Abstract
Environmental heterogeneity can drive patterns of functional trait variation and lead to the formation of locally adapted ecotypes. Plant ecotypes are often differentiated by suites of correlated root and shoot traits that share common genetic, developmental, and physiological relationships. For instance, although plant water loss is largely governed by shoot systems, root systems determine water access and constrain shoot water status. To evaluate the genetic basis of root and shoot trait divergence, we developed a recombinant inbred population derived from mesic and xeric ecotypes of the perennial grass Panicum hallii. Our study sheds light on the genetic architecture underlying the relationships between root and shoot traits. We identified several genomic “hotspots” which control suites of correlated root and shoot traits, thus indicating genetic coordination between plant organ systems in the process of ecotypic divergence. Genomic regions of colocalized quantitative trait locus (QTL) for the majority of shoot and root growth related traits were independent of colocalized QTL for shoot and root resource acquisition traits. The allelic effects of individual QTL underscore ecological specialization for drought adaptation between ecotypes and reveal possible hybrid breakdown through epistatic interactions. These results have implications for understanding the factors constraining or facilitating local adaptation in plants.
Keywords: adaptation, ecotype, genetic architecture, quantitative trait locus, root architecture, pleiotropy, epistasis, recombinant inbred line
Introduction
Adaptations to abiotic stress have been implicated as driving factors in ecological speciation (Stebbins, 1952; Lexer and Fay, 2005), where populations have diverged across a number of traits, exhibit different niche characteristics, and eventually become reproductively isolated (Clausen, 1951; Lowry, 2012; Yardeni et al., 2016). Local adaptation to soil water availability is an especially important driver of plant evolution (Stebbins, 1952; Rajakaruna, 2004; Kooyers et al., 2015) and can impose strong natural selection on populations, leading to the formation of ecotypes that are differentially adapted to xeric and mesic habitats (Joly et al., 1989; Kumar et al., 2008). Xeric and mesic ecotypes are often characterized by the divergence of common suites of morphological and phenological traits (Clausen, 1951; Lowry, 2012) related to maintaining water status and tolerating drought (Chapin et al., 1993; Markesteijn and Poorter, 2009; Juenger, 2013).
While leaf and shoot traits are important drivers of adaptation to drought (Carmo-Silva et al., 2009; Juenger, 2013), the properties of root systems determine plant water access and can place constraints on shoot water status (Price et al., 2002; Hund et al., 2009). Shoot traits may be related to root traits through genetic correlation (Bouteillé et al., 2012) or be dependent upon root traits through resource allocation tradeoffs (Hammer et al., 2009), including changes in carbon allocation between root and shoot systems (Hummel et al., 2010). Higher root mass ratio (RMR) increases water foraging capability to maintain plant water status, which can be accomplished by allocating more resources toward roots (Knight et al., 2006) or by inhibiting above ground growth (Hendriks et al., 2015). Specific leaf area (SLA, the ratio of leaf area to leaf dry mass) and specific root length (SRL, the ratio of root length to root dry mass) are both important plant traits linked to resource acquisition (Reich, 2014; Cheng et al., 2016) and SRL is typically thought of as the below ground analog of SLA (Eissenstat et al., 2000; Reich, 2014). These traits are often positively correlated (Withington et al., 2006; Reich, 2014; Valverde-Barrantes et al., 2017) and associated with rapid growth (Pérez-Harguindeguy et al., 2013; Reich, 2014) – where an acquisitive root strategy (high SRL) can be aided by an acquisitive leaf strategy (high SLA; Pérez-Ramos et al., 2013). Despite evidence that root and shoot trait covariance is an important driver of plant adaptation, few studies have documented how combinations of specific shoot and root traits generate locally adapted ecotypes. The genetic basis of such trait complexes and the implications of recombining adaptive shoot and root traits in hybrids are poorly understood.
Quantitative genetic analyses and the mapping of quantitative trait loci (QTL) permit exploration of the genetic basis of trait correlations and trait divergence (Fishman et al., 2002; Lovell et al., 2015; Milano et al., 2016). Importantly, by simultaneously analyzing multiple traits, QTL mapping can infer the loci and genetic interactions that drive ecological trait correlations. Functional traits with a high degree of correlation that underlie divergence can result from pleiotropy through shared developmental genetics or genetic linkage (Via and Hawthorne, 2005; Lovell et al., 2013) as a result of correlational selection (Brodie et al., 1995). For example, colocalized QTL for root and shoot traits including root biomass, root volume, shoot biomass and plant height have been identified in a wheat recombinant inbred line (RIL) population (Iannucci et al., 2017) likely resulting from pleiotropy or tightly physically linked genes. Overall, there is growing evidence for substantial genetic variation in root system architecture and root/shoot relationships. However, the loci driving these trait correlations and the degree to which these patterns impact plant productivity are largely unknown.
Panicum hallii is a small, self-fertilizing, C4 perennial bunch grass native to North America that occurs across a large geographical range comprised of diverse habitats and climates. Average annual precipitation ranges from 127 cm per year on the eastern border of its distribution to 13 cm per year on the west. P. hallii occurs as two distinct ecotypes (xeric upland and mesic lowland) that are classified as separate varieties, P. hallii var. hallii (hereafter referred to as hallii) and P. hallii var. filipes (hereafter referred to as filipes). Hallii is typically found in xeric upland habitats with shallow, dry, calcareous and rocky soils in the American southwest and northern Mexico; while filipes occurs in mesic lowland areas on clay and silt soils mostly along the Gulf Coast Plain of Texas and Mexico (Gould, 1975; Waller, 1976). The xeric upland ecotype, hallii, is smaller in stature and overall size than the mesic lowland ecotype filipes: with smaller leaves, fewer tillers, earlier flowering time, fewer flowers per inflorescence, but larger seed size and seed mass (Waller, 1976; Lowry et al., 2013). This is consistent with its polyploid relative, Panicum virgatum (an important biofuel candidate), where upland ecotypes are typically smaller, flower earlier (Lowry et al., 2014a) and have less leaf area (McMillan, 1965) than lowland ecotypes. Previous analyses of shoot traits in a F2 population of P. hallii (Lowry et al., 2014b) demonstrated that a few large-effect loci drove multivariate shoot trait divergence between hallii and filipes, and complete genomes has been assembled and compared (Lovell et al., 2018). Here, we investigate the genetic architecture of multidimensional root phenotypic traits and their relationship with shoots to develop a more complete picture of the adaptive differences between these ecotypes.
In this study, we cross xeric and mesic ecotypes of P. hallii, to generate a population of RIL at the F7 generation and subsequently constructed a new genetic map based on whole genome re-sequencing. We utilized extensive phenotyping of root and shoot traits and a quantitative genetic approach to identify the genetic architecture of trait relationships and their divergence among ecotypes. We discovered shared QTL clusters involved in genetic correlations between root and shoot growth related traits that were independent of QTL clusters for carbon allocation and phenology related traits. The allelic effects of individual QTL underscore ecological specialization for drought adaptation between hallii and filipes and reveal possible hybrid breakdown through epistatic interactions.
Materials and Methods
Morphological Shoot and Root Phenotyping Under Greenhouse Conditions
We developed a population of RILs derived from a cross of hallii and filipes and constructed a genetic map from whole genome re-sequencing (see Supplementary Appendix 1). Raw sequence data was deposited in the NCBI short read archive (see Supplementary Table 5). Seedlings of 174 F7 RILs and the two parental genotypes were planted to 6 × 30 cm Cone-Tainers (Stuewe and Sons, Tangent, OR, United States) filled with Field and Fairway Profile (The Turf Trade, NJ, United States) media. Plants were grown in a completely randomized block design within three blocks on a single bench at the University of Texas greenhouse (see Supplementary Appendix 2). Plants were harvested within three days of a common developmental stage defined as when a fully expanded flag leaf with a visible ligule was observable on any tiller with an emerging panicle. Harvest dates across the population ranged from 27 to 51 days after germination. The tiller height, leaf length and area of the flag leaf of the main tiller were measured and tiller number was counted at the time of harvest. Total root number was counted and then the root system was spread out in a clear acrylic water filled tray and scanned at a 600 dpi resolution using an EPSON Scanner (Model 12000XL, Epson America, Inc., San Jose, CA, United States) calibrated for use with WinRhizo Pro 2015 root image analysis software (Regent Instruments Inc., Canada). Leaf, shoot and root tissue was dried and weighed to obtain biomass. SLA was calculated for each plant as the ratio of leaf area to dry mass (Supplementary Appendix 2).
Root trait data was obtained from scans using WinRhizo Pro 2015 software and included total root length (cm), total root volume (cm3), and average root diameter (mm). Specific root length (SRL; cm g-1), root tissue density (RTD, g cm-3), and RMR were calculated for each plant (Supplementary Appendix 2).
Data and QTL Analysis
Data analyses centered on fitting linear mixed models and considered RIL genotype as a fixed effect (proc mixed, SAS) for the measured phenotypic traits. Block was also included as a fixed effect covariate when it had a significant impact on measured traits (emergence day, specific root length and root diameter). The SAS procedure PROC CORR was used to calculate genetic correlation coefficients of traits based on RIL line means. Broad-sense trait heritability was calculated using h2boot software using one-way ANOVA among inbred RILs with 1000 bootstrap runs (Phillips and Arnold, 1999). Trait divergence between parental lines was evaluated with a t-test in SAS.
The majority of the measured traits were continuously distributed with relatively strong multivariate structure based on pairwise correlational analyses. As such, we also used genetic principal component analysis (PCA) to obtain a multidimensional overview of shoot and root trait variation and integration. PCA was performed on the trait means of each line for the following phenotypic variables: emergence day, tiller number, root number, root biomass, shoot biomass, root diameter, root tissue density, specific root length, specific leaf area, tiller height, leaf length, root volume and total root length. PCA was completed using SAS with the proc princomp function. The first three principal components that together explained 75% of total variation were retained for QTL analysis.
Quantitative trait locus mapping was completed in R using the R/qtl package (Broman and Sen, 2009) on the RIL breeding values as described above (Supplementary Table 1). When quantitative trait data distributions were not normally distributed, data was log (emergence day, tiller number) or square root (shoot biomass) transformed. Two functions were used to determine the position of QTL and to conduct the calculation of estimates for additive effects and epistasis (an additive-by-additive interaction between quantitative trait loci) (script1). The scantwo function with 1000 permutations was used to calculate penalties for main effect and interactions for each phenotypic trait, and the stepwise QTL function was used to conduct a forward-backward search and account for epistasis with a maximum of 6 QTL (at least two QTL peaks in addition to those detected with the scanone function) that optimized the penalized LOD score criterion. Threshold values for type 1 error rates were set at alpha = 0.05 for all traits based on permutation. 1.5 LOD drop intervals of QTL were calculated using the qtlStats function (Lovell, 2018). In addition, QTL analysis was performed on the first three principal components following the above procedure.
Confirming Root and Shoot Biomass QTL in a Field Study
To further confirm and evaluate major QTL detected in our greenhouse study, we conducted a follow up field experiment on a focal QTL during the 2016 growing season. Ten RILs homozygous at the shared QTL region for root and shoot biomass were selected for this experiment (5 with filipes alleles and 5 with hallii alleles). Eight biological replicates of each selected RIL line and eight replicates of the two parental genotypes were planted on May 10, 2016 under both restrictive and well-watered irrigation treatments [(10 RILs + 2 parents) × 8 biological replicates × 2 irrigation levels = 192 plants; see Supplementary Appendix 2]. Plants were harvested toward the end of the summer growing season in August. Shoots were separated from roots, dried at 55°C for 4 days before weighing for biomass. Trait values more extreme than 1.5 × the interquartile range were removed as outliers prior to analysis. For statistical analysis, we used linear mixed models with proc mixed in SAS. The main effect for the model was genotype at the focal QTL (filipes or hallii alleles at the marker position), treatment and genotype-by-treatment interaction. RIL line was used as a random effect to control for background genetic variance.
Results
Heritable Shoot and Root Trait Differences Between Mesic and Xeric Ecotypes
The RIL parents representing mesic and xeric ecotypes of Panicum hallii (HAL2 and FIL2) had significantly different shoot and root trait mean values (Table 1). The xeric genotype, HAL2, had 2.3-fold earlier first panicle emergence (t values at 5 dfs and P values; t = 2.87, P = 0.035), 3.3-fold less shoot biomass (t = 4.39, P = 0.007) and 2.8-fold less root biomass (t = 3.08, P = 0.028), 1.8-fold shorter plant height (t = 3.43, P = 0.018), 2.2-fold shorter leaf length (t = 6.3, P = 0.001), 2-fold shorter total root length (t = 3.29, P = 0.022), 2.5-fold lower total root volume (t = 3.41, P = 0.02), and 1.3-fold increased specific root length (t = -2.5, P = 0.05) relative to the mesic genotype FIL2 (Table 1).
Table 1.
FIL2 and HAL2 root and shoot trait value means with SE and t-statistics; and RIL root and shoot trait value means, range, and broad-sense heritability (H2) with SE.
| Phenotypic Trait | FIL2 | HAL2 | t | P-value | RIL mean | RIL range | H2±SE |
|---|---|---|---|---|---|---|---|
| Panicle Emergence (day) | 9.25 1.19 | 4.00 1.38 | 2.87 | 0.035 | 7.01 1.74 | 1.00–18.33 | 0.51 0.05 |
| Shoot Biomass (g) | 4.74 0.49 | 1.41 0.57 | 4.39 | 0.007 | 1.65 0.33 | 0.29–4.74 | 0.59 0.05 |
| Tiller Number (count) | 6.25 0.48 | 5.00 0.56 | 1.68 | 0.150 | 6.00 0.83 | 3.00–14.50 | 0.50 0.05 |
| SLA (cm2g-1) | 325.62 18.15 | 382.77 20.96 | –2.06 | 0.094 | 381.58 33.17 | 264.67–499.36 | 0.18 0.08 |
| Plant Height (cm) | 21.18 1.82 | 11.63 2.11 | 3.43 | 0.018 | 12.57 1.56 | 4.30–23.65 | 0.63 0.04 |
| Leaf Length (cm) | 30.77 1.72 | 14.23 1.98 | 6.30 | 0.001 | 15.66 1.46 | 4.75–24.27 | 0.66 0.04 |
| Root Biomass (g) | 1.38 0.18 | 0.51 0.21 | 3.08 | 0.028 | 0.54 0.10 | 0.12–1.60 | 0.58 0.06 |
| Root Number (count) | 14.00 0.97 | 8.33 1.11 | 3.84 | 0.012 | 8.87 1.39 | 2.50–15.00 | 0.38 0.05 |
| SRL (cm g-1) | 10.14 0.85 | 13.37 0.98 | –2.50 | 0.055 | 12.27 1.11 | 6.12–17.95 | 0.43 0.06 |
| RTD (g cm-3) | 0.06 0.01 | 0.05 0.01 | 1.31 | 0.247 | 0.05 0.01 | 0.03–0.08 | 0.39 0.07 |
| Root Diameter (mm) | 0.46 0.01 | 0.44 0.02 | 1.27 | 0.259 | 0.45 0.01 | 0.37–0.55 | 0.37 0.05 |
| Root Volume (cm3) | 2.43 0.28 | 0.98 0.32 | 3.41 | 0.019 | 1.00 0.17 | 0.26–2.90 | 0.56 0.05 |
| Root Length (m) | 1.37 0.14 | 0.67 0.16 | 3.29 | 0.022 | 0.65 0.11 | 0.12–1.64 | 0.59 0.04 |
| RMR (ratio) | 0.22 0.01 | 0.27 0.01 | –3.44 | 0.018 | 0.25 0.02 | 0.16–0.39 | 0.34 0.09 |
t-statistics given at 5 degrees of freedom with statistically significant P-values indicated in bold text.
We estimated broad-sense trait heritability (H2) as the proportion of observed phenotypic variance due to genetic differences among RILs in the population. In the RIL population, all measured traits were heritable, with H2 ranging from 18 to 66% for shoot traits and from 34 to 60% for root traits (bootstrap based significance, in all cases P < 0.001). The most heritable traits were leaf length (66%), plant height (64%), shoot biomass (60%), root length (60%) and root biomass (58%; Table 1). Transgressive segregation, where the range of recombinant phenotypes extends beyond the range of parental values (Rieseberg et al., 1999), was found among the majority of traits except shoot biomass, plant height, leaf length, root biomass and root number, where FIL2 had trait values that were the highest or close to the highest of population wide values, while HAL2 values were generally in the middle of the population trait distribution (Table 1).
Many shoot and root phenotypic traits showed remarkably strong genetic correlations in the RIL population (Supplementary Table 2). For example, shoot and root biomass (r = 0.92, P < 0.0001), tiller and root number (r = 0.67, P < 0.001), shoot biomass and root volume (r = 0.91, P < 0.0001), and shoot biomass and total root length (r = 0.90, P < 0.001) were all positively genetically correlated. We performed PCA to characterize the multivariate structure of our data. The first three PCA axes explained 75% of the overall trait variance. Principal component one (PC1; 45.5% variance explained) was composed of general plant size traits (shoot biomass, root biomass, number of tillers, number of roots, tiller height, leaf length, root volume and root length). Principal component two (PC2; 16.5%) was mainly composed of root resource acquisition traits (SRL, root diameter and root tissue density). Principal component three (PC3; 12.6%) was composed of carbon acquisition and allocation traits (SLA, RMR and panicle emergence; Supplementary Table 3 and Supplementary Figure 1).
QTL Underscore Root and Shoot Trait Divergence Between Hallii and Filipes
Given high H2 values, it is not surprising that QTL were detected for all measured traits. A total of 32 QTL were identified for 14 phenotypic traits: two QTL for one phenological trait, 14 QTL for five shoot traits and 16 QTL for eight root traits (Table 2, Figure 1, and Supplementary Figure 2). QTL for all traits showed additive effects in the direction of parental divergence, except for one of three QTL for tiller number, one of four QTL for root diameter, and one of three QTL for SRL. Filipes alleles had later panicle emergence and increased trait values for plant size related traits, including: emergence day, root number, root tissue density, root biomass, shoot biomass, tiller height, leaf length and root volume. Hallii alleles increased trait values associated with water acquisition (SRL) and carbon acquisition and allocation (RMR, SLA).
Table 2.
Main and epistatic effects of QTL for the Panicum hallii RIL population.
| Phenotype | Chr | Peak (cM) | 1.5 Lod Interval | LOD | % var | Effect | SE | Positive allele donor | QTL Cluster (CL) |
|---|---|---|---|---|---|---|---|---|---|
| Panicle Emergence | 5 | 52.1 | 40–59 | 4.59 | 9.85 | –0.044 | 0.009 | filipes | CL5.1 |
| (day) | 7 | 80.0 | 31–83 | 4.31 | 9.2 | –0.039 | 0.008 | filipes | CL7.2 |
| Shoot Biomass | 5 | 58.6 | 56–60 | 7.43 | 14.8 | –0.044 | 0.007 | filipes | CL5.1 |
| (g) | 5 | 136.0 | 128–142 | 5.08 | 9.82 | –0.031 | 0.007 | filipes | CL5.3 |
| 9 | 66.1 | 60–71 | 4.78 | 9.19 | –0.027 | 0.005 | filipes | CL9.1 | |
| Epi5:5 | 2.86 | 5.36 | 0.027 | 0.007 | |||||
| Tiller Number | 3 | 40.5 | 38–48 | 7.23 | 14.74 | –0.054 | 0.009 | filipes | |
| (count) | 5 | 137.0 | 128–142 | 3.47 | 6.73 | –0.037 | 0.009 | filipes | CL5.3 |
| 7 | 73.6 | 46–81 | 4.84 | 9.56 | 0.039 | 0.008 | hallii | CL7.2 | |
| SLA | 5 | 13.3 | 0–26 | 3.15 | 5.25 | 9.772 | 2.543 | hallii | |
| (cm2g-1) | 7 | 66.0 | 60–74 | 8.56 | 15.37 | 16.394 | 2.494 | hallii | CL7.2 |
| 8 | 19.8 | 16–23 | 8.33 | 14.90 | 16.077 | 2.484 | hallii | ||
| Tiller Height | 5 | 76.0 | 74–77 | 6.16 | 13.34 | –1.765 | 0.320 | filipes | CL5.2 |
| (cm) | 6 | 83.9 | 69–88 | 3.82 | 8.05 | –1.096 | 0.256 | filipes | |
| Leaf Length | 2 | 89.7 | 76–96 | 4.28 | 8.56 | –1.19 | 0.264 | filipes | |
| (cm) | 7 | 43.6 | 35–64 | 4.39 | 8.80 | –1.293 | 0.283 | filipes | CL7.1 |
| 9 | 63.4 | 59–75 | 3.41 | 6.76 | –0.985 | 0.246 | filipes | CL9.1 | |
| Root Biomass | 5 | 58.6 | 56–60 | 8.81 | 18.61 | –0.012 | 0.002 | filipes | CL5.1 |
| (g) | 5 | 136.0 | 135–142 | 8 | 16.71 | –0.010 | 0.002 | filipes | CL5.3 |
| Epi5:5 | 4.61 | 9.21 | 0.008 | 0.002 | |||||
| Root Number | 3 | 88.0 | 69–104 | 6.18 | 13.9 | –1.08 | 0.199 | filipes | |
| (count) | 5 | 125.7 | 125–130 | 5.36 | 11.94 | –0.81 | 0.196 | filipes | CL5.3 |
| Epi3:5 | 2.79 | 5.99 | 0.73 | 0.202 | |||||
| SRL (cm g-1) | 1 | 91.5 | 82–94 | 5.3 | 11.02 | 0.66 | 0.131 | hallii | CL1.1 |
| 3 | 18.8 | 17–36 | 5.16 | 10.7 | 0.78 | 0.156 | hallii | CL3.1 | |
| 7 | 44.7 | 34–49 | 3.16 | 6.4 | –0.55 | 0.145 | filipes | CL7.1 | |
| RTD (g cm-3) | 1 | 6.3 | 0–20 | 3.15 | 7.9 | –0.001 | 0.0004 | filipes | |
| Root Diameter | 1 | 86.0 | 82–94 | 4.73 | 8.68 | –0.009 | 0.002 | filipes | CL1.1 |
| (mm) | 3 | 34.2 | 30–36 | 5.36 | 9.91 | –0.011 | 0.002 | filipes | CL3.1 |
| 5 | 71.9 | 66–75 | 3.78 | 6.84 | 0.010 | 0.002 | hallii | CL5.2 | |
| 8 | 47.9 | 43–52 | 4.65 | 8.50 | –0.009 | 0.002 | filipes | ||
| Root Volume | 5 | 58.6 | 56–63 | 3.96 | 8.85 | –0.134 | 0.030 | filipes | CL5.1 |
| (cm3) | 5 | 117.2 | 109–142 | 3.07 | 6.77 | –0.119 | 0.032 | filipes | CL5.3 |
| Root Length (cm) | 5 | 58.6 | 44–138 | 3.12 | 7.85 | –0.82 | 21.29 | filipes | CL5.1,2,3 |
| RMR (ratio) | 7 | 67.0 | 62–74 | 6.36 | 15.34 | 0.0137 | 0.002 | hallii | CL7.2 |
Chr, chromosome; Peak, cM (centimorgan) position of the QTL peak; LOD, logarithm of odds; % var, percent of variance; SE, one standard error; SLA, specific leaf area; SRL, specific root length; RTD, root tissue density; RMR, root mass ratio; Epi, epistasis.
FIGURE 1.
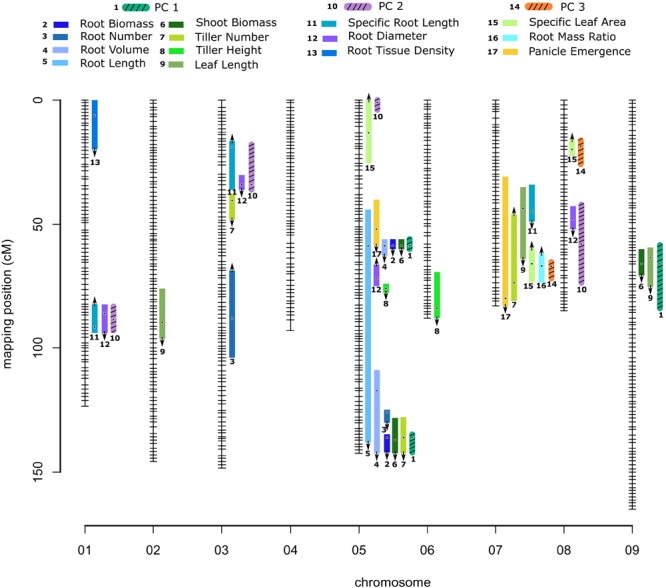
Genetic map of the Panicum hallii RIL population with location of trait QTL. Colored bars indicate 1.5-LOD drop confidence intervals. Location of dots within the bars is the location of QTL peaks. Arrow represents the direction of additive effect, with up or down arrows indicating that the hallii allele increases or decreases the trait value.
The additive effects of each QTL explained from 5.25 to 15.4% of phenotype variation for shoot traits, and from 5.9 to 18.6% for root traits (Table 2). Of these 32 QTL, eight QTL occupied unique positions in the genome: root tissue density on chr1, leaf length on chr2, tiller number on chr3, root number on chr3, SLA on chr5 and chr8, tiller height on chr6, and root diameter on chr8. As expected, three of these single QTL were also identified by principle component QTL (Supplementary Table 4 and Figure 1). The confidence intervals of all other QTL are shared or colocalized with at least one other QTL.
Trait-Specific QTL Cluster Into Genomic “Hotspots”
We identified three major and five minor clusters of root and shoot trait QTL occurring over five different chromosomes (Table 2 and Figure 1). Here we identify QTL clusters (CL) by chromosome and numerical order from the telomere for each chromosome. As expected, we found that positions of QTL for principle components were highly indicative of the locations of QTL clusters for the traits loading on particular PC axes (Supplementary Table 4, Figure 1, and Supplementary Figure 2).
Quantitative trait locus for PC1 localized to three genomic clusters of QTL for plant size traits. CL9.1 contains shoot biomass and leaf length QTL. CL5.1 contains root biomass, shoot biomass, root volume, total root length and panicle emergence QTL. CL5.3 contains root biomass, shoot biomass, root volume, total root length, tiller number and root number QTL. A separate QTL pair for tiller height and root diameter not identified with PC1 lies between these two large clusters. PC2 QTL localized with one of two genomic clusters of QTL for root resource acquisition traits. CL1.1 and 3.1 both contain SRL and root diameter traits. PC3 QTL localized to a single genomic cluster (CL7.2) related to carbon allocation traits. CL7.2 contains panicle emergence day, leaf length, number of tillers, RMR and SLA. Near this PC3 associated QTL is a minor cluster (CL7.1) of leaf length and SRL (Table 2, Supplementary Table 4, and Figure 1).
Four pairwise epistatic interactions, where the effect of one QTL depends on the allelic state of an unlinked QTL, were detected (Table 2, Supplementary Table 4, and Figure 2). Three QTL from cluster CL5.3 (shoot biomass, root biomass and PC1) interacted with other QTL for these traits located in CL5.1. In addition, the root number QTL from CL5.3 interacted with the root number QTL on chr3. Individuals that possess the hallii allele for these QTL at CL5.3 mask the positive effects of their interactive QTL.
FIGURE 2.
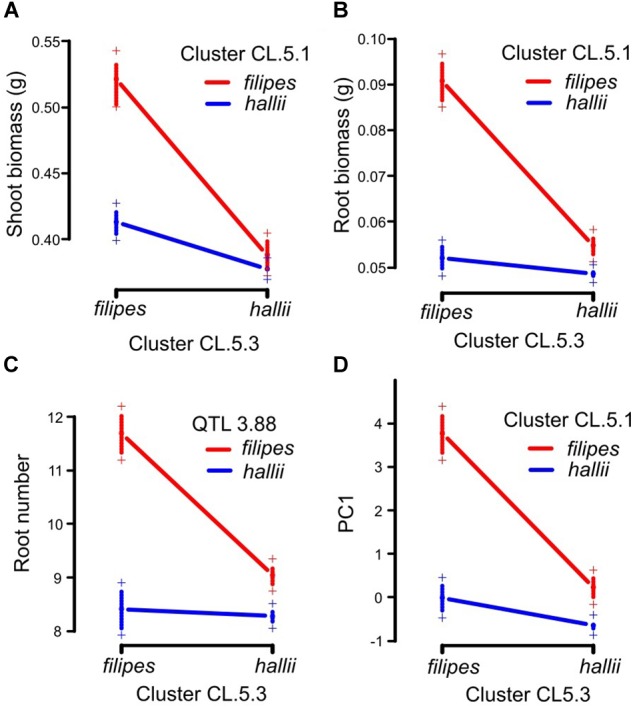
Pairwise epistatic QTL in the P. hallii RIL population. Plotted points indicate two-locus genotype means ± 1SE for the two loci containing root biomass between CL.5.1 and CL.5.3 (A), shoot biomass between CL.5.1 and CL.5.3 (B), root number between QTL 3.88 and CL.5.3 (C) and PC1 between CL.5.1 and CL5.3 (D).
A Major Pleotropic Effect QTL Is Confirmed in the Field
To confirm the effects of QTL observed in a controlled greenhouse study, we phenotyped two sets of RILs homozygous for different parental alleles at the loci for shoot and root biomass (CL5.2) in a field experiment. While the magnitude of increased biomass for lines with filipes alleles at the selected QTL observed in the field is 24% less for the root biomass and 11% less for the shoot biomass relative to the greenhouse, the effects are significant and in the same direction as those observed in the greenhouse. Field grown lines with filipes parental alleles produced 1.9-fold more root biomass (P = 0.0024) and 2.7-fold more shoot biomass (P = 0.0002) relative to field grown lines with hallii parental alleles (Figure 3). In addition, the HAL2 parental line showed a 1.8-fold increase trend in RMR (P = 0.09) over the FIL2 parental line under field conditions compared to the 1.2-fold difference observed in the greenhouse (P = 0.018). There were no significant differences between the irrigation treatments or the interaction of treatment by genotype for RILs or the parental genotypes. However, root biomass showed a 1.2-fold increase trend under the dry treatment relative to the wet treatment (P = 0.08).
FIGURE 3.

Mean ± 1SE of shoot biomass (A) and root biomass (B) for field grown P. hallii RILs homozygous for either filipes or hallii parental alleles at shoot and root biomass QTL located in cluster CL5.1. Picture of field grown RILs homozygous at CL5.1 for filipes allele (top row) and hallii allele (bottom row) (C).
Discussion
Ecotypes are often differentiated by suites of correlated root and shoot traits that may share common genetic and developmental architectures as a result of adaptive differentiation. One of our major findings was several genomic “hotspots” of colocalized QTL for multiple shoot and root traits. This is consistent with a previous study of a P. hallii F2 population covering a suite of ecotype differentiating shoot trait QTL which clustered on chr5 (Lowry et al., 2014b). In addition to confirming this important locus, we discovered additional root traits linked to this region along with additional regions of clustered loci for root and shoot traits. Colocalized QTL controlling traits such as root biomass, shoot biomass, among others, has also been shown in RIL populations of wheat and sorghum (Mace et al., 2012; Iannucci et al., 2017). These findings indicate that specific loci can shape both shoot and root morphological traits, through tight linkage of several genes controlling individual traits or a single pleiotropic gene that controls several traits.
PC1 QTL localized to three genomic regions controlling several size related root and shoot traits (shoot biomass, root biomass, root volume, and other). We found that the hallii allele had additive effects in the direction of ecotype divergence and contributed to smaller root and shoot phenotypes in every case compared to the filipes allele. This finding is consistent with the global pattern observed in angiosperm plants whose shoot and root biomass are positively correlated (Enquist and Niklas, 2002) and with other studies on perennial grasses where total biomass is decreased under water limited conditions (Baruch, 1994; Weißhuhn et al., 2011; Tozer et al., 2017). Importantly, we show that one of the main growth QTL effects is robust to the environment and persists under natural field conditions.
In addition to differences in absolute size, there are expected differences in carbon acquisition and allocation between xeric and mesic ecotypes. PC3 resulted from cluster of carbon allocation and phenology related traits (SLA, RMR, tiller number, and panicle emergence). Plants with hallii alleles had greater SLA, RMR, tiller number, and faster panicle emergence. Thinner leaves (high SLA) have lower carbon cost and are associated with increased photosynthetic capacity (Reich et al., 1997; Cornelissen et al., 2003). Increased RMR helps to maintain plant water status and productivity under drought (Comas et al., 2013). Faster flowering time along with greater tiller number allows for rapid production of seeds when resources are available for short time periods. These factors combined may indicate that hallii employs a fast acquisitive strategy for drought escape; acquiring nutrients rapidly and flowering quickly to enter a dormant state before periods of summer drought. Acquisitive shoot and root strategies have been associated with fast growth strategies and summer dormancy in other perennial grasses (Balachowski et al., 2016). This contrasts with the lower SLA, and RMR of the mesic filipes, which may employ a slow strategy of thicker longer lasting leaves, larger more persistent roots, and abundant above ground foliage. This common genetic control of ecotype differentiating traits involving shoot and root organs suggests that these factors evolved in tandem. Alternatively, we found a relatively weak genetic correlation between SLA and SRL, which are important plant traits linked to resource acquisition (Reich, 2014; Cheng et al., 2016) and associated with fast growth (Pérez-Harguindeguy et al., 2013; Reich, 2014). Each of these traits had three independent QTL. Thus, divergence of these traits is likely due to independent loci which become structured across ecotypes as a result of strong directional or correlational selection. In this case, our crossing scheme was able to largely decouple these traits through recombination.
Observed pairwise epistatic interactions for root biomass, shoot biomass and root number showed that hallii alleles mask the effects of filipes alleles in all cases. When lines are homozygous for hallii parental alleles at CL5.3, it contributes to smaller phenotypes for these traits, regardless of the genotype at their respective interactive QTL. This suggests that the CL5.3 loci could include a pleiotropic gene with major effect that controls the development of multiple shoot and root size related traits. Natural populations of P. hallii ecotypes are largely homozygous, thus these linked QTL likely work together in a positive direction and contribute to the phenotypic trait correlations that underlie ecotype divergence. The observed epistasis in the RIL population could be involved in ecological speciation (Burke and Arnold, 2001), and these interactions in hybrid plants could be deleterious and impact survivorship by undermining synergistic trait relationships. For example, the combination of reduced root and shoot size effected by hallii alleles may be desirable in xeric environments, but deleterious in natural hybrids or under the higher competition mesic environments that filipes inhabits.
Greenhouse Detected Genetic Correlations Confirmed Under Field Conditions
There is persistent concern that effects observed in greenhouse studies are not representative of plant performance in natural or agronomic environments. Although greenhouse and growth chambers may be able to replicate a wide range of temperature and light conditions, other differences between these artificial and natural environments can be significant. Furthermore, greenhouse studies are often conducted on very young plants and in smaller than optimal pots, which can significantly alter root architectures compared to natural environments. Several recent studies have highlighted how differences in conditions between glasshouse and natural settings can affect the mapping of genetic architectures for various plant traits (Poorter et al., 2012; reviewed in Lovell et al., 2016).
We sought to overcome this concern by confirming the glasshouse detected genetic architecture of two of our chief traits of interest (root biomass and shoot biomass) in selected RILs and parental genotypes in a field setting at full plant maturity. In the RILs, we found that our glasshouse observed QTL were confirmed. For the parental lines, we found that RMR differences between the xeric and mesic ecotypes nearly doubled under field conditions as compared to the glasshouse study. This suggests that adaptive allocation of biomass to roots increases with plant age and can also be constrained by pot limitations in the glasshouse. More importantly, these results provide credence to the assumption that our glasshouse study is predictive of plant performance in a natural setting. Future studies with P. hallii should explore the genetic architecture of shoot:root traits over multiple perennial seasons in additional field studies. These data will help to clarify the lifetime fitness consequences of allocation strategies and potential ecological tradeoffs that arise in natural habitats.
Conclusion
In the process of ecotype formation, populations can diverge across many functional traits and exhibit different niche characteristics, which requires coordination between plant organ systems. Root traits are involved in adaptive differentiation to abiotic stresses by their direct effects on water acquisition, and through correlation, tradeoffs or constraints with shoot traits (Hammer et al., 2009; Mace et al., 2012). Our study sheds light on the genetic architecture underlying the relationships between root and shoot traits involved in ecotype divergence of P. hallii and demonstrates that some correlated traits are under common genetic control as a result of QTL colocalization and interaction, while other traits are controlled by independent loci. We found several genomic hotspots relating to multiple root and shoot traits and a striking pattern of epistatic interaction impacting overall plant growth. Further insight into the molecular basis of these loci will be an important step in understanding the genetic coordination and ecological importance of root and shoot systems involved in ecotype divergence.
Data Availability
The raw sequencing data was deposited at NCBI, phenotypic data is included as Supplementary Material.
Author Contributions
AK and TJ designed the experiments. AK and JB conducted the experiments. JJ and JS conducted the Panicum hallii genome assembly. XW, YY, JJ, and JS performed sequencing of the RIL population. JL created the genomic map. AK, JL, JB, and TJ contributed to statistical analysis. AK wrote the first draft of the manuscript, JL, JB, and TJ contributed to revisions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank M. Donahue, J. Shih, L. Mayer, S. Faries, D. Miller, and B. Campitelli for their help phenotyping in the greenhouse and field experiments; and J. Heiling, B. Whitaker, and M. Stuke for help in generating RILs. Seeds for hallii and filipes parental lines were originally provided by the Lady Bird Johnson Wildflower Center and J. L. Reilly, respectively; and the original cross of this material was created by D. Lowry. We also thank B. Campitelli for discussion and comments that aided in the improvement of this manuscript.
Funding. This research was supported by the DOE Office of Science, Office of Biological and Environmental Research (BER), grant no. DE-SC0008451 to TJ. Additional funding came from an NSF Plant Genome Research Program Grant (IOS-0922457) to TJ and an NSF postdoctoral fellowship (IOS-1402393) to JL. The work conducted by the United States Department of Energy Joint Genome Institute is supported by the Office of Science of the United States Department of Energy under Contract No. DE-AC02-05CH11231.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00366/full#supplementary-material
References
- Balachowski J. A., Bristiel P. M., Volaire F. A. (2016). Summer dormancy, drought survival and functional resource acquisition strategies in California perennial grasses. Ann. Bot. 118 357–368. 10.1093/aob/mcw109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch Z. (1994). Responses to drought and flooding in tropical forage grasses. I. Biomass allocation, leaf growth and mineral nutrients. Plant Soil 164 87–96. 10.1007/BF00010114 [DOI] [Google Scholar]
- Bouteillé M., Rolland G., Balsera C., Loudet O., Muller B. (2012). Disentangling the intertwined genetic bases of root and shoot growth in arabidopsis. PLoS One 7:e32319. 10.1371/journal.pone.0032319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie E. D., Moore A. J., Janzen F. J. (1995). Visualizing and quantifying natural selection. Trends Ecol. Evol. 10 313–318. 10.1016/S0169-5347(00)89117-X [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen S. (2009). A Guide to QTL Mapping With R/Qtl. New York, NY: Springer; 10.1007/978-0-387-92125-9 [DOI] [Google Scholar]
- Burke J. M., Arnold M. L. (2001). Genetics and the fitness of hybrids. Annu. Rev. Genet. 35 31–52. 10.1146/annurev.genet.35.102401.085719 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva A. E., Francisco A., Powers S. J., Keys A. J., Ascensão L., Parry M. A. J., et al. (2009). Grasses of different C4 subtypes reveal leaf traits related to drought tolerance in their natural habitats: changes in structure, water potential, and amino acid content. Am. J. Bot. 96 1222–1235. 10.3732/ajb.0800224 [DOI] [PubMed] [Google Scholar]
- Chapin S., Autumn K., Pugntairet F. (1993). Evolution of suites of traits in response to environmental stress. Am. Nat. 142 S78–S92. 10.1086/285524 [DOI] [Google Scholar]
- Cheng J., Chu P., Chen D., Bai Y. (2016). Functional correlations between specific leaf area and specific root length along a regional environmental gradient in Inner Mongolia grasslands. Funct. Ecol. 30 985–997. 10.1111/1365-2435.12569 [DOI] [Google Scholar]
- Clausen J. (1951). Stages in the Evolution of Plant Species. New York, NY: Cornell University Press. [Google Scholar]
- Comas L. H., Becker S. R., Cruz V. M. V., Byrne P. F., Dierig D. A. (2013). Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:442. 10.3389/fpls.2013.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen J. H. C., Lavorel S., Garnier E., Díaz S., Buchmann N., Gurvich D. E., et al. (2003). A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 51:335 10.1071/BT02124 [DOI] [Google Scholar]
- Eissenstat D. M., Wells C. E., Yanai R. D., Whitbeck J. L. (2000). Building roots in a changing environment: implications for root longevity. New Phytol. 147 33–42. 10.1046/j.1469-8137.2000.00686.x [DOI] [Google Scholar]
- Enquist B. J., Niklas K. J. (2002). Global allocation rules for patterns of biomass partitioning in seed plants. Science 295 1517–1520. 10.1126/science.1066360 [DOI] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Willis J. H. (2002). Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56 2138–2155. 10.1111/j.0014-3820.2002.tb00139.x [DOI] [PubMed] [Google Scholar]
- Gould F. W. (1975). The Grasses of Texas. College Station, TX: Texas A&M University Press. [Google Scholar]
- Hammer G. L., Dong Z., McLean G., Doherty A., Messina C., Schussler J., et al. (2009). Can changes in canopy and/or root system architecture explain historical maize yield trends in the U.S. corn belt? Crop Sci. 49 299–312. 10.2135/cropsci2008.03.0152 [DOI] [Google Scholar]
- Hendriks P. W., Kirkegaard J. A., Lilley J. M., Gregory P. J., Rebetzke G. J. (2015). A tillering inhibition gene influences root-shoot carbon partitioning and pattern of water use to improve wheat productivity in rainfed environments. J. Exp. Bot. 67 327–340. 10.1093/jxb/erv457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I., Pantin F., Sulpice R., Piques M., Rolland G., Dauzat M., et al. (2010). Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol. 154 357–372. 10.1104/pp.110.157008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund A., Ruta N., Liedgens M. (2009). Rooting depth and water use efficiency of tropical maize inbred lines, differing in drought tolerance. Plant Soil 318 311–325. 10.1007/s11104-008-9843-6 [DOI] [Google Scholar]
- Iannucci A., Marone D., Russo M. A., Vita P., De Miullo V., Ferragonio P., et al. (2017). Mapping QTL for root and shoot morphological traits in a durum wheat × T. dicoccum Segregating Population at Seedling Stage. Int. J. Genom. 3 1–17. 10.1155/2017/6876393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly R. J., Adams W. T., Stafford S. G. (1989). Phenological and morphological responses of mesic and dry site sources of coastal Douglas-fir to water deficit. For. Sci. 35 987–1005. [Google Scholar]
- Juenger T. E. (2013). Natural variation and genetic constraints on drought tolerance. Curr. Opin. Plant Biol. 16 274–281. 10.1016/j.pbi.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Knight C. A., Vogel H., Kroymann J., Shumate A., Witsenboer H., Mitchell-Olds T. (2006). Expression profiling and local adaptation of Boechera holboellii populations for water use efficiency across a naturally occurring water stress gradient. Mol. Ecol. 15 1229–1237. 10.1111/j.1365-294X.2006.02818.x [DOI] [PubMed] [Google Scholar]
- Kooyers N. J., Greenlee A. B., Colicchio J. M., Oh M., Blackman B. K. (2015). Replicate altitudinal clines reveal that evolutionary flexibility underlies adaptation to drought stress in annual Mimulus guttatus. New Phytol. 206 152–165. 10.1111/nph.13153 [DOI] [PubMed] [Google Scholar]
- Kumar A., Bernier J., Verulkar S., Lafitte H. R., Atlin G. N. (2008). Breeding for drought tolerance: direct selection for yield, response to selection and use of drought-tolerant donors in upland and lowland-adapted populations. Field Crops Res. 107 221–231. 10.1016/j.fcr.2008.02.007 [DOI] [Google Scholar]
- Lexer C., Fay M. F. (2005). Adaptation to environmental stress: a rare or frequent driver of speciation? J. Evol. Biol. 18 893–900. 10.1111/j.1420-9101.2005.00901.x [DOI] [PubMed] [Google Scholar]
- Lovell J. T. (2018). qtlTools: Data Processing AND Plotting in Association with R/qtl. Available at: https://rdrr.io/github/jtlovell/qtlTools/. [Last accessed March 05 2018]. [Google Scholar]
- Lovell J. T., Jenkins J., Lowry D. B., Mamidi S., Sreedasyam A., Weng X., et al. (2018). The genomic landscape of molecular responses to natural drought stress in Panicum hallii. Nat. Commun. 9:5213. 10.1038/s41467-018-07669-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell J. T., Juenger T. E., Michaels S. D., Lasky J. R., Platt A., Richards J. H., et al. (2013). Pleiotropy of FRIGIDA enhances the potential for multivariate adaptation. Proc. R. Soc. B Biol. Sci. 280:1043. 10.1098/rspb.2013.1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell J. T., Mullen J. L., Lowry D. B., Awole K., Richards J. H., Sen S., et al. (2015). Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell. 27 969–983. 10.1105/tpc.15.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell J. T., Shakirov E. V., Schwartz S., Lowry D. B., Aspinwall M. J., Taylor S. H., et al. (2016). Promises and challenges of eco-physiological genomics in the field: tests of drought responses in switchgrass. Plant Phys. 172 734–748. 10.1104/pp.16.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B. (2012). Ecotypes and the controversy over stages in the formation of new species. Biol. J. Linn. Soc. 106 241–257. 10.1111/j.1095-8312.2012.01867.x [DOI] [Google Scholar]
- Lowry D. B., Behrman K. D., Grabowski P., Morris G. P., Kiniry J. R., Juenger T. E. (2014a). Adaptations between ecotypes and along environmental gradients in Panicum virgatum. Am. Nat. 183 682–692. 10.1086/675760 [DOI] [PubMed] [Google Scholar]
- Lowry D. B., Hernandez K., Taylor S. H., Meyer E., Logan T. L., Barry K. W., et al. (2014b). The genetics of divergence and reproductive isolation between ecotypes of Panicum hallii. New Phytol. 205 402–414. 10.1111/nph.13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Purmal C. T., Juenger T. E. (2013). A population genetic transect of Panicum hallii (Poaceae). Am. J. Bot. 100 592–601. 10.3732/ajb.1200379 [DOI] [PubMed] [Google Scholar]
- Mace E. S., Singh V., van Oosterom E. J., Hammer G. L., Hunt C. H., Jordan D. R. (2012). QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theor. Appl. Genet. 124 97–109. 10.1007/s00122-011-1690-9 [DOI] [PubMed] [Google Scholar]
- Markesteijn L., Poorter L. (2009). Seedling root morphology and biomass allocation of 62 tropical tree species in relation to drought- and shade-tolerance. J. Ecol. 97 311–325. 10.1111/j.1365-2745.2008.01466.x [DOI] [Google Scholar]
- McMillan C. (1965). Ecotypic differentiation within 4 North American Prairie grasses. II. Behavioral variation within transplanted community fractions. Am. J. Bot. 52 55–65. 10.2307/2439975 [DOI] [Google Scholar]
- Milano E. R., Lowry D. B., Juenger T. E. (2016). The genetic basis of upland/lowland ecotype divergence in Switchgrass (Panicum virgatum). G3 6 3561–3570. 10.1534/g3.116.032763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Harguindeguy N., Diaz S., Garnier E., Lavorel S., Poorter H., Jaureguiberry P., et al. (2013). New Handbook for standardized measurment of plant functional traits worldwide. Aust. J. Bot. 61 167–234. 10.1071/BT12225 [DOI] [Google Scholar]
- Pérez-Ramos I. M., Volaire F., Fattet M., Blanchard A., Roumet C. (2013). Tradeoffs between functional strategies for resource-use and drought-survival in Mediterranean rangeland species. Environ. Exp. Bot. 87 126–136. 10.1016/j.envexpbot.2012.09.004 [DOI] [Google Scholar]
- Phillips P. C., Arnold S. J. (1999). Hierarchical comparison of genetic variance-covariance matrices. I. Using the flury hierarchy. Evolution 53 1506–1515. 10.2307/2640896 [DOI] [PubMed] [Google Scholar]
- Poorter H., Bühler J., Van Dusschoten D., Climent J., Postma J. A. (2012). Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct. Plant Biol. 39 839–850. 10.1071/FP12049 [DOI] [PubMed] [Google Scholar]
- Price A. H., Steele K. A., Gorham J., Bridges J. M., Moore B. J., Evans J. L., et al. (2002). Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes. I. Root distribution, water use and plant water status. Field Crop. Res. 76 11–24. 10.1016/S0378-4290(02)00012-6 [DOI] [Google Scholar]
- Rajakaruna N. (2004). The edaphic factor in the origin of plant species. Int. Geol. Rev. 46 471–478. 10.2747/0020-6814.46.5.471 [DOI] [Google Scholar]
- Reich P. B. (2014). The world-wide “fast-slow” plant economics spectrum: a traits manifesto. J. Ecol. 102 275–301. 10.1111/1365-2745.12211 [DOI] [Google Scholar]
- Reich P. B., Walters M. B., Ellsworth D. S. (1997). From tropics to tundra: global convergence in plant functioning. Proc. Natl. Acad. Sci. U. S. A. 94 13730–13734. 10.1073/pnas.94.25.13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Archer M. A., Wayne R. K. (1999). Transgressive segregation, adaptation and speciation. Heredity 83 363–372. 10.1038/sj.hdy.6886170 [DOI] [PubMed] [Google Scholar]
- Stebbins G. L. (1952). Aridity as a stimulus to plant evolution. Am. Nat. 86 33–44. 29073895 [Google Scholar]
- Tozer A. K. N., Carswell K., Griffiths W. M., Crush J. R., Cameron C. A., Chapman D. F., et al. (2017). Growth responses of diploid and tetraploid perennial ryegrass (Lolium perenne) to soil-moisture deficit, defoliation and a root-feeding invertebrate. Crop Pasture Sci. 68 632–642. 10.1071/CP17154 [DOI] [Google Scholar]
- Valverde-Barrantes O. J., Freschet G. T., Roumet C., Blackwood C. B. (2017). A worldview of root traits: the influence of ancestry, growth form, climate and mycorrhizal association on the functional trait variation of fine-root tissues in seed plants. New Phytol. 215 1562–1573. 10.1111/nph.14571 [DOI] [PubMed] [Google Scholar]
- Via S., Hawthorne D. J. (2005). Back to the future: genetic correlations, adaptation and speciation. Genetica 123 147–156. 10.1007/s10709-004-2731-y [DOI] [PubMed] [Google Scholar]
- Waller F. R. (1976). A Biosystematic Study of Panicum Section Diffusa (Poaceae)in North America. [dissertation] [College Station (TX)]: TexasA&M University. [Google Scholar]
- Weißhuhn K., Auge H., Prati D. (2011). Geographic variation in the response to drought in nine grassland species. Basic Appl. Ecol. 12 21–28. 10.1016/j.baae.2010.11.005 [DOI] [Google Scholar]
- Withington J. M., Reich P. B., Oleksyn J., Eissenstat D. M. (2006). Comparisons of structure and life span in roots and leaves among temerate trees. Ecol. Monogr. 76 381–397. 10.1890/0012-9615(2006)076[0381:COSALS]2.0.CO;2 [DOI] [Google Scholar]
- Yardeni G., Tessler N., Imbert E., Sapir Y. (2016). Reproductive isolation between populations of Iris atropurpurea is associated with ecological differentiation. Ann. Bot. 118 971–982. 10.1093/aob/mcw139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data was deposited at NCBI, phenotypic data is included as Supplementary Material.


