Abstract
Most studies of environmental adaptations in plants have focused on either biotic or abiotic stress factors in an attempt to understand the defense mechanisms of plants against individual stresses. However, in the natural ecosystem, plants are simultaneously exposed to multiple stresses. Stress-tolerant crops developed in translational studies based on a single stress often fail to exhibit the expected traits in the field. To adapt to abiotic stress, recent studies have identified the need for interactions of plants with various microorganisms. These findings highlight the need to understand the multifaceted interactions of plants with biotic and abiotic stress factors. The endoplasmic reticulum (ER) is an organelle that links various stress responses. To gain insight into the molecular integration of biotic and abiotic stress responses in the ER, we focused on the interactions of plants with RNA viruses. This interaction points toward the relevance of ER in viral pathogenicity as well as plant responses. In this mini review, we explore the molecular crosstalk between biotic and abiotic stress signaling through the ER by elaborating ER-mediated signaling in response to RNA viruses and abiotic stresses. Additionally, we summarize the results of a recent study on phytohormones that induce ER-mediated stress response. These studies will facilitate the development of multi-stress-tolerant transgenic crops in the future.
Keywords: ABA, ER stress, heat stress, osmotic stress, RNA virus, SA
Introduction
Being sessile organisms, plants rely on their interactions with various organisms to adapt to environmental changes. Most terrestrial plants establish symbiotic relationships with microorganisms such as fungi and bacteria (Parniske, 2008; Finkel et al., 2017). Interactions of plants with these microorganisms play an important role not only in protecting plants against various pathogens but also in defense against abiotic stresses (Lau and Lennon, 2012). Plant molecular biologists have been studying plant responses to individual environmental stresses for a long time, and attempts have been made to apply the results of these studies to agriculture. However, in the field, plants are exposed to a variety of stresses simultaneously, and the responses of plants to these stresses are often different from those predicted in the laboratory (Pandey et al., 2015). Rapid ongoing global climate change is increasing the need to study plant responses to simultaneous stresses.
The endoplasmic reticulum (ER) is one of the largest, most functionally complex, and architecturally variable organelles discovered in eukaryotic cells (Schuldiner and Schwappach, 2013). It is a highly dynamic and complex cytoplasmic membrane system composed of two structurally distinct subdomains: the nuclear envelope enclosing the nucleus and an interconnected network called the peripheral ER, comprising a series of flattened sacs and tubules (Voeltz et al., 2002; Westrate et al., 2015). The ER is a central organelle that regulates stress responses in both plant and animal cells (Ellgaard and Helenius, 2003; Schroder and Kaufman, 2005). Stresses affecting protein folding lead to ER stress, which is communicated to the nucleus via the unfolded protein response (UPR), a cellular homeostatic response to ER stress (Fontes et al., 1991; Ellgaard and Helenius, 2003). Although the molecular mechanism of ER stress in plants is not as well understood as in animals, the expansion and diversity of ER stress-related genes revealed by genome sequencing of various plant species suggests that plants use more ER stress responses to adapt to the environment than animals (Liu and Howell, 2010b; Howell, 2017).
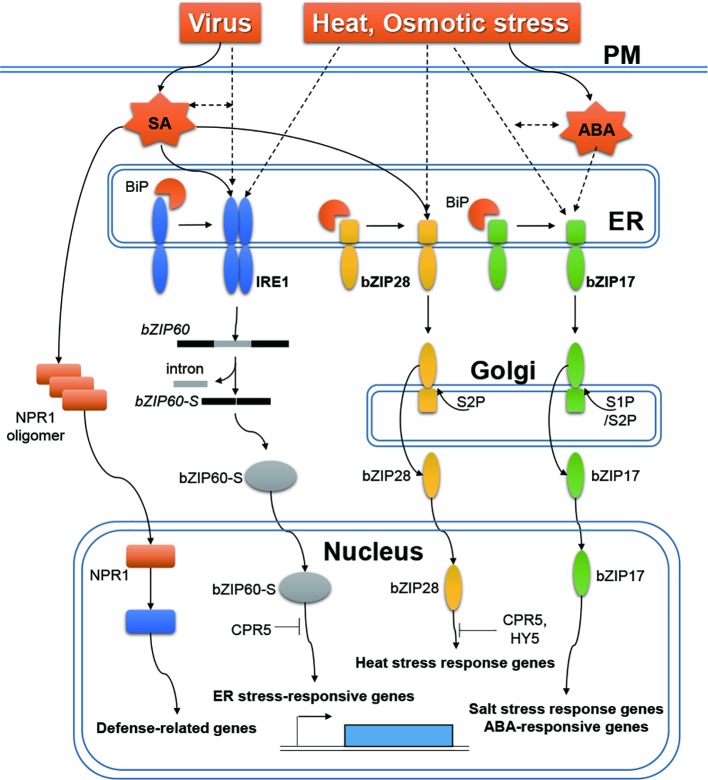
In plants, two main types of ER stress sensors, which regulate different UPR signaling pathways, have been identified: ER membrane-associated basic leucine zipper (bZIP) transcription factors, bZIP28 (Srivastava et al., 2014) and bZIP17 (Liu et al., 2008), and an ER resident transmembrane protein, inositol-requiring enzyme 1 (IRE1) (Koizumi et al., 2001) (Figure 1). Under unstressed normal conditions, bZIP17/28 are retained in the ER by their association with the binding protein (BiP), which is a master regulator of UPR. Under stress conditions, when unfolded proteins accumulate, BiP is sequestered away and released from bZIP17/28 (Gao et al., 2008; Srivastava et al., 2014). bZIP transcription factors are then transported from the ER to the Golgi apparatus, where they are proteolytically cleaved. The cytosol-facing regions of bZIP17/28 are then transported from the Golgi apparatus to the nucleus, to upregulate the expression of stress response genes, and to restore ER homeostasis (Liu et al., 2007a; Liu and Howell, 2010a). The IRE1 harbors both kinase and ribonuclease domains (Koizumi et al., 2001). In Arabidopsis (Arabidopsis thaliana), activated IRE1 splices bZIP60 mRNA, resulting in a frame shift and yielding a bZIP60 variant targeted to the nucleus (Deng et al., 2011; Hayashi et al., 2012, 2016; Liu and Howell, 2016). In the nucleus, bZIP60 plays a role in expression of ER stress response-related genes.
Figure 1.
ER stress responses to RNA viruses, heat and osmotic stress, and stress-related hormones in plants. Under biotic/abiotic conditions, BiP dissociates from IRE1 to bind unfolded proteins that accumulate in the ER. Activated IRE1 splices bZIP60 mRNA (bZIP60), resulting in a frame shift that generates a bZIP60 variant (bZIP60-S) targeted to the nucleus. When BiP is sequestered away, bZIP17/28 transcription factors are released from the ER to the Golgi apparatus, where they are cleaved. The cytosol-facing regions of bZIP17/28 are transported from the Golgi apparatus to the nucleus. After translocation, bZIP transcription factors upregulate the expression of stress response genes. Expression of bZIP60 is increased by viral infection. SA induces IRE1-mediated splicing of bZIP60 mRNA and proteolytic processing of bZIP28. SA regulates the expression of stress response genes via an NPR1-depenedent pathway. ABA promotes proteolytic processing of bZIP17. Dashed arrows indicate yet-uncharacterized molecular pathways.
In this mini review, we summarize ER stress responses in plants to RNA viruses, abiotic stresses, and stress-related hormones to understand plant cell perception of simultaneous biotic and abiotic stresses and the responses to these stresses through the ER.
Exploitation of ER Membranes by RNA Viruses
The majority of plant viruses that contains a positive-sense RNA genome and amplifies RNA in specific membrane-associated regions of host organelles are called replication complexes (RCs). Viral RCs play an important role in the enrichment of host cellular components for viral replication and prevent the activation of specific host defense mechanisms triggered by double-stranded RNA (dsRNA) intermediates of viral replication (Heinlein, 2015; Romero-Brey and Bartenschlager, 2016). Over the past decade, visualization of viral RC formation using advanced cell fluorescence imaging and microscopy techniques has enhanced our understanding of the process of virus replication in plant cells and the host factors involved (Jin et al., 2018). Although cell membrane origin and replication components and processes vary among plant RNA viruses, the RCs associated with plant RNA viruses can be divided into two types: double membrane vesicles and spherules/invaginations, as in animal viruses. This suggests that the process of viral infection is evolutionarily conserved among plant and animal viruses (Jin et al., 2018).
The ER is connected to the membrane of most other cellular organelles and the plasma membrane through membrane contact sites (MCSs) (Pérez-Sancho et al., 2016). Recent research suggests that inter-organelle communications via ER-driven MCSs helps communicate stress signals faster and more accurately than via vesicles or molecular diffusion (Pérez-Sancho et al., 2016). In plants, various membrane-bound organelles have been targeted as viral RC sites by one or more viral species, although the significance of interactions between these viruses and host cell organelles remains unclear (Grangeon et al., 2012). Tomato bushy stunt virus (TBSV) replicates on peroxisomes but is capable of forming viral RCs on the ER in the absence of peroxisomes, suggesting that a particular organelle is not a limiting factor for plant RNA virus infection (Jonczyk et al., 2007; Chuang et al., 2014). However, a particular type of lipid has been shown to affect viral RC formation and function. For example, the phosphatidylcholine (PC) content is high at sites of viral RCs of Brome mosaic virus (BMV), and blockade of PC synthesis inhibits BMV replication both in the native host barley and in the alternative host yeast, indicating that PC is important for BMV replication (Zhang et al., 2016). In addition, experiments using plants, yeast, and artificial phospholipid vesicles revealed that phosphatidylethanolamine is required at the site of TBSV replication (Xu and Nagy, 2015). Further research is needed to understand the effect of virus-specific lipid requirement on the host immune response, lipid metabolism in ER, and inter-organelle communication.
ER Stress Signaling Induced by Virus Infection
Overexpression of viral proteins and changes in the lipid membrane structure during virus infection induce ER stress and UPR signaling by perturbing protein homeostasis in both plant and animal hosts (Rutkowski and Kaufman, 2004; Zhang and Wang, 2012). The importance of ER stress signaling for infection of animal viruses has been highlighted in recent years (Li et al., 2015). In particular, much work has focused on mammalian viral proteins that interact directly with components of the UPR signaling pathway to inhibit or induce this response, thereby improving viral replication or transmission in mammalian cells (Blázquez et al., 2014). More recently, increasing evidence indicates that perturbation of the lipid bilayer composition also activates UPR signaling through IRE1 and PERK independently of its effect on protein folding homeostasis in the ER lumen (Volmer et al., 2013; Halbleib et al., 2017). IRE1 and PERK mutants lacking the luminal domain, which is required to sense unfolded protein stress, are activated by deprivation of inositol in yeast and by treatment with saturated fatty acids in mammalian cells (Volmer et al., 2013; Halbleib et al., 2017). Although the molecular mechanism by which UPR sensors recognize lipid bilayer stress has not been elucidated, the hepatitis C and West Nile viruses, which belong to the family Flaviviridae, induce lipid-dependent UPR signaling through nonstructural viral proteins to facilitate viral replication and to avoid host immune surveillance (Leier et al., 2018).
The molecular components involved in ER stress and UPR signaling were identified later in plants than in animals. Consequently, associations between plant RNA virus infections and ER stress responses were mostly investigated by measuring the expression levels of ER stress-related genes such as BiPs and protein disulfide isomerase and calreticulin (Zhang and Wang, 2012). Overexpression of several plant viral proteins such as Turnip mosaic virus (TuMV) 6K2, Potato virus X (PVX) triple gene block protein 3 (TGBp3), Rice black-streak dwarf virus P10, and Garlic virus X p11 induces the expression of ER stress response-related genes in plants (Heinlein, 2015; Lu et al., 2016; Howell, 2017). These proteins also bind to the ER membrane and enhance expression of the bZIP60 and BiP genes in tobacco (Nicotiana benthamiana) and Arabidopsis (Ye et al., 2011, 2012, 2013). The Arabidopsis bZIP60 knockout mutants show a low viral titer when infected with PVX and TuMV (Zhang et al., 2015). These viruses associate with different host subcellular organelles; TuMV forms viral RCs with the peripheral membrane of peroxisomes or chloroplasts, while PVX associates with the ER. These results suggest that activation of ER stress by viral proteins through bZIP60 benefits pathogenesis of plant viruses regardless of their RC-associated subcellular organelle. In addition, eukaryotic translation elongation factor 1 alpha (eEF1A) has been known to interact with viral RNA-dependent RNA polymerase of potyviruses, tobamoviruses, and tombusviruses, which is essential for viral replication (Nishikiori et al., 2006; Yamaji et al., 2006; Thivierge et al., 2008; Li et al., 2010). A recent report demonstrated that the UPR is activated in soybean during Soybean mosaic virus (SMV) infection and that this promotes accumulation of the virus (Luan et al., 2016). The authors showed that eEF1A, which interacts with the P3 protein of SMV, plays an important role in UPR activation and viral replication, although the molecular mechanism by which eEF1A activates the UPR is unclear (Luan et al., 2016). The P3 protein is known to reside in ER as a virulence factor of SMV, but its exact biochemical function is unknown (Chowda-Reddy et al., 2011). IRE1 activates splicing of bZIP60 in plants; therefore, it would be interesting to determine whether plant viral proteins targeting ER, similar to animal viral proteins, disturb ER lipid homeostasis and utilize the ER stress response through IRE1–bZIP60 signaling to support viral RCs.
Although the mutual regulatory mechanisms of UPR and programmed cell death (PCD) are well known in animals, it is not yet clear how plant UPR is linked to pathogen-induced cell death (Shore et al., 2011). In mammalian cells, a protein kinase R (PKR)-like ER kinase (PERK) induces PCD by phosphorylating the α-subunit of eukaryotic initiation factor 2 (eIF-2α), which prevents general protein synthesis and ultimately induces apoptosis (Liu et al., 2015b). However, few studies have investigated the induction of cell death by ER stress in plants. The existence of PERK, an ER stress sensor, in plants remains controversial, and the relevance of PERK-mediated phosphorylation of eIF-2α to the general plant defense response, including that against virus infection, has not yet been clarified (Zhang and Wang, 2012). A genome-wide search for eIF-2α kinases suggests that Arabidopsis and rice (Oryza sativa) plants lack a mammalian PERK homolog and possess only a homolog of the yeast general control non-derepressible-2 (GCN2) (Zhang et al., 2008). Indeed, phosphorylation of eIF-2α by AtGCN2 in Arabidopsis was observed under stress conditions and unlike in animals, it was not observed during viral infection (Zhang et al., 2008). Silencing of the gene encoding p58IPK, a putative plant ortholog of the mammalian PKR inhibitor, causes cell death following viral infection in tobacco (Bilgin et al., 2003). In animals, p58IPK acts as an inhibitor of the antiviral protein PKR, which induces apoptosis by phosphorylating eIF-2α upon recognition of viral dsRNA (Tan et al., 1998). Bilgin et al. (2003) demonstrated a direct interaction between tobacco p58IPK and helicases of TMV and Tobacco etch virus and an increase in eIF-2α phosphorylation in p58IPK-silenced plants. These results suggest the presence of a PKR- or PERK-like kinase-mediated pathway for eIF-2α phosphorylation, although so far no PKR- or PERK-like kinases have been found in plants using sequence similarity searches with mammalian counterparts.
ER Stress Signaling in Response to Abiotic Stress
Heat Stress
Over the past several decades, extensive studies have elucidated several molecular mechanisms of plant responses to high temperature, mainly focusing on flower development, circadian clock modulation, and immune response (Liu et al., 2015a). Although great progress has been achieved in the identification of molecular mechanisms of plant thermotolerance, how plants sense and transduce the heat signal remains unknown. Recently, it was reported that phytochromes, which sense the ratio of red to far-red light, function as thermosensors. Indeed, phytochrome-null plants display a constitutive warm temperature response (Jung et al., 2016). In addition, elongated hypocotyl 5 (HY5), a bZIP transcription factor and a downstream component of phytochrome-mediated light signaling, negatively regulates UPR by competing with bZIP28, which upregulates the expression of stress response genes (Nawkar et al., 2017). These reports shed light on the molecular mechanism of crosstalk between UPR and thermal sensing, mediated by HY5, which positively mediates light signaling but negatively regulates UPR gene expression.
The most detrimental effect of heat shock is the accumulation of unfolded proteins in both the cytosol and ER. Therefore, unfolded protein sensors in the ER and cytosol are proposed to play an essential role in thermotolerance. One of the UPR components, bZIP28, contributes to the upregulation of heat responsive genes, leading to heat tolerance of Arabidopsis (Gao et al., 2008). Knockout mutants of bZIP28 are sensitive to high temperature, suggesting an essential role of UPR in the general heat stress response and thermotolerance (Gao et al., 2008). Chromatin immunoprecipitation coupled with high-throughput sequencing revealed 133 putative direct targets of bZIP28 in Arabidopsis seedlings subjected to heat stress (Zhang S.S. et al., 2017). Another UPR component, IRE1, is also reported to be involved in heat stress response in Arabidopsis. Heat activated IRE1 splices bZIP60 mRNA, which is required for the upregulation of BiP3 in response to ER stress (Deng et al., 2011). Furthermore, IRE1 regulates the stress transcriptome by degrading various mRNAs (Mishiba et al., 2013; Maurel et al., 2014). Therefore, phytochromes are major regulators of heat stress response and thermotolerance in plants through the adjustment of downstream molecular components and different types of ER stress sensors.
Osmotic Stress
Osmotic stress caused by drought and high salinity has a major impact on plant growth and crop production, which highlights the importance of osmotic stress tolerance in plants. Drought and high salinity elicit many common and interactive downstream effects such as high levels of abscisic acid (ABA) (Nambara and Marion-Poll, 2005) and stress-responsive gene expression (Zhu, 2002). In Arabidopsis, the salt stress signaling pathway is reported to resemble an ER stress response (Liu et al., 2007b). Salt treatment activates the Golgi apparatus-resident site-1 protease (S1P) and cleaves ER membrane-associated transcription factor bZIP17. The released cytosol-facing region of bZIP17 is translocated to the nucleus, where it activates the expression of salt stress response genes. In this pathway, the activated bZIP17 transcription factor upregulates the expression of downstream genes, which desensitize ABA signaling (Zhou et al., 2015). Another major UPR component, BiP, plays an important role in osmotic stress tolerance in soybean (Glycine max) and tobacco plants via an unknown mechanism (Alvim et al., 2001; Valente et al., 2009). Expression profile analyses of soybean plants treated with ER stress inducers (tunicamycin/azidothymidine) or an osmotic stress inducer (polyethylene glycol) suggest a link between ER stress and osmotic stress pathway (Irsigler et al., 2007). In wheat (Triticum aestivum), the expression of BiP is upregulated during osmotic stress-related cell death caused by tauroursodeoxycholic acid, an apoptosis inhibitor (Zhang L. et al., 2017).
In addition to the ER-resident proteins, NAC (NAM/ATAF1/2/CUC2) domain-containing transcription factors are also receiving attention in osmotic stress response. Rice (151), soybean (152), and Arabidopsis (117) harbor a large family of NAC domain-containing proteins that are involved in multiple stress responses (Le et al., 2011; Nakashima et al., 2012; Shao et al., 2015). In soybean, GmNAC81 (also known as GmNAC6) has been identified as a component of ER stress- and osmotic stress-induced cell death response (Costa et al., 2008; Faria et al., 2011); this cell death response is synergistically activated by GmNAC30 and GmNAC81 (Mendes et al., 2013).
ER Stress Induced by Stress-Related Hormones
Salicylic Acid (SA)
SA is a key signaling component of both local defense response at infection sites and systemic resistance (Vernooij et al., 1994; Klessig et al., 2000). Functional crosstalk between SA and ER stress was first observed in Arabidopsis (Wang et al., 2005). Treatment of Arabidopsis with SA alters the level of many ER proteins required for protein folding and secretion, including BiP2 (Wang et al., 2005) and BiP3 (Pajerowska-Mukhtar et al., 2012). SA-induced expression of some of the ER stress-related genes is regulated by the heat shock factor-like transcription factor, TL1-binding transcription factor 1 (TBF1), which is genetically dependent on the non-expressor of pathogenesis-related genes 1 (NPR1), a master regulator of SA signaling (Wang et al., 2005; Pajerowska-Mukhtar et al., 2012). Because TBF1 regulates only selected ER genes, it was initially suggested that plants have evolved a specific mechanism for regulating SA-induced expression of ER stress-related genes (Pajerowska-Mukhtar et al., 2012). It was later shown that SA induces these genes via two main UPR signaling pathways: proteolytic processing of bZIP28 and IRE1-mediated splicing of bZIP60 mRNA (Nagashima et al., 2014). However, because it is unlikely that SA directly inhibits protein folding in the ER, the mechanism of activation of the UPR pathway by SA remains unclear. In Arabidopsis, constitutive expresser of pathogenesis-related genes 5 (CPR5), a plant-specific growth and stress regulator, acts as a negative modulator of SA in the early signal transduction steps, downstream of pathogen recognition and upstream of SA (Bowling et al., 1997). Recently, CPR5 was reported to act as a negative regulator of bZIP28 and bZIP60 through protein–protein interactions (Meng et al., 2017). Consequently, CPR5 suppresses the function of ER stress-induced bZIP28 and IRE1-bZIP60 pathways in the homeostatic control of SA-mediated plant growth (Meng et al., 2017).
ABA
ABA regulates many aspects of plant growth and development and plays a central role in the response to heat and osmotic stress (Shinozaki and Yamaguchi-Shinozaki, 2000; Raghavendra et al., 2010; Hauser et al., 2011; Huang et al., 2016). For example, in Arabidopsis, high temperature treatment upregulates ABA biosynthesis-related genes and downregulates ABA degradation-related genes (Toh et al., 2008). Similarly, in rice, heat treatment increases the level of ABA (Wu et al., 2016). Despite the increasing number of reports on ABA and heat stress, only a few studies have been conducted to investigate the effect of ABA on ER stress. Arabidopsis plants overexpressing the maize ortholog of Arabidopsis bZIP17 (ZmbZIP17) upregulates the expression of ER stress response genes. ER stress inducers such as dithiothreitol and tunicamycin induce ZmbZIP17 and its translocation to the nucleus (Yang et al., 2013). In addition, ZmbZIP17 interacts with ABA-responsive cis-elements in the promoter of ABA-responsive genes in yeast. Considering these reports, it is very likely that bZIP17 is involved in ABA-mediated ER stress response.
The mechanism of activation of bZIP17, which triggers ABA signaling in response to ER stress, has been elucidated in a study on seed germination in Arabidopsis; the authors showed that the Golgi apparatus-resident site-2 protease (S2P) cleaves and activates bZIP17, thus regulating downstream target genes that encode negative regulators of ABA signaling (Zhou et al., 2015). The level of ABA is also elevated by salt stress, which induces a signaling cascade involving the processing of bZIP17 by S1P, translocation of bZIP17 to the nucleus, and upregulation of salt stress genes (Nambara and Marion-Poll, 2005; Liu et al., 2007b). However, whether bZIP17-mediated ABA response is directly linked to the ER stress response remains unclear. It is possible that salt or ABA treatments promote protein misfolding, and S1P/S2P-mediated bZIP17 processing plays an essential role in desensitizing the plant to ER stress.
Conclusion and Perspectives
Changes in the agricultural environment, such as changes in temperature and water availability caused by climate change, can cause enormous reductions in crop yields via increased biological stress (Pandey et al., 2017). Global warming, for example, increases temperature stress in plants while at the same time increasing insect populations, which results in the spread of insect-borne viruses and their expansion to new host areas (Bebber, 2015). To help crops withstand newly emerging stresses, it is important to understand the mechanisms that plants have evolved to counteract various stress factors. Here, we have summarized recent advances in our understanding of ER responses to RNA viruses, abiotic stresses, and hormone responses (Figure 1). Of the three known ER stress sensors, IRE1 and bZIP28 are both involved in ER stress responses to viral infection and abiotic stress, whereas bZIP17 appears to be only an abiotic stress-specific sensor. Prasch and Sonnewald (2013) have shown that Arabidopsis plants exposed simultaneously to multiple stresses, such as heat, drought, and TuMV infection, have different responses to those of plants exposed to only one stress. For instance, defense-related genes induced by viral infection were not observed in Arabidopsis plants exposed to three stresses, but the ER stress response, which was not observed during TuMV infection, was induced (Prasch and Sonnewald, 2013). These results suggest that the mechanisms by which plants adapt to both external and internal stresses must be elucidated to understand the molecular adaptation of plants to multiple external stresses.
Author Contributions
JMP designed the outline of the article and C-JP and JMP wrote the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Bioedit Ltd for critically reviewing the paper and for language assistance.
Footnotes
Funding. This work was supported by the KRIBB Initiative Program and the Basic Research Program of the National Research Foundation of Korea (NRF-2017R1D1A1B03032215 to C-JP and NRF-2017R1A2B4012820 to JMP) funded by the Ministry of Science and ICT.
References
- Alvim F. C., Carolino S. M., Cascardo J. C., Nunes C. C., Martinez C. A., Otoni W. C., et al. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126, 1042–1054. 10.1104/pp.126.3.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebber D. P. (2015). Range-expanding pests and pathogens in a warming world. Annu. Rev. Phytopathol. 53, 335–356. 10.1146/annurev-phyto-080614-120207 [DOI] [PubMed] [Google Scholar]
- Bilgin D. D., Liu Y., Schiff M., Dinesh-Kumar S. P. (2003). P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev. Cell 4, 651–661. [DOI] [PubMed] [Google Scholar]
- Blazquez A. B., Escribano-Romero E., Merino-Ramos T., Saiz J. C., Martin-Acebes M. A. (2014). Stress responses in flavivirus-infected cells: activation of unfolded protein response and autophagy. Front. Microbiol. 5:266. 10.3389/fmicb.2014.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling S. A., Clarke J. D., Liu Y., Klessig D. F., Dong X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. 10.1105/tpc.9.9.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowda-Reddy R. V., Sun H., Chen H., Poysa V., Ling H., Gijzen M., et al. (2011). Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype soybean. Mol. Plant Microbe Interact 24, 37–43. 10.1094/MPMI-07-10-0158 [DOI] [PubMed] [Google Scholar]
- Chuang C., Barajas D., Qin J., Nagy P. D. (2014). Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog. 10:e1003944. 10.1371/journal.ppat.1003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. D., Reis P. A., Valente M. A., Irsigler A. S., Carvalho C. M., Loureiro M. E., et al. (2008). A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J. Biol. Chem. 283, 20209–20219. 10.1074/jbc.M802654200 [DOI] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J. X., Srivastava R., Rothstein S. J., Howell S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 108, 7247–7252. 10.1073/pnas.1102117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191. 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- Faria J. A., Reis P. A., Reis M. T., Rosado G. L., Pinheiro G. L., Mendes G. C., et al. (2011). The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 11, 129. 10.1186/1471-2229-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel O. M., Castrillo G., Herrera Paredes S., Salas González I., Dangl J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. 10.1016/j.pbi.2017.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes E. B., Shank B. B., Wrobel R. L., Moose S. P., Gr O. B., Wurtzel E. T., et al. (1991). Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell 3, 483–496. 10.1105/tpc.3.5.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Brandizzi F., Benning C., Larkin R. M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 105, 16398–16403. 10.1073/pnas.0808463105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R., Jiang J., Laliberté J.-F. (2012). Host endomembrane recruitment for plant RNA virus replication. Curr. Opin. Virol. 2, 683–690. 10.1016/j.coviro.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib K., Pesek K., Covino R., Hofbauer H. F., Wunnicke D., Hanelt I., et al. (2017). Activation of the Unfolded Protein Response by Lipid Bilayer Stress. Mol. Cell 67, 673–684, e678. 10.1016/j.molcel.2017.06.012 [DOI] [PubMed] [Google Scholar]
- Hauser F., Waadt R., Schroeder J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. 10.1016/j.cub.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Wakasa Y., Ozawa K., Takaiwa F. (2016). Characterization of IRE1 ribonuclease-mediated mRNA decay in plants using transient expression analyses in rice protoplasts. New Phytol. 210, 1259–1268. 10.1111/nph.13845 [DOI] [PubMed] [Google Scholar]
- Hayashi S., Wakasa Y., Takaiwa F. (2012). Functional integration between defence and IRE1-mediated ER stress response in rice. Sci. Rep. 2:670. 10.1038/srep00670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M. (2015). Plant virus replication and movement. Virology 479–480, 657–671. 10.1016/j.virol.2015.01.025 [DOI] [PubMed] [Google Scholar]
- Howell S. H. (2017). When is the unfolded protein response not the unfolded protein response? Plant Sci. 260, 139–143. 10.1016/j.plantsci.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Huang Y. C., Niu C. Y., Yang C. R., Jinn T. L. (2016). The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 172, 1182–1199. 10.1104/pp.16.00860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irsigler A. S., Costa M. D., Zhang P., Reis P. A., Dewey R. E., Boston R. S., et al. (2007). Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics 8:431. 10.1186/1471-2164-8-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Cao X., Wang X., Jiang J., Wan J., Laliberté J.-F., et al. (2018). Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front. Plant Sci. 9:57. 10.3389/fpls.2018.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk M., Pathak K. B., Sharma M., Nagy P. D. (2007). Exploiting alternative subcellular location for replication: Tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362, 320–330. 10.1016/j.virol.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Jung J. H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Klessig D. F., Durner J., Noad R., Navarre D. A., Wendehenne D., Kumar D., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. U. S. A. 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N., Martinez I. M., Kimata Y., Kohno K., Sano H., Chrispeels M. J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 172, 949–962. [PMC free article] [PubMed] [Google Scholar]
- Lau J. A., Lennon J. T. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. U. S. A. 109, 14058–14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T., Nishiyama R., Watanabe Y., Mochida K., Yamaguchi-Shinozaki K., Shinozaki K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276. 10.1093/dnares/dsr015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leier H. C., Messer W. B., Tafesse F. G. (2018). Lipids and pathogenic flaviviruses: An intimate union. PLoS Pathog. 14:e1006952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Kong L., Yu X. (2015). The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit. Rev. Microbiol. 41, 150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pogany J., Tupman S., Esposito A. M., Kinzy T. G., Nagy P. D. (2010). Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Feng L., Li J., He Z. (2015). Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 6:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010a). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22, 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010b). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2016). Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 211, 418–428. [DOI] [PubMed] [Google Scholar]
- Liu Z., Lv Y., Zhao N., Guan G., Wang J. (2015). Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 6, e1822–e1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R., Che P., Howell S. H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 19, 4111–4119. 10.1105/tpc.106.050021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R., Che P., Howell S. H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909. 10.1111/j.1365-313X.2007.03195.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R., Howell S. H. (2008). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31, 1735–1743. 10.1111/j.1365-3040.2008.01873.x [DOI] [PubMed] [Google Scholar]
- Lu Y., Yin M., Wang X., Chen B., Yang X., Peng J., et al. (2016). The unfolded protein response and programmed cell death are induced by expression of garlic virus X p11 in Nicotiana benthamiana. J. Gen. Virol. 97, 1462–1468. 10.1099/jgv.0.000460 [DOI] [PubMed] [Google Scholar]
- Luan H., Shine M. B., Cui X., Chen X., Ma N., Kachroo P., et al. (2016). The Potyviral P3 Protein Targets Eukaryotic Elongation Factor 1A to Promote the Unfolded Protein Response and Viral Pathogenesis. Plant Physiol. 172, 221–234. 10.1104/pp.16.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M., Chevet E., Tavernier J., Gerlo S. (2014). Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 39, 245–254. 10.1016/j.tibs.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Mendes G. C., Reis P. A., Calil I. P., Carvalho H. H., Aragao F. J., Fontes E. P. (2013). GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. U. S. A. 110, 19627–19632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Ruberti C., Gong Z., Brandizzi F. (2017). CPR5 modulates salicylic acid and the unfolded protein response to manage tradeoffs between plant growth and stress responses. Plant J. 89, 486–501. 10.1111/tpj.13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K., Nagashima Y., Suzuki E., Hayashi N., Ogata Y., Shimada Y., et al. (2013). Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. U. S. A. 110, 5713–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y., Iwata Y., Ashida M., Mishiba K., Koizumi N. (2014). Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 55, 1772–1778. 10.1093/pcp/pcu108 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. 10.1146/annurev.arplant.56.032604.144046 [DOI] [PubMed] [Google Scholar]
- Nawkar G. M., Kang C. H., Maibam P., Park J. H., Jung Y. J., Chae H. B., et al. (2017). HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 2084–2089. 10.1073/pnas.1609844114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikiori M., Dohi K., Mori M., Meshi T., Naito S., Ishikawa M. (2006). Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80, 8459–8468. 10.1128/JVI.00545-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K. M., Wang W., Tada Y., Oka N., Tucker C. L., Fonseca J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112. 10.1016/j.cub.2011.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Irulappan V., Bagavathiannan M. V., Senthil-Kumar M. (2017). Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 8:537. 10.3389/fpls.2017.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Ramegowda V., Senthil-Kumar M. (2015). Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms. Front. Plant Sci. 6:723. 10.3389/fpls.2015.00723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6:763. 10.1038/nrmicro1987 [DOI] [PubMed] [Google Scholar]
- Pérez-Sancho J., Tilsner J., Samuels A. L., Botella M. A., Bayer E. M., Rosado A. (2016). Stitching organelles: organization and function of specialized membrane contact sites in plants. Trends Cell Biol. 26, 705–717. 10.1016/j.tcb.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Prasch C. M., Sonnewald U. (2013). Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in Signaling networks. Plant Physiol. 162, 1849–1866. 10.1104/pp.113.221044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A. S., Gonugunta V. K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. 10.1016/j.tplants.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. (2016). Endoplasmic reticulum: thefavorite intracellular niche for viral replication and assembly.. Viruses 8. 10.3390/v8060160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., Kaufman R. J. (2004). A trip to the ER: coping with stress. Trends Cell Biol. 14, 20–28. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. (2005). The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789. 10.1146/annurev.biochem.73.011303.074134 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Schwappach B. (2013). From rags to riches–the history of the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2389–2391. [DOI] [PubMed] [Google Scholar]
- Shao H., Wang H., Tang X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. 10.3389/fpls.2015.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. 10.1016/S1369-5266(00)00067-4 [DOI] [PubMed] [Google Scholar]
- Shore G. C., Papa F. R., Oakes S. A. (2011). Signaling cell death from the endoplasmic reticulum stress response. Curr. Opin. Cell Biol. 23, 143–149. 10.1016/j.ceb.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Deng Y., Howell S. H. (2014). Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front. Plant Sci. 5:59. 10.3389/fpls.2014.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. L., Gale M. J., Jr., Katze M. G. (1998). Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol. Cell Biol. 18, 2431–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge K., Cotton S., Dufresne P. J., Mathieu I., Beauchemin C., Ide C., et al. (2008). Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377, 216–225. 10.1016/j.virol.2008.04.015 [DOI] [PubMed] [Google Scholar]
- Toh S., Imamura A., Watanabe A., Nakabayashi K., Okamoto M., Jikumaru Y., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146, 1368–1385. 10.1104/pp.107.113738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente M. A., Faria J. A., Soares-Ramos J. R., Reis P. A., Pinheiro G. L., Piovesan N. D., et al. (2009). The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot. 60, 533–546. 10.1093/jxb/ern296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij B., Uknes S., Ward E., Ryals J. (1994). Salicylic acid as a signal molecule in plant-pathogen interactions. Curr. Opin. Cell Biol. 6, 275–279. 10.1016/0955-0674(94)90147-3 [DOI] [PubMed] [Google Scholar]
- Voeltz G. K., Rolls M. M., Rapoport T. A. (2002). Structural organization of the endoplasmic reticulum. EMBO Rep. 3, 944–950. 10.1093/embo-reports/kvf202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R., Van Der Ploeg K., Ron D. (2013). Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U. S. A. 110, 4628–4633. 10.1073/pnas.1217611110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Weaver N. D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. 10.1126/science.1108791 [DOI] [PubMed] [Google Scholar]
- Westrate L. M., Lee J. E., Prinz W. A., Voeltz G. K. (2015). Form follows function: the importance of endoplasmic reticulum shape. Annu. Rev. Biochem. 84, 791–811. 10.1146/annurev-biochem-072711-163501 [DOI] [PubMed] [Google Scholar]
- Wu C., Cui K., Wang W., Li Q., Fahad S., Hu Q., et al. (2016). Heat-induced phytohormone changes are associated with disrupted early reproductive development and reduced yield in rice. Sci. Rep. 6:34978. 10.1038/s41598-016-0031-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Nagy P. D. (2015). RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc. Natl. Acad. Sci. U. S. A. 112, E1782–E1791. 10.1073/pnas.1418971112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji Y., Kobayashi T., Hamada K., Sakurai K., Yoshii A., Suzuki M., et al. (2006). In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347, 100–108. 10.1016/j.virol.2005.11.031 [DOI] [PubMed] [Google Scholar]
- Yang Y. G., Lv W. T., Li M. J., Wang B., Sun D. M., Deng X. (2013). Maize membrane-bound transcription factor Zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol. 54, 2020–2033. 10.1093/pcp/pct142 [DOI] [PubMed] [Google Scholar]
- Ye C. M., Chen S., Payton M., Dickman M. B., Verchot J. (2013). TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol. Plant Pathol. 14, 241–255. 10.1111/mpp.12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Dickman M. B., Whitham S. A., Payton M., Verchot J. (2011). The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156, 741–755. 10.1104/pp.111.174110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C. M., Kelly V., Payton M., Dickman M. B., Verchot J. (2012). SGT1 is induced by the potato virus X TGBp3 and enhances virus accumulation in Nicotiana benthamiana. Mol. Plant 5, 1151–1153. 10.1093/mp/sss026 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen H., Brandizzi F., Verchot J., Wang A. (2015). The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 11:e1005164. 10.1371/journal.pgen.1005164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang A. (2012). Virus-induced ER stress and the unfolded protein response. Front. Plant Sci. 3:293. 10.3389/fpls.2012.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xin Z., Yu X., Ma C., Liang W., Zhu M., et al. (2017). Osmotic stress induced cell death in wheat is alleviated by tauroursodeoxycholic acid and involves endoplasmic reticulum stress-related gene expression. Front. Plant Sci. 8:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Kanyuka K., Parry M. A., Powers S. J., Halford N. G. (2008). GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. J. Exp. Bot. 59, 3131–3141. 10.1093/jxb/ern169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. S., Yang H., Ding L., Song Z. T., Ma H., Chang F., et al. (2017). Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in arabidopsis. Plant Cell 29, 1007–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang Z., Chukkapalli V., Nchoutmboube J. A., Li J., Randall G., et al. (2016). Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc. Natl. Acad. Sci. 113:E1064. 10.1073/pnas.1605871113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. F., Sun L., Valdes A. E., Engstrom P., Song Z. T., Lu S. J., et al. (2015). Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in arabidopsis. New Phytol. 208, 188–197. 10.1111/nph.13436 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]