Abstract
Gastropod molluscs are among the most diverse and abundant animals in the oceans, and are successful colonizers of terrestrial and freshwater environments. Past phylogenetic efforts to resolve gastropod relationships resulted in a range of conflicting hypotheses. Here, we use phylogenomics to address deep relationships between the five major gastropod lineages—Caenogastropoda, Heterobranchia, Neritimorpha, Patellogastropoda and Vetigastropoda—and provide one congruent and well-supported topology. We substantially expand taxon sampling for outgroups and for previously underrepresented gastropod lineages, presenting new transcriptomes for neritimorphs and patellogastropods. We conduct analyses under maximum-likelihood, Bayesian inference and a coalescent-based approach, accounting for the most pervasive sources of systematic errors in large datasets: compositional heterogeneity, site heterogeneity, heterotachy, variation in evolutionary rates among genes, matrix completeness, outgroup choice and gene tree conflict. We find that vetigastropods and patellogastropods are sister taxa, and that neritimorphs are the sister group to caenogastropods and heterobranchs. We name these two major unranked clades Psilogastropoda and Angiogastropoda, respectively. We additionally provide the first genomic-scale data for internal relationships of neritimorphs and patellogastropods. Our results highlight the need for reinterpreting the evolution of morphological and developmental characters in gastropods, especially for inferring their ancestral states.
Keywords: gastropod phylogeny, transcriptomes, sequence heterogeneity, Psilogastropoda, Angiogastropoda, Mollusca
1. Introduction
Gastropods are one of the most diverse clades of marine animals [1], and the only mollusc group to successfully colonize terrestrial environments. With an extant diversity of many tens of thousands of described species, gastropods also have a high degree of morphological disparity—snails, limpets and slugs with enormous variation in shell shape, coloration and size—and inhabit all kinds of environments and depths. Gastropods have embryonic spiral cleavage, an array of developmental modes (direct and indirect, with more than one type of larva), and undergo torsion of the body during development. Five main lineages are currently recognized: Caenogastropoda (e.g. cowries, whelks, conchs, cones), Heterobranchia (e.g. bubble snails, sea slugs, sea hares, most terrestrial snails and slugs), Neritimorpha (nerites), Patellogastropoda (true limpets) and Vetigastropoda (e.g. abalones, keyhole limpets, turban snails, top shells).
Early classifications included members of the vetigastropods, patellogastropods and neritimorphs in the Archaeogastropoda [2,3]. With the first numerical cladistic analysis of morphological data, patellogastropods were recovered as the sister group to all other gastropods, which were united in the clade Orthogastropoda [4,5]. The sister group relationship of the most diverse lineages, the heterobranchs and caenogastropods into the clade Apogastropoda, has been consistently recovered in most morphological and molecular analyses. Other than that, almost all possible topologies for gastropod relationships have been proposed (for a historical review, see [6]). Early molecular studies had mixed success in recovering even the well-established monophyly of gastropods or some of the main lineages [7–11]. Mitogenomic efforts have also produced discordant results [12–14], but recently have recovered a topology congruent with orthogastropods [15]. The first transcriptomic analyses of the group were able to reject several of the historically proposed hypotheses, including the clade Orthogastropoda [16]. However, different methods still resulted in contrasting topologies, and three hypotheses remain [16]. The major uncertainty is the position of Neritimorpha, which is recovered either as the sister group to Apogastropoda or as the sister group to Patellogastropoda and Vetigastropoda, in this case forming the traditional Archaeogastropoda. The third remaining hypothesis has vetigastropods as the sister lineage to all other gastropods [16].
Although the most diverse gastropod lineages were well sampled in the transcriptomic analyses of Zapata et al. [16], the dataset had only one species of Patellogastropoda and two of Neritimorpha, which are crucial for the proper rooting of the gastropod tree. As the three remaining hypotheses differ in their rooting, better outgroup sampling is another key necessary improvement. Furthermore, several biases known to be present in large genomic datasets have not been accounted for in the phylogenetic methods used so far to resolve gastropod relationships. Heterogeneity in the stationary frequency of amino acids among samples is one such issue that can artificially group taxa that are actually not closely related based on convergent amino acid composition [17]. Within-site rate variation through time (heterotachy) is another likely violation [18]. Some genes with slow rates of evolution (e.g. ribosomal protein genes) have also been shown to bias phylogenetic inference [19,20], while genes with fast rates and high levels of saturation can cause long-branch attraction [15,21]. An additional model violation comes from gene tree discordance, not accounted for by concatenation methods, that can be caused by incomplete lineage sorting and be particularly relevant in areas of the tree with short internal branches [22–24], such as the radiation of crown gastropods during the Ordovician [16,25]. More commonly considered issues include rate heterogeneity between sites and missing data.
Our goal was to resolve between the three remaining hypotheses for the early divergences of gastropods. We present an extended sampling of Neritimorpha and Patellogastropoda by producing new transcriptomes, and complement the dataset with the latest published gastropod transcriptomes. We further increase representation for the closest outgroups—bivalves, scaphopods and cephalopods—sampling all of the major lineages within each of these mollusc clades. We employ a variety of methods and models with strategic gene subsampling to account for the most widespread potential sources of systematic error in large datasets, namely compositional heterogeneity, site heterogeneity, heterotachy, variation in evolutionary rates among genes, matrix completeness, outgroup choice and gene tree conflict.
2. Methods
(a). Sampling and sequencing
We sequenced the transcriptomes of 17 species, mostly patellogastropods and neritimorphs, and combined them with published transcriptome sequences from 39 other gastropods and 18 mollusc outgroups, for a total of 74 terminals. All new data and selected published sequences are paired-end Illumina reads. New samples were fixed in RNAlater (Invitrogen) or flash frozen in liquid nitrogen. RNA extraction and mRNA isolation were done with the TRIzol Reagent and Dynabeads (Invitrogen). Libraries were prepared with the PrepX RNA-Seq Library kit using the Apollo 324 System (Wafergen). Quality control of mRNA and cDNA was done with a 2100 Bioanalyzer, a 4200 TapeStation (Agilent) and the Kapa Library Quantification kit (Kapa Biosystems). Samples were pooled in equimolar amounts and sequenced in the Illumina HiSeq 2500 platform (paired end, 150 bp) at the Bauer Core Facility at the Harvard University. New sequences were deposited in the NCBI Sequence Read Archive (BioProject PRJNA508436, SRA SRR8318344–SRR8318360); voucher information, library indexes and assembly statistics are available in electronic supplementary material, table S1.
(b). Transcriptome assembly
Both new and previously published transcriptomes were assembled de novo; a detailed pipeline, scripts and assemblies are available in the electronic supplementary material. Raw reads were cleaned with RCorrector [26] and Trim Galore! [27], removing unfixable reads (as identified by RCorrector), Wafergen library adapters and reads shorter than 50 bp. Filtered reads were compared against a set of mollusc ribosomal RNAs and mitochondrial DNA and removed with Bowtie2 v. 2.2.9 [28]. This set was created from the well-curated databases SILVA [29] (18S and 28S rRNAs), AMIGA [30] (mtDNA) and from GenBank [31] (5S and 5.8S rRNAs), and is also deposited in the electronic supplementary material. Reads were assembled into transcripts with Trinity v. 2.3.2 [32,33] (–SS_lib_type FR for our new strand-specific data generated with Wafergen kits; precise information was not available from published data, so the default non-strand-specific mode was used for reads downloaded from SRA). A second run of Bowtie2 was done on the assemblies, before removing transcripts with sequence identity higher than 95% with CD-HIT-EST v. 4.6.4 [34,35]. Transcripts were then translated to amino acids with TransDecoder v. 3.0 [33], and the longest isoform of each gene was retained with a custom python script (choose_longest_iso.py). The completeness of the assemblies was evaluated with BUSCO v. 3.0.2 by comparison with the Metazoa database [36].
(c). Matrix construction
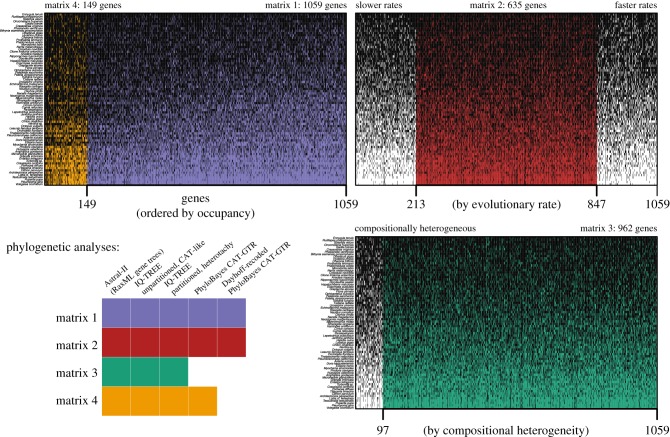
We built four matrices to account for extreme evolutionary rates, amino acid composition heterogeneity and different levels of matrix completeness. Scripts, gene content for each matrix and alignment files are available in the electronic supplementary material. Orthology assignment of the peptide assemblies was done with OMA v. 2.0 [37]. We then used a custom python script (selectslice.py) to select all orthogroups for which at least half of the terminals were represented (50% taxon occupancy), resulting in a matrix with 1059 genes (matrix 1) (figure 1). Each orthogroup was aligned with MAFFT v. 7.309 [38], and the alignment ends were trimmed to remove positions with more than 80% missing data with a custom bash script (trimEnds.sh). To avoid possible biases, saturation and long-branch attraction, matrix 2 was built by removing from matrix 1 the 20% slowest and the 20% fastest evolving genes, as calculated with TrimAl [39], for a final size of 635 genes (figure 1). Matrix 3 is the subset of 962 genes from matrix 1 that are homogeneous regarding amino acid composition. Homogeneity for each gene was determined with a simulation-based test from the python package p4 [17,40], with a custom script modified from Laumer et al. [41] (p4_compo_test.py) and a conservative p-value of 0.1. Finally, a subset of 149 genes with 70% taxon occupancy constitutes matrix 4 (figure 1). For inference methods that require concatenation, genes were concatenated using Phyutility [42]. We further reduced composition heterogeneity in matrices 1 and 2 by recoding amino acids into the six Dayhoff categories [43] with a custom bash script (recdayhoff.sh).
Figure 1.
Matrices and phylogenetic methods used to infer gastropod relationships. With 50% taxon occupancy, matrix 1 is the largest, with 1059 genes. Matrix 4 is the subset of the best sampled 149 genes, with 70% taxon occupancy. Genes and species are sorted with the best sampling on the upper left. Matrix 2 is the subset of 635 genes after ordering all genes by evolutionary rate and removing the 20% slowest and 20% fastest evolving genes. Matrix 3 includes the 962 genes that are homogeneous in amino acid composition; genes are ordered by p-value of the homogeneity test. Black cells indicate genes present for each species. See Methods for details. (Online version in colour.)
All of our matrices include a dense outgroup sampling from the closest mollusc relatives. However, most previous molecular gastropod phylogenies have sampled only a couple outgroup species and/or very distantly related molluscs. To test the effect of such limited outgroup sampling, we built four extra datasets based on the largest matrix 1, each containing all gastropods plus only one of the other mollusc classes from our complete set (bivalves, scaphopods, cephalopods or polyplacophorans).
(d). Phylogenetic analyses
Amino acid matrices were used for phylogenetic inference with a coalescent-based approach in Astral-II v. 4.10.12 [44], with maximum likelihood (ML) in IQ-TREE MPI v. 1.5.5 [45–47] and with Bayesian inference in PhyloBayes MPI v. 1.7a [48]. The two Dayhoff-recoded matrices were analysed in PhyloBayes (figure 1). Full details and scripts are explained in a custom pipeline in the electronic supplementary material. For the coalescent-based method, gene trees were inferred with RAxML v. 8.2.10 [49] (-N 10 -m PROTGAMMALG4X) and then used as input for Astral-II for species tree estimation. For each concatenated matrix, we inferred the best ML tree with two strategies: a gene-partitioned analysis with model search including LG4 mixture models and accounting for heterotachy (-bb 1500 -sp partition_file -m MFP+MERGE -rcluster 10 -madd LG4 M,LG4X -mrate G,R,E); and a non-partitioned analysis with model search also including the C10 to C60-profile mixture models [50] (ML variants of the Bayesian CAT model [51]) (-bb 1500 -m MFP+MERGE -rcluster 10 -madd LG4 M,LG4X,LG+C10,LG+C20,LG+C30,LG+C40,LG+C50,LG+C60 -mrate G,R,E). The search for the models LG+C60 (matrices 1 and 3) and LG+C50 (matrix 1) required more memory than available, and these models were disregarded for the respective matrices. Outgroup test datasets were analysed with the ML profile mixture model. PhyloBayes was run with the CAT-GTR model on a subset of the concatenated alignments (matrices 1, 2 and 4), discarding constant sites to speed up computation. Tree figures were edited with the R package ggtree [52].
3. Results and discussion
(a). Main gastropod relationships
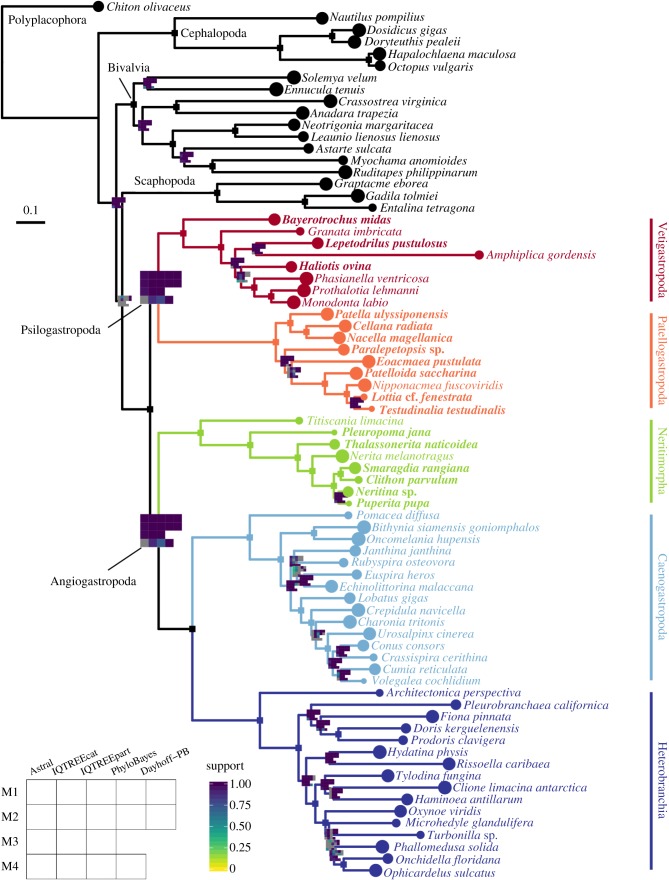
Our main goal was to resolve the deep nodes of the gastropod tree and distinguish between three hypotheses of the relationships among its five main lineages. All but one of our inference methods and matrices congruently support a clade uniting Vetigastropoda with Patellogastropoda, and Neritimorpha as the sister group to Apogastropoda (figure 2). The only exception is the coalescent-based analysis on the smallest dataset of 149 genes (Astral, matrix 4), in which these two key nodes were left unresolved (all tree files are available in the electronic supplementary material). Accordingly, the few analyses with lower support on these nodes also refer to the smaller matrix 4, which is unsurprising given that it comprises fewer informative sites in concatenated analyses and fewer genes in the coalescent-based analysis [53]. In summary, the resulting topology is congruent based on an array of analyses testing for the major common sources of systematic error in phylogenomic datasets, including gene tree discordance, compositional heterogeneity, heterotachy, site heterogeneity, variation in evolutionary rates and missing data.
Figure 2.
Gastropod phylogeny inferred from the largest matrix (M1) with ML and a profile mixture model (IQTREEcat). A single square marks branches where all analyses had full support; branches where at least one analysis had less than full support are marked with a plot, coloured in a continuous scale according to support value, from 0 to 1. Grey squares in the plots represent splits that were absent in a given analysis. New transcriptomes are represented in bold. M1–M4, matrices 1–4; IQTREEpart, ML partitioned analysis; Dayhoff-PB, Bayesian analysis on a matrix recoded according to the six Dayhoff categories. See Methods for details. (Online version in colour.)
To explore the signal of genes with heterogeneous amino acid composition, we used the ML and coalescent-based methods to infer trees for the set of 97 genes that failed the p4 homogeneity test (trees available in the electronic supplementary material). Interestingly, in the ML partitioned analysis that had a simpler site heterogeneity model, Patellogastropoda was recovered as the sister group to all other lineages. This is possibly the most commonly used strategy for phylogenetic inference, highlighting the risks of not accounting for high complexity in sequence data, in this case, site and composition heterogeneity combined. Even with exclusively heterogeneous genes, analyses that do not rely on a concatenated matrix (coalescent-based) or that consider more complex models of site heterogeneity (ML with a profile mixture model) still recovered the same relationships of figure 2.
Our enriched taxon representation ensured that all major lineages within each of the closest outgroups (scaphopods, bivalves and cephalopods) were represented, mitigating issues of long-branch attraction to the outgroups (figure 2). However, most previous molecular studies of gastropods had limited outgroup representation, often resulting in long branches leading to the ingroup [9,10,14,15]. We tested the sensitivity of our results to restricted outgroup sampling by limiting outgroups to just one mollusc class at a time in matrix 1 (trees available in the electronic supplementary material). Datasets with only cephalopods, only bivalves and even with the single polyplacophoran resulted in the same topology of figure 2. Only the dataset restricted to scaphopods produced a different topology, finding patellogastropods as the sister group to all other gastropods, but with low support. Scaphopods and patellogastropods have respectively the longest internal branch among outgroups and gastropods, pointing to an effect of long branch attraction. These results highlight the importance of maximizing outgroup sampling when targeting hard and ancient nodes.
Our inferred topology for gastropod relationships (figure 2) has been previously recovered by a few molecular [16,54] and total evidence [6] analyses, with numerous alternatives proposed in the literature (e.g. [5,6,10,12,13,15,53]), even within the same studies. With 17 analyses (combinations of four subsampled matrices, two data types—amino acids and Dayhoff recoding—and four inference methods/models), for the first time, we find strong congruence towards this single topology for deep gastropod relationships. With that we reject the clade Archaeogastropoda, proposed almost a century ago by Thiele [2], which united Neritimorpha, Vetigastropoda and Patellogastropoda. Although this grouping had given way to other predominant hypotheses along the years (e.g. Eogastropoga versus Orthogastropoda divergence), this classification is still used in the organization of malacology and paleontology collections of many natural history museums, and was one of the three resulting hypotheses from the transcriptomic study of Zapata et al. [16].
The close relationship of neritimorphs and apogastropods had already been recognized based on early developmental characters [55,56], such as the time of formation of the 4d blastomere (mesentoblast). In these groups, the differentiation of this key embryonic cell, which gives rise to the mesoderm in spiralians [57–59], is accelerated, happening at an earlier cell stage than in vetigastropods and patellogastropods [55,56]. Other traits shared by neritimorphs and apogastropods include complex reproductive anatomy, internal fertilization and encapsulated eggs, which hatch into a feeding veliger larva or directly into a juvenile [6,25,60–62]. By contrast, vetigastropods and patellogastropods are mostly broadcast spawners, with embryos that develop in the plankton into non-feeding larvae, first as a trochophore that later gives rise to a veliger [6,25,60,62,63]. Character states shared by patellogastropods and vetigastropods have historically been interpreted as plesiomorphic based on the phylogenetic hypothesis in which patellogastropods were the sister group to all other gastropods, or due to a misguided notion that these are ‘primitive’ taxa [5,55]. In the light of a sister group relationship between Patellogastropoda and Vetigastropoda, it is not possible to confidently infer which were the ancestral gastropod conditions without an extensive comparative analysis. Sampling of morphological and developmental data from a larger diversity of gastropods and especially their closest outgroups—bivalves and scaphopods—will be needed to reinterpret their evolution under the framework presented here (figure 2).
Although exceptions exist in such diverse clades, we use the most general features of the reproductive strategy and early life history of gastropods, irrespective of their ancestral state, to name the two major lineages in figure 2: Psilogastropoda, new taxon, from the Greek psilos meaning bare, naked. This is the most inclusive clade containing Vetigastropoda and Patellogastropoda, but not Neritimorpha, Caenogastropoda or Heterobranchia, therefore also accounting for stem taxa. The name represents the unprotected nature of the gametes, which are released in the water for external fertilization, and of the embryos and larvae that develop in the plankton, exposed to the environment. Angiogastropoda, new taxon, from the Greek angeion meaning vessel, capsule. It is the most inclusive clade containing Neritimorpha, Caenogastropoda and Heterobranchia, but not psilogastropods. The name reflects the enclosed nature of the embryo after internal fertilization, which is encapsulated during early development, followed by either direct development or a late stage veliger larva hatching from the egg.
We then propose the adjusted classification of Gastropoda (table 1). Important questions that remain regarding major gastropod relationships include the position of Cocculiniformia and Neomphalina, smaller deep sea clades that have been considered somehow related to vetigastropods, neritimorphs, patellogastropods or as independent branches in the gastropod tree [60]. They are yet to be sampled in a phylogenomic analyses, and we therefore keep their independent status relative to the other major lineages, but note that future phylogenomic studies could reveal either one as part of psilogastropods or angiogastropods.
Table 1.
Higher level classification of the extant Gastropoda proposed here. We follow [64] in not presenting the authority of high level names because some of them have a taxonomic composition that differs substantially from that of the original author.
| classification proposed here |
|---|
| Class Gastropoda |
| Psilogastropoda, new taxon |
| Patellogastropoda |
| Vetigastropoda |
| Angiogastropoda, new taxon |
| Neritimorpha |
| Apogastropoda |
| Caenogastropoda |
| Heterobranchia |
| Incertae sedis |
| Neomphalina |
| Cocculiniformia |
Regarding overall mollusc relationships, we recover a well-supported clade of gastropods, bivalves and scaphopods in all analyses; however, as in previous phylogenomic efforts [65,66], relationships between these three groups are unstable (figure 2). The Dayhoff datasets and most of the ML analyses with the profile mixture model result in a clade of gastropods and scaphopods; while most coalescent-based trees recover a clade of bivalves and scaphopods; and finally, the ML partitioned analyses produce a clade of gastropods and bivalves. Perhaps a way ahead to resolve such hard nodes will be to use other types of data, such as genomic rearrangements and presence/absence of genes from complete genomes.
(b). A note about convergence in PhyloBayes
While PhyloBayes runs converged on the Dayhoff-recoded datasets presented here, analyses on the more complex amino acid matrices did not converge for all parameters. The problem was especially pronounced for the large matrices (a summary table with convergence metrics for all analyses is given in the electronic supplementary material). We observed that some convergence issues were due to small differences between chains regarding the position of one or few derived terminals within the outgroups or within apogastropods, whose relationships were not the goal of this study. We suspect this may be caused by a problem in topology proposals for these derived nodes, leading some of the chains to get stuck in local maxima. One example comes from the Dayhoff analysis of matrix 1: the initial two chains seemed to be very far from topological convergence (maxdiff = 1) even after more than 20 000 generations. Upon closer inspection, both trees were basically indistinguishable, with the only variation being the position of Charonia or Crepidula as the sister group to Neogastropoda. Removal of either one of the two terminals from the treelist files with a custom script (remove_terminal_treelist.py) resulted in the same converged topology (tree files in the electronic supplementary material). For that particular analysis, we ran two additional independent chains that converged without presenting this issue. This behaviour was recently discussed [67], and perhaps has been underreported in the literature.
(c). Relationships within gastropod lineages
This is the first genomic-scale dataset for Patellogastropoda and Neritimorpha. Internal relationships of patellogastropods have presented incongruent results even among studies using the same type of data (reviewed in [60,68]). We consistently recover Nacellidae (Cellana, Nacella) as the sister group of Patellidae (figure 2), a clade originally supported by some of the earliest morphological [69] and mitochondrial phylogenies [70]. Nacellids have also been placed either as a grade at the base of the tree [71] or closer to Lottiidae [72], and the current taxonomic classification has Nacellidae in the superfamily Lottioidea [64,73]; our results indicate the family should be transferred to Patelloidea. Another interesting finding regards Eoacmaea, which had gained family and superfamily status due to being recovered as the sister taxon to all other patellogastropods with mitochondrial markers [72]. None of our results recover this position, but rather indicate that the genus is either part of Lottiidae (most ML and Bayesian results), which was its original assignment, or is sister group to the Lottioidea families Neolepetopsidae (Paralepetopsis) and Lottiidae (Patelloida, Nipponacmea, Lottia, Testudinalia) (coalescent-based trees and one ML tree) (figure 2).
Neritimorphs had mostly congruent phylogenies recovered from 28S rRNA [74] and mitogenomes [75]. Our reconstruction supports the same topology, with Neritopsoidea (Titiscania) as the sister group to all other neritimorphs, followed by the divergence between Helicinoidea (Pleuropoma) and Neritoidea (figure 2). Within the latter, we recover a monophyletic Neritidae as the sister group of Phenacolepadidae (Thalassonerita). The nested position of Smaragdia inside Neritininae disagrees with the current classification of the genus in its own subfamily [64,73].
Vetigastropoda and Heterobranchia had similar taxon representation as in Zapata et al. [16] (with newly sequenced replacements for some vetigastropod families). As expected, the relationships are the same, and highlight the need for future studies focused on each group, given the uncertain position of Haliotis in Vetigastropoda, and low resolution of internal relationships of panpulmonates in Heterobranchia (figure 2). Our results contrast with recent mitogenomic analyses of vetigastropods, which recovered a monophyletic group of Seguenzioidea (Granata), Lepetodriloidea (Lepetodrilus) and Haliotoidea (Haliotis) [14,76,77].
We substantially increased sampling of Caenogastropoda by adding the latest published transcriptomes of eight families. Despite that, caenogastropods are the most diverse gastropod lineage, with over a hundred families, and the following results are still limited in sampling. We recover a monophyletic Neogastropoda; its internal relationships differ from a molecular study with denser taxon sampling [78], in that we find Buccinoidea (Cumia, Volegalea) closer to Conoidea (Conus, Crassispira) than to Muricoidea (Urosalpinx). We also recover a monophyletic Truncatelloidea (Bithynia, Oncomelania) as the sister group to all other Hypsogastropoda (figure 2). The relative position of Tonnoidea (Charonia) and Calyptraeoidea (Crepidula) regarding Neogastropoda is unclear; nonetheless, the close relationship between Tonnoidea, Neogastropoda and also Stromboidea (Lobatus) agrees with previous molecular studies [78,79]. The branching pattern of the closest relatives of neogastropods reveals a paraphyletic Littorinimorpha.
Acknowledgements
We are grateful to James Reimer for kindly providing support for fieldwork in Okinawa. We also thank Vanessa Knutson, Shawn Miller, Hin Boo Wee, Kristen Soong and Taku Ohara for help in the field. Don Colgan (Australian Museum) and Lluis Cardona (University of Barcelona) each donated a specimen. The computations in this paper were done on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. We thank software developers and community forums for help with software, in particular Brian Haas (Trinity), Adrian Altenhoff (OMA) and Bui Quang Minh (IQ-TREE). The manuscript was greatly improved by comments from Juan Moles, Bruno de Medeiros and two anonymous reviewers. We thank Bruno de Medeiros for many discussions and help with scripts.
Data accessibility
New transcriptomes were deposited in the NCBI Sequence Read Archive (BioProject PRJNA508436). Electronic supplementary materials are deposited in Harvard Dataverse: https://doi.org/10.7910/DVN/O85KLQ. Scripts are available from: https://github.com/tauanajc/Cunha_Giribet_2019_ProcRSocB.
Authors' contributions
T.J.C. and G.G. conceived the study, collected and identified specimens. T.J.C. carried out laboratory work, analysed the data and drafted the manuscript. Both authors improved the manuscript and gave final approval for publication.
Competing interests
The authors have no competing interests.
Funding
Collection of most new biological material used in this paper was made possible by the MCZ Putnam Expeditions Grants. Laboratory work and sequencing was funded by internal funds from the MCZ and the Faculty of Arts and Sciences, Harvard University. T.J.C. received a student research grant from the Society of Systematic Biologists, a doctoral stipend from the Department of Organismic and Evolutionary Biology at Harvard University and a Faculty for the Future Fellowship from the Schlumberger Foundation. Published as open access by a grant from the Wetmore Colles Fund.
References
- 1.Appeltans W, et al. 2012. The magnitude of global marine species diversity. Curr. Biol. 22, 2189–2202. ( 10.1016/j.cub.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 2.Thiele J. 1929. Handbook of Systematic Malacology. Part 1. Translation of: Handbuch der systematischen Weichtierkunde. Washington, DC: Smithsonian Institution Libraries, National Science Foundation. [Google Scholar]
- 3.Haszprunar G. 1988. On the origin and evolution of major gastropods group, with special reference to the streptoneura. J. Mollus. Stud. 54, 367–441. ( 10.1093/mollus/54.4.367) [DOI] [Google Scholar]
- 4.Ponder WF, Lindberg DR. 1996. Gastropod phylogeny—challenges for the 90s. In Origin and evolutionary radiation of the mollusca (ed. Taylor JD.), pp. 135–154. Oxford, UK: Oxford University Press. [Google Scholar]
- 5.Ponder WF, Lindberg DR. 1997. Towards a phylogeny of gastropod molluscs: an analysis using morphological characters. Zool. J. Linn. Soc. 119, 83–265. ( 10.1006/zjls.1996.0066) [DOI] [Google Scholar]
- 6.Aktipis SW, Giribet G, Lindberg DR, Ponder WF. 2008. Gastropoda, an overview and analysis. In Phylogeny and evolution of the Mollusca (eds Ponder WF, Lindberg DR), pp. 201–237. Berkeley, CA: University of California Press. [Google Scholar]
- 7.Harasewych MG, Adamkewicz SL, Blake JA, Saudeck D, Spriggs T, Bult CJ. 1997. Phylogeny and relationship of pleurotomariid gastropods (Mollusca: Gastropoda): an assessment based on partial 18S rDNA and cytochrome c oxydase I sequences. Mol. Mar. Biol. Biotech. 6, 1–20. [PubMed] [Google Scholar]
- 8.Harasewych MG, Adamkewicz SL, Plassmeyer M, Gillevet PM. 1998. Phylogenetic relationships of the lower Caenogastropoda (Mollusca, Gastropoda, Architaenioglossa, Campaniloidea, Cerithioidea) as determined by partial 18S rDNA sequences. Zool. Scr. 27, 361–372. ( 10.1111/j.1463-6409.1998.tb00467.x) [DOI] [Google Scholar]
- 9.Colgan DJ, Ponder WF, Eggler PE. 2000. Gastropod evolutionary rates and phylogenetic relationships assessed using partial 28S rDNA and histone H3 sequences. Zool. Scr. 29, 29–63. ( 10.1046/j.1463-6409.2000.00021.x) [DOI] [Google Scholar]
- 10.Colgan DJ, Ponder WF, Beacham E, Macaranas JM. 2003. Gastropod phylogeny based on six segments from four genes representing coding or non-coding and mitochondrial or nuclear DNA. Mollusc. Res. 23, 123–148. ( 10.1071/MR03002) [DOI] [Google Scholar]
- 11.McArthur AG, Harasewych MG. 2003. Molecular systematics of the major lineages of the Gastropoda. In Molecular systematics and phylogeography of mollusks (eds Lydeard C, Lindberg DR), pp. 140–160. Washington, DC: Smithsonian Books. [Google Scholar]
- 12.Grande C, Templado J, Zardoya R. 2008. Evolution of gastropod mitochondrial genome arrangements. BMC Evol. Biol. 8, 61 ( 10.1186/1471-2148-8-61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams ST, Foster PG, Littlewood DTJ. 2014. The complete mitochondrial genome of a turbinid vetigastropod from MiSeq Illumina sequencing of genomic DNA and steps towards a resolved gastropod phylogeny. Gene 533, 38–47. ( 10.1016/j.gene.2013.10.005) [DOI] [PubMed] [Google Scholar]
- 14.Uribe JE, Kano Y, Templado J, Zardoya R. 2015. Mitogenomics of Vetigastropoda: insights into the evolution of pallial symmetry. Zool. Scr. 45, 145–159. ( 10.1111/zsc.12146) [DOI] [Google Scholar]
- 15.Uribe JE, Irisarri I, Templado J, Zardoya R. 2019. New patellogastropod mitogenomes help counteracting long-branch attraction in the deep phylogeny of gastropod mollusks. Mol. Phylogenet. Evol. 133, 12–23. ( 10.1016/j.ympev.2018.12.019) [DOI] [PubMed] [Google Scholar]
- 16.Zapata F, Wilson NG, Howison M, Andrade SCS, Jorger KM, Schrodl M, Goetz FE, Giribet G, Dunn CW. 2014. Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc. R. Soc. B 281, 20141739 ( 10.1098/rspb.2014.1739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster PG. 2004. Modeling compositional heterogeneity. Syst. Biol. 53, 485–495. ( 10.1080/10635150490445779) [DOI] [PubMed] [Google Scholar]
- 18.Lopez P, Casane D, Philippe H. 2002. Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 19, 1–7. ( 10.1093/oxfordjournals.molbev.a003973) [DOI] [PubMed] [Google Scholar]
- 19.Whelan NV, Kocot KM, Moroz LL, Halanych KM. 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl Acad. Sci. USA 112, 5773–5778. ( 10.1073/pnas.1503453112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannon JT, Vellutini BC, Smith J, Ronquist F, Jondelius U, Hejnol A. 2016. Xenacoelomorpha is the sister group to Nephrozoa. Nature 530, 89–93. ( 10.1038/nature16520) [DOI] [PubMed] [Google Scholar]
- 21.Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 31, 2963–2984. ( 10.1093/molbev/msu235) [DOI] [PubMed] [Google Scholar]
- 22.Degnan JH, Rosenberg NA. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340. ( 10.1016/j.tree.2009.01.009) [DOI] [PubMed] [Google Scholar]
- 23.Salichos L, Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497, 327–331. ( 10.1038/nature12130) [DOI] [PubMed] [Google Scholar]
- 24.Tian Y, Kubatko LS. 2017. Expected pairwise congruence among gene trees under the coalescent model. Mol. Phylogenet. Evol. 106, 144–150. ( 10.1016/j.ympev.2016.09.023) [DOI] [PubMed] [Google Scholar]
- 25.Fryda J, Nützel A, Wagner PJ. 2008. Paleozoic Gastropoda. In Phylogeny and evolution of the Mollusca (eds Ponder WF, Lindberg DR), pp. 239–270. Berkeley, CA: University of California Press. [Google Scholar]
- 26.Song L, Florea L. 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 4, 48 ( 10.1186/s13742-015-0089-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krueger F.2018. Trim Galore. See https://github.com/FelixKrueger/TrimGalore .
- 28.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. ( 10.1093/nar/gks1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feijão PC, Neiva LS, Azeredo-Espin AML, Lessinger AC. 2006. AMiGA: the arthropodan mitochondrial genomes accessible database. Bioinformatics 22, 902–903. ( 10.1093/bioinformatics/btl021) [DOI] [PubMed] [Google Scholar]
- 31.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res. 44, D67–D72. ( 10.1093/nar/gkv1276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. ( 10.1093/bioinformatics/btl158) [DOI] [PubMed] [Google Scholar]
- 35.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. ( 10.1093/bioinformatics/bts565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 37.Altenhoff AM, et al. 2018. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 46, D477–D485. ( 10.1093/nar/gkx1019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster PG.2018. p4 (Python package). See http://p4.nhm.ac.uk/tutorial/tut_compo.html .
- 41.Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, Giribet G. 2018. Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. eLife 7, 1–32. ( 10.7554/eLife.36278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24, 715–716. ( 10.1093/bioinformatics/btm619) [DOI] [PubMed] [Google Scholar]
- 43.Dayhoff MO, Schwartz RM, Orcutt BC. 1978. A model of evolutionary change in proteins. In Atlas of protein sequence and structure (ed. Dayhoff MO.), pp. 345–352. National Biomedical Research Foundation. [Google Scholar]
- 44.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31, i44–i52. ( 10.1093/bioinformatics/btv234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. ( 10.1038/nmeth.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chernomor O, von Haeseler A, Minh BQ.. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65, 997–1008. ( 10.1093/sysbio/syw037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615. ( 10.5061/dryad.c459h) [DOI] [PubMed] [Google Scholar]
- 49.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quang LS, Gascuel O, Lartillot N. 2008. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24, 2317–2323. ( 10.1093/bioinformatics/btn445) [DOI] [PubMed] [Google Scholar]
- 51.Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. ( 10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 52.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. ( 10.1111/2041-210X.12628) [DOI] [Google Scholar]
- 53.Molloy EK, Warnow T. 2018. To include or not to include: the impact of gene filtering on species tree estimation methods. Syst. Biol. 67, 285–303. ( 10.1093/sysbio/syx077) [DOI] [PubMed] [Google Scholar]
- 54.Castro LR, Colgan DJ. 2010. The phylogenetic position of Neritimorpha based on the mitochondrial genome of Nerita melanotragus (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 57, 918–923. ( 10.1016/j.ympev.2010.08.030) [DOI] [PubMed] [Google Scholar]
- 55.van den Biggelaar JAM, Haszprunar G. 1996. Cleavage patterns and mesentoblast formation in the Gastropoda: an evolutionary perspective. Evolution 50, 1520–1540. ( 10.1111/j.1558-5646.1996.tb03925.x) [DOI] [PubMed] [Google Scholar]
- 56.Lindberg DR, Guralnick RP. 2003. Phyletic patterns of early development in gastropod molluscs. Evol. Dev. 5, 494–507. ( 10.1046/j.1525-142X.2003.03055.x) [DOI] [PubMed] [Google Scholar]
- 57.Lambert JD. 2008. Mesoderm in spiralians: the organizer and the 4d cell. J. Exp. Zool. Part B: Mol. Dev. Evol. 310B, 15–23. ( 10.1002/jez.b.21176) [DOI] [PubMed] [Google Scholar]
- 58.Hejnol A. 2010. A twist in time—the evolution of spiral cleavage in the light of animal phylogeny. Integr. Comp. Biol. 50, 695–706. ( 10.1093/icb/icq103) [DOI] [PubMed] [Google Scholar]
- 59.Lambert JD. 2010. Developmental patterns in spiralian embryos. Curr. Biol. 20, R72–R77. ( 10.1016/j.cub.2009.11.041) [DOI] [PubMed] [Google Scholar]
- 60.Lindberg DR. 2008. Patellogastropoda, Neritimorpha, and Cocculinoidea: the low-diversity gastropod clades. In Phylogeny and evolution of the Mollusca (eds Ponder WF, Lindberg DR), pp. 271–296. Berkeley, CA: University of California Press. [Google Scholar]
- 61.Ponder WF, Colgan DJ, Healy JM, Nutzel A, Simone LRL, Strong EE. 2008. Caenogastropoda. In Phylogeny and evolution of the mollusca (eds Ponder WF, Lindberg DR), pp. 331–383. Berkeley, CA: University of California Press. [Google Scholar]
- 62.Giese AC, Pearse JS (eds). 1977. Reproduction of marine invertebrates. Volume IV molluscs: gastropods and cephalopods. New York, NY: Academic Press. [Google Scholar]
- 63.Geiger DL, Nützel A, Sasaki T. 2008. Vetigastropoda. In Phylogeny and evolution of the Mollusca (eds Ponder WF, Lindberg DR), pp. 297–330. Berkeley, CA: University of California Press. [Google Scholar]
- 64.Bouchet P, Rocroi J-P, Hausdorf B, Kaim A, Kano Y, Nützel A, Parkhaev P, Schrödl M, Strong EE. 2017. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia 61, 1–526. ( 10.4002/040.061.0201) [DOI] [Google Scholar]
- 65.Kocot KM, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477, 452–456. ( 10.1038/nature10382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SCS, Rouse GW, Giribet G, Dunn CW. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–369. ( 10.1038/nature10526) [DOI] [PubMed] [Google Scholar]
- 67.Laumer CE. 2018. Inferring ancient relationships with genomic data: a commentary on current practices. Integr. Comp. Biol. 58, 623–639. ( 10.1093/icb/icy075) [DOI] [PubMed] [Google Scholar]
- 68.Nakano T, Sasaki T. 2011. Recent advances in molecular phylogeny, systematics and evolution of patellogastropod limpets. J. Mollusc. Stud. 77, 203–217. ( 10.1093/mollus/eyr016) [DOI] [Google Scholar]
- 69.Sasaki T. 1998. Comparative anatomy and phylogeny of the Recent Archaeogastropoda (Mollusca: Gastropoda). Univ. Tokyo Bull. 38, 1–223. [Google Scholar]
- 70.Nakano T, Ozawa T. 2004. Phylogeny and historical biogeography of limpets of the order Patellogastropoda based on mitochondrial DNA sequences. J. Mollusc. Stud. 70, 31–41. ( 10.1093/mollus/70.1.31) [DOI] [Google Scholar]
- 71.Yoon SH, Kim W. 2007. 18S ribosomal DNA sequences provide insight into the phylogeny of patellogastropod limpets (Mollusca: Gastropoda). Mol. Cells 23, 64–71. [PubMed] [Google Scholar]
- 72.Nakano T, Ozawa T. 2007. Worldwide phylogeography of limpets of the order Patellogastropoda: molecular, morphological and palaeontological evidence. J. Mollusc. Stud. 73, 79–99. ( 10.1093/mollus/eym001) [DOI] [Google Scholar]
- 73.WoRMS. 2018. World register of marine species. See http://www.marinespecies.org
- 74.Kano Y, Chiba S, Kase T. 2002. Major adaptive radiation in neritopsine gastropods estimated from 28S rRNA sequences and fossil records. Proc. R. Soc. B 269, 2457–2465. ( 10.1098/rspb.2002.2178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uribe JE, Colgan DJ, Castro LR, Kano Y, Zardoya R. 2016. Phylogenetic relationships among superfamilies of Neritimorpha (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 104, 21–31. ( 10.1016/j.ympev.2016.07.021) [DOI] [PubMed] [Google Scholar]
- 76.Lee H, Samadi S, Puillandre N, Tsai MH, Dai CF, Chen WJ. 2016. Eight new mitogenomes for exploring the phylogeny and classification of Vetigastropoda. J. Mollusc. Stud. 82, 534–541. ( 10.1093/mollus/eyw027) [DOI] [Google Scholar]
- 77.Wort EJG, Fenberg PB, Williams ST. 2017. Testing the contribution of individual genes in mitochondrial genomes for assessing phylogenetic relationships in Vetigastropoda. J. Mollusc. Stud. 83, 123–128. ( 10.1093/mollus/eyw044) [DOI] [Google Scholar]
- 78.Zou S, Li Q, Kong L. 2011. Additional gene data and increased sampling give new insights into the phylogenetic relationships of Neogastropoda, within the caenogastropod phylogenetic framework. Mol. Phylogenet. Evol. 61, 425–435. ( 10.1016/j.ympev.2011.07.014) [DOI] [PubMed] [Google Scholar]
- 79.Colgan DJ, Ponder WF, Beacham E, Macaranas J. 2007. Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca). Mol. Phylogenet. Evol. 42, 717–737. ( 10.1016/j.ympev.2006.10.009) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
New transcriptomes were deposited in the NCBI Sequence Read Archive (BioProject PRJNA508436). Electronic supplementary materials are deposited in Harvard Dataverse: https://doi.org/10.7910/DVN/O85KLQ. Scripts are available from: https://github.com/tauanajc/Cunha_Giribet_2019_ProcRSocB.