Abstract
Soil detritivores such as Collembola impact plant growth, tissue nutrient concentration and gene expression. Using a model system with pedunculate oak (Quercus robur) microcuttings that display a typical endogenous rhythmic growth with alternating shoot (SF) and root flushes (RF), we investigated the transcriptomic response of oak with and without mycorrhiza (Piloderma croceum) to the presence of Collembola (Protaphorura armata), and linked it to changes in resource allocation by pulse labelling the plants with 13C and 15N. Collembola impacted Gene Ontology (GO) terms as well as plant morphology and elemental ratios with the effects varying markedly with developmental phases. During SF Collembola increased GO terms related to primary growth and this was mirrored in increased 13C and 15N excess in aboveground plant compartments. During RF, Collembola increased GO terms related to plant secondary metabolism and physical fortification. Further, Collembola presence resulted in an increase in plant defence-related GO terms suggesting that Collembola in the rhizosphere prime oak shoots against the attack by fungi or herbivores. Notably, the impact of Collembola on growth, resource allocation and oak gene expression was modified by presence of P. croceum. The results indicate that oaks clearly react to the presence of Collembola in the rhizosphere and respond in a complex way by changing the expression of genes of both primary and secondary metabolism, and this resulted in concomitant changes in plant morphology and physiology.
Keywords: Collembola, Quercus robur, plant nutrition, stable isotopes, plant defence, Gene Ontology
1. Introduction
Detritivore animals form part of the biota responsible for the recycling of plant litter and therefore contribute to the provisioning of nutrients to plants. Thereby, they indirectly impact plant growth and plant performance. However, detritivores also modify plant growth by a number of other mechanisms including modifying soil structure, grazing on rhizosphere microorganisms, altering plant–mycorrhiza symbiosis and changing root pathogen infections [1–3].
The great majority of plants are colonized by mycorrhizal fungi [4] which, by exploiting nutrients in the rhizosphere via extra-radical hyphae, foster the uptake of nutrients. Via their hyphal network, mycorrhizal fungi are embedded into the belowground food web and interact with detritivore animals. Grazing by fungal feeding soil invertebrates may detrimentally affect mycorrhizal fungi, but, on the other side, plants may benefit from nutrients made available, e.g. in animal excrements.
Collembola, as major soil detritivores reaching densities of more than 100 000 individuals per square metre [5], have been shown to graze on hyphae and mycelial mats of ectomycorrhizal fungi thereby impacting plant–mycorrhiza interrelationships [6,7]. Further, Collembola have been shown to increase the supply of nutrients to plants due to processing of detritus [8,9]. Overall, in the great majority of studies the presence of Collembola caused an increase in plant nutrient uptake and plant tissue nutrient concentration, and thereby fostered plant growth and performance [10–15], suggesting that the Collembola-mediated increase in nutrient mobilization outweighs their detrimental effect via grazing on mycorrhizal fungi. In fact, there is evidence that moderate grazing on fungal hyphae by fungal feeding soil invertebrates increases fungal productivity and functionality due to liberating nutrients from senescent hyphae thereby fostering fungal regrowth [16,17]. Further, there is evidence that Collembola preferentially graze on saprotrophic rather than mycorrhizal fungi [2,9], thereby favouring the competitive strength of mycorrhiza, mycorrhizal nutrient capture and plant growth. However, non-nutrient effects may also contribute to Collembola-mediated changes in mycorrhizal functioning and plant growth. For example, Collembola have been shown to alter root morphology without changing plant nutrient concentrations [18,19]. To disentangle nutrient- and non-nutrient-based effects of Collembola on plant growth and elucidate the role of mycorrhiza in Collembola–plant interactions, laboratory experiments are needed, manipulating mycorrhiza and Collembola independently and investigating the transcriptional response of plants to these manipulations. Further, for understanding plant responses, detailed analysis of plant growth as well as plant carbon and nutrient allocation are needed. Unfortunately, experiments combining these approaches are lacking. The only study investigating the transcriptional response of plants to the presence of Collembola focused on Arabidopsis thaliana, a non-mycorrhizal plant species [20]. The study showed that the presence of Collembola modified gene expression with both genes of primary and secondary metabolic pathways being altered. Other studies using earthworms and dung beetles also showed that detritivore–plant interactions change plant secondary metabolite synthesis and defence characteristics of plants against herbivores [21–24]. However, only herbaceous plants have been considered in these studies, while detritivore-mediated changes in gene expression patterns in trees have not been studied until today. This is surprising because detritivore animals reach maximum density in forests and therefore are likely to interact with trees and the mycorrhizal fungi with which they are associated [14].
To disentangle detritivore–mycorrhiza interactions and their effects on the transcriptional and growth response of trees we established an experimental model system including an ectomycorrhizal tree, Quercus robur (L.), the ectomycorrhizal fungal species Piloderma croceum (J. Erikss. & Hjortst. Strain 729; DSM-4924) and the Collembola species Protaphorura armata (Tullberg). Quercus robur is among the most common European tree species and of significant economic importance. Piloderma croceum is a widespread ectomycorrhizal fungal species in deciduous forests and P. armata is a widespread soil living (euedaphic) Collembola species common in the rhizosphere of plants. To unravel morphological, nutritional and gene expression changes in Q. robur due to the presence of P. croceum and P. armata we established a full factorial experiment varying the presence of mycorrhiza and Collembola. As oak displays rhythmic growth with alternating shoot (SF) and root flushes (RF) we investigated both of these growth phases (cf. [25]). The study formed part of the TrophinOak project (http://www.trophinoak.de) aiming at investigating in a comprehensive way the transcriptomic response of Q. robur to biological interactors including mutualists and antagonists [26–28]. Further, using 13C and 15N labelling, the influence of P. armata and P. croceum on carbon and nitrogen allocation during the rhythmic growth of Q. robur was investigated. As the response of Q. robur to the mycorrhizal fungus P. croceum has been investigated in detail [29,30], we focus on effects of P. armata and its interaction with P. croceum in the present study. We hypothesized that (i) gene expression patterns of oaks are altered in the presence of Collembola with the patterns varying between SF and RF, and including both genes involved in primary and secondary metabolism; (ii) these changes are attenuated in presence of mycorrhiza, assuming that both Collembola and mycorrhiza improve plant nutrient supply; (iii) Collembola-mediated changes in the expression of genes involved in plant primary metabolism reflect respective changes in plant carbon and nitrogen uptake, i.e. upregulation of genes related to plant growth are associated by increased incorporation of 13C and 15N; and (iv) Collembola-mediated changes in the expression of genes involved in plant secondary metabolism reflect upregulation of genes involved in plant defence against herbivores.
2. Material and methods
2.1. Quercus robur
The present study and other studies in the framework of the TrophinOak consortium [26,28–30] employed clonally propagated pedunculate oak microcuttings (DF159). Propagating and rooting of microcuttings was performed at the Helmholtz Center for Environmental Research (UFZ; Halle, Germany) as described previously [25,31]. The root system of single microcuttings was implanted into Petri dishes (12 × 12 cm) filled with sterilized soil with the shoots growing outside the microcosms. The soil was taken from an oak forest stand at the Dölauer Heide close to Halle/Saale, Saxony Anhalt, Germany (51.51016° N, 11.91291° E). The Oh and upper Ah layers of the soil were taken, homogeneously mixed by sieving (5 mm mesh), air-dried, mixed with sand (1 : 1, v/v), packed in 500 ml aliquots and sent for γ-radiation for sterilization (50 kGy; Beta-Gamma-Service, Wiehe, Germany). The soil/sand mixture contained 4.88 ± 0.11% carbon, had a C-to-N ratio of 19.3 and a pH of 4.12. Sterilized soil aliquots were stored at 8°C and sterility was tested before use by plating on Lysogeny broth agar.
The experiment was set up in a climate chamber at 23°C, relative air humidity of 75% and a photosynthetic photon flux density of 180 µmol m−2 s−1 at long day conditions (16/8 h). A total of 129 oak microcuttings were used, which under the specified conditions establish an endogenous rhythmic growth as described in Herrmann et al. [25]. High relative humidity reduces plant stress, but does not improve mycorrhization of oak microcuttings [21]. However, although not fully mycorrhized, oak microcuttings treated in that way show typical effects of mycorrhizal plants, i.e. increased growth, photosynthesis and stress resistance [11,28,29,31].
2.2. Piloderma croceum
The ectomycorrhizal fungus P. croceum (Piloderma) was reared and cultivated at 20°C in darkness in Petri dishes containing Modified Melin-Norkrans agar (for details see [25]). Piloderma croceum is a widespread mycorrhizal fungus preferentially colonizing deciduous trees such as pedunculated oak [25,31]. Microcosms were inoculated with Piloderma as described in Tarkka et al. [28]. In brief, the Piloderma inoculum was produced using a substrate mixture of vermiculite (675 ml), sphagnum peat (75 ml), and Melin-Norkrans liquid medium (300 ml) modified by Marx [32] without carbohydrates and with 1/10 strength for phosphorus and nitrogen as described in Herrmann et al. [25] using a 48-day-old liquid fungal culture reared in 100 ml glass flasks at 20°C in darkness with agitation (100 r.p.m.). The inoculum was incubated in darkness at 20°C for four weeks and used for mycorrhiza establishment. Mycorrhization was checked after five weeks using a dissecting microscope and showed that the inoculation was successful.
2.3. Protaphorura armata
The Collembola species P. armata (Protaphorura) was taken from laboratory cultures established from field populations close to Darmstadt (Germany) in 2002. Protaphorura armata is widespread in Europe and preferentially colonizes the mineral soil and rhizosphere of plants (euedaphic species). Cultures were kept on a mixture of sterilized potting soil and clay pellets (3 : 1) at 14°C in darkness and fed with moistened baker's yeast. Collembola treatments received 90 P. armata individuals which were added six weeks after transplantation of microcuttings into the microcosms. Collembola were added after the mycorrhizal inoculum to allow mycorrhizal fungi to establish without being grazed, i.e. to ensure effective mycorrhization of the microcuttings.
2.4. 13C and 15N labelling
Twenty-four hours before harvest, plants were transferred into a plexiglas chamber and labelled using a mobile 13CO2 labelling system controlling CO2 concentration and air humidity (set to 400 µl l−1 and 70%, respectively) in the chamber (see [26,31]). CO2 in ambient air was removed and replaced by CO2 containing 8.3 ± 0.2 atom% 13CO2 (mean ± s.d.) (Eurisotop, Saarbrücken, Germany). Thereby, Q. robur microcuttings were exposed to the 13CO2-enriched atmosphere over a complete light period of 16 h and the CO2 concentration adjusted to 400 ± 2 ml l−1 as described in Herrmann et al. [30]. 15N labelling was performed 72 h before harvest using 98 atom% 15NH415NO3 (Sigma, Darmstadt, Germany); 0.1 mg 15NH415NO3 dissolved in 5 ml sterile distilled water was injected into the rhizosphere under sterile conditions.
2.5. Experimental procedure
The experiment was set up in a full factorial design with the factors Piloderma (with and without) and Protaphorura (with and without) allowing to inspect effects of individual organisms but also their interaction. Microcosms were incubated in a climate chamber at the conditions specified above for 8 weeks and then destructively sampled. During incubation the position of individual microcosms was changed in a random way twice a week. Depending on the rhythmic growth phase of microcuttings at harvest, individual experimental systems were grouped into SF and RF. The grouping was based on the opening of individual buds and the anthocyane colouring of leaves as described in more detail in Herrmann et al. [25]. Microcosms with microcuttings which could not be ascribed unequivocally to either of these phases were discarded.
From the initial 129 microcuttings a total of 78 reached the root or shoot flush stage at harvest. Of those, 49 microcuttings were assigned to RF and 29 to SF, the remaining microcuttings were discarded. As defined in Herrmann et al. [25] RF microcuttings are characterized by rush bud stage B which is associated by maximum root growth, and SF microcuttings are characterized by stage D which is associated by maximum leaf expansion. Petri dishes of the 78 microcosms were opened and microcuttings were purged from soil. Harvest started early in the morning (08.00) and lasted until the afternoon (17.00). Microcosms were processed sequentially in random order with handling of individual microcosms and placement of plant tissue into N2 lasting a maximum of 5 min. Microcuttings were cut into five compartments: source leaves (i.e. fully expanded leaves with net export of photosynthates to growing parts of the plant), sink leaves (i.e. not fully expanded leaves with proliferating cells during plant growth), stem, principal root and lateral roots. The principal root comprised the most developed root from which lateral roots branched. The compartments were weighed and the length of stems was measured. Based on total plant biomass at harvest and duration of incubation the relative growth rate of microcuttings was calculated assuming linear growth. Then, the compartments were wrapped into aluminium foil, frozen in liquid nitrogen and stored at −80°C. Some plant compartments (leaves and lateral roots) were pooled before 13C and 15N analysis and RNA extraction to obtain enough material for the analyses. Each pool included 2–5 individual plant compartments (see electronic supplementary material, table S2).
2.6. RNA assay
Transcriptome analysis was based on pooled leaf samples; a total of 19 samples were used (see electronic supplementary material, table S2). RNA extraction was performed using the MasterPure Plant RNA Purification Kit (Epicentre Technologies Corporation, Madison, WI, USA). According to the manufacturer protocol 50 mg of leaf tissue were used for RNA extraction. Unfortunately, there was not enough tissue material to also perform transcriptome analysis of roots. Quality controls were conducted using formaldehyde-agarose gels, Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). The extracted RNA was used to accomplish 100 bp paired-end libraries. RNA was sequenced using an Illumina HiSeq 2000 at Beijing Genomics Institute, Hong Kong, China.
2.7. Read processing and analysis of differential gene expression
Reads were processed following Tarkka et al. [28]. Briefly, low-quality sequences and sequencing artefacts were removed with SeqClean (http://sourceforge.net/projects/seqclean/files/) and low-quality sequencing ends were trimmed with a custom Java script. Short sequences (less than 50 bp) and sequences lacking paired-end information were discarded. The processed Illumina reads were aligned against the reference transcriptome OakContigDF159.1 [28] by Bowtie [33] and quantified by RSEM [34]. The mapping rate onto the OakContigDF159 reference was 76.5–80.4%. Fold changes in gene expression were calculated by pairwise comparisons using the edgeR function [35] implemented in the Bioconductor package [36]. In these comparisons, negative binomial models are fitted to the transcript abundancies determined by RSEM. Contigs were considered differentially expressed when the Benjamini–Hochberg adjusted p-value of this fit was less than 1%. Blast2GO was used to get a description for each contig based on up to 20 hits against NCBI NR database in a blastx search (E-value 1 × 10−5). Protein sequences from Arabidopsis thaliana TAIR database were downloaded to perform a blastx search of DF159.1 and to assign homologue proteins from A. thaliana to each contig. Only hits with an E-value of at least 1 × 10−5 were taken into account. The best A. thaliana protein hit for each oak contig was determined by taking the A. thaliana protein exhibiting the largest per cent identity to the contig in the local alignment. Gene Ontology (GO) [37] enrichment analysis was performed with the Bioconductor package GOseq [38]. GOseq performs a statistical test based on a hypergeometric distribution to determine if in a given list of DE tags (e.g. genes or contigs) tags assigned to a certain category (e.g. GO terms) are significantly enriched, i.e. if they occur more frequently than expected by chance. Thereby, GOseq adjusts the estimation of the p-value for tag-length. We used the capability of GOseq to perform enrichment analyses for a second type of categories, protein families (Pfam). The OakContigDF159.1 reference library, GO annotations as well as best blast hits of each contig have been deposited at www.trophinoak.de. This publication will focus on results of treatments containing Collembola; straight Piloderma effects are described in Herrmann et al. [29].
2.8. Carbon and nitrogen allocation analysis
For 13C and 15N analysis of the compartments of oak microcuttings, the plant material was dried, milled, weighed into tin capsules (1.50–1.75 mg) and stored in a desiccator until analysis (plant material contained pools (leaves and lateral roots) or individual plants (stems and principal roots); for pools see electronic supplementary material, table S2). For 13C and 15N analysis of Collembola, 15–20 individuals of P. armata, equivalent to 50–200 µg dry weight, were transferred into tin capsules, dried at 60°C for 24 h and stored in a desiccator until analysis.
Samples were analysed with a combined system consisting of an elemental analyser (NA 1500, Carlo Erba, Milan, Italy) and a mass spectrometer (MAT 251, Finnigan, Bremen, Germany) [39]. The precision of the measurement is 0.1 delta per mil for 13C and 0.2 delta per mil for 15N. Stable isotope abundance is expressed as atom% excess calculated as 13C (%) = [13C/(12C + 13C)]/100 and 15N (%) = [15N/(14N + 15N)]/100. For 13C PD belemnite (PDB) and for 15N atmospheric N was used as primary standard. Acetanilide (C8H9NO; Merck, Darmstadt, Germany) was used for internal calibration.
2.9. Statistical analysis
13C and 15N values of plant tissue (i.e. lateral root, principal root, sink leaf, source leaf, stem) as well as plant biomass, relative growth rate and stem length were analysed using three-factorial general linear model (GLM), accounting for the non-balanced design, with the factors Stage (RF and SF), Protaphorura (with and without) and Piloderma (with and without). Significance level was set to p < 0.05. Statistical analyses were carried out using SAS (SAS Institute Inc., Cary, NC, USA). As not all plant compartments could be analysed because of lack of enough material in each of the treatments, the number of replicates of the individual variables analysed differed and is given in the legends of figures 1 and 2.
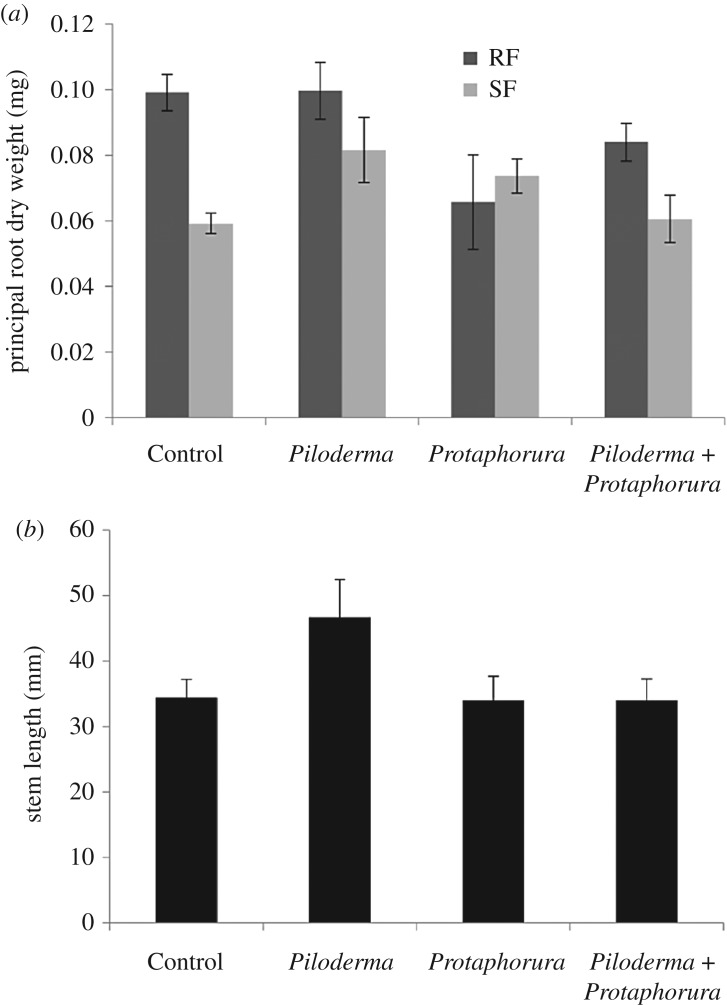
Figure 1.
Effects of Piloderma, Protaphorura and Stage on (a) principal root dry weight during root (RF) and shoot flush (SF) (Control n = 9, Piloderma n = 15, Protaphorura n = 6 and Protaphorura + Piloderma n = 20 in RF, and Control n = 9, Piloderma n = 7, Protaphorura n = 6 and Protaphorura + Piloderma n = 5 in SF treatments), and (b) on stem length (pooled for RF and SF) (Control n = 18, Piloderma n = 22, Protaphorura n = 12 and Protaphorura + Piloderma n = 25).
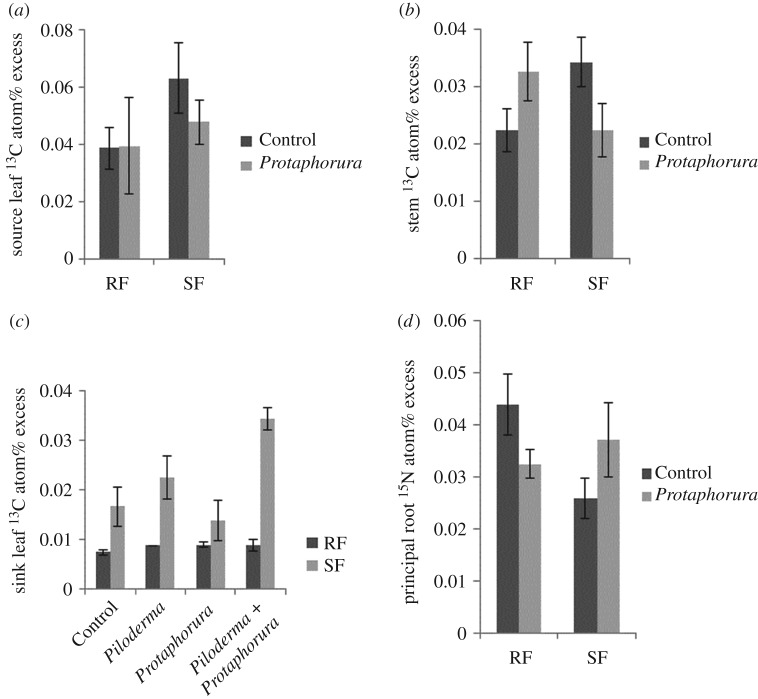
Figure 2.
Effects of (a) Protaphorura on source leaf 13C atom% excess during root (RF) and shoot flush (SF) (pooled for Piloderma) (RF: Control n = 6; Protaphorura n = 4; SF: Control n = 5; Protaphorura n = 6); 13C and 15N atom% excess in plant compartments, (b) effects of Protaphorura on stem 13C atom% excess during RF and SF (pooled for Piloderma) (RF: Control n = 6, Protaphorura n = 4; SF: Control n = 5, Protaphorura n = 4), (c) effects of Protaphorura and Piloderma and their combination on sink leaf 13C atom% excess during RF and SF (RF: Control n = 5, Piloderma n = 6, Protaphorura n = 3, Protaphorura + Piloderma n = 6; SF: Control n = 5, Piloderma n = 3, Protaphorura n = 3, Protaphorura + Piloderma n = 6), and (d) effects of Protaphorura on principal root 15N atom% excess (pooled for Piloderma) (RF: Control n = 6, Protaphorura n = 3; SF: Control n = 5, Protaphorura n = 3).
3. Results
3.1. Morphology
Protaphorura significantly affected principal root biomass, but the effect varied with growth stage of the microcuttings and presence of Piloderma (significant Protaphorura × Stage × Piloderma interaction; F1,70 = 4.01, p = 0.0492; figure 1a; for complete results of statistical analyses see electronic supplementary material, table S1). Protaphorura reduced principal root biomass during RF, with the effect being less pronounced in presence of Piloderma. In contrast, during SF Protaphorura increased principal root biomass, with the effect being less pronounced in presence of Piloderma. Additionally, presence of Protaphorura significantly affected stem length, but the effect varied with the presence of Piloderma (significant Protaphorura × Piloderma interaction; F1,70 = 4.13, p = 0.046; figure 1b). Protaphorura reduced stem length during RF, but the effect was less pronounced in presence of Piloderma. In contrast, during SF Protaphorura increased stem length, but again the effect was less pronounced in presence of Piloderma. Further, the presence of Protaphorura uniformly reduced relative growth rate independent of stage and presence of Piloderma by on average 9.8% (F1,53 = 5.81, p = 0.019).
3.2. 13C and 15N uptake
The presence of Protaphorura significantly affected source leaf 13C atom% excess, but the effect varied with growth stage of the microcuttings (significant Protaphorura × Stage interaction; F1,30 = 4.82, p = 0.036; figure 2a); during RF Protaphorura slightly increased source leaf 13C atom% excess, whereas during SF it was strongly reduced. Further, Protaphorura significantly affected stem 13C atom% excess, but the effect varied with stage (significant Protaphorura × Stage interaction; F1,31 = 7.58, p = 0.010; figure 2b); during RF Protaphorura increased stem 13C atom%, whereas during SF it was decreased. Furthermore, sink leaf 13C atom% excess was increased by Protaphorura, but the effect varied with the presence of Piloderma and with stage (significant Protaphorura × Piloderma × Stage interaction; F1,26 = 4.83, p = 0.037; figure 2c); the effect was restricted to SF and was most pronounced in presence of both Protaphorura and Piloderma.
In addition to aboveground plant compartments, Protaphorura significantly affected 15N atom% excess of principal roots, but again the effect varied with stage (significant Protaphorura × Stage interaction; F1,29 = 4.73, p = 0.038; figure 2d). Protaphorura reduced principal root 15N atom% excess during RF, but increased it during SF. Independent of the presence of Protaphorura, stem 15N atom% excess was increased during RF as compared to SF by 63% (F1,31 = 9.17, p = 0.005). Similarly, in lateral roots, 15N atom% excess was increased during RF as compared to SF by 27% (F1,22 = 7.42, p = 0.012) (data not shown).
3.3. Differential expression profiles
In the Protaphorura treatment 467 and 120 contigs were differentially expressed in leaves during SF and RF, respectively. Of the differentially expressed contigs during SF, 410 were enriched in upregulated and 57 were enriched in downregulated contigs; respective numbers during RF were 65 and 55. In the combined treatment with Protaphorura and Piloderma, 1004 and 104 contigs were differentially expressed in leaves during SF and RF, respectively. Of the differentially expressed contigs during SF, 591 were enriched in upregulated and 413 in downregulated contigs; respective numbers during RF were 46 and 58.
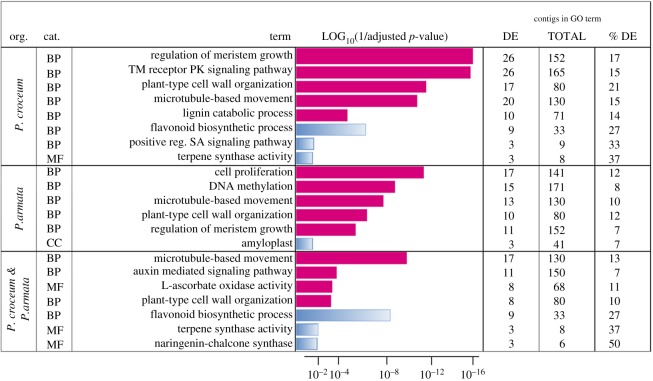
A number of enriched GO terms mirrored changes in oak gene expression levels in sink leaves during SF, and this was true for each pairwise comparison, i.e. Control versus Piloderma, Control versus Protaphorura and Control versus Combined (figure 3; electronic supplementary material, table S3). In each of the three treatments, the GO terms microtubule-based movement and plant-type cell wall organization were enriched in upregulated genes, as was the GO term regulation of meristem growth in Control versus Piloderma and Control versus Protaphorura treatments. By contrast, the term flavonoid biosynthesis was enriched in downregulated genes in the treatments Control versus Piloderma and Control versus Combined. Apart from this, the treatments showed specific patterns of GO enrichment. In presence of Piloderma, mainly growth- and development-related GO terms were enriched in upregulated contigs, as well as GO terms transmembrane receptor protein tyrosine kinase signaling pathway and lignin catabolism, whereas GO terms salicylic acid mediated signaling pathway and terpene synthase activity were enriched in downregulated contigs. In presence of Protaphorura, GO terms of cell proliferation and DNA methylation were enriched in upregulated contigs during SF, but the GO term amyloplasts was enriched in downregulated contigs. In the combined treatment, enriched GO terms included auxin mediated signaling pathway and L-ascorbate oxidase activity. By contrast, the GO terms oxidation-reduction process, terpene synthase activity and naringenin-chalcone synthase activity were enriched in downregulated contigs. In the Protaphorura treatment, two defence-related contigs were enriched in upregulated contigs during SF in leaves. These included the enhanced disease susceptibility 5 contig, which is connected to defence responses including salicylic acid mediated pathway, jasmonic acid mediated pathway and response to chitin, and a contig related to a chalcone-flavanone isomerase family protein, which forms part of the flavonoid biosynthesis (table 1). In the combined treatment with Protaphorura and Piloderma, no enriched contigs related to defence were present. Generally, gene-expression in the combined treatment was least diverse.
Figure 3.
Results of Gene Ontology terms over-representation analysis (FDR < 0.01). Gene Ontology (GO) terms enriched in up- and down-regulated contigs from sink leaves at shoot flush (SF) of plants treated with Piloderma croceum, Protaphorura armata, or from combined treatment are shown, with GO terms related to upregulated (red) and downregulated (blue) contigs. BP marks GO category biological process, MF molecular function and CC cellular compartment, and significance levels are marked by column lengths with maximum column length at p = 1 × 10−16. The absolute numbers and percentages of differentially expressed contigs under each GO term are indicated. TM, transmembrane; PK, protein kinase; SA, salicylic acid. Note that plant growth-related GO terms regulation of meristem growth and microtubule based movement are enriched in upregulated contigs at all three interaction types.
Table 1.
Defence- and secondary-metabolism-related contigs with consistent enrichment or depletion. Pairwise comparisons Control-Pilo (Piloderma treatment), Control-Prot (Protaphorura treatment) and Control-Combined (Combined treatment) in leaves of oak microcuttings during two developmental stages, i.e. shoot flush (SF) and root flush (RF). Significant enrichment (orange) was determined by edgeR with a threshold Benjamini-corrected p-value of 0.01, indicated by FDR.
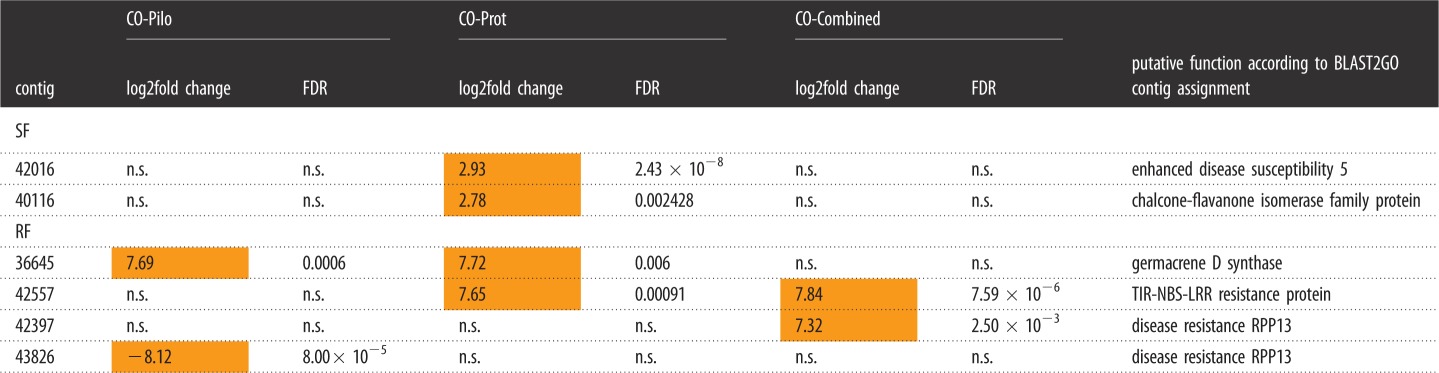 |
Only a few GO terms were enriched in source leaves during RF (electronic supplementary material, table S3). In presence of Protaphorura, GO terms L-ascorbate oxidase activity, laccase activity and secondary cell wall biogenesis were enriched in upregulated contigs. During RF, acid phosphatase transcripts were upregulated in all three treatments, germacrene D synthase by Piloderma and Protaphorura, and TIR-NBS-LRR resistance protein by Protaphorura and combined treatment. Further, there was a highly upregulated contig in presence of Protaphorura which potentially is related to defence, disease resistance RPP13-like protein 1, and interestingly, the abundance of a closely related transcript was downregulated by Piloderma (table 1). Related to phenylpropanoid metabolism, hydroxycinnamoyl-coenzyme A transcripts were upregulated by Piloderma, and according to GO enrichment analysis, cell-wall-related transcripts were upregulated by Protaphorura encoding two laccases, cellulose synthases, a beta-1,4-xylosyltransferase and a proline-rich cell wall protein.
4. Discussion
Collembola are among the most widespread and abundant soil arthropods modifying plant performance in a multitude of ways. Modifications in plant performance at least in part are assumed to be due to Collembola interacting with rhizosphere microorganisms, in particular fungi including mycorrhiza [7,9,40,41]. The only study investigating changes in plant gene expression as affected by Collembola used Arabidopsis thaliana and showed that Collembola affected both plant primary and secondary metabolism [20]. Conforming to the finding that Collembola also affect plant secondary compounds, effects of Collembola on plant growth have been shown to propagate into the herbivore system affecting e.g. aphid reproduction [12,42]. The present study for the first time addressed the combined effect of Collembola and mycorrhizal fungi using a woody plant model system and targeted both changes in plant gene expression as well as plant carbon and nitrogen incorporation. As hypothesized, Collembola indeed altered plant gene expression, and impacted both plant carbon and nitrogen incorporation. Notably, this was true in presence and absence of mycorrhiza. Also, as hypothesized, the effect of Collembola on plant gene expression, and plant carbon and nutrient incorporation varied markedly between developmental stages of oaks, i.e. SF and RF.
We used P. armata as model Collembola species, which is widespread in forests but also agricultural systems in Europe. It represents euedaphic Collembola species which preferentially colonize the mineral soil and rhizosphere of plants. As Collembola are morphologically and trophically diverse and also colonize the soil surface (epedaphic species), their interaction with plants may be more diverse than indicated by investigating only a single species. However, representing euedaphic Collembola species, the effects caused by P. armata are likely to be typical for a wide range of Collembola species colonizing the rhizosphere of plants. Piloderma croceum, our model species of ectomycorrhizal fungi, is a broad host range ectomycorrhizal fungus and common mutualist of both conifer and hardwood species, typically occurring in boreal and temperate forests. The effects of P. armata and its interaction with P. croceum, as found with our model system, therefore are likely to be also of relevance in the field.
4.1. Shoot flush
Protaphorura significantly increased root biomass and also root 15N atom% excess during SF. Since oak roots are not growing during SF, roots mainly function in supplying aboveground plant compartments with nutrients and water. Supporting this scenario a GO term related to water channel activity was increased in each of the treatments during SF and this was most pronounced in the combined treatment with Protaphorura and Piloderma. This suggests that both Collembola and mycorrhiza fostered water and nutrient uptake in oak roots. Notably, this is in line with earlier findings that Collembola and mycorrhiza interact in fostering plant nitrogen uptake [14,19]. Additionally, in presence of Protaphorura the GO term nutrient reservoir activity was enriched in upregulated contigs in leaves, suggesting that Protaphorura stimulated the mobilization of nutrients from storage pools to support leaf development. Notably, the enrichment in the nutrient reservoir activity GO term was most pronounced in the combined treatment with Protaphorura and Piloderma, again suggesting that Collembola and mycorrhiza complement each other in fostering plant nutrition. Collembola-mediated changes in root growth and N incorporation are in line with earlier findings that Collembola alter root morphology [18,19] and nitrogen uptake by plants from soil organic matter [43,44]. The increased root biomass and root 15N atom% excess during SF in presence of Protaphorura and the observed increase in GO terms suggest that Collembola increased the availability of nutrients thereby triggering increased water and nitrogen uptake by oak microcuttings. This then triggered increased mobilization of nitrogen from plant reservoirs and transfer of both nitrogen taken up by plants and mobilized from plant reservoirs into sink leaves, thereby fostering plant growth.
In fact, during SF, presence of Protaphorura resulted in enrichment of upregulated contigs related to plant growth in leaves, such as cell proliferation and regulation of cell cycle; further, the Piloderma-mediated increase in 13C atom% excess in sink leafs during SF was more pronounced in presence of Protaphorura. Earlier studies also reported Collembola to increase plant growth [12,14,43,44]. Concomitant with the reduction in 13C atom% in source leaves and stems in presence of Collembola during SF, Protaphorura increased allocation of carbon resources from source to sink leaves thereby fostering leaf growth and expansion. Again, this supports the above scenario that the Collembola-mediated increase in plant nitrogen uptake triggered plant nutrient mobilization and plant growth.
Similar to leaves and stems, Protaphorura also affected carbon allocation to roots during SF. Complementary to the enrichment of upregulated contigs regarding the nutrient reservoir activity GO term, presence of Protaphorura led to enrichment of the GO term amyloplast formation, supporting the assumption that oak microcuttings increased investment in sink leaf development during SF. Further, Protaphorura reduced 13C atom% excess in source leaves and stems during SF, suggesting that carbon storage, predominantly occurring in source leaves, was reduced by Protaphorura. This indicates that the Collembola-mediated increase in the allocation of resources from roots to shoots fostered the investment of carbon by plants into growth rather than into storage. Supporting this conclusion, earlier studies showed that Collembola increase nitrogen concentration in plants and shift plant biomass towards shoots [43–45]. This again supports our conclusion that Collembola-mediated increase in plant nutrient concentrations triggers oak microcuttings to shift resource allocation from storage to growth via allocating carbon resources into actively growing sink leaves. Overall, the results suggest that Collembola-mediated changes in plant resource uptake and allocation as well as gene expression patterns are triggered by Collembola indirectly impacting plants via increasing nutrient availability, which probably resulted from Collembola grazing on rhizosphere microorganisms and thereby liberating nutrients bound in microbial biomass [46,47]. However, in addition to these nutrient-based effects, Collembola may also have triggered changes in plant performance via plants sensing chemical or mechanical cues of Collembola and modifying gene expression patterns [20,48,49].
In addition to genes related to plant primary metabolism, Protaphorura also altered the expression of defence-related contigs during SF, such as the enhanced disease susceptibility 5 contig. This contig is related to the salicylic acid mediated pathway, jasmonic acid mediated pathway and response to chitin. Additionally, a defence-related contig related to chalcone-flavanone isomerase family protein forming part of flavonoid biosynthesis was enriched in upregulated contigs by Protaphorura during SF. Plant defence is known to be induced by herbivores including those feeding on roots [50,51]. However, induced defence has the disadvantage that plants may suffer from damage before the defence is in place. For preventing damage, plants may use environmental cues providing information on potential or upcoming attacks [52]. Due to the enrichment of upregulated contigs related to defence, Protaphorura probably primed oak seedlings against potential attacks by herbivores. Priming may be related to plants physically sensing the presence of rhizosphere arthropods, suggesting that discrimination of different arthropod taxa by plants is limited. Indeed, it has been shown that plants sensitively respond to mechanostimulation and alter gene expression patterns after contact [53,54]. Previous studies performed in the framework of the TrophinOak project demonstrated that the expression of defence genes also is elicited in SF by Streptomyces sp. and plant parasitic nematodes [27,55]. Similar to the present study, these responses were attenuated in presence of Piloderma. In the case of the bacterium, the priming-like response led to diminished powdery mildew symptoms. However, the response of oak microcuttings in the present study may also be based on Protaphorura feeding on plant roots, which occasionally occurs, but does not detrimentally affect plant growth [19]. Further, plants may also sense cues of the cuticle of Collembola, potentially chitin. In fact, the contig enhanced disease susceptibility 5 is assumed to be related to the response of plants to chitin. Notably, sensing of other soil invertebrates than root herbivores may be advantageous as they may function as vectors for pathogenic microorganisms [56,57].
4.2. Root flush
In contrast to SF, Protaphorura significantly reduced 15N atom% excess of principal roots and reduced their biomass during RF. Biomass reduction of roots might be due to root feeding and in fact, P. fimata, a closely related species of P. armata, has been shown to switch diet from litter resources in soil to feeding on fine roots of Zea mays (L.) if available [58]. More intensive feeding on roots by Collembola during RF than during SF may reflect that during RF oaks are investing in root growth and therefore allocate plant resources, including compounds of high nutritional value such as sugars and amino acids, into roots.
Due to the low number of DE contigs, only a few GO terms in leaves were over-represented in source leaves at RF. Upregulated contigs related to cell wall formation suggests that during RF Protaphorura stimulated the establishment of mechanic barriers of plants and plant structural fortification, contrasting the stimulation of plant investment into growth during SF. Supporting this conclusion, presence of Protaphorura increased stem 13C atom% excess during RF. Similarly, Scheu et al. [59] showed that Collembola increase plant tissue C concentrations, suggesting that plant structural cell wall constituents were enhanced. Increased investment into structural components may explain why Protaphorura reduced shoot length during RF; notably, reduced shoot length in presence of Collembola has been reported previously [60]. Supporting the assumption that Protaphorura altered the investment of oaks into plant defence during RF, their presence led to an enrichment of contigs related to defence, namely tir-nbs-lrr resistance protein related to a wide spectrum of defence responses against antagonists. The above-mentioned reduction in root 15N concentrations and root biomass as well as the enrichment of a defence-related contig probably was due to Protaphorura feeding on roots (see above), resulting in increased defence-related GO terms.
The contrasting effects of Protaphorura during RF as compared to SF reflect the very different gene expression patterns of oak during these phases [29] and suggest that oaks very differently respond to environmental cues including interacting biota during the two allocation and developmental phases RF and SF. Further, during SF and RF Protaphorura were interacting with mycorrhiza regarding gene expression and nutrient uptake, with mycorrhiza attenuating the effect of Protaphorura. This is in line with our second hypothesis.
5. Conclusion
Collembola in the rhizosphere of oak seedlings significantly impacted plant gene expression patterns, plant morphology and plant carbon and nitrogen incorporation. Notably, effects of Protaphorura markedly varied during allocation phases, i.e. SF and RF. During both SF and RF, Protaphorura modified the expression of growth-specific genes which was mirrored in increased 13C and 15N plant tissue nutrient concentrations. In SF Protaphorura presence resulted in an enrichment of GO terms related to plant growth and plant primary metabolism. In contrast, during RF presence of Protaphorura led to an enrichment of GO terms related to physical plant fortification and plant secondary growth. In addition to plant growth and primary metabolism, presence of Protaphorura induced the expression of defence-related GO terms, suggesting that they primed the defence against herbivores during SF and RF. Notably, Collembola triggered alterations in gene expression patterns and plant allocation in non-mycorrhizal plants, but they also interacted with mycorrhiza in altering plant performance with the effects of both varying markedly between plant growth phases. The results suggest that oaks recognize the presence of Collembola and respond by increasing the allocation of carbon into growth and by preparing against herbivore attacks in particular during SF. In contrast, during RF Protaphorura stimulated plant fortification and secondary growth. Overall, the results document that both plant gene expression and allocation patterns can only be understood by considering the multitude of biotic interactors including root associated soil invertebrates. Focusing on soil microarthropods the results document that plant performance and defence gene expression is not only modified by herbivores above and below ground, but also by detritivore animals in soil, highlighting the role of the decomposer system for plant performance and aboveground food webs.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
This study was funded by the German Science Foundation (DFG; SCHE 376/26-1) and was part of the ‘TrophinOak’ project supported by the Helmholtz Centre for Environmental Research (UFZ, Halle). We thank Elke Schulz for technical support in analysing soil parameters. We thank Ines Krieg and Barbara Krause for establishment of the Microsoms with P. croceum.
Data accessibility
All data needed for replicating this work are provided with this article and the included electronic supplementary material.
Authors' contributions
M.G. conducted the molecular laboratory work, participated in the data analysis, carried out the statistical analyses and drafted the manuscript. M.B. and L.F. did the bio-informatics for further analysis of the sequencing data, the Read processing and analysis of differential gene expression. F.K. improved the protocol of the RNA extraction and gave practical assistance during the quantification of the RNA assay. TEEG constructed the mobile 13CO2 labelling system and provided assistance during the labelling process. S.H. established the oak microcutting system including the mycorrhizal fungi, provided the oak microcuttings and participated in the design of the study. M.T. supervised the RNA assay, the Read processing and analysis of differential gene expression and the evaluation of the differential expression profiles. F.B. participated in the design of the study and helped draft the manuscript. S.S. participated in the design of the study, helped drafting the manuscript, revised and finalized the manuscript. All authors gave final approval for publication.
Competing interests
We do not have competing interests.
Funding
This study was funded by the German Science Foundation (DFG; SCHE 376/26-1) and was part of the ‘TrophinOak’ project supported by the Helmholtz Centre for Environmental Research (UFZ, Halle).
References
- 1.Friberg H, Lagerlof J, Ramert B. 2005. Influence of soil fauna on fungal plant pathogens in agricultural and horticultural systems. Biocontrol Sci. Technol. 15, 641–658. ( 10.1080/09583150500086979) [DOI] [Google Scholar]
- 2.Gormsen D, Olsson PA, Hedlund K. 2004. The influence of collembolans and earthworms on AM fungal mycelium. Appl. Soil Ecol. 27, 211–220. ( 10.1016/j.apsoil.2004.06.001) [DOI] [Google Scholar]
- 3.Scheu S. 2001. Plants and generalist predators as links between the below-ground and above-ground system. Basic Appl. Ecol. 2, 3–13. ( 10.1078/1439-1791-00031) [DOI] [Google Scholar]
- 4.Smith SE, Read DJ. 1996. Mycorrhizal symbiosis, 2nd edn London, UK: Academic Press. [Google Scholar]
- 5.André HM, Noti M-I, Lebrun P. 1994. The soil fauna: the other last biotic frontier. Biodivers. Conserv. 3, 45–56. ( 10.1007/BF00115332) [DOI] [Google Scholar]
- 6.Ek H, Sjögren M, Arnebrant K, Söderström B. 1994. Extramatrical mycelial growth, biomass allocation and nitrogen uptake in ectomycorrhizal systems in response to collembolan grazing. Appl. Soil Ecol. 1, 155–169. ( 10.1016/0929-1393(94)90035-3) [DOI] [Google Scholar]
- 7.Sawahata T, Narimatsu M. 2007. Abundance of Collembola (Insecta) inhabiting the hyphal mat of an ectomycorrhizal fungus, Sarcodon scabrosus, in a Pinus densiflora forest. Mycoscience 48, 63–65. ( 10.1007/S10267-006-0322-8) [DOI] [Google Scholar]
- 8.Chamberlain PM, Bull ID, Black HIJ, Ineson P, Evershed RP. 2006. Collembolan trophic preferences determined using fatty acid distributions and compound-specific stable carbon isotope values. Soil Biol. Biochem. 38, 1275–1281. ( 10.1016/j.soilbio.2005.09.022) [DOI] [Google Scholar]
- 9.Tiunov AV, Scheu S. 2005. Arbuscular mycorrhiza and Collembola interact in affecting community composition of saprotrophic microfungi. Oecologia 142, 636–642. ( 10.1007/s00442-004-1758-1) [DOI] [PubMed] [Google Scholar]
- 10.Eisenhauer N, Sabais ACW, Schonert F, Scheu S. 2010. Soil arthropods beneficially rather than detrimentally impact plant performance in experimental grassland systems of different diversity. Soil Biol. Biochem. 42, 1418–1424. ( 10.1016/j.soilbio.2010.05.001) [DOI] [Google Scholar]
- 11.Harris KK, Boerner REJ. 1990. Effects of belowground grazing by collembolan on growth, mycorrhizal infection, and P uptake of Geranium robertianum. Plant Soil 129, 203–210. ( 10.1007/BF00032414) [DOI] [Google Scholar]
- 12.Ke X, Scheu S. 2008. Earthworms, Collembola and residue management change wheat (Triticum aestivum) and herbivore pest performance (Aphidina: Rhopalosiphum padi). Oecologia 157, 603–617. ( 10.1007/s00442-008-1106-y) [DOI] [PubMed] [Google Scholar]
- 13.Ladygina N, et al. 2010. Additive and interactive effects of functionally dissimilar soil organisms on a grassland plant community. Soil Biol. Biochem. 42, 2266–2275. ( 10.1016/j.soilbio.2010.08.027) [DOI] [Google Scholar]
- 14.Lussenhop J, BassiriRad H. 2005. Collembola effects on plant mass and nitrogen acquisition by ash seedlings (Fraxinus pennsylvanica). Soil Biol. Biochem. 37, 645–650. ( 10.1016/j.soilbio.2004.08.021) [DOI] [Google Scholar]
- 15.Mitschunas N, Wagner M, Filser J. 2006. Evidence for a positive influence of fungivorous soil invertebrates on the seed bank persistence of grassland species. J. Ecol. 94, 791–800. ( 10.1111/j.1365-2745.2006.01146.x) [DOI] [Google Scholar]
- 16.Bengtsson G, Hedlund K, Rundgren S. 1993. Patchiness and compensatory growth in a fungus Collembola system. Oecologia 93, 296–302. ( 10.1007/BF00317684) [DOI] [PubMed] [Google Scholar]
- 17.Crowther TW, Bear AD. 2012. Impacts of grazing soil fauna on decomposer fungi are species-specific and density-dependent. Fugal Ecol. 5, 277–281. ( 10.1016/j.funeco.2011.07.006) [DOI] [Google Scholar]
- 18.Endlweber K, Scheu S. 2006. Effects of Collembola on root properties of two competing ruderal plant species. Soil Biol. Biochem. 38, 2025–2031. ( 10.1016/j.soilbio.2006.01.004) [DOI] [Google Scholar]
- 19.Endlweber K, Scheu S. 2007. Interactions between mycorrhizal fungi and Collembola: effects on root structure of competing plant species. Biol. Fertil. Soils 43, 741–749. ( 10.1007/s00374-006-0157-7) [DOI] [Google Scholar]
- 20.Endlweber K, Krome K, Welzl G, Schäffner AR, Scheu S. 2011. Decomposer animals induce differential expression of defence and auxin-responsive genes in plants. Soil Biol. Biochem. 43, 1130–1138. ( 10.1016/j.soilbio.2010.11.013) [DOI] [Google Scholar]
- 21.Blouin M, Zuily-Fodil Y, Pham-Thi A-T, Laffray D, Reversat G, Pando A, Tondoh J, Lavelle P. 2005. Belowground organism activities affect plant aboveground phenotype, inducing plant tolerance to parasites. Ecol. Lett. 8, 202–208. ( 10.1111/j.1461-0248.2004.00711.x) [DOI] [Google Scholar]
- 22.Lohmann M, Scheu S, Müller C. 2009. Decomposers and root feeders interactively affect plant defence in Sinapis alba. Oecologia 160, 289–298. ( 10.1007/s00442-009-1306-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Megías AG, Müller C. 2010. Root herbivores and detritivores shape above-ground multitrophic assemblage through plant-mediated effects. J. Anim. Ecol. 79, 923–931. ( 10.1111/j.1365-2656.2010.01681.x) [DOI] [PubMed] [Google Scholar]
- 24.Wurst S, Dugassa-Gobena D, Langel R, Bonkowski M, Scheu S. 2004. Combined effects of earthworms and vesicular-arbuscular mycorrhiza on plant and aphid performance. New Phytol. 163, 169–176. ( 10.1111/j.1469-8137.2004.01106.x) [DOI] [PubMed] [Google Scholar]
- 25.Herrmann S, Munch J-C, Buscot F. 1998. A gnotobiotic culture system with oak microcuttings to study specific effects of mycobionts on plant morphology before, and in the early phase of, ectomycorrhiza formation by Paxillus involutus and Piloderma croceum. New Phytol. 138, 203–212. ( 10.1046/j.1469-8137.1998.00105.x) [DOI] [PubMed] [Google Scholar]
- 26.Angay O, Fleischmann F, Recht S, Herrmann S, Matyssek R, Oßwald W, Buscot F, Grams TEE. 2014. Sweets for the foe—effects of nonstructural carbohydrates on the susceptibility of Quercus robur against Phytophthora quercina. New Phytol. 203, 1282–1290. ( 10.1111/nph.12876) [DOI] [PubMed] [Google Scholar]
- 27.Kurth F, Mailänder S, Bönn M, Feldhahn L, Herrmann S, Große I, Buscot F, Schrey SD, Tarkka MT. 2014. Streptomyces-induced resistance against oak powdery mildew involves host plant responses in defense, photosynthesis, and secondary metabolism pathways. Mol. Plant Microbe Interact. 27, 891–900. ( 10.1094/MPMI-10-13-0296-R) [DOI] [PubMed] [Google Scholar]
- 28.Tarkka MT, et al. 2013. OakContigDF159.1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis. New Phytol. 199, 529–540. ( 10.1111/nph.12317) [DOI] [PubMed] [Google Scholar]
- 29.Herrmann S, Recht S, Boenn M, Feldhahn L, Angay O, Fleischmann F, Tarkka MT, Grams TEE, Buscot F. 2015. Endogenous rhythmic growth in oak trees is regulated by internal clocks rather than resource availability. J. Exp. Bot. 66, 7113–7127. ( 10.1093/jxb/erv408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrmann S, et al. 2016. Endogenous rhythmic growth, a trait suitable for the study of interplays between multitrophic interactions and tree development. Perspect. Plant Ecol. Evol. Syst. 19, 40–48. ( 10.1016/j.ppees.2016.02.003) [DOI] [Google Scholar]
- 31.Buscot F, Herrmann S. 2004. At the frontier between Basidiomycotes and plants: reciprocal interactions between mycorrhiza formation and root development in an in vitro system with oaks and Hymenomycetes. In Frontiers in Basidiomycote mycology (eds Agerer R, Piepenbring M, Blanz P), pp. 361–376. Eching, Germany: IHW-Verlag. [Google Scholar]
- 32.Marx DK. 1969. The influence of ectotrophic mycorrhizal fungi on the resistance of pine roots to pathogenic infections. I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology 59, 153–163. [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 ( 10.1186/gb-2009-10-3-r25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 ( 10.1186/1471-2105-12-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentleman RC, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 ( 10.1186/gb-2004-5-10-r80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris MA, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32D, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, 2–12. ( 10.1186/gb-2010-11-2-r14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reineking A, Langel R. 1993. 15N, 13C-on-line measurements with an elemental analyser (Carlo Erba, NA 1500), a modified trapping box and a gas isotope mass spectrometer (Finnigan, MAT 251). Environ. Health Stud. 29, 169–174. [Google Scholar]
- 40.Bakonyi G, Posta K, Kiss I, Fábián M, Nagy P, Nosek JN. 2002. Density-dependent regulation of arbuscular mycorrhiza by collembolan. Soil Biol. Biochem. 34, 661–664. ( 10.1016/S0038-0717(01)00228-0) [DOI] [Google Scholar]
- 41.Gange A. 2000. Arbuscular mycorrhizal fungi, Collembola and plant growth. Trends Ecol. Evol. 15, 369–372. ( 10.1016/S0169-5347(00)01940-6) [DOI] [PubMed] [Google Scholar]
- 42.Schütz K, Bonkowski M, Scheu S. 2008. Effects of Collembola and fertilizers on plant performance (Triticum aestivum) and aphid reproduction (Rhopalosiphum padi). Basic Appl. Ecol. 9, 182–188. ( 10.1016/j.baae.2006.07.003) [DOI] [Google Scholar]
- 43.Bardgett RD, Chan KF. 1999. Experimental evidence that soil fauna enhance nutrient mineralization and plant nutrient uptake in montane grassland ecosystems. Soil Biol. Biochem. 31, 1007–1014. ( 10.1016/S0038-0717(99)00014-0) [DOI] [Google Scholar]
- 44.Partsch S, Milcu A, Scheu S. 2006. Decomposer (Lumbricidae, Collembola) affect plant performance in model grassland of different diversity. Ecology 87, 2548–2558. ( 10.1890/0012-9658(2006)87[2548:DLCAPP]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 45.Haase J, Brandl R, Scheu S, Schädler M. 2008. Above- and belowground interactions are mediated by nutrient availability. Ecology 89, 3072–3081. ( 10.1890/07-1983.1) [DOI] [PubMed] [Google Scholar]
- 46.Scheu S, Setälä H. 2002. Multitrophic interactions in decomposer communities. In Multitrophic level interactions (eds Tscharntke T, Kawkins BA), pp. 223–264. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 47.Wardle DA, Yeates GW, Williamson WM, Bonner KI, Barker GM. 2004. Linking aboveground and belowground communities: the indirect influence of aphid species identity and diversity on a three trophic level soil food web. Oikos 107, 283–294. ( 10.1111/j.0030-1299.2004.13523.x) [DOI] [Google Scholar]
- 48.Phillips DA, Ferris H, Cook DR, Strong DR. 2003. Molecular control points in rhizosphere food webs. Ecology 84, 816–826. ( 10.1890/0012-9658(2003)084[0816:MCPIRF]2.0.CO;2) [DOI] [Google Scholar]
- 49.Soler R, Van der Putten WH, Harvey JA, Vet LEM, Dicke M, Bezemer TM. 2012. Root herbivore effects on aboveground multitrophic interactions: patterns, processes and mechanisms. J. Chem. Ecol. 38, 755–767. ( 10.1007/s10886-012-0104-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bezemer TM, van Dam NM. 2005. Linking aboveground and belowground interactions via induced plant defenses. Trends Ecol. Evol. 11, 617–624. ( 10.1016/j.tree.2005.08.006) [DOI] [PubMed] [Google Scholar]
- 51.Ogallo JL, McClure MA. 1996. Systemic acquired resistance and susceptibility to root-knot nematodes in tomato. Phytopathology 86, 498–501. ( 10.1094/Phyto-86-498) [DOI] [Google Scholar]
- 52.Frost CJ, Mescher MC, Carlson JE, De Moraes CM. 2008. Plant defense priming against herbivores: getting ready for a different battle. Plant Physiol. 146, 818–824. ( 10.1104/pp.107.113027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coutand C. 2010. Mechanosensing and thigmomorphogenesis, a physiological and biomechanical point of view. Plant Sci. 179, 168–182. ( 10.1016/j.plantsci.2010.05.001) [DOI] [Google Scholar]
- 54.Lee D, Polisensky DH, Braam J. 2005. Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. New Phytol. 165, 429–444. ( 10.1111/j.1469-8137.2004.01238.x) [DOI] [PubMed] [Google Scholar]
- 55.Maboreke HR, Feldhahn L, Bönn M, Tarkka MT, Buscot F, Herrmann S, Menzel R, Ruess L. 2016. Transcriptome analysis in oak uncovers a strong impact of endogenous rhythmic growth on the interaction with plant-parasitic nematodes. BMC Genomics 17, 627–643. ( 10.1186/s12864-016-2992-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dromph K. 2003. Collembolans as vectors of entomopathogenic fungi. Pedobiologia 47, 245–256. ( 10.1078/0031-4056-00188) [DOI] [Google Scholar]
- 57.Thimm T, Hoffmann A, Borkott H, Munch JC, Tebbe CC. 1998. The gut of the soil microarthropod Folsomia candida (Collembola) is a frequently changeable but selective habitat and a vector for microorganisms. Appl. Environ. Microbiol. 64, 2660–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endlweber K, Ruess L, Scheu S. 2009. Collembola switch diet in presence of plant roots thereby functioning as herbivores. Soil Biol. Biochem. 41, 1151–1154. ( 10.1016/j.soilbio.2009.02.022) [DOI] [Google Scholar]
- 59.Scheu S, Theenhaus A, Jones TH. 1999. Links between the detritivore and the herbivore system: effects of earthworms and Collembola on plant growth and aphid development. Oecologia 119, 541–551. ( 10.1007/s004420050817) [DOI] [PubMed] [Google Scholar]
- 60.Cole L, Staddon PL, Sleep D, Bardgett RD. 2004. Soil animals influence microbial abundance, but not plant-microbial competition for organic nitrogen. Funct. Ecol. 18, 631–640. ( 10.1111/j.0269-8463.2004.00894.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed for replicating this work are provided with this article and the included electronic supplementary material.