Abstract
Leukemogenic potential of MLL fusion with the coiled-coil domain-containing partner genes and the downstream target genes of this type of MLL fusion have not been clearly investigated. In this study, we demonstrated that the coiled-coil–four-helix bundle structure of EB1 that participated in the MLL/EB1 was required for immortalizing mouse bone marrow (BM) cells and producing myeloid, but not lymphoid, cell lines. Compared to MLL/AF10, MLL/EB1 had low leukemogenic ability. The MLL/EB1 cells grew more slowly owing to increased apoptosis in vitro and induced acute monocytic leukemia with an incomplete penetrance and longer survival in vivo. A comparative analysis of transcriptome profiling between MLL/EB1 and MLL/AF10 cell lines revealed that there was an at least two-fold difference in the induction of 318 genes; overall, 51.3% (163/318) of the genes were known to be bound by MLL, while 15.4% (49/318) were bound by both MLL and MLL/AF9. Analysis of the 318 genes using Gene Ontology–PANTHER overrepresentation test revealed significant differences in several biological processes, including cell differentiation, proliferation/programmed cell death, and cell homing/recruitment. The Ets1 gene, bound by MLL and MLL/AF9, was involved in several biological processes. We demonstrated that Ets1 was selectively upregulated by MLL/EB1. Short hairpin RNA knockdown of Ets1 in MLL/EB1 cells reduced the expression of CD115, apoptosis rate, competitive engraftment to BM and spleen, and incidence of leukemia and prolonged the survival of the diseased mice. Our results demonstrated that MLL/EB1 upregulated Ets1, which controlled the balance of leukemia cells between apoptosis and BM engraftment/clonal expansion.
Novelty and impact of this study
The leukemogenic potential of MLL fusion with cytoplasmic proteins containing coiled-coil dimerization domains and the downstream target genes of this type of MLL fusion remain largely unknown. Using a retroviral transduction/transplantation mouse model, we demonstrated that MLL fusion with the coiled-coil–four-helix bundle structure of EB1 has low leukemogenic ability; Ets1, which is upregulated by MLL/EB1, plays a critical role in leukemic transformation by balance between apoptosis and BM engraftment/clonal expansion.
Abbreviations: aa, amino acid; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; AMoL, acute monocytic leukemia; APC, allophycocyanin; BD, DNA-binding domain; BM, bone marrow; CBC, complete blood cell; CC, coiled-coil; cDNA, complementary DNA; CFC, colony forming capacity; CTD, C-terminal domain; DAPI, 4′,6-diamidino-2-phenylindole; FHB, four-helix bundle; GM-CSF, granulocyte-monocyte colony stimulating factor; H&E, hematoxylin and eosin; IL, interleukin; ip, intraperitoneally; PB, peripheral blood; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PE, phycoerythrin; PI, propidium iodide; RT, reverse transcription; SCF, stem cell factor; shRNA, short hairpin RNA; WBC, white blood cell; 5-FU, 5-florouracil
Introduction
Chromosomal rearrangements involving the MLL (recently renamed KMT2A, also called ALL1, HTRX1, and HRX) gene at 11q23 are detected in patients with acute lymphoid and myeloid leukemias.1 In general, MLL rearrangements are predictive of a poor clinical outcome.[2], [3] To date, 135 MLL rearrangements have been found in leukemia patients, in whom 94 MLL partner genes have been identified.4 Half of the MLL partner genes encode nuclear proteins, and the rest encode cytoplasmic proteins. Among the nuclear partner genes, AF4 (AFF1), AF9 (MLLT3), ENL (MLLT1), AF10 (MLLT10), or ELL accounts for ~80% of cases.[4], [5], [6], [7] Structural/functional studies revealed MLL fusion with the transcriptional effector domains of the nuclear proteins involved the recruitment of DOT1L-containing complexes which epigenetically modify the MLL target genes and aberrantly sustain their expression.8 The most well-known MLL and MLL fusion target genes are 5′ HOXA (HOXA7, HOXA9, HOXA10) and MEIS1. Evidence indicates that overexpression of Hoxa9 and Meis1 induces leukemic transformation.9 Recently, Xu et al. (2016) performed chromatin immunoprecipitation sequencing analysis and identified 5233 MLL1- and 3140 MLL/AF9-binding genes, with 1369 joint targets including 5′ Hoxa and Meis1 genes, in the mouse MLL/AF9 cells. They also demonstrated that MLL/AF9-binding sites are enriched for the sequences of embryonic or T-cell transcription factors, whereas MLL1 is preferentially recruited to the chromatin regions that are enriched for the consensus sequences of ETS family transcription factors, including ETS1, PU.1, ERG, and ETV1.10 The transcription factors of ETS family are involved in stem cell development, cell proliferation, survival, and tumorigenesis.11
Leukemic transformation induced by MLL fusion with the cytoplasmic fusion partners is less understood. Evidence shows that the coiled-coil dimerization domain of AF1p (EPS15), GAS7, and gephyrin or the self-association domain of AF6 (MLLT4) was required for MLL fusion-mediated leukemia development.[12], [13], [14], [15] MLL/AF1p and MLL/GAS7 also activated homeobox genes13; however, Hoxa7 and Hoxa9 were not required for the MLL/GAS7-induced leukemic transformation.16 Thus, the downstream target genes that participate in such a type of MLL fusion-induced leukemic transformation remain to be elucidated.
We previously identified a novel MLL fusion partner gene, EB1 (MAPRE1), from a patient having de novo pro-B acute lymphoblastic leukemia (ALL).17 MLL exhibits 36 exons (NM_005933.3) and EB1 exhibits 7 exons (NM_012325.3). The resultant MLL/EB1 fusion protein is composed of MLL exons 1 to 8 (aa 1-1362) fused to EB1 exons 6 to 7 (aa 200-268). EB1 is a microtubule plus-end trafficking protein and is involved in cytoskeleton and spindle formation.18 An examination of the EB1 crystal structure revealed a coiled-coil–four-helix bundle structure at its carboxy-terminal end, and the coiled-coil domain from amino acid [aa] 211 to aa 229 conferred a homo-dimerization activity.[19], [20] In this study, we performed retroviral transduction/transplantation and structural/functional assays to investigate the leukemogenic potential of MLL/EB1. We also compared transcriptome profiling between the MLL/EB1 and MLL/AF10 cell lines as well as identified and investigated the role of the Ets1 gene, which is selectively upregulated by MLL/EB1 during MLL/EB1-induced leukemic transformation.
Materials and Methods
Plasmid Construction
Total RNA was prepared from patient leukemia cells using Trizol reagent (Gibco BRL, Gaithersburg, MD). Complementary DNA (cDNA) was synthesized from the total RNA by reverse transcription (RT) using oligo-dT and Superscript II reverse transcriptase (Life Technologies, Rockville, MD). The gene fragments of truncated EB1 (tEB1, encoding aa 200-268 of EB1), truncated MLL (tMLL, encoding aa 1-1362 of MLL), MLL/EB1 (encoding aa 1-1362 of MLL and aa 200-268 of EB1), MLL/EB1∆αBC (encoding aa 1-1362 of MLL and aa 200-234 of EB1), and MLL/EB1∆αA (encoding aa 1-1362 of MLL and aa 235-268 of EB1) (Figure 1, A and C) were amplified from a cDNA by performing polymerase chain reaction (PCR) using the Expand Long Template PCR system (Roche Applied Science, Mannheim, Germany). The amplified gene fragments were cloned into the EcoRI and XhoI sites of the retroviral vector pMSCVneo (Clontech, Palo Alto, CA). Construction of pMSCVneo carrying MLL/AF10(OM-LZ) has been described elsewhere. For performing the luciferase reporter assay, the promoter region of Ets1 (nucleotide [nt] −703 to +293 around the transcription start site of Ets1 variant 1, NM_011808) (Supplementary Figure 1A) was amplified using PCR and cloned into the KpnI and BglII sites of the luciferase vector pGL4.20 (Promega, Madison, WI) to generate PEts1-luc. The promoter region of human HOXA7 (nt −1988 to +21 around the transcription start site of HOXA7, NM_006896.3) was PCR-amplified and cloned into the KpnI and SacI sites of the luciferase vector pGL2-basic (Promega) to generate PHOXA7-luc. Furthermore, the promoter regions of HOXA9 (nt −988 to −87 near the transcription start site of HOXA9, NM_152739.3) and MEIS1 (nt −1430 to +453 around the transcription start site of MEIS1, NM_002398.2) were cloned into the XhoI and HindIII sites of pGL2-basic to generate PHOXA9-luc and PMEIS1-luc, respectively. The fidelity of the cloned gene fragments was confirmed by DNA sequencing.
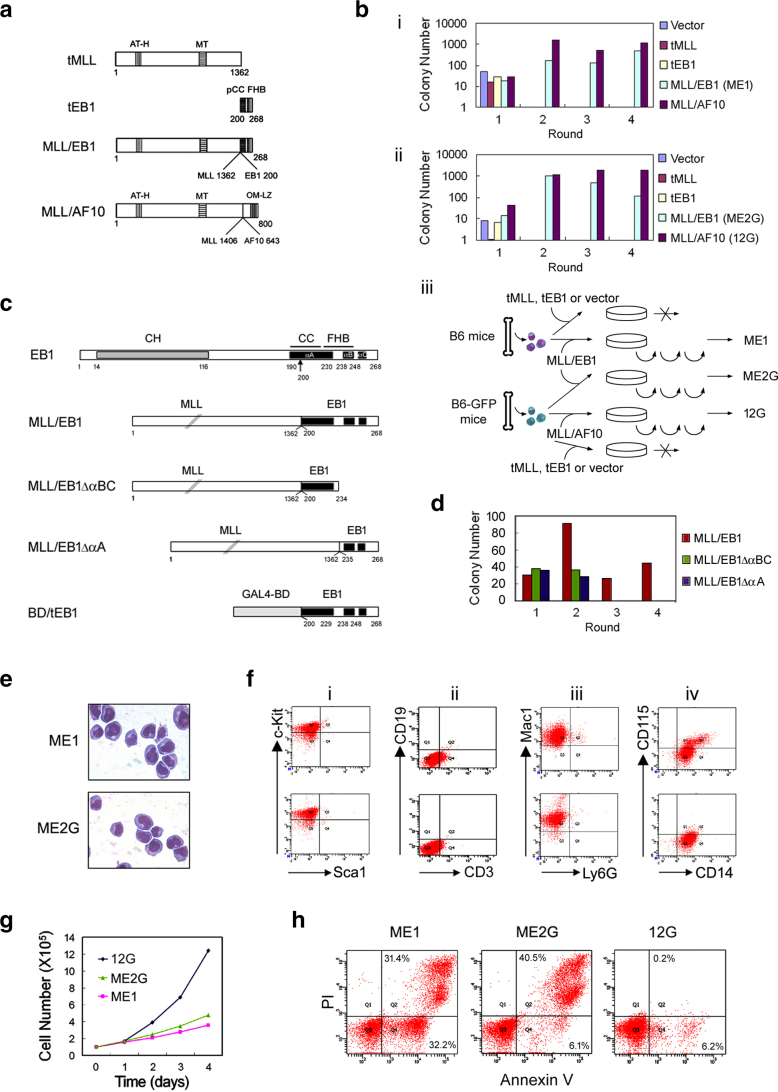
Figure 1.
Establishment and characterization of the MLL/EB1-immortalized cell lines. (A) Schematic representations of truncated MLL (tMLL), truncated EB1 (tEB1), MLL/EB1, and MLL/AF10(OM-LZ) fusion proteins. AT-H, AT hooks; MT, DNA methyltransferase domain; pCC, partial coiled coil domain; FHB, four-helix bundle; OM, octapeptide motif; LZ, leucine zipper. The numbers indicate the amino acid of each protein. (B) Transduction efficiency of the blank retrovirus (Vector) or retroviruses expressing tMLL, tEB1, MLL/EB1, and MLL/AF10(OM-LZ) in two independent experiments using B6 (i) or B6-GFP mice BM cells (ii). The bars indicate the colonies generated per 3 × 104 transduced murine BM cells. MLL/AF10(OM-LZ) was used as a positive control. ME1 and ME2G were MLL/EB1-immortalized cell lines generated by adaptation of the cells, which were pooled from the fourth round of plating, to liquid culture (iii). 12G was the MLL/AF10(OM-LZ)-immortalized cell line (iii). (C) Schematic representation of EB1, MLL/EB1, MLL/EB1∆αBC, and MLL/EB1∆αA. CH, calponin homology domain; CC, coiled coil domain; FHB, four-helix bundle; αA, αB, and αC, the three α-helices of EB1. (D) Transduction efficiency of the retroviruses expressing truncated and fusion genes. The bars indicate the colonies generated per 1 × 104 transduced mouse BM cells. (E) Liu staining of the MLL/EB1-immortalized cell lines cultured in the medium containing IL3 (oil immersion, ×1000). (F) Immunophenotypic analysis of the MLL/EB1-immortalized cell lines [upper panel (ME1), lower panel (ME2G)]. (G) Growth curve of the MLL/EB1-immortalized cell lines (ME1 and ME2G) and MLL/AF10(OM-LZ)-immortalized cell line (12G) in the medium containing IL3. (H) Flow cytometry analysis of live and apoptotic cells by staining with Annexin V and propidium iodide (PI).
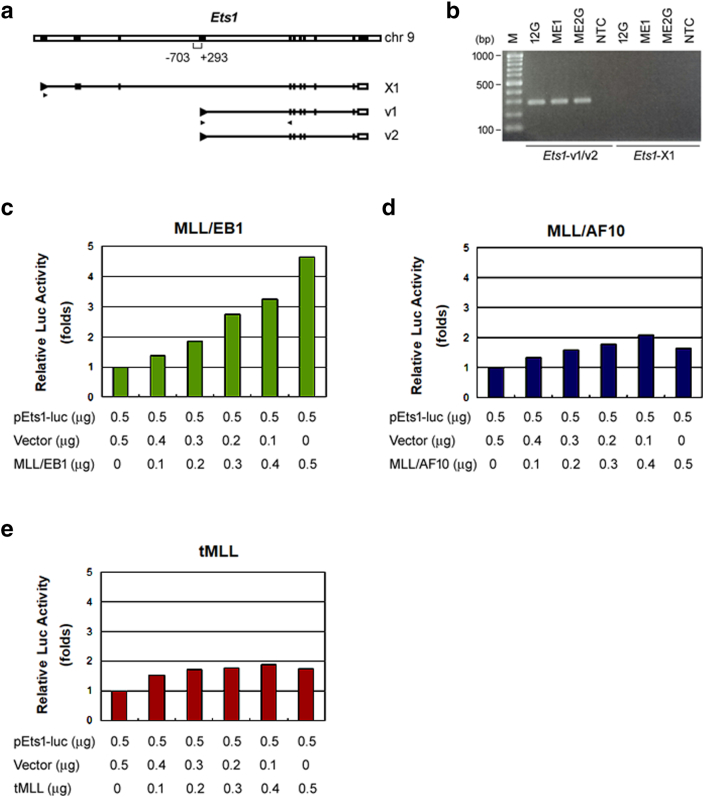
Supplementary Figure 1.
MLL/EB1 transactivates mouse Ets1 proximal promoter. (a) Schematic representation of mouse Ets1 gene and its transcript variants. Variants 1 and 2 (v1/v2) are generated from the proximal promoter and variant X1 is generated from the distal promoter. Black boxes indicate exons. A promoter region of ~1-kb around the transcription start site of v1/v2 (nucleotides -703 to +293) was cloned to generate a luciferase reporter plasmid, pEts1-luc. Triangles under the transcripts indicate the locations of primer sets used to amplify v1/v2 or X1. (b) RT-PCR analysis detected PCR products amplified from Ets1 transcripts v1/v2, but not X1, in the 12G, ME1, and ME2G cells. (c-e) Fold change in luciferase activity of the 32Dcl3 cells transfected with pEts1-luc reporter plasmid plus the empty vector or a gradient amount of vector-expressing tMLL, MLL/EB1 or MLL/AF10(OM-LZ). Error bars indicate SD of mean. Results are representative of three independent experiments.
In Vitro Retroviral Transduction/Replating and In Vivo Leukemogenesis Assays
The retroviral production, retroviral transduction of murine BM cells, and intraperitoneal (ip) injection of immortalized BM cell lines into mice were performed as described previously21 with some modifications. The titer of virus was determined using NIH3T3 cells, and the viral titer of MLL/EB1 was around 2 × 105/ml. Briefly, the BM cells were prepared from C57BL/6J-Tg(Pgk1-EGFP) 03N male mice (GFP-B6, National Laboratory Animal Center, Taiwan) or C57BL/6J (B6) male mice 5 days after injecting them with 5-florouracil (5-FU; 150 μg/g). BM cells were transduced with retroviruses at an MOI of 1 and selected by replating four times in a methylcellulose medium (M3231, Stem Cell Technologies, Vancouver, BC, Canada) containing lymphoid differentiation-inducing cytokines [20 ng/ml stem cell factor (SCF), 10 ng/ml interleukin 3 (IL3), 10 ng/ml IL7, and 100 ng/ml flt3 ligand] or containing myeloid differentiation-inducing cytokines [20 ng/ml SCF and 10 ng/ml each of IL3, IL6, and granulocyte-monocyte colony stimulating factor (GM-CSF)]. Cells pooled from the colonies grown during the fourth round of plating were adapted to liquid culture [1× RPMI medium containing IL3 (10 ng/ml)]. To determine in vivo leukemogenic potential, 1 × 106 immortalized cells or 1.5 × 105-3 × 105 MLL fusion-transduced BM cells were ip injected into sublethally irradiated (a dose of 5.25 Gy total-body γ-irradiation) male B6 mice. To monitor leukemia development, peripheral blood (PB) was collected from the transplanted mice every week for complete blood count (CBC) analysis using a hemocytometer (Hemavet 950, Drew Scientific, Oxford, CT).
Phenotypic Analyses
The hematological lineage of mouse BM cells and immortalized cell lines was determined using flow cytometry (FACS-Calibur or CantoII, Becton-Dickinson, Mountain View, CA), which employed phycoerythrin (PE)-Mac1, PE-CD115, PE-CD19, PE-B220, PE-Cy7-c-kit, allophycocyanin (APC)-Sca1, APC-CD3, APC-CD14 (eBioscience, San Diego, CA), and APC-Ly6G (Miltenyi Biotec, Auburn, CA, USA). The liver, spleen, and lymph node collected from the moribund mice were fixed in buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) using the standard technique. Stamp specimens of the tissues, PB smears, and BM cytospin preparations were stained with Liu reagents (Handsel Technologies, Taipei, Taiwan).
Cell Growth and Apoptosis Analyses
Cell proliferation was evaluated by counting cell numbers every day or by performing WST assay (BioVision, Mountain View, CA). For cell apoptosis analysis, cells were stained with Pacific Blue-conjugated Annexin V (Biolegend, San Diego, CA) and propidium iodide (Annexin V apoptosis detection kit, BD Pharmingen, San Diego, CA) according to the manufacturer's instruction. The stained cells were analyzed immediately using a flow cytometer (FACS-Calibur; Becton-Dickinson).
Luciferase Reporter Assay
32Dcl3 cells, a murine myeloid leukemia cell line, were transfected with pMSCVneo-expressing plasmid [empty vector, tEB1 (for homeobox genes only), tMLL, MLL/EB1 or MLL/AF10] and promoter-luciferase vector (pEts1-luc, pHOXA7-luc, pHOXA9-luc, or pMEIS1-luc) using LT1 liposome (Mirus, Madison, WI). pRL-TK (Promega) was used as an internal control (for homeobox genes only). Cells were harvested at 24 hours after transduction, and luciferase activity was determined using the dual-luciferase assay kit (Promega) and a luminometer (GLOMA X 20/20, Promega).
Microarray Analysis and Gene Ontology–PANTHER Enrichment Analysis
The total RNA of the cells was amplified, labeled, and hybridized to the mouse genome 430 2.0 Array chips (Affymetrix, Santa Clara, CA) by following the manufacturer's instructions (this procedure was performed by the staff of the Genomic Medicine Research Core Laboratory at Chang Gung Memorial Hospital, Linkou, Taiwan). Microarray data are available at NCBI GEO website (accession number: GSE82156). Differentially expressed genes with a fold change of at least two-fold between MLL/EB1 and MLL/AF10 cell lines were classified into functional groups of biological process using an online Gene Ontology (GO) enrichment analysis–PANTHER classification system (GO database released 2018-09-06) (http://geneontology.org/page/go-enrichment-analysis). Statistical significance was evaluated using the Fisher's exact test and corrected by Bonferroni correction for multiple testing. The biological process categories of GO having P values of <.05 were considered to be statistically significant.
RT-PCR and Quantitative RT-PCR (RT-qPCR) Analyses
To determine the gene expression level, quantitative PCRs were performed in triplicate using SYBR Green PCR mastermix and analyzed using the ABI Prism 7500 system (Applied Biosystems, Foster City, CA). Primer sets used in PCR were 5′-ATC CAG CTG TGG CAG TTT CTT and 5′-CAC GGC TCA GTT TCT CAT AAT for Ets1, 5′-CTG AGT CAG AAG CCC TTC GAC and 5′-CCC AGA CCA AAG GCT GTA GCC for CD115, 5′-CTG AAG GGG GCC TGT ATG TG and 5′-CAT TCT TCG ATT TTG TCT GC for Cybb, 5′-CCC ACA GGC AGC ACA GTG GAC and 5′-GGA GGC CGA GGA GGA CCA GG for CD68, and 5′-TTC ACC ACC ATG GAG AAG GC and 5′-GGC ATG GAC TGT GGT CAT GA for Gapdh. Gene expression was normalized by calculating the difference between threshold cycle numbers of Gapdh and target genes (∆Ct) for each sample. The difference in gene expression level relative to control cells was obtained by calculating the difference between ∆Ct of the control and target cell lines (∆∆Ct). Fold change was calculated as 2−∆∆Ct.
To detect which Ets1 transcription variant was expressed in the cell lines, RT-PCR was performed using the following primer sets: 5′-CGG ACT GGC TGG GCG CGC AC and 5′-GGA GTT AAC AGC GGG ACA TCT for variants 1 and 2 and 5′-ACC CGT CTC TGC AGC AAA TTA and 5′-GGA GTT AAC AGC GGG ACA TCT for variant X1.
Gene Knockdown by shRNA
Lentivirus vector (pLKO_TRC025) and lentivirus that expressed shRNA against mouse Ets1 (TRCN0000042639 and TRCN0000042642) were obtained from the National RNAi Core Facility Platform (Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan). Cells were infected with lentivirus in the presence of polybrene (4 μg/ml) for 3 hours; the medium was then replaced with fresh RPMI complete medium, and the cells were cultured for another 24 hours. The infected cells were selected by adding puromycin (2 μg/ml) over a total of 2 weeks. Gene knockdown efficiency in the infected cells was evaluated using RT-qPCR, Western blot, and immunofluorescence labeling.
Immunofluorescence Labeling and Western Blot Analyses
Cytospin preparations of leukemia cells were fixed, permeabilized, and incubated with rabbit polyclonal anti-Ets1 antibody (Santa Cruz Biotechnology, CA); subsequently, the preparations were incubated with FITC-conjugated affinity purified goat anti-rabbit IgG (Jackson Immuno Research, West Grove, PA) and stained with 4′,6-diamidino-2-phenylindole (DAPI). The slides were viewed under a fluorescence microscope BX61 (Olympus, Melville, NY). Western blot analysis was performed as described previously.22 The primary antibody against mouse β-actin was obtained from Sigma (St Louis, MO). The secondary antibodies, namely, horseradish peroxidase–conjugated anti-mouse IgG and anti-rabbit IgG, were obtained from Jackson Immuno Research Laboratories (West Grove, PA).
In Vivo Competitive Engraftment/Clonal Expansion Assay
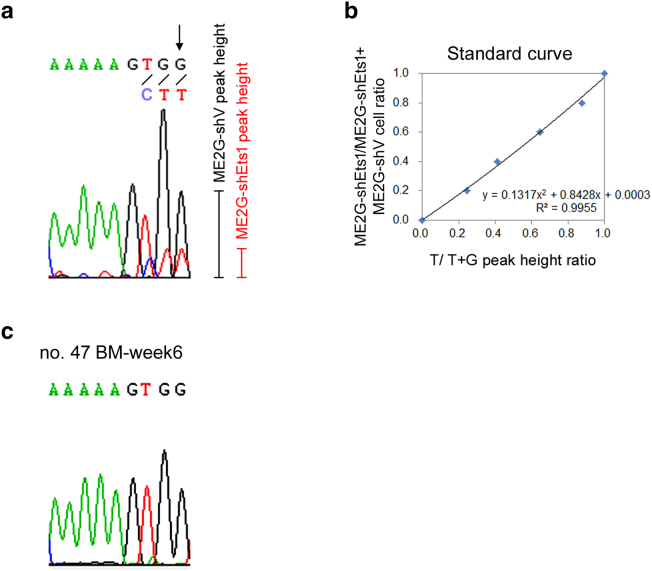
To generate a standard curve of cell ratio vs. peak height ratio, ME2G-shEts1 cells and ME2G-shV cells were mixed in ratios of 10:0, 8:2, 6:4, 4:6, 2:8, and 0:10. Genomic DNAs prepared from the cell mixtures were used to amplify the shRNA region via nested PCR. The first round of PCR was performed using the primer set 5′-ACA AAA TAC GTG ACG TAG and 5′-CTG TTG CTA TTA TGT CTA C, whereas the second round of PCR was performed using the primer set 5′-TGG ACT ATC ATA TGC TTA CCG and 5′-CTG TTG CTA TTA TGT CTA C. The PCR products were sequenced using the PCR primer 5′-CTG TTG CTA TTA TGT CTA C, and the peak height of the 99th nucleotide (T for ME2G-shEts1 and G for ME2G-shV; Supplementary Figure 3A) was measured. The relationship between cell ratio (ME2G-shEts1/ ME2G-shEts1+ ME2G-shV) and nucleotide peak height ratio (T/T + G) was used to generate the standard curve (Supplementary Figure 3B). In vivo competitive engraftment/clonal expansion assays were performed as described previously.23 The mice were sacrificed at 2, 4, 6, 8, and 10 weeks after transplantation (3 mice for each time point). Genomic DNAs were extracted from the initial cell mixture or mice BM and splenic cells, and subsequently nested PCR-DNA sequencing was performed. The cell ratios of ME2G-shEts1/ME2G-shEts1 + ME2G-shV in the samples were determined by aligning T/T + G peak height ratio to the standard curve.
Supplementary Figure 3.
MLL/EB1-shEts1 leukemia cells lost competitive growth advantage in vivo, but not in vitro. (a) A representative DNA sequencing electropherogram of a PCR product of the shRNA region amplified from a mixture of ME2G-shV and ME2G-shEts1cells. The peak heights of the arrow pointed at nucleotide G/T (G from ME2G-shV and T from ME2G-shEts1) were measured. (b) A standard curve of nucleotide peak height ratio (allele burden) vs. cell ratio was determined by PCR-DNA sequencing of ME2G-shV and ME2G-shEts1 mixtures at varying ratios. (c) A representative DNA sequencing electropherogram of BM cells from a recipient mice ip injected with a mixture of ME2G-shV and ME2G-shEts1 cells (1:1) at 6 weeks after transplantation.
Ethical Requirements
All animal experiments were approved by the Animal Research Committee of Chang Gung Memorial Hospital and performed in accordance with its guidelines.
Statistical Analyses
Survival analyses were conducted according to the Kaplan-Meier method. Differences in survival were assessed using the log-rank test. Gene expression level was examined via the paired Student's t test. P values of <.05 were considered to be statistically significant.
Results
MLL/EB1 Expression Immortalizes Mouse BM cells in the Medium Supplemented with Myeloid Differentiation-Inducing Cytokines
To determine the transformation capacity of MLL/EB1, we transduced 5-FU–treated BM cells with MSCV carrying tMLL, tEB1, MLL/EB1, or MLL/AF10(OM-LZ) (Figure 1A) and subsequently evaluated colony-forming capacity (CFC) via a serial replating assay. Our results of three independent experiments showed that BM cells transduced with MLL/EB1, tEB1, tMLL, or MLL/AF10(OM-LZ) were unable to form colonies when plated for the second time in the methylcellulose medium supplemented with lymphoid differentiation-inducing cytokines (IL3, IL7, Flt3 ligand). On the contrary, in the medium containing myeloid differentiation-inducing cytokines (IL3, IL6, SCF, and GM-CSF), MLL/EB1- and MLL/AF10(OM-LZ)-transduced BM cells formed colonies all four rounds of plating in two independent experiments, whereas tMLL- and tEB1-transduced BM cells did not show any proliferation potential in the second round of plating (Figure 1, B-i and ii). The MLL/EB1-transduced BM cells had a lower CFC than that of MLL/AF10(OM-LZ)-transduced BM cells at rounds three and four of plating (Figure 1B).
The carboxy-terminal end of EB1 consists of the three α-helices, αA (aa 190-230), αB (aa 238-248), and αC (aa 251-254) (Figure 1C). The αA and αB helices form a coiled coil (CC)–four-helix bundle (FHB) structure (Figure 1C), which mediates EB1 homo-dimerization.20 To investigate whether both CC (aa 200-230) and FHB were required for MLL/EB1-mediated BM cell transformation, the two constructs αA-deleted MLL/EB1 (MLL/EB1∆αA) and αB-αC-deleted MLL/EB1 (MLL/EB1∆αBC) were generated (Figure 1C). Data of retroviral transduction/replating assay showed that the BM cells transduced with both types of deletion constructs ran out of proliferation potential in the second round of plating (Figure 1D), suggesting that both CC and FHB are required for MLL/EB1-induced cell transformation.
Two cell lines, ME1 and ME2G, were generated from the independent retroviral transduction/replating assays (Figure 1E). Both ME1 and ME2G cells showed a blastic morphology with a high nucleus-to-cytoplasm ratio in the medium containing IL3 (Figure 1E). Immunophenotypic analysis of ME1 and ME2G cells revealed the following expression patterns for both cell types: c-kit hematopoietic progenitor cell marker expression without Sca1 hematopoietic stem cell marker expression (Figure 1, F-i); dim CD3 T cell marker expression without CD19 B lymphocyte marker expression (Figure 1, F-ii); Mac1 myeloid lineage marker expression without Ly6G granulocyte marker expression (Figure 1, F-iii); and CD14 monocytic marker expression, with only 17.7% of the ME1 cells expressing monocytic lineage marker CD115 (Figure 1, F-iv); this observation indicated that MLL/EB1 mediated the immortalization of mouse BM cells along the monocytic lineage.
Compared with 12G cells, MLL/AF10(OM-LZ)-immortalized cells, ME1, and ME2G cells had lower growth rates in the IL3-containing medium (Figure 1G). The high percentages of cells in the early and late apoptotic stages [32.2% and 31.4% (ME1) and 6.1% and 40.5% (ME2G) vs. 6.2% and 0.2% (12G)] (Figure 1H) indicated that MLL/EB1 had a higher potential to induce apoptosis than MLL/AF10(OM-LZ).
MLL/EB1 Induced Acute Monocytic Leukemia with Low Penetrance and Longer Survival
To assess the in vivo leukemogenic potential of MLL/EB1, 5-FU–treated BM cells were retrovirally transduced with MLL/EB1 or MLL/AF10(OM-LZ), followed by a direct ip injection of the cells into B6 mice. The mouse injected with the MLL/AF10(OM-LZ)-transduced BM cells developed MPN-like myeloid leukemia within 12 weeks, whereas none of the mice injected with the MLL/EB1-transduced BM cells developed leukemia [a total of 7 mice, 2 were injected with a dose of 1.5 × 105 BM cells/mouse (observation time 54 weeks) and 5 were injected with a dose of 3 × 105 BM cells/mouse (observation time 2 years)]. These results suggested that MLL/EB1 had a lower leukemogenic potential than MLL/AF10(OM-LZ).
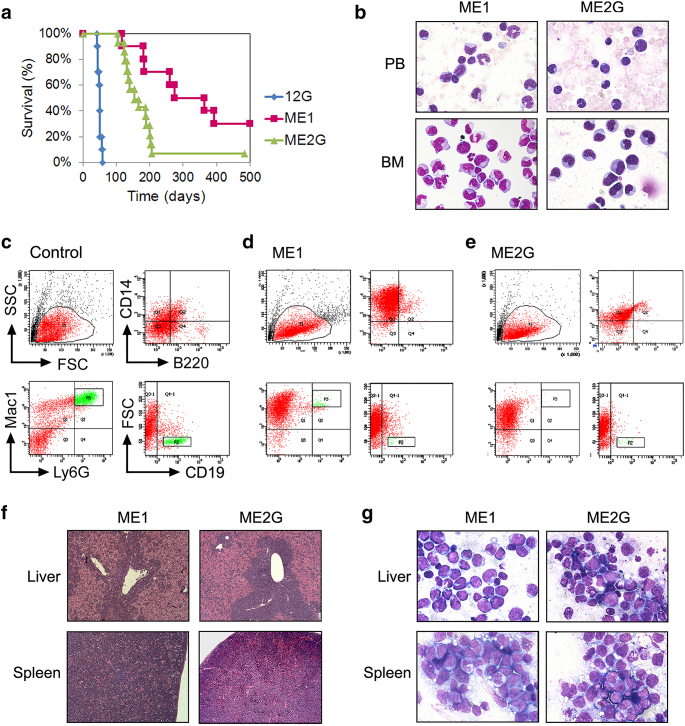
We subsequently assessed the leukemogenic potential of ME1, ME2G, and control (12G) cell lines. Seven of the 10 (70%) mice injected with ME1 cells and 13 of the 14 (93%) mice injected with ME2G cells developed leukemia (Table 1 and Figure 2A). Leukemic ME1 and ME2G mice had significantly longer median survivals than the leukemic 12G mice [260 (range 116-393) days and 155 (range 104-206) days vs. 50 days21] (12G vs. ME1 P < .005; 12G vs. ME2G P < .0001; ME1 vs. ME2G P < .05) (Figure 2A). Most of the leukemic mice showed hyperleukocytosis, but ME1 mice had a lower median white blood cell (WBC) count than the ME2G mice (14.6 × 103/μl vs. 81.5 × 103/μl) before moribund (Table 1). The time from the appearance of leukocytosis in PB to moribund was usually <3 weeks. ME1 and ME2G mice had 43.3% and 82.3% immature forms/blasts in BM, respectively (Table 1 and Figure 2B). Immunophenotypic analysis of the mouse BM cells revealed lower levels of CD19+ /B220+ B-lymphocytes and Ly6G+ granulocytes in ME1 and ME2G mice than those in control mice, indicating suppression of B-cell lymphopoiesis and granulopoiesis of the recipient BM by ME1/ME2G cells (Figure 2, C-E). The leukemic cells of ME1 mice expressed brilliant Mac1 and CD14, which were moderately expressed in ME2G mice (Figure 2, D-E). The cytological and immunophenotypic findings showed that the leukemic cells of ME1 and ME2G mice had varied levels of monocytic maturation. Both ME1 and ME2G mice developed anemia (57.1% vs. 53.8%) and thrombocytopenia (85.7% vs. 76.9%) at similar rates (Table 1). All of the leukemic ME1 and ME2G mice had hepatosplenomegaly (Table 1). Compared with the ME2G mice, ME1 mice had a lower frequency of enlarged lymph nodes but a higher frequency of ascites (Table 1). Pathological examination showed that the periportal region of the liver was infiltrated with leukemia cells; the architecture of the red and white pulps of the spleen was disrupted by leukemia cells (Figure 2F). Liu-staining of the stamp smears showed that both the liver and spleen were infiltrated with leukemic blasts (Figure 2G). According to the Bethesda proposals for the classification of murine nonlymphoid hematopoietic neoplasms,24 MLL/EB1 induced acute monocytic leukemia (AMoL), namely, M5b-like for ME1 mice and M5a-like for ME2G mice.
Table 1.
Summary of the Phenotypic Features of Mice Transplanted with MLL/EB1 and Ets1-Knockdown MLL/EB1 Cell Lines
| Cell Line | ME1 | ME2G | ME2G-shV | ME2G-shEts1 |
|---|---|---|---|---|
| Mouse number | 10 | 14 | 10 | 10 |
| Sick mouse number (%) | 7 (70%) | 13 (93%) | 9 (90%) | 4 (40%) |
| Median survival (range) days | 260 (116-393) | 155 (104-206) | 99 (85-120) | 264 (213-265) |
| Median BM blasts/immature form (range) | 43.3% (8.4%-49.3%) | 82.3% (68.9%-95.3%) | >90% | >90% |
| Median WBC (range) × 103/μl | 14.6 (7.2-52.2) | 81.5 (33.9-161.5) | 67.0 (30.5-118.9) | 4.4 (1.2-6.3) |
| Anemia (%) | 4/7 (57.1%) | 7/13 (53.8%) | 2/7 (28.6%) | 2/3 (66.7%) |
| Thrombocytopenia (%) | 6/7 (85.7%) | 10/13 (76.9%) | 4/7 (57.1%) | 2/3 (66.7%) |
| Hepatosplenomegaly (%) | 7/7 (100%) | 13/13 (100%) | 9/9 (100%) | 4/4 (100%) |
| Enlarged lymph node (%) | 3/7 (42.9%) | 10/13 (76.9%) | 9/9 (100%) | 4/4 (100%) |
| Ascites (%) | 3/7 (42.9%) | 1/13 (7.7%) | 1/9 (11.1%) | 0/4 (0%) |
| Diagnosis | AMoL (M5b-like) | AMoL (M5a-like) | AMoL (M5a-like) | AML |
Figure 2.
Characteristics of the leukemia mice transplanted with the MLL/EB1-immortalized cell lines. (A) Survival curves of the sublethally irradiated mice which were ip injected with ME1 or ME2G cells. Mice injected with 12G cells were used as the positive control (data has been shown in ref. 21). (B) Liu staining of PB smears or cytospin preparations of BM cells from the representative leukemic mice (oil immersion, ×1000). (C-E) Flow cytometric analysis of hematopoietic subsets in the BM cells of control mouse (C), representative (D) ME1- and (E) ME2G-leukemic mice. (F) Histological sections of the liver and spleen of the representative leukemic mice (H&E staining, ×100). (G) Stamp smears of the liver and spleen of the representative leukemic mice (Liu staining, oil immersion, ×1000).
MLL/EB1 Cells Had Higher Ets1 Expression Level
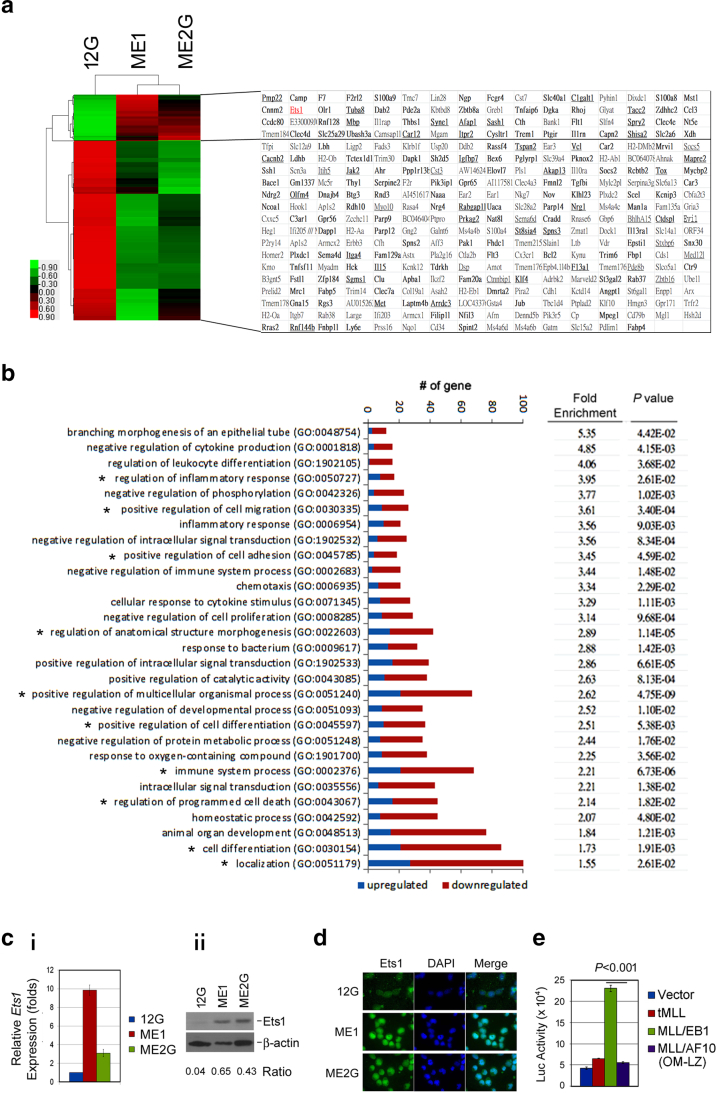
The leukemic transformation induced by MLL/EB1 was different from that induced by MLL/AF10(OM-LZ). To identify MLL/EB1-specific downstream target gene(s) involved in leukemic transformation, we compared the transcriptome between 12G and ME1/ME2G cell lines on the basis of cDNA microarray data. Sixty-four upregulated (≥2-fold) and 254 downregulated (≤0.5-fold) genes were identified in both ME1 and ME2G as opposed to the 12G cells (Figure 3A). Among these 318 differentially expressed genes, 163 genes (49 upregulated and 114 downregulated) have been reported to be MLL-bound genes, and 49 (13 upregulated and 36 downregulated) were joint targets of MLL1 and MLL/AF910 (Figure 3A). The GO biological process terms that were overrepresented in the 318 genes included several terms related to cell proliferation/apoptosis, namely, “negative regulation of cell proliferation (GO:0008285),” “regulation of programmed cell death (GO:0043067)”; cell differentiation, namely, “regulation of leukocyte differentiation (GO:1902105),” “positive regulation of cell differentiation (GO:0045597),” “cell differentiation (GO:0030154)”; cell homing/engraftment, namely, “positive regulation of cell migration (GO:0030335),” “positive regulation of cell adhesion (GO:0045785),” “chemotaxis (GO:0006953)”; and inflammatory response, namely, “regulation of inflammatory response (GO:0050727),” “inflammatory response (GO:0006954),” “immune system process (GO:0002376)” (Figure 3B). The enrichment in these biological processes is consistent with the observations that ME1/ME2G and 12G cells were different in terms of their leukemogenic potential as well as cell differentiation and proliferation ability.
Figure 3.
Ets1 is overexpressed in MLL/EB1 cell lines. (A) Heat-map and list of the 318 genes differentially expressed in the transcriptome profiling of MLL/EB1 cell lines (ME1 and ME2G) in comparison with the MLL/AF10 cell line (12G). Hierarchical clustering was performed and heat-map was obtained using Cluster 3.0 and TreeView1.1.6r4. Raw values were log2-transformed and centered relative to the median. Red indicates that the level of gene expression is higher than the median, and green indicates that the level is lower than the median. The known MLL1-bound genes are indicated in bold type; genes that were jointly bound to both MLL1 and MLL/AF9 are underlined. (B) Enriched biological process Gene Ontology (GO) terms for the 318 differentially expressed genes. All GO terms listed on the bar plot show significant enrichment (P < .05). Fold enrichment refers to the number of genes represented in each category relative to the random expression of all genes in the human genome. Bars indicate gene counts; blue and red bars indicate the upregulated and downregulated genes, respectively. The GO biological processes involving Ets1 gene are indicated by *. (C) RT-qPCR (i) and Western blot (ii) analyses of Ets1 expression level in 12G, ME1, and ME2G cells. Error bars indicate the standard deviation (SD) of mean. (D) Immunofluorescence staining of 12G, ME1, and ME2G cells with Ets1 (green, left column) and DAPI (blue, middle column). The merged images were shown in the right column. (E) Luciferase reporter analysis. 32Dcl3 cells were transduced with Ets1 promoter-reporter construct, PEts1-luc, and retrovirus empty vector or vector-carrying tMLL, MLL/EB1, or MLL/AF10(OM-LZ) plasmid DNA. Graph shows a representative result from four individual assays. Error bars represent SD.
Among the 64 upregulated genes, Ets1, which is bound by MLL1 and MLL/AF9, was seen to be involved in the GO biological processes of inflammatory/immune system, cell migration/adhesion, differentiation, and programmed cell death (Figure 3B). The RT-qPCR and Western blot analyses confirmed that Ets1 expression level was higher in ME1/ME2G than in 12G cells (Figure 3, C-i and ii). Immunofluorescence labeling illustrated the presence of more nuclear Ets1 protein in ME1/ME2G cells than in 12G cells (Figs. 3D). To determine whether the high expression level of Ets1 was induced by MLL/EB1 fusion, we performed luciferase reporter assay. Transcription of Ets1 is controlled by two promoters (Supplementary Figure 1A). The proximal promoter generates Ets1 variants 1 and 2, whereas the distal promoter generates variant X1. RT-PCR analysis revealed that the variant X1 was not expressed in our leukemic cell lines (Supplementary Figure 1B). Therefore, only a region that spans the transcription start site of variant 1/2 (nt −703 to +293) (Supplementary Figure 1A) was cloned into a luciferase reporter plasmid. Our results showed that the Ets1 promoter-driven luciferase activity was upregulated by MLL/EB1 but not by MLL/AF10(OM-LZ) or tMLL (Figure 3E and supplementary Figs. 1, C-E), suggesting that Ets1 was selectively upregulated by MLL/EB1.
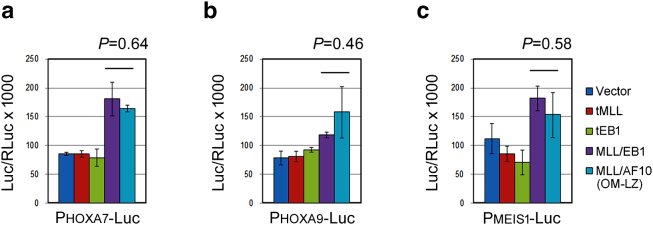
The 318 differentially expressed genes did not include 5′ Hoxa (Hoxa7, Hoxa9, Hoxa10) and Meis1 (Figure 3A). We performed luciferase reporter assays and demonstrated that HOXA7, HOXA9, and MEIS1 promoter-driven luciferase activities were upregulated by both MLL/EB1 and MLL/AF10(OM-LZ) (Supplementary Figs. 2, A-C), indicating that MLL/EB1, like MLL/AF10 and other MLL fusions, activated 5′ HOXA and MEIS1 genes.
Supplementary Figure 2.
Both MLL/EB1 and MLL/AF10(OM-LZ) transactivate human 5′ HOXA and MEIS1 promoters. The luciferase reporter construct driven by the indicated promoters (a) HOXA7, (b) HOXA9, or (c) MEIS1 were transduced into 32Dcl3 cells with a retrovirus empty vector or vector-carrying tMLL, tEB1, MLL/EB1, or MLL/AF10(OM-LZ) plasmid DNA. The luc/RLuc ×1000 value was calculated as the final transcriptional activation activity. Data are mean±SE of three independent replicates.
Knockdown of Ets1 Expression Reduced Cell Monocytic Differentiation and Apoptosis
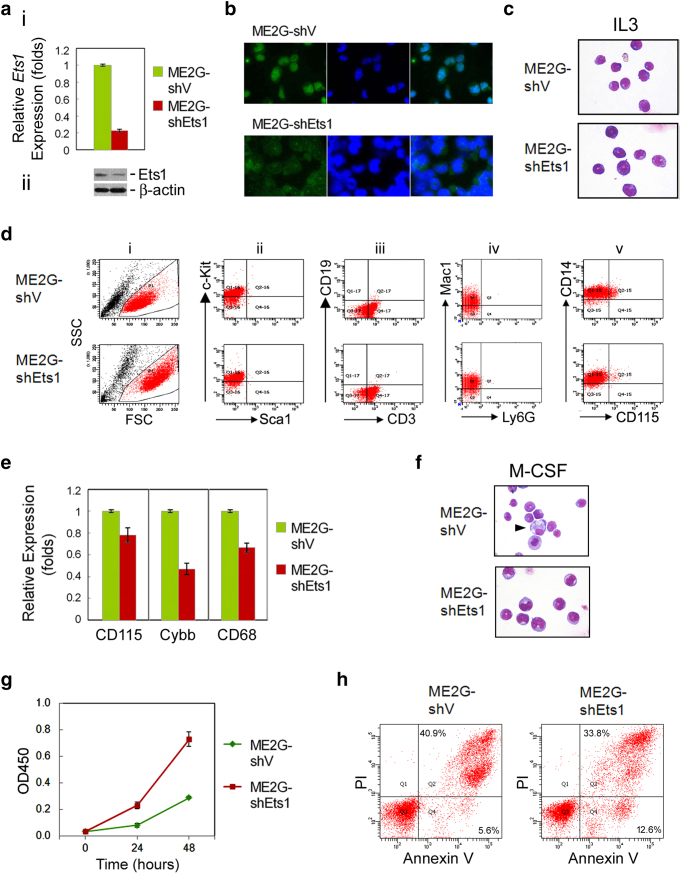
To investigate the role of Ets1 in the monocytic differentiation and apoptosis of MLL/EB1 cells, we transduced two lentivirus-based shRNAs against Ets1 into ME1 and ME2G cells to generate Ets1-knockdown cell lines. RT-qPCR analysis showed that, as opposed to the cells transduced with empty lentiviruses, Ets1 expression levels were reduced to 29% (ME1-shEts1) and 23% (ME2G-shEts1) owing to transduction with shRNA TRCN0000042639 (Figure 4, A-i). The second shRNA clone TRCN0000042642 failed to generate Ets1-knockdown cell lines. Because of the low penetrance of leukemia (please see below), we did not further analyze ME1-shEts1 cells. Reduction of Ets1 protein level in ME2G-shEts1 cells was confirmed by Western blot analysis (Figure 4, A-ii). Immunofluorescence labeling further confirmed that the level of Ets1 protein was substantially decreased in the nuclei of ME2G-shEts1 cells (Figure 4B). Both ME2G-shV and ME2G-shEts1 cells had a blastic morphology; however, ME2G-shEts1 cells were enlarged (Figure 4C). ME2G-shEts1 cells showed high forward and side light scatters, consistent with their large size (Figure 4, D-i). The expression of the hematopoietic lineage markers, including Sca1, c-Kit, CD19, CD3, Mac1, Ly6G, and CD14, in ME2G-shV and ME2G-shEts1 cell lines remained the same as that in their parental ME2G cells (Figures 4D and 1D). However, ME2G-shEts1 cells had decreased expression of the monocytic lineage marker CD115 (Figure 4, D-v), as confirmed by RT-qPCR analysis (Figure 4E). Moreover, the expression levels of mature monocytic markers Cybb and CD68 were also decreased in ME2G-shEts1 cells (Figure 4E). M-CSF treatment induced the generation of macrophage-like cells from ME2G-shV but not ME2G-shEts1 cells, which continued to have a blastic morphology (Figure 4F). These results indicated that knockdown of Ets1 expression in MLL/EB1 cells reduced monocytic differentiation and blocked M-CSF–induced macrophage development.
Figure 4.
Characteristics of Ets1-knockdown MLL/EB1 cells. (A) RT-qPCR (i) and Western blot (ii) analyses of Ets1 expression level in the lentivirus-transduced ME2G cells. ME2G-shEts1, ME2G cells transfected with lentivirus expressing-shRNA-targeted Ets1 (TRCN0000042639). ME2G-shV, ME2G cells transfected with blank lentivirus. Error bars indicate SD of mean. (B) Immunofluorescence staining of ME2G-shV and ME2G-shEts1 cells. Cells were stained with anti-Ets1 (left column) or DAPI (middle column), and a merged image (right column). (C) Cytological and (D) immunophenotypic characteristics of ME2G-shV and ME2G-shEts1 cells. (E) RT-qPCR analysis of CD115, Cybb, and CD68 expression in ME2G-shV and ME2G-shEts1 cells. (F) Liu staining of ME2G-shV and ME2G-shEts1 cells cultured in the medium containing M-CSF (oil immersion, ×1000). (G) Growth curve and (H) cell apoptosis analyses of ME2G-shV and ME2G-shEts1 cells cultured in the medium containing IL3. Error bars indicate SD of mean (G).
ME2G-shEts1 cells had a higher growth rate (Figure 4G) and a lower percentage of cells at the late apoptotic stage (33.8% vs. 40.9%) than ME2G-shV cells (Figure 4H), indicating that knockdown of Ets1 expression reduces the frequency of MLL/EB1 cell apoptosis.
Knockdown of Ets1 Expression in MLL/EB1 Cells Impaired Cell Engraftment to BM/Spleen and Leukemia Development
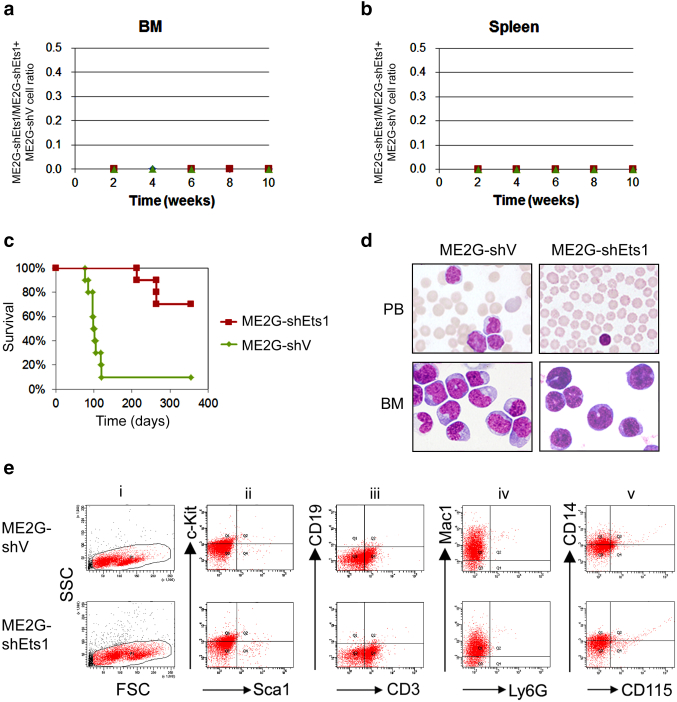
To investigate the role of Ets1 in MLL/EB1-mediated leukemia development, we first compared the in vivo competitive engraftment/clonal expansion activity between ME2G-shEts1 and ME2G-shV cells. By aligning with the standard curve for cell ratio vs. allele burden (Supplementary Fig. 3, A and B), we showed that ME2G-shEts1 cells were poorly engrafted/expanded in the BM and spleen of the recipient mice because the shRNA region of none of the ME2G-shEts1 cells was amplified from the samples collected at 2, 4, 6, 8, and 10 weeks after transplantation (Figure 5, A-B and Supplementary Figure 3C). These results indicated that knockdown of Ets1 expression severely impaired in vivo engraftment activity of MLL/EB1 cells despite reducing the frequency of cell apoptosis.
Figure 5.
Characteristics of leukemic mice transplanted with Ets1-knockdown MLL/EB1 cells. (A-B) In vivo competitive engraftment/clonal expansion of the ME2G-shV and ME2G-shEts1 cells in the BM (A) and spleen (B) of recipient mice. ME2G-shEts1 and ME2G-shV cells were initially mixed at a 1:1 ratio (by cell number). (C) Survival curves of the sublethally irradiated mice ip injected with ME2G-shV or ME2G-shEts1 cells. (D) Liu staining of PB smears (upper panel) or cytospin preparations of BM cells (lower panel) from representative leukemic mice (oil immersion, ×1000). (E) Representative BM flow cytometric analysis of hematopoietic subsets in the ME2G-shV- (upper panel) or ME2G-shEts1- (lower panel) leukemic mice.
We also performed a BM transplantation assay to determine the leukemogenesis of Ets1-knockdown MLL/EB1 cells. None of the six mice injected with ME1-shEts1 cells developed AML, whereas one of the six mice injected with ME1-shV developed AML before the experiment ended (335 days). On the other hand, only 4/10 (40%) mice injected with ME2G-shEts1 cells developed AML, whereas 9/10 (90%) mice transplanted with ME2G-shV cells developed AMoL (Table 1 and Figure 5C). The median survival of the leukemic ME2G-shEts1 mice was significantly longer than leukemic ME2G-shV mice (264 days vs. 99 days, P < .01) (Table 1). The PB of ME2G-shV mice at moribund stage showed hyperleukocytosis with a median WBC count (comprising lymphocytes and leukemia blasts) of 67.0 × 103/μl, whereas the leukemic ME2G-shEts1 mice showed pancytopenia or a normal WBC count, with a median WBC count (exclusively comprising lymphocytes) of 4.4 × 103/μl (Figure 5D upper panel and Table 1). At moribund stage, >90% of the BM cells from ME2G-shV and ME2G-shEts1 mice were immature forms/blasts (Table 1), but ME2G-shEts1 mice had a larger size of blast cells with a minimal CD115 expression rate (Figure 5D, E-i and E-v). Both ME2G-shV and ME2G-shEts1 mice showed a rare occurrence of CD19+ B lymphocytes and Ly6G+ granulocytes (Figure 5, E-iii and iv), indicating that Ets1-knockdown ME2G cells retain the ability to suppress recipient B-cell lymphopoiesis and granulopoiesis in vivo. Compared with leukemic ME2G-shV mice, leukemic ME2G-shEts1 mice had a higher incidence rate of anemia (66.7% vs. 28.6%) but a similar rate of thromobocytopenia (66.7% vs. 57.1%); however, both mice had the same degree of hepatosplenomegaly (100% vs. 100%) and lymph node enlargement (100% vs. 100%) (Table 1). One of the ME2G-shV mice had ascites (11.1% vs. 0%) (Table 1). Taken together, our results demonstrated that the knockdown of Ets1 expression reduced MLL/EB1-induced leukemia development and prolonged the survival of leukemic mice in vivo.
Discussion
Previous studies have revealed that the carboxy-terminal end of EB1 contained a coiled-coil–four-helix bundle structure that conferred dimerization activity.[19], [20] In this study, we demonstrated that MLL fusion between both the coiled-coil and four-helix bundle structure is required for facilitating the immortalization of murine BM cells. Our results further supported that the N-terminal MLL fused to the coiled-coil dimerization domain is leukemogenic.[12], [13] However, while MLL/AF10-transduced BM cells (1.5 × 105 /mouse) induced MPN-like myeloid leukemia in the recipient mice, MLL/EB1-transduced BM cells (1.5 × 105-3 × 105 BM cells/mouse) failed to induce leukemia. The retroviral transduction/replating assay also showed lower CFC of MLL/EB1-transduced cells than that of MLL/AF10(OM-LZ)-transduced cells. These results suggested that the leukemogenic potential of MLL/EB1 was lower than that of MLL/AF10.
Our patient with MLL/EB1 was diagnosed as having pro-B ALL, but the transduction of MLL/EB1 did not immortalize murine BM cells in the medium supplemented with lymphoid differentiation-inducing cytokines. Recently, it had been reported that BM stroma was required for normal B-cell development and lymphoid malignancies25; hence, there could be a possibility that our in vitro culture condition was not enough to sustain the growth of lymphoid leukemia cells. On the other hand, a recent report had shown that MLL fusions could target multipotent hematopoietic stem and progenitor cells to induce mixed-lineage leukemia as well as lineage switch from B-ALL to AML or vice versa in acute leukemia patients with MLL rearrangements.26 Unfortunately, our study patient left the hospital without undergoing treatment17; therefore, follow-up samples were not available to assess whether linage conversion occurred in the patient.
The low leukemogenic potential and various cell differentiation- and apoptosis-associated characteristics of MLL/EB1 cells in comparison with MLL/AF10(OM-LZ) cells implicated that there may be different target genes downstream of MLL/EB1. Comparative transcriptome analysis identified 318 differentially expressed genes between MLL/EB1 cells and MLL/AF10(OM-LZ) cells. The enrichment of the genes involved in the biological processes of cell differentiation, proliferation/apoptosis, homing/recruitment, and immune response was consistent with our experimental observations. From the differentially expressed genes, we identified Ets1, a known MLL and MLL/AF9-bound gene, which was selectively upregulated by MLL/EB1 and played critical roles in MLL/EB1-mediated leukemic transformation. Ets1 gene encodes a widely expressed transcription factor involving in many biological processes, including embryogenesis, vasculogenesis, angiogenesis, proliferation, hematopoiesis, and tumorigenesis.[27], [28], [29], [30] In hematopoiesis, Ets1 was involved in decisions governing the fate of B-/T-lymphocytic cells and megakaryocyte/erythrocyte progenitor cells as well as in the development of natural killer cells, granulocytes, and macrophages.[31], [32], [33], [34], [35] Ets1/Ets2 and METS/DP103 function cooperatively in linking terminal macrophage differentiation to cell cycle arrest.33 Our results demonstrated that the knockdown of Ets1 expression in MLL/EB1 cells reduced monocytic gene expression, blocked M-CSF–induced terminal macrophage differentiation, and decreased the frequency of cell apoptosis. These results indicated that Ets1 in MLL/EB1 leukemia cells, like in macrophages, linked macrophage differentiation and cell cycle arrest (cell apoptosis). On the other hand, the knockdown of Ets1 expression in MLL/EB1 cells strongly hampered cell engraftment to BM/spleen and leukemia development and consequently prolonged the survival of leukemic mice. These results uncovered the dual roles of Ets1; it was found to not only attenuate leukemia cell growth but also promote tumorigenesis. Similar observation was reported that Ets1 controls balance between cell invasion and growth in breast cancer.36
Gene amplification and missense mutations of ETS1 have been detected in patients with marginal zone B cell lymphoma and diffuse large B cell lymphoma (7.9%-20.8%),[37], [38], [39] but no direct involvement of Ets1 was reported in leukemia. More AML samples showing MLL fusion with dimerization domain-containing partner genes will be needed to evaluate whether ETS1 overexpression is common in such type of AML patients. Moreover, Thiel et al. (2010) proposed that wild-type MLL is required for MLL fusion-induced leukemogenesis.40 The molecular mechanism governing the selective upregulation of Ets1 by MLL in association with MLL/EB1 and the tumorigenic role of Ets1 in MLL/EB1-induced leukemic development remain to be explored.
The following are the supplementary data related to this article.
Acknowledgement
This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 103-2320-B-182A-005), and Chang Gung Memorial Hospital, Taiwan (CMRPG3B1421-3, CMRPG 3E0301-3, CMRPG3E1391-3). We thank Ms. Yu-Ju Lin and Mr. Jun-Wei Huang for technical assistance in all the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contributor Information
Jen-Fen Fu, Email: cgfujf@cgmh.org.tw.
Lee-Yung Shih, Email: sly7012@adm.cgmh.org.tw.
References
- 1.Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Behm FG, Downing JR, Hancock ML, Shurtleff SA, Ribeiro RC, Head DR, Mahmoud HH, Sandlund JT, Furman WL. 11q23/MLL rearrangement confers a poor prognosis in infants with acute lymphoblastic leukemia. J Clin Oncol. 1994;12:909–915. doi: 10.1200/JCO.1994.12.5.909. [DOI] [PubMed] [Google Scholar]
- 3.Rubnitz JE, Link MP, Shuster JJ, Carroll AJ, Hakami N, Frankel LS, Pullen DJ, Cleary ML. Frequency and prognostic significance of HRX rearrangements in infant acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1994;84:570–573. [PubMed] [Google Scholar]
- 4.Meyer C, Burmeister T, Groger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MS. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32:273–284. doi: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daser A, Rabbitts TH. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin Cancer Biol. 2005;15:175–188. doi: 10.1016/j.semcancer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 7.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 8.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisig BB, Garcia-Cuellar MP, Winkler TH, Slany RK. The oncoprotein MLL-ENL disturbs hematopoietic lineage determination and transforms a biphenotypic lymphoid//myeloid cell. Oncogene. 2003;22:1629–1637. doi: 10.1038/sj.onc.1206104. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Li L, Xiong J, denDekker A, Ye A, Karatas H, Liu L, Wang H, Qin ZS, Wang S, Dou Y. MLL1 and MLL1 fusion proteins have distinct functions in regulating leukemic transcription program. Cell Discov 2016;2: 16008. [DOI] [PMC free article] [PubMed]
- 11.Kar A, Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit Rev Biochem Mol Biol. 2013;48:522–543. doi: 10.3109/10409238.2013.838202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.So CW, Karsunky H, Passegue E, Cozzio A, Weissman IL, Cleary ML. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–171. doi: 10.1016/s1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 13.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 14.Eguchi M, Eguchi-Ishimae M, Greaves M. The small oligomerization domain of gephyrin converts MLL to an oncogene. Blood. 2004;103:3876–3882. doi: 10.1182/blood-2003-11-3817. [DOI] [PubMed] [Google Scholar]
- 15.Liedtke M, Ayton PM, Somervaille TC, Smith KS, Cleary ML. Self-association mediated by the Ras association 1 domain of AF6 activates the oncogenic potential of MLL-AF6. Blood. 2010;116:63–70. doi: 10.1182/blood-2009-09-243386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 17.Fu JF, Hsu HC, Shih LY. MLL is fused to EB1 (MAPRE1), which encodes a microtubule-associated protein, in a patient with acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2005;43:206–210. doi: 10.1002/gcc.20174. [DOI] [PubMed] [Google Scholar]
- 18.Su LK, Qi Y. Characterization of human MAPRE genes and their proteins. Genomics. 2001;71:142–149. doi: 10.1006/geno.2000.6428. [DOI] [PubMed] [Google Scholar]
- 19.Rehberg M, Graf R. Dictyostelium EB1 is a genuine centrosomal component required for proper spindle formation. Mol Biol Cell. 2002;13:2301–2310. doi: 10.1091/mbc.E02-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu JF, Hsu CL, Shih LY. MLL/AF10(OM-LZ)-immortalized cells expressed cytokines and induced host cell proliferation in a mouse bone marrow transplantation model. Int J Cancer. 2010;126:1621–1629. doi: 10.1002/ijc.24867. [DOI] [PubMed] [Google Scholar]
- 22.Fu JF, Yen TH, Chen Y, Huang YJ, Hsu CL, Liang DC, Shih LY. Involvement of Gpr125 in the myeloid sarcoma formation induced by cooperating MLL/AF10(OM-LZ) and oncogenic KRAS in a mouse bone marrow transplantation model. Int J Cancer. 2013;133:1792–1802. doi: 10.1002/ijc.28195. [DOI] [PubMed] [Google Scholar]
- 23.Fu J-F, Liang S-T, Huang Y-J, Liang K-H, Yen T-H, Liang D-C, Shih L-Y. Cooperation of MLL/AF10(OM-LZ) with PTPN11 activating mutation induced monocytic leukemia with a shorter latency in a mouse bone marrow transplantation model. Int J Cancer. 2017;140:1159–1172. doi: 10.1002/ijc.30515. [DOI] [PubMed] [Google Scholar]
- 24.Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, Carter JS, de Coronado S, Downing JR, Fredrickson TN. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 25.Sangaletti S, Tripodo C, Portararo P, Dugo M, Vitali C, Botti L, Guarnotta C, Cappetti B, Gulino A, Torselli I. Stromal niche communalities underscore the contribution of the matricellular protein SPARC to B-cell development and lymphoid malignancies. Oncoimmunology. 2014;3 doi: 10.4161/onci.28989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorantes-Acosta E, Pelayo R. Lineage switching in acute leukemias: a consequence of stem cell plasticity? Bone Marrow Res. 2012;2012:406796. doi: 10.1155/2012/406796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:1–21. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi: 10.1016/j.semcancer.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res. 2013;119:1–61. doi: 10.1016/B978-0-12-407190-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrett-Sinha LA. Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci. 2013;70:3375–3390. doi: 10.1007/s00018-012-1243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bories J-C, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 32.Barton K, Muthusamy N, Fischer C, Ting C-N, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 33.Klappacher GW, Lunyak VV, Sykes DB, Sawka-Verhelle D, Sage J, Brard G, Ngo SD, Gangadharan D, Jacks T, Kamps MP. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell. 2002;109:169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 34.Lulli V, Romania P, Morsilli O, Gabbianelli M, Pagliuca A, Mazzeo S, Testa U, Peschle C, Marziali G. Overexpression of Ets-1 in human hematopoietic progenitor cells blocks erythroid and promotes megakaryocytic differentiation. Cell Death Differ. 2006;13:1064–1074. doi: 10.1038/sj.cdd.4401811. [DOI] [PubMed] [Google Scholar]
- 35.Lulli V, Romania P, Riccioni R, Boe A, Lo-Coco F, Testa U, Marziali G. Transcriptional silencing of the ETS1 oncogene contributes to human granulocytic differentiation. Haematologica. 2010;95:1633–1641. doi: 10.3324/haematol.2010.023267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furlan A, Vercamer C, Bouali F, Damour I, Chotteau-Lelievre A, Wernert N, Desbiens X, Pourtier A. Ets-1 controls breast cancer cell balance between invasion and growth. Int J Cancer. 2014;135:2317–2328. doi: 10.1002/ijc.28881. [DOI] [PubMed] [Google Scholar]
- 37.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flossbach L, Holzmann K, Mattfeldt T, Buck M, Lanz K, Held M, Moller P, Barth TF. High-resolution genomic profiling reveals clonal evolution and competition in gastrointestinal marginal zone B-cell lymphoma and its large cell variant. Int J Cancer. 2013;132:E116–E127. doi: 10.1002/ijc.27774. [DOI] [PubMed] [Google Scholar]
- 40.Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA. MLL-AF9–induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]