnight. For paraffin infiltration, Paraplast® (Sigma, USA) was gradually added to histoclear solution at 60 °C. Finally, seeds were stored in 100% melted paraffin for 24 h at 60 °C. The paraffin-infiltrated samples were embedded in an embed block and cut into 10 μm sections with an HM 340 E Rotary Microtome (Microm Lab, Germany). The sections were stained with 1% safranin O solution (1% safranin O and 30% ethanol) and examined under CX31 Microscope (Olympus, Japan).
Quantification of GABA Content
Fifty milligrams of finely ground brown rice powder was homogenized with 1 ml of 0.01 N HCl prepared in 25% acetonitrile and centrifuged at 13,000 rpm for 3 min. The supernatant was filtered through a 0.2 μm syringe filter. The clear supernatant was derivatized using EZ:FAAST Kit (Phenomenex, USA) and subjected to gas chromatography (GC) using a GC-2010 gas chromatograph (GC-2010, Japan) equipped with a flame ionization detector (FID) and a ZB-5 capillary column (30 m × 0.25 mm; 0.25 μm diameter) for the analysis of GABA. Each sample was tested four times.
Allelism Test and Sequence Analysis
F1 and F2 seeds derived from reciprocal crosses among HC, and each of the three mutants, le, ge, and ges, were used for genetic analysis. F2 seeds produced from each cross were classified according to the embryo size, and chi-square (χ2) test was performed to detect statistically significant differences. Two overlapping DNA fragments spanning the GE ORF (Nagasawa et al. 2013) were PCR amplified and sequenced with ABI Prism 3730 XL DNA Analyzer (PE Applied Biosystems, USA). The sequences of HC and three giant embryo mutants were aligned using CodonCode Aligner software (CodonCode Corporation, USA) and analyzed. Co-segregation test was conducted to detect SNP using CAPS and dCAPS markers (Additional file 1: Table S1).
Map-Based Cloning of LE
F2 seeds derived from a cross between le mutant and Hangangchal 1 were classified into mutant type and wild type based on the embryo size. Twenty mutant type F2 seeds and 20 wild type F2 seeds were selected to prepare DNA bulks. Mutant and wild DNA bulks were subjected to BSA using 66 sequence-tagged-site (STS) markers (Michelmore et al. 1991; Chin et al. 2007). For fine mapping the LE gene, 397 F2 and 857 F3 plants were used, and seven new STS markers were designed using Primer3 version 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0) based on polymorphisms between sequences of indica and japonica rice (https://blast.ncbi.nlm.nih.gov and http://gramene.org). Primer sequences used in this study are listed in Additional file 1: Table S1.
Candidate Gene Analysis
To identify nucleotide polymorphisms in candidate genes, DNA fragments of genes were PCR amplified from HC and le mutant using gene-specific primers (Additional file 1: Table S1) and purified using Gel & PCR Purification Kit (Inclone, Korea). The purified PCR products were cloned into pGEM-T Easy vector (Promega, USA) and transformed into Escherichia coli strain DH5α. Plasmid DNA was isolated from the transformed colonies using Plasmid MiniPrep Kit (Inclone, Korea) and sequenced using an ABI Prism 3730 XL DNA Analyzer (PE Applied Biosystems, USA). DNA sequencing data were aligned using CodonCode Aligner software (CodonCode Corporation, USA).
Multiple Sequence Alignment and Phylogenetic Analysis
The amino acid sequences with high similarity to LE were downloaded from the Universal Protein Resource (UniProt, http://www.uniprot.org). Multiple sequence alignment was carried out using Probcons, and background color shading was applied with Jalview using the percent identity scheme. Details of protein sequences used in this study are provided in Additional file 2: Figure S1. Transmembrane motifs were predicted using TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from leaf, stem, and root tissues collected at heading time, and from seeds 5 and 20 DAP, using Hybrid-R™ RNA Purification Kit (GeneAll, Korea). All RNA samples were treated with RNase-free Recombinant DNase Ι (Takara Bio, Japan) to eliminate genomic DNA contamination. First-strand cDNA was synthesized using M-MLV reverse transcriptase (Promega, USA). qRT-PCR was performed using SYBR Premix Ex Taq (Takara, Japan) on a CFX96™ Real-Time PCR Detection System (Bio-Rad, USA), according to the manufacturer’s instructions. Primers used for qRT-PCR analysis are listed in Additional file 1: Table S1. Expression levels of LE were normalized relative to UBIQUITIN5, a housekeeping gene.
GUS Staining
GUS staining was carried out as previously described (Jefferson et al. 1987). The GUS staining solution was vacuum-infiltrated into various organ samples for 10 min. All samples were submerged in the staining solution and incubated overnight at 37 °C. This was followed by incubation in 90% ethanol overnight at room temperature to bleach the chlorophyll.
Vector Construction and Transformation
For the generation of RNAi and proLE::GUS transgenic plants, the 3′ untranslated region (3′ UTR; + 6925 to + 7215 bp) and promoter region (− 2135 to − 1 bp) of LE were PCR amplified and cloned into pH7GWIWG2(II) and pHGWFS7, respectively, using Gateway® BP and LR Clonase™ II enzyme mixes (Invitrogen, USA). The final constructs were introduced into the japonica cv. Dongjin via Agrobacterium-mediated transformation using the strain LBA4404 as described (Nishimura et al. 2007) with slight modifications. Primers used for cloning PCR products are listed in Additional file 1: Table S1.
Additional files
Table S1. Agronomic traits of HC three giant embryo mutants. DTH, days to heading; CL, culm length; PL, panicle length; PN, panicle number; SPP, spikelet per panicle; SF, spikelet fertility; GL, grain length; GW, grain width; LWR, length-width ratio. The means followed by lowercase letters are significantly different (p < 0.05, ANOVA) according to Duncan test. (XLSX 10 KB)
Figure S1. Types of segregated F2 seed derived from cross-combinations of le/ge (a) and le/ges (b). (PDF 96 KB)
Figure S2. Multiple sequence alignment of LE orthologs. Asterisk represents the location of amino acid substitution caused by one base deletion in le mutant. Substituted amino acid residues by frameshift are highlighted by red background shading. Predicted transmembrane motifs and the C3HC4-type RING finger domain are underlined with blue and red lines, respectively. Conserved residues in the C3HC4-type RING finger domain are enclosed within black open boxes. Os, Oryza sativa; Ta, Triticum aestivum; Hv, Hordeum vulgare; Zm, Zea mays; At, Arabidopsis thaliana; Br, Brassica rapa; Gm, Glycine max. (PDF 531 KB)
Figure S3. Co-segregation test on F2 individuals and various rice genotypes. a Co-segregation test on F2 individuals derived from the cross between HC/le mutant. b Co-segregation test on other varieties and O.nivara. 1, Hwacheong; 2, Hwaseonchal, 3. Ilpum, 4. Unkwang, 5. Hapcheon, 6. Nipponbare, 7. Dasan, 8. Hangangchal 1ho, 9. IR36, 10. IR64, 11. IR56, 12. IR21015, 13. O.nivara. (PDF 23 KB)
Information of the primers used in this study. (XLSX 11 KB)
Information of LE orthologs used in multiple sequence alignment. (XLSX 9 KB)
Acknowledgements
We thank Dr. Joohyun Lee (Konkuk University) for kind advice on map-based cloning of LE gene.
Funding
This study was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ013165) of the Rural Development Administration, Korea.
Availability of Data and Materials
All data supporting the conclusions of this article are provided within the article and its additional files (Additional file 3: Table S1, Additional file 4: Figure S1, Additional file 5: Figure S2, Additional file 6: Figure S3, Additional file 1: Table S2. Additional file 2: Table S3, respectively).
Abbreviations
- BSA
Bulked segregant analysis
- CL
Culm length
- DAP
Days after pollination
- EER
Embryo to endosperm ratio
- EL
Embryo length
- EMW
Embryo weight
- ENW
Endosperm weight
- GE
Giant embryo
- GL
Grain length
- GUS
β-glucuronidase
- GW
Grain width
- HC
Hwacheong
- LE
Large embryo
- LWR
Length to width ratio
- NILs
Near-isogenic lines
- ORF
Open reading frame
- qRT PCR
Quantitative real time PCR
- RNAi
dsRNA-mediated interference
- TGW
Thousand-grain weight
Authors’ Contributions
GL designed and performed the experiments, and wrote the manuscript. RP, YL and BK provided assistance and advice on experiments. JS, DL, SJ, and ZJ provided assistance on the experiments. CL and JHC improved the manuscript. HJK supervised the research and improved the manuscript. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gileung Lee, Email: leegileung@gmail.com.
Rihua Piao, Email: p2h78@163.com.
Yunjoo Lee, Email: han14@hanmail.net.
Backki Kim, Email: uptfamily@hanmail.net.
Jeonghwan Seo, Email: rightseo@hotmail.com.
Dongryung Lee, Email: bok4jang@hanmail.net.
Su Jang, Email: oryzasativalinne@gmail.com.
Zhuo Jin, Email: jinzhuo0116@gmail.com.
Choonseok Lee, Email: seokkch@gmail.com.
Joong Hyoun Chin, Email: jhchin@sejong.ac.kr.
Hee-Jong Koh, Email: heejkoh@snu.ac.kr.
References
- Cheung M-Y, Zeng N-Y, Tong S-W, Wing-Yen Li F, Zhao K-J, Zhang Q, Sai-Ming Sun S, Lam H-M. Expression of a RING-HC protein from rice improves resistance to Pseudomonas syringae pv. Tomato DC3000 in transgenic Arabidopsis thaliana. J Exp Bot. 2007;58(15–16):4147–4159. doi: 10.1093/jxb/erm272. [DOI] [PubMed] [Google Scholar]
- Chin JH, Kim JH, Jiang W, Chu SH, Woo MO, Han L, Brar D, Koh HJ. Identification of subspecies-specific STS markers and their association with segregation distortion in rice (Oryza sativa L.) J Crop Sci Biotechnol. 2007;10(3):175–184. [Google Scholar]
- Chung SI, Kim TH, Rico CW, Kang MY. Effect of instant cooked giant embryonic rice on body fat weight and plasma lipid profile in high fat-fed mice. Nutrients. 2014;6(6):2266. doi: 10.3390/nu6062266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SI, Lo LMP, Nam SJ, Jin X, Kang MY. Antioxidant capacity of giant embryo rice Seonong 17 and Keunnunjami. J Adv Agric Technol. 2016;3:94–98. [Google Scholar]
- Chung SI, Rico CW, Lee SC, Kang MY. Hypolipidemic and body fat–lowering effects of giant embryo brown rice (Seonong 17 and Keunnunjami) in high-fat-fed mice. Cereal Chemistry J. 2014;91(1):18–22. doi: 10.1094/CCHEM-04-13-0062-R. [DOI] [Google Scholar]
- Fan JL, Quan S, Orth T, Awai C, Chory J, Hu JP. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiology. 2005;139(1):231–239. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MA, McCouch SR, Hall RD. Not just a grain of rice: the quest for quality. Trends Plant Sci. 2009;14(3):133–139. doi: 10.1016/j.tplants.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Gopala Krishna AG, Prabhakar JV, Sen DP. Effect of degree of milling on tocopherol content of rice bran. J Food Sci Technol. 1984;21(4):222–224. [Google Scholar]
- Han SJ, Kwon SW, Chu S-H, Ryu SN. A new rice variety ‘Keunnunjami’, with high concentrations of Cyanidin 3-glucoside and Giant embryo. Korean J Breed Sci. 2012;44(2):185–189. [Google Scholar]
- Hardtke CS, Okamoto H, Stoop-Myer C, Deng XW. Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8) Plant J. 2002;30(4):385–394. doi: 10.1046/j.1365-313X.2002.01298.x. [DOI] [PubMed] [Google Scholar]
- Hong H-C, Kim Y-G, Choi Y-H, Yang S-J, Lee K-S, Lee J-H, Jung O-Y, Yang C-I, Cho Y-C, Choi I-S, Baek M-K, Kim M-K, Yea J-D, Hwang H-G, Roh J-H, Kim S-L, Choi H-C, Lee Y-T, Lee S-H. A Medium-Maturing, Giant-embryo, and Germination Brown Rice Cultivar ‘Keunnun’. Korean J Breed Sci. 2012;44(2):160–164. [Google Scholar]
- Hong S-K, Aoki T, Kitano H, Satoh H, Nagato Y. Phenotypic diversity of 188 rice embryo mutants. Dev Genet. 1995;16(4):298–310. doi: 10.1002/dvg.1020160403. [DOI] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng TL, Shih YJ, Ho PT, Lai CC, Lin YW, Wang CS, Sung JM. γ-Oryzanol, tocol and mineral compositions in different grain fractions of giant embryo rice mutants. J Sci Food Agric. 2012;92(7):1468–1474. doi: 10.1002/jsfa.4728. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Taramino G, Itoh J-I, Allen J, Sato Y, Hong S-K, Yule R, Nagasawa N, Kojima M, Kusaba M, Sakakibara H, Sakai H, Nagato Y. PLASTOCHRON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. Plant J. 2009;58(6):1028–1040. doi: 10.1111/j.1365-313X.2009.03841.x. [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TH. Manufacturing method of aseptic packing cooked Rice using specialty Rice with embryo buds. KR patent. 2013. pp. 10–1545056. [Google Scholar]
- Kim JY, Seo WD, Park DS, Jang KC, Choi KJ, Kim SY, Oh SH, Ra JE, Yi G, Park SK, Hwang UH, Song YC, Park BR, Park MJ, Kang HW, Nam MH, Han SI. Comparative studies on major nutritional components of black waxy rice with giant embryos and its rice bran. Food Sci Biotechnol. 2013;22(1):121–128. doi: 10.1007/s10068-013-0057-1. [DOI] [Google Scholar]
- Kim KH, Heu MH, Park SZ, Koh H-J. New mutants for endosperm and embryo characters in Rice. Korean J Crop Sci. 1991;36(3):197–203. [Google Scholar]
- Kim KH, Park SZ, Koh HJ, Heu MH (1992) New mutants for endosperm and embryo characters in rice: two dull endosperms and a giant embryo. Proc SABRAO Int Symp ‘Impact of Biological Research on Agricultural Productivity:125–131
- Koh H, Won Y, Heu M, Park S (1993) Nutritional and agronomic characteristics of super-giant embryo mutant in rice. Korean J Crop Sci 38(6):537–544
- Koh HJ, Heu MH, McCouch SR. Molecular mapping of the ge (s) gene controlling the super-giant embryo character in rice (Oryza sativa L.) Theor Appl Genet. 1996;93(1–2):257–261. doi: 10.1007/BF00225754. [DOI] [PubMed] [Google Scholar]
- Liu K, Wang L, Xu Y, Chen N, Ma Q, Li F, Chong K. Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta. 2007;226(4):1007–1016. doi: 10.1007/s00425-007-0548-5. [DOI] [PubMed] [Google Scholar]
- Ma K, Xiao J, Li X, Zhang Q, Lian X. Sequence and expression analysis of the C3HC4-type RING finger gene family in rice. Gene. 2009;444(1–2):33–45. doi: 10.1016/j.gene.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Maeda H, Nemoto H, Iida S, Ishii T, Nakagawa N, Hoshino T, Sakai M, Okamoto M, Shinoda H, Yoshida T. A New Rice Variety with Giant Embryos, “Haiminori&rdquo. Breed Sci. 2001;51(3):211–213. doi: 10.1270/jsbbs.51.211. [DOI] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked Segregant analysis - a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Hibara K, Heppard EP, Vander Velden KA, Luck S, Beatty M, Nagato Y, Sakai H. GIANT EMBRYO encodes CYP78A13, required for proper size balance between embryo and endosperm in rice. Plant J. 2013;75(4):592–605. doi: 10.1111/tpj.12223. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Aichi I, Matsuoka M. A protocol for agrobacterium-mediated transformation in rice. Nat Protocols. 2007;1(6):2796–2802. doi: 10.1038/nprot.2006.469. [DOI] [PubMed] [Google Scholar]
- Pepper AE, Chory J. Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics. 1997;145(4):1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata M, Seno M, Matsusaka H, Takahashi K, Nakamura Y, Yamagata Y, Angeles ER, Mochizuki T, Kumamaru T, Sato M, Enomoto A, Tashiro K, Kuhara S, Satoh H, Yoshimura A. Development and evaluation of rice giant embryo mutants for high oil content originated from a high-yielding cultivar ‘Mizuhochikara’. Breed Sci. 2016;66(3):425–433. doi: 10.1270/jsbbs.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Iwata N. Linkage analysis in rice. On three mutant loci for endosperm properties, ge (giant embryo), du-4 (dull endosperm-4) and flo-1 (floury endosperm-1) Japan J Breed. 1990;40(Suppl 2):268–269. [Google Scholar]
- Satoh H, Omura T. New endosperm mutations induced by chemical mutagens in Rice Oryza sativa L. Japanese J Breed. 1981;31(3):316–326. doi: 10.1270/jsbbs1951.31.316. [DOI] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci U S A. 2003;100(16):9626–9631. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Ohtani T, Iida S, Sunohara Y, Matsushita K, Maeda H, Tanetani Y, Kawai K, Kawamukai M, Kadowaki K-i. Development of CoQ10-enriched rice from giant embryo lines. Breed Sci. 2009;59(3):321–326. doi: 10.1270/jsbbs.59.321. [DOI] [Google Scholar]
- Tanaka K, Ogawa M, Kasai Z. The Rice scutellum. II. A comparison of Scutellar and Aleurone Electron-dense particles by transmission Electron microscopy including energy-dispersive X-ray analysis. Cereal Chemistry. 1977;54(3):684–689. [Google Scholar]
- Taramino G, Allen J, Hong S-K, Nagasawa N, Nagato Y, Sakai H. Mapping of GOLIATH, a new gene controlling embryo size in rice. Rice Genet Newsl. 2003;20:24–26. [Google Scholar]
- Tsuge T, Inagaki N, Yoshizumi T, Shimada H, Kawamoto T, Matsuki R, Yamamoto N, Matsui M. Phytochrome-mediated control of COP1 gene expression in rice plants. Mol Gen Genomics. 2001;265(1):43–50. doi: 10.1007/s004380000396. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J Biol Chem (26) 1993;268:19626–19631. [PubMed] [Google Scholar]
- Wang G, Wang H, Zhu J, Zhang J, Zhang X, Wang F, Tang Y, Mei B, Xu Z, Song R. An expression analysis of 57 transcription factors derived from ESTs of developing seeds in maize (Zea mays) Plant Cell Rep. 2010;29(6):545–559. doi: 10.1007/s00299-010-0843-7. [DOI] [PubMed] [Google Scholar]
- Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, Ronald PC, Song WY. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell. 2006;18(12):3635–3646. doi: 10.1105/tpc.106.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Gao M, Yin X, Liu J, Xu Y, Zeng L, Li Q, Zhang S, Wang J, Zhang X, He Z. Control of rice embryo development, shoot apical meristem maintenance, and grain yield by a novel cytochrome p450. Mol Plant. 2013;6(6):1945–1960. doi: 10.1093/mp/sst107. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hu P, Tang S, Zhao H, Wu D. Comparative studies on major nutritional components of rice with a giant embryo and a Normal embryo. J Food Biochem. 2005;29(6):653–661. doi: 10.1111/j.1745-4514.2005.00039.x. [DOI] [Google Scholar]
- Zhang P, Allen WB, Nagasawa N, Ching AS, Heppard EP, Li H, Hao X, Li X, Yang X, Yan J, Nagato Y, Sakai H, Shen B, Li J. A transposable element insertion within ZmGE2 gene is associated with increase in embryo to endosperm ratio in maize. Theor Appl Genet. 2012;125(7):1463–1471. doi: 10.1007/s00122-012-1926-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. 2007;19(6):1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Agronomic traits of HC three giant embryo mutants. DTH, days to heading; CL, culm length; PL, panicle length; PN, panicle number; SPP, spikelet per panicle; SF, spikelet fertility; GL, grain length; GW, grain width; LWR, length-width ratio. The means followed by lowercase letters are significantly different (p < 0.05, ANOVA) according to Duncan test. (XLSX 10 KB)
Figure S1. Types of segregated F2 seed derived from cross-combinations of le/ge (a) and le/ges (b). (PDF 96 KB)
Figure S2. Multiple sequence alignment of LE orthologs. Asterisk represents the location of amino acid substitution caused by one base deletion in le mutant. Substituted amino acid residues by frameshift are highlighted by red background shading. Predicted transmembrane motifs and the C3HC4-type RING finger domain are underlined with blue and red lines, respectively. Conserved residues in the C3HC4-type RING finger domain are enclosed within black open boxes. Os, Oryza sativa; Ta, Triticum aestivum; Hv, Hordeum vulgare; Zm, Zea mays; At, Arabidopsis thaliana; Br, Brassica rapa; Gm, Glycine max. (PDF 531 KB)
Figure S3. Co-segregation test on F2 individuals and various rice genotypes. a Co-segregation test on F2 individuals derived from the cross between HC/le mutant. b Co-segregation test on other varieties and O.nivara. 1, Hwacheong; 2, Hwaseonchal, 3. Ilpum, 4. Unkwang, 5. Hapcheon, 6. Nipponbare, 7. Dasan, 8. Hangangchal 1ho, 9. IR36, 10. IR64, 11. IR56, 12. IR21015, 13. O.nivara. (PDF 23 KB)
Information of the primers used in this study. (XLSX 11 KB)
Information of LE orthologs used in multiple sequence alignment. (XLSX 9 KB)
Data Availability Statement
All data supporting the conclusions of this article are provided within the article and its additional files (Additional file 3: Table S1, Additional file 4: Figure S1, Additional file 5: Figure S2, Additional file 6: Figure S3, Additional file 1: Table S2. Additional file 2: Table S3, respectively).