Abstract
A novel alcohol dehydrogenase from Bartonella apis (BaADH) was heterologous expressed in Escherichia coli. Its biochemical properties were investigated and used to catalyze the synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate ((S)-CHBE), which is a chiral intermediate of the cholesterol-lowering drug atorvastatin. The purified recombinant BaADH displayed 182.4 U/mg of the specific activity using ethyl 4-chloroacetoacetate as substrate under the conditions of 50 °C in pH 7.0 Tris–HCl buffer. It was stable in storage buffers of pH 7 to 9 and retains up to 96.7% of the initial activity after 24 h. The Km and Vmax values of BaADH were 0.11 mM and 190.4 μmol min−1 mg−1, respectively. Synthesis of (S)-CHBE catalyzed by BaADH was performed with a cofactor regeneration system using a glucose dehydrogenase, and a conversion of 94.9% can be achieved after 1 h reaction. Homology modeling and substrate docking revealed that a typical catalytic triad is in contact with local water molecules to form a catalytic system. The results indicated this ADH could contribute to the further enzymatic synthesis of (S)-CHBE.
Keywords: Alcohol dehydrogenase, Expression, Characterization, Cofactor regeneration, Ethyl (S)-4-chloro-3-hydroxybutyrate
Introduction
Alcohol dehydrogenase (ADH) is an important biocatalyst for the manufacture of high optical purity chiral alcohols. It is classified as short chain dehydrogenase/reductase (SDR) family [1], which can catalyze the reduction of various carbonyl compounds to the corresponding chiral hydroxy derivatives. Among these chiral products, the high optical purity chiral alcohol ethyl (S)-4-chloro-3-hydroxybutyrate ((S)-CHBE) is an important intermediate for various chiral drugs including cholesterol-lowering drugs, antidepressants and anticancer drugs [2].
Therefore, an economical and efficient method for synthesizing high purity (S)-CHBE has great significance. A chemical method using a ruthenium catalyst for producing (S)-CHBE by asymmetric synthesis under high pressure has been developed, but the expensive catalyst and lower optical purity (92%) of product limit its industrial application. Compared with chemical production, biocatalysis has the advantages of high selectivity, mild reaction environment and simple operating conditions which made enzymatic production of CHBE become an important environmentally sustainable alternative [3]. The preparation of (S)-CHBE through reduction of relatively inexpensive ethyl 4-chloroacetoacetate (COBE) catalyzed by ADH or carbonyl reductase is the most promising method in enzymatic synthesis [4]. Due to the generally high optical purity (more than 99%) of the product, ADH has received much attention in the past few years [5]. To date, few ADHs with high enantioselectivities for highly efficient conversion of COBE have been discovered, while their low enzymatic activity is an obstacle to the industrial applications [6, 7].
To economically produce these chiral alcohols, the cofactors must be recycled because of their expense [8, 9]. The construction of a coenzyme regeneration system by coupling between enzymes is a potential cost-saving method [10–12]. Although there are several reports regarding enzymatic synthesis of (S)-CHBE, NADPH was employed as the coenzyme [13], which is more expensive but less stable than NADH [14]. To reduce the cost, reductase could be engineered to exchange the coenzyme preference from NADP+ to NAD+ [15]. Therefore, highly active NADH-dependence alcohol dehydrogenases play essential role in the industrial production of chiral alcohols.
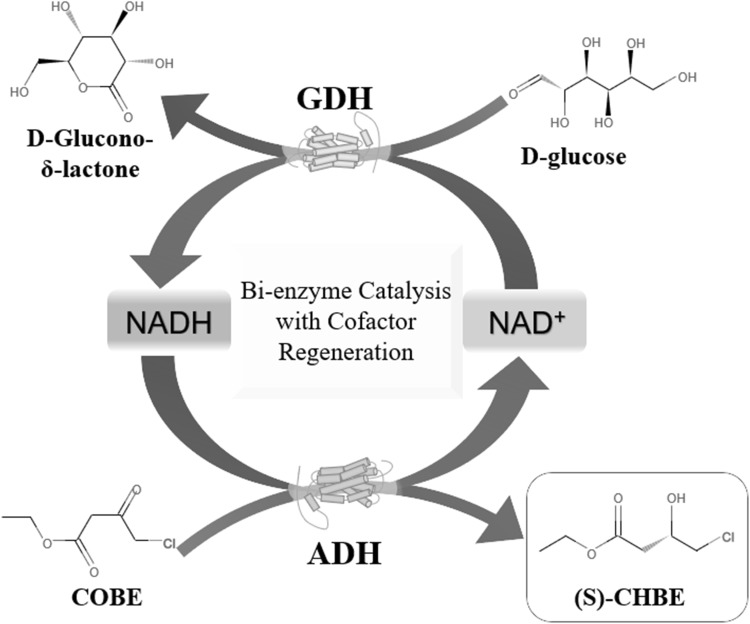
In the present work, an ADH gene from Bartonella apis was cloned and purified after heterologous expression in Escherichia coli. The enzyme was characterized and explored the possibility to produce (S)-CHBE with a cofactor regeneration system using glucose dehydrogenase to enzymatically regenerate cofactor NADH (Scheme 1).
Scheme 1.
Enzymatic synthesis of (S)-CHBE using BaADH and BsGDH for cofactor regeneration
Materials and Methods
Materials
Bartonella apis (DSM: 29779) was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunsweig, Germany). Restriction enzymes, DNA polymerase and primers were obtained from Sangon Biotech Co., Ltd. (Shanghai, China). Genomic DNA extraction kits, E. coli DH5α and BL21 (DE3) were purchased from Beyotime Bio (Haimen, China). T4 DNA ligase and pGEM-T Easy vector were obtained from Promega Biotech Co., Ltd (Beijing, China). The pET-28a(+) expression vector and the nickel–nitrilotriacetic acid (Ni–NTA) super-flow column were supplied by Qiagen (Hilden, Germany). All cofactors and other chemicals, unless otherwise indicated, were obtained from Sigma-Aldrich (St. Louis, MO, USA).
BaADH Gene Cloning
Primers were designed according to DNA sequences of the BaADH gene (GenBank: AQT47207) from B. apis: upstream, 5′-CGCCATATGGCACGTGTAGCAGGTAAGGTTGCAATT-3′, and downstream, 5′-CCGCTCGAGTTATTGTGCTGTGTAGCCACC ATCAAT-3′ (NdeI and XhoI sites are marked with the underlines respectively). The BaADH gene was amplified from the B. apis genome by polymerase chain reaction (PCR) using primers and dNTPs. The PCR product with the NdeI and XhoI sites brought by the upstream and downstream primers was ligated into the pGEM-T easy vector by DNA ligase. And the recombinant plasmid was transformed into E. coli DH5α. The resulted plasmid was submitted for sequencing by Sangon Biotech Co., Ltd. to eliminate the possibility of gene mutation. Then the pGEM-T plasmid was digested with NdeI and XhoI, and the released ADH gene was subcloned into the vector pET-28a(+). The harvested recombinant plasmid was transformed into E. coli BL21 (DE3) to complete the construction of the expression host.
Expression and Purification of BaADH
The recombinant E. coli cells harboring BaADH gene were cultured in LB medium with 50 mg l−1 kanamycin (37 °C, 200 rpm) and the induction of protein expression started by the addition of 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG) when the OD600 reached to 0.8. After 6 h of induction at 15 °C, the cells were collected through centrifugation at 6000×g for 5 min and resuspended in binding buffer (0.1 M Tris–HCl, 0.1 M NaCl, pH 7.0). Lysozyme was added to the suspension to 1 mg ml−1 and incubated on ice for 30 min, then the cells were disrupted by sonication for 5 min. The cell debris suspending in the lysate was separated by centrifugation at 10,000×g for 15 min and then the purification was implemented. The Ni–NTA ultra-flow column equilibrated with binding buffer was used to bind the enzyme protein in the supernatant, then the unbound protein was washed away by washing buffer, and finally the elution buffer was used to release BaADH protein from the column. The protein was quantified with Bradford method [16] and the purified enzyme protein was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
Enzyme Assay
The enzyme solution (containing 5 μg of purified BaADH) was added to a 1 cm3 quartz cuvette containing a premix (0.2 M pH 7.0 Tris-HCl, 50 mM COBE, 10 mM NADH) at 25 °C. The change in absorbance at 340 nm of the reaction mixture caused by the decrease in NADH concentration can be detected by an ultraviolet–visible spectrophotometer. One unit of BaADH was defined as the amount of enzyme oxidizing 1 μmol of NADH per minute under assay conditions mentioned above.
Effects of Temperature and pH on the BaADH Activity
The best catalytic temperature of BaADH was determined at the ranging of temperature from 25 to 55 °C. To determine the activity of BaADH at different temperatures, the pre-mixed reaction solution was heated to a certain temperature and the enzyme solution was added to initiate the reaction. The favorite buffer and the top suitable pH of BaADH were evaluated in different pH solutions: 0.2 M citrate buffers (pH 4.0–6.0), phosphate buffers (pH 6.0–7.0), Tris-HCl buffer (pH 7.0–8.5) and glycine-NaOH buffers (pH 8.5–10.0).
Effects of Metal Ions and Surfactants on the BaADH Activity
Metal ions including Mg2+, Ca2+, Li+, Zn2+, Mn2+, Ni2+, Mo2+ and ethylenediaminetetraacetic acid (EDTA) were added to the pH 7.0 Tris-HCl buffer and brought to a concentration of 5 mM. Tween 80 or Triton X-100 was dissolved in buffer to form 5% (v/v) surfactant solutions. These reaction mixtures containing metal ions and surfactants were used to determine the activity of BaADH under the standard conditions. Reactions without metal ions or surfactant were used as the control.
Effects of Substrate Concentration on the BaADH Activity
The BaADH activity was measured to assess the effect of different COBE concentrations. The concentration of the coenzyme NADH was set to 0.2 mM. The apparent Michaelis–Menten constant (Km) and maximum reaction rate (Vmax) values of BaADH were evaluated at 25 °C and pH 7.0 with the COBE concentrations ranging from 0.03 mM to 3 mM and calculated win nonlinear fitting.
Thermal Stability and pH Stability of BaADH
To investigate the thermal stability of BaADH, the enzyme solution was incubated in a water bath from the temperature 25 to 45 °C for different time ranging from 5 min to 8 h, then cooled on ice and measured in form of reactive activity at enzyme assay conditions.
To evaluate the pH stability of BaADH, the enzyme solution was diluted in different pH buffers. The initial specific activity of BaADH in each sample was set as 100%, and the activity after storage for 24 h was measured at 4 °C. The remained activity of BaADH was calculated as an index for pH stability.
Homology Modeling and Docking of the BaADH
A similar homologous sequence was searched for as a template from the BaADH sequence using BLAST. ESPript-3.0 and Clustal-W were used to align different DNA sequences. The alcohol dehydrogenase crystal structure from Lactobacillus kefir (PDB: 1JVZ) was chosen as the template. The structure of COBE was downloaded from Zinc and the crystal structure of NADH was downloaded from PDB. The substrate is docked with the active sites of coenzyme and ADH, and the interaction between local water molecules and active sites is performed by AutoDock4.
Bioconversion of COBE to (S)-CHBE with BaADH
BaADH and a glucose dehydrogenase from Bacillus sp. (BsGDH) were coupled to construct an enzyme coupling system to achieve continuous regeneration of NADH. (S)-CHBE was synthesized in this optimized NADH regeneration system constructed by pH 7.0 Tris-HCl buffer (0.2 M), NADH (0.1 mM), NAD+ (0.1 mM), glucose (100 mM), COBE (100 mM) and BaADH/BsGDH mixture (50 μg ml−1). To maximize the catalytic activity of the BaADH and BsGDH, the ratio of the two enzymes was optimized from 1:4 to 4:1. To improve the solubility of COBE, 10% (v/v) choline chloride–glycerol (ChCl–Gly) was added as an ionic solvent. The reaction temperature was 35 °C with a continuous rotating at 30 rpm and the sample was withdrawn after reaction for 8 h. The glucose conversion was measured by the cysteine-sulfuric acid-carbazole method [17] and the COBE conversion was calculated accordingly.
Results and Discussion
Identification of the BaADH Gene and its Heterologous Expression
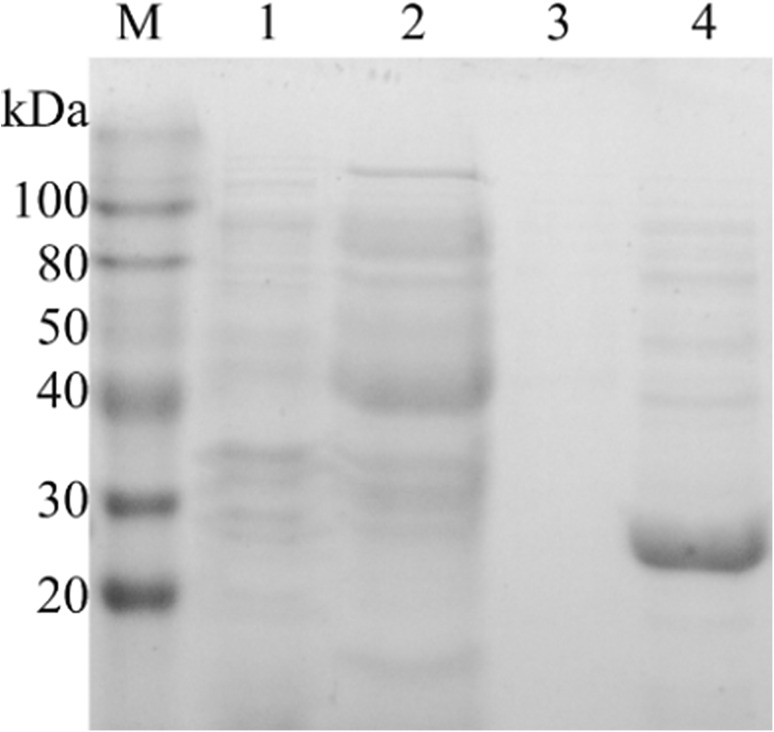
Gene sequencing showed the BaADH gene encoding 254 amino acids was 762 bp. The encoded peptide chain had a calculated molecular weight of 27.19 kDa. This amino acid sequence was compared to other reported enzymes by multiple sequence alignment, showing different degrees of identity with other functional enzymes. The Blast results found it is classified as one of short-chain dehydrogenases. Compared with other reductases from Candida parapsilosis [18], Kefir lactic [19], Lactobacillus reuteri [20], the cloned ADH was found to have a 47, 45 and 44% identity respectively, and had a relatively lower identity (24%) with carbonyl reductase from Kluyveromyces [21]. Alignment of amino acid sequences revealed that BaADH was a member of short-chain dehydrogenase family. Although the amino acid sequence similarity in this enzyme family was not very high, they all showed a conserved catalytic site (Tyr154). After induction with IPTG for 6 h, the BaADH protein was successfully expressed in E. coli. The crude and purified enzymes were analyzed by SDS-PAGE. The over-expressed BaADH was clearly displayed in lane 4 (Fig. 1). The band was estimated 27 kDa, which was consistent with the predicted molecular weight.
Fig. 1.
SDS-PAGE analysis of expressed BaADH (lane M: protein marker, lane 1: precipitation, lane 2: flow-through, lane 3: elute with 50 mM imidazole elution buffer, lane 4: elute with 250 mM imidazole elution buffer)
Characterization of BaADH
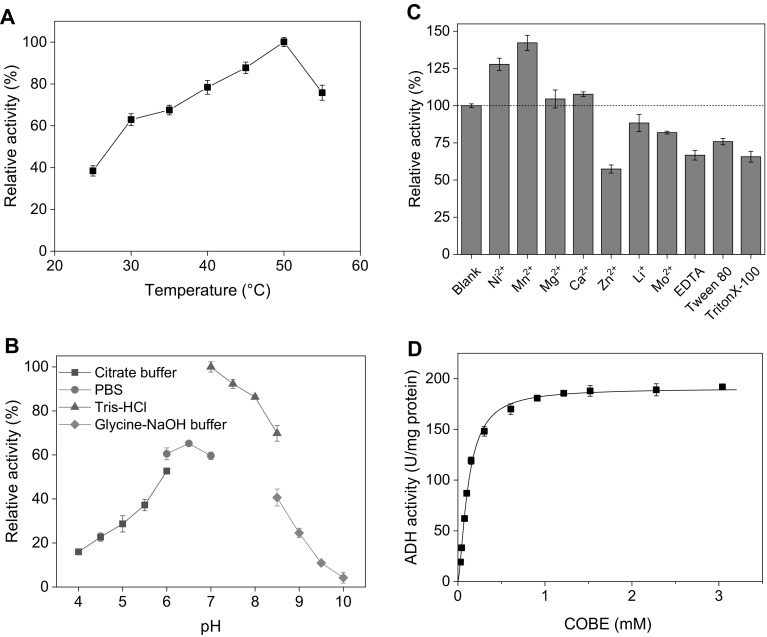
The effect on the BaADH activity of temperature was shown in Fig. 2a in the range of 25–55 °C. The maximal activity was observed at 50 °C and BaADH could maintain relative activity above 60% over a wide temperature range (30–55 °C). When the temperature was raised to 55 °C, the enzyme activity was around 76% of the initial activity. The possible reason for the decreased relative activity is that the spatial structure change of the enzyme at high temperature [22]. Several short-chain dehydrogenases including carbonyl reductase and alcohol dehydrogenase could catalyze conversion of COBE to (S)-CHBE. Exception of the carbonyl reductase from Pichia stipitis showed the highest activity at 30 °C [23], most of the enzymes have an optimal temperature between 45 and 50 °C, which is the same as BaADH.
Fig. 2.
Effect of a temperature, B pH, c metal ions and surfactant and d substrate concentration on the activity of BaADH
Figure 2b showed the effect of pH on the BaADH activity. The BaADH showed the highest catalytic activity at pH 7.0, and maintained over 60% of relative activity in the range of pH 6 to 8. In order to use these enzymes highly efficient, cofactor regeneration system should be coupled [24, 25]. Generally, many NADH-dependent dehydrogenases, including glucose dehydrogenase, has an optimal pH between 7 and 8 [26, 27]. Compared to other short-chain dehydrogenases, the optimum pH of BaADH is the closest to the optimum pH of BsGDH (pH 8.0), which is beneficial to the regeneration of the cofactor NADH [28].
Effects of metal ions, EDTA and surfactants on the BaADH activity were explored and are shown in Fig. 2c. The blank group with no metal ions or surfactants was set as 100%. It can be found that most metal ions shown in the figure had little influence on the BaADH activity, while Ni2+ and Mn2+ increased the activities of BaADH by 32.1% and 48.0%, respectively. The activation of some dehydrogenases by Mn2+ and Ni2+ has been reported in some studies. The addition of Ni2+ restored 65% of activity of glucose dehydrogenase inhibited by EDTA, while Mn2+ increased the activity of dehydrogenase from Klebsiella pneumonia by 4.6 times [29]. However, the activity of BaADH was reduced to 56.1% in the presence of Zn2+. In addition, EDTA and Tween 80 reduced the activities of BaADH by 39% and 24%, respectively.
To check the effect of COBE concentration on BaADH activity, different substrate concentrations were set up from 30 μM to 3 mM (Fig. 2d). The Michaelis constant (Km) and maximum reaction rate (Vmax) of BaADH were calculated to be 0.11 mM and 190.4 μmol min−1 mg−1 with non-linear regression, respectively. It can be found that the Vmax of the purified enzyme in this study is close to the Vmax of the carbonyl reductase from P. stipitis, and exceeds the other enzymes. Compared with the other ADH, BaADH has the smallest Km, which means BaADH has the highest catalytic efficiency and binding specificity to COBE.
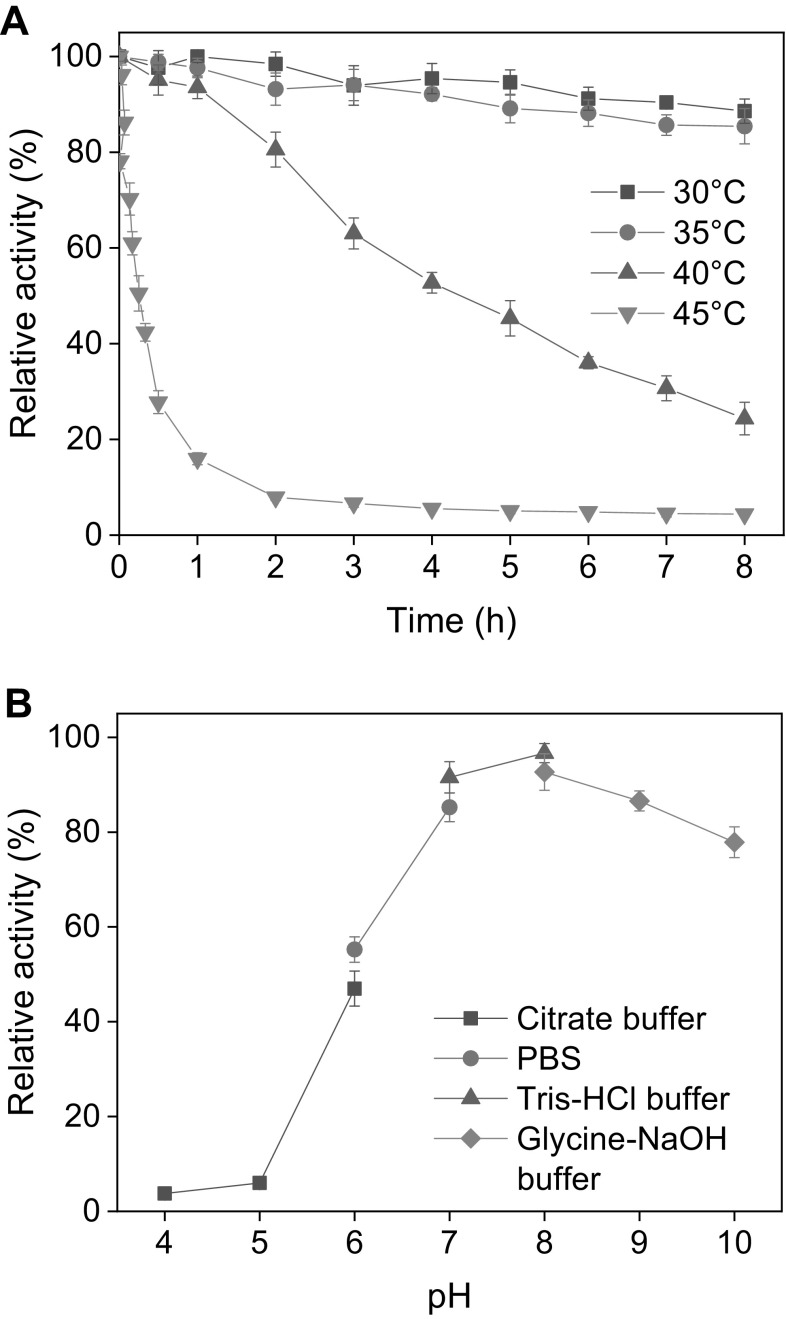
Thermal Stability and pH Stability
The thermal stability of BaADH was studied with the half-lives at different temperatures from 30 to 45 °C (Fig. 3a). BaADH retained more than 85% of initial activity at 30 and 35 °C during the 8-hour incubation period, and the half-lives at 30 and 35 °C were determined as 35 h and 24.9 h, respectively. It has been reported that most carbonyl reductases exhibit significant instability when incubated at 40 °C or above. It has been reported that most carbonyl reductases exhibit significant instability when incubated at temperatures of 40 °C or above. However, the alcohol dehydrogenase is relatively stable at 40 °C and the half-life of BaADH is determined to be 4.9 h, about 2.2 times that of carbonyl reductase from Candida parapsilosis [2].
Fig. 3.
The thermal stability (a) and pH stability (b) of BaADH
The pH stability of BaADH was measured with the residual activity after storing at 4 °C for 24 h in buffers of pH 4 to 10 (Fig. 3b). The enzyme was found to be stable in pH 8.0 Tris-HCl buffer and retained 96.7% initial activity. A similar optimal preservation pH was found in ADH from Yeast (pH 7.0) [30]. However, the difference was that BaADH still showed good stability in alkaline buffer with pH greater than 8, and maintained more than 75% of initial activity in the pH range of 8–10, while the ADH from Yeast was left with only 5% initial activity after storage in pH 8 buffer.
Homology Modeling and Docking of BaADH
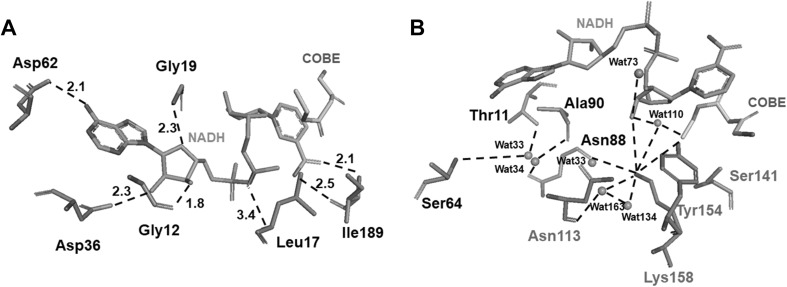
Homology modeling of BaADH was performed by finding the most suitable template. As shown in the Fig. 4a, Asp62 contact with the adenine group of NADH (2.1 Å), Asp36, Gly19 and Gly12 bind the ribose moiety with 2.3, 2.3 and 1.8 Å hydrogen bonds respectively. In addition, the Leu17 approaches to the phosphate group with a hydrogen bond with a length of 3.4 Å which was close to the adenine group of NADH. The Ile189 contacts with the nicotinamide with 2.5 Å and 2.1 Å hydrogen bonds. In Fig. 4b, Asn113, Lys158, Tyr154 and Ser141 contact with the local water molecules (Wat33, Wat163, Wat134 and Wat110) and form a catalytic system. The Lys158/Tyr154 couple was relatively close to Ser141 to form a typical catalytic triad. Both the triad and substrate (COBE) are in close hydrogen bonding distance to the Wat 110.
Fig. 4.
Homology modeling and molecular docking of BaADH. The mode of binding of the catalytic group to COBE and NADH (a) and local water molecules (b) in the putative catalytic site of BaADH. Hydrogen bonds are shown as black dashed lines and distances are indicated by Å
The interactions between cofactor and enzyme suggested that the NADH could be stabilized by several residues and the substrate (COBE). In the catalytic triad which contains Tyr154/Lys158 couple and Ser141 residue, the Tyr154 residue was the catalytic residue and is positioned near the substrate COBE [31]. The Ser141 has a hydroxyl side chain which is critical to fix the substrate and could tune the pKa value of the catalytic residue (Tyr154) to match that of substrate. These three residues contact with the water molecules closely, and the proton could be effectively shared. In addition, coupled with the Wat163 and Wat134, a “proton relay system” composed of a catalytic Tyr154 and the Asn113 residue was formed. This system could allow the rapid transfer of proton from solvent to the active site. This could be the reason for high catalytic activity of BaADH.
(S)-CHBE Synthesis using ADH in a Cofactor Regeneration System
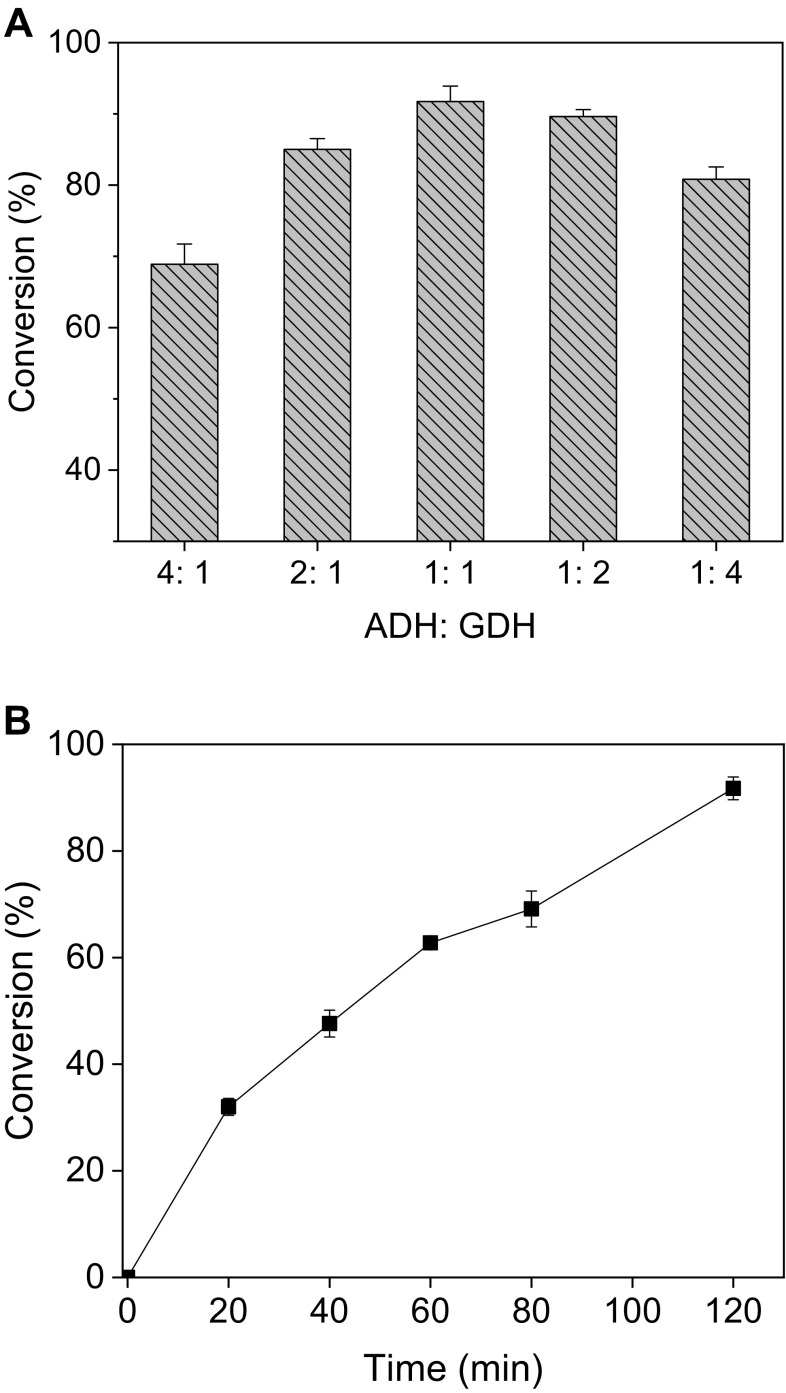
To explore the possible industrial applications of BaADH, it was used to synthesize (S)-CHBE with a coenzyme NADH regeneration system. In this system, ADH reduces COBE to (S)-CHBE while converting NADH to the oxidized coenzyme NAD+, while the glucose dehydrogenase is responsible for reducing NAD+ to NADH and consuming glucose to accumulate glucono-δ-lactone. To increase the conversion rate and maximize the enzyme utilization, the two enzymes of different mass ratios were coupled and the conversion of COBE was shown in Fig. 5a. When the ratio of the two enzymes was set to 1:1 (Fig. 5b), around 94.9% of the substrate was converted to S-CHBE within 2 h. When two different enzymes are catalyzed under the same reaction conditions, the ratio of enzyme addition may have a non-negligible effect on the conversion rate, and the optimization of enzyme ratio helps to increase the conversion rate and maximize the enzyme utilization [32, 33]. For example, in the NAD+ regeneration system comprising glycerol dehydrogenase, when the enzyme ratio was adjusted from 1:1 to 1:2, the yield of dihydroxyacetone was increased by 8% [26].
Fig. 5.
Effect of BaADH: BsGDH ratio on the enzymatic synthesis of (S)-CHBE (a) and the time profile of CHBE production at the ratio of 1:1 (BaADH: BsGDH) (b)
Conclusions
A novel gene encoding ADH was successfully cloned and expressed. The purified BaADH showed relatively high enzyme activity and good storage stability in a wide pH range. The high specific activity of BaADH to COBE indicates its possible application for production of (S)-CHBE. The efficiency of producing (S)-CHBE was demonstrated by coupling BaADH with a highly active glucose dehydrogenase to establish a coenzyme regeneration system.
Acknowledgements
This work was supported by Brain Pool Grant (NRF-2018H1D3A2001746) by National Research Foundation of Korea (NRF) to work at Konkuk University. This paper was written as part of Konkuk University’s research support program for its faculty on sabbatical leave in 2018.
Compliance with Ethical Standards
Conflict of interest
The authors have no financial conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vipin C. Kalia, Email: vckaliaku@gmail.com
Jung-Kul Lee, Email: jkrhee@konkuk.ac.kr.
Ye-Wang Zhang, Email: zhangyewang@ujs.edu.cn.
References
- 1.Kallberg Y, Oppermann U, Jörnvall H, Persson B. Short-chain dehydrogenase/reductase (SDR) relationships: a large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 2010;11:636–641. doi: 10.1110/ps.26902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You ZY, Liu ZQ, Zheng YG. Characterization of a newly synthesized carbonyl reductase and construction of a biocatalytic process for the synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate with high space-time yield. Appl Microbiol Biotechnol. 2013;98:1671–1680. doi: 10.1007/s00253-013-5042-3. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Pai CC, Kwok WH, Guo RW, Au-Yeung TTL, Yeung CH, Chan ASC. Studies on the rhodium- and ruthenium-catalyzed asymmetric hydrogenation of α-dehydroamino acids using a family of chiral dipyridylphosphine ligand (P-Phos) Tetrahedron Asymmetry. 2003;14:987–992. doi: 10.1016/S0957-4166(03)00098-3. [DOI] [Google Scholar]
- 4.An M, Cai P, Yan M, Hao N, Wang S, Liu H, Li Y, Xu L. A novel reductase from Candida albicans for the production of ethyl (S)-4-chloro-3-hydroxybutanoate. Biosci Biotechnol Biochem. 2012;76:1210–1212. doi: 10.1271/bbb.120048. [DOI] [PubMed] [Google Scholar]
- 5.Inoue K, Makino Y, Dairi T, Itoh M. Gene cloning and expression of Leifsonia alcohol dehydrogenase (LSADH) involved in asymmetric hydrogen-transfer bioreduction to produce (R)-form chiral alcohols. J Agric Chem Soc Jpn. 2006;70:418–426. doi: 10.1271/bbb.70.418. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Wang H, Chen L, Wu K, Xie J, Wei D. Efficient production of ethyl (R)-4-chloro-3-hydroxybutanoate by a novel alcohol dehydrogenase from Lactobacillus curieae S1L19. J Mol Catal B Enzym. 2016;134:51–60. doi: 10.1016/j.molcatb.2016.09.010. [DOI] [Google Scholar]
- 7.Kumar P, Patel SKS, Lee JK, Kalia VC. Extending the limits of Bacillus for novel biotechnological applications. Biotechnol Adv. 2013;31:1543–1561. doi: 10.1016/j.biotechadv.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Liu CH, Li XQ, Jiang XP, Zhuang MY, Zhang JX, Bao CH, Zhang YW. Preparation of functionalized graphene oxide nanocomposites for covalent immobilization of NADH oxidase. Nanosci Nanotechnol Lett. 2016;8:164–167. doi: 10.1166/nnl.2016.2102. [DOI] [Google Scholar]
- 9.Kumar P, Ray S, Patel SKS, Lee JK, Kalia VC. Bioconversion of crude glycerol to polyhydroxyalkanoate by Bacillus thuringiensis under non-limiting nitrogen conditions. Int J Biol Macromol. 2015;78:9–16. doi: 10.1016/j.ijbiomac.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, Kim IW, Choi JH, Khera E, Wen F, Lee JK. Repeated production of L-xylulose by an immobilized whole-cell biocatalyst harboring L-arabinitol dehydrogenase coupled with an NAD(+) regeneration system. Biochem Eng J. 2015;96:23–28. doi: 10.1016/j.bej.2014.12.017. [DOI] [Google Scholar]
- 11.Moon HJ, Tiwari MK, Singh R, Kang YC, Lee JK. Molecular determinants of the cofactor specificity of ribitol dehydrogenase, a short-chain dehydrogenase/reductase. Appl Environ Microbiol. 2012;78:3079–3086. doi: 10.1128/AEM.07751-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao H, Khera E, Lee JK, Wen F. Immobilization of multi-biocatalysts in alginate beads for cofactor regeneration and improved reusability. JoVE J Vis Exp. 2016;110:e53944. doi: 10.3791/53944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray S, Kalia VC. Microbial cometabolism and polyhydroxyalkanoate co-polymers. Indian J Microbiol. 2017;57:39–47. doi: 10.1007/s12088-016-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Q, Ouyang PK, Ying HJ. A review—biosynthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate ester: recent advances and future perspectives. Appl Microbiol Biotechnol. 2011;89:513–522. doi: 10.1007/s00253-010-2942-3. [DOI] [PubMed] [Google Scholar]
- 15.Morikawa S, Nakai T, Yasohara Y, Nanba H, Kizaki N, Hasegawa J. Highly active mutants of carbonyl reductase S1 with inverted coenzyme specificity and production of optically active alcohols. Biosci Biotechnol Biochem. 2005;69:544–552. doi: 10.1271/bbb.69.544. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhang YW, Jeya M, Lee JK. L-Ribulose production by an Escherichia coli harboring l-arabinose isomerase from Bacillus licheniformis. Appl Microbiol Biotechnol. 2010;87:1993–1999. doi: 10.1007/s00253-010-2600-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu ZQ, Ye JJ, Shen ZY, Hong HB, Yan JB, Lin Y, Chen ZX, Zheng YG, Shen YC. Upscale production of ethyl (S)-4-chloro-3-hydroxybutanoate by using carbonyl reductase coupled with glucose dehydrogenase in aqueous-organic solvent system. Appl Microbiol Biotechnol. 2015;99:2119–2129. doi: 10.1007/s00253-014-6245-y. [DOI] [PubMed] [Google Scholar]
- 19.Xu Z, Jing K, Liu Y, Cen P. High-level expression of recombinant glucose dehydrogenase and its application in NADPH regeneration. J Ind Microbiol Biotechnol. 2007;34:83–90. doi: 10.1007/s10295-006-0168-2. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y, Hou C, Lan M, Ming Y, Yan W, Qinting H, Jian L, Lin X, Yong C, Jian X. Biosynthesis of (S)-4-chloro-3-hydroxybutanoate ethyl using Escherichia coli co-expressing a novel NADH-dependent carbonyl reductase and a glucose dehydrogenase. Bioresour Technol. 2010;101:8911–8914. doi: 10.1016/j.biortech.2010.01.063. [DOI] [PubMed] [Google Scholar]
- 21.Hiroaki Y, Kazuya M, Norihiro K, Akinobu M, Nobuyoshi E, Yoshinori K. A novel NADH-dependent carbonyl reductase from Kluyveromyces aestuarii and comparison of NADH-regeneration system for the synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate. J Agric Chem Soc Jpn. 2004;68:638–649. doi: 10.1271/bbb.68.638. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Wang AR, Jiang XP, Zhang JX, Zhang YW. Preparation of expoxy-functionalized magnetic nanoparticles for immobilization of glycerol dehydrogenase. J Nanosci Nanotechnol. 2018;18:4852–4857. doi: 10.1166/jnn.2018.15266. [DOI] [PubMed] [Google Scholar]
- 23.Cao H, Mi L, Ye Q. Purification and characterization of a novel NADH-dependent carbonyl reductase from Pichia stipitis involved in biosynthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate. Bioresour Technol. 2011;102:1733–1739. doi: 10.1016/j.biortech.2010.08.072. [DOI] [PubMed] [Google Scholar]
- 24.Nie Y, Xu Y, Hu QS, Xiao R. A new strategy to improve the efficiency and sustainability of Candida parapsilosis catalyzing deracemization of (R, S)-1-phenyl-1,2-ethanediol under non-growing conditions: increase of NADPH availability. J Microbiol Biotechnol. 2009;19:65–71. [PubMed] [Google Scholar]
- 25.Xu MQ, Wang SS, Li LN, Gao J, Zhang YW, Xu MQ, Wang SS, Li LN, Gao J, Zhang YW. Combined cross-linked enzyme aggregates as biocatalysts. Catalysts. 2018;8:460. doi: 10.3390/catal8100460. [DOI] [Google Scholar]
- 26.Kalia VC, Lal S, Rashmi, Chauhan A, Bhattacharyya G. In Silico reconstitution of novel routes for microbial plastic. In: Kalia VC, editor. Microbial factories: biodiversity, biopolymers, bioactive molecules. New Delhi: Springer; 2015. pp. 299–315. [Google Scholar]
- 27.Zhuang MY, Jiang XP, Ling XM, Xu MQ, Zhu YH, Zhang YW. Immobilization of glycerol dehydrogenase and NADH oxidase for enzymatic synthesis of 1,3-dihydroxyacetone with in situ cofactor regeneration. J Chem Technol Biotechnol. 2018;93:2351–2358. doi: 10.1002/jctb.5579. [DOI] [Google Scholar]
- 28.Jiang XP, Lu TT, Liu CH, Ling XM, Zhuang MY, Zhang JX, Zhang YW. Immobilization of dehydrogenase onto epoxy-functionalized nanoparticles for synthesis of (R)-mandelic acid. Int J Biol Macromol. 2016;88:9–17. doi: 10.1016/j.ijbiomac.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 29.Li FL, Shi Y, Zhang JX, Gao J, Zhang YW. Cloning, expression, characterization and homology modeling of a novel water-forming NADH oxidase from Streptococcus mutans ATCC 25175. Int J Biol Macromol. 2018;113:1073–1079. doi: 10.1016/j.ijbiomac.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Kim TS, Patel SKS, Selvaraj C, Jung WS, Pan CH, Kang YC, Lee JK. A highly efficient sorbitol dehydrogenase from Gluconobacter oxydans G624 and improvement of its stability through immobilization. Sci Rep. 2016;6:33438. doi: 10.1038/srep33438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 32.Ray S, Kalia VC. Biological significance of degradation of polyhydroxyalkanoates. In: Kalia VC, Kumar P, editors. Microbial applications Vol.1: bioremediation and bioenergy. Cham: Springer; 2017. pp. 125–139. [Google Scholar]
- 33.Ray S, Sharma R, Kalia VC. Co-utilization of crude glycerol and biowastes for producing polyhydroxyalkanoates. Indian J Microbiol. 2018;58:33–38. doi: 10.1007/s12088-017-0702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]