Abstract
Diffusion tensor imaging (DTI) studies in early Parkinson's disease (PD) to understand pathologic changes in white matter (WM) organization are variable in their findings. Evaluation of different analytic techniques frequently employed to understand the DTI-derived change in WM organization in a multisite, well-characterized, early stage PD cohort should aid the identification of the most robust analytic techniques to be used to investigate WM pathology in this disease, an important unmet need in the field. Thus, region of interest (ROI)-based analysis, voxel-based morphometry (VBM) analysis with varying spatial smoothing, and the two most widely used skeletonwise approaches (tract-based spatial statistics, TBSS, and tensor-based registration, DTI-TK) were evaluated in a DTI dataset of early PD and Healthy Controls (HC) from the Parkinson's Progression Markers Initiative (PPMI) cohort. Statistical tests on the DTI-derived metrics were conducted using a nonparametric approach from this cohort of early PD, after rigorously controlling for motion and signal artifacts during DTI scan which are frequent confounds in this disease population. Both TBSS and DTI-TK revealed a significantly negative correlation of fractional anisotropy (FA) with disease duration. However, only DTI-TK revealed radial diffusivity (RD) to be driving this FA correlation with disease duration. HC had a significantly positive correlation of MD with cumulative DaT score in the right middle-frontal cortex after a minimum smoothing level (at least 13mm) was attained. The present study found that scalar DTI-derived measures such as FA, MD, and RD should be used as imaging biomarkers with caution in early PD as the conclusions derived from them are heavily dependent on the choice of the analysis used. This study further demonstrated DTI-TK may be used to understand changes in DTI-derived measures with disease progression as it was found to be more accurate than TBSS. In addition, no singular region was identified that could explain both disease duration and severity in early PD. The results of this study should help standardize the utilization of DTI-derived measures in PD in an effort to improve comparability across studies and time, and to minimize variability in reported results due to variation in techniques.
Keyword: Neuroscience
1. Introduction
Parkinson's disease (PD) is a progressive neurodegenerative disorder characterized by a variety of clinical symptoms such as tremor, rigidity, and abnormalities of gait and posture (Magrinelli et al., 2016). Motor symptoms in PD have classically been correlated with loss of dopaminergic neurons in the substantia nigra (SN) pars compacta (Alexander, 2004). In addition to dopamine-related motor dysfunction, however, multiple nondopaminergic deficits in PD affecting cognitive (Kudlicka et al., 2011; Yarnall et al., 2013) and other neuropsychiatric domains have been reported that result in non-motor symptoms including depression (Marsh, 2013), apathy (Kaji and Hirata, 2011), and cognitive impairment including dementia (Aarsland et al., 2017; Meireles and Massano, 2012; Watson and Leverenz, 2010; Yarnall et al., 2013). Furthermore, diffuse pathologic progression has been posited to underlie these non-motor features, including cortical lesions observed in the later stages of disease (Goedert et al., 2013) as well as other non-dopaminergic anatomic involvement (Hawkes et al., 2010) even in the pre-motor stage of PD. Neuroimaging techniques such as positron emission tomography (PET) and single-photon emission computed tomography (SPECT) have been used to aid diagnosis, assess potential new therapies, and monitor disease progression in PD (Politis, 2014). MRI, however, is ubiquitously present, logistically more feasible, potentially easier to standardize, avoids radiation exposure, and has the potential to provide simultaneous structural, functional, and perfusion investigation into PD, unlike PET and SPECT.
Identification of an MRI-derived imaging biomarker early in PD will not only improve understanding of the disease, but also allow for monitoring of disease progression as well as understanding response to treatment (Lang and Mikulis, 2009), which in turn is vital for the conduct of symptomatic and disease-modifying clinical trials (Walsh, 2016). Three complementary MRI techniques involving anatomical, diffusion, and functional changes have been widely investigated to aid early diagnosis and to assess the dynamics of disease progression in PD (Brooks, 2010). Specifically, resting-state functional MRI (fMRI) has shown that PD induces functional alterations (Prodoehl et al., 2014) involving sensorimotor (Tessitore et al., 2014), visual (Holroyd and Wooten, 2006), and basal ganglia networks (Rolinski et al., 2015). Furthermore, recent studies investigating the temporal dynamics of resting-state fMRI networks in PD have shown dynamic functional brain disorganization of major resting state networks, including in PD with and without mild cognitive impairment (Cordes et al., 2018; Díez-Cirarda et al., 2018; Meszlényi et al., 2017; Zhuang et al., 2018). However, fMRI findings can be vulnerable both to acquisition parameters (Prodoehl et al., 2014) as well as to medication status of the patient (Krajcovicova et al., 2012; Tessitore et al., 2012). Anatomical MRI employing voxel-based morphometry (VBM) (Eidelberg, 2011; Fioravanti et al., 2015) and deformation-based morphometry (DBM) (Borghammer et al., 2010; Zeighami et al., 2015) has revealed cortical atrophy in PD involving various cortical and subcortical regions which were correlated with disease duration (Eidelberg, 2011) and degree of motor impairment (Zeighami et al., 2015). Despite these important findings, conflicting reports from fMRI and anatomic MRI studies complicate their understanding and applicability to further investigation of PD. Further, most studies using anatomical MRI have shown correlations between cortical volumes and cognitive status but no discernible volumetric changes in early PD participants. Detection of neuromelanin, which is a characteristic pigment of dopaminergic substantia nigra (SN), using anatomical MRI have been shown to discriminate PD patients and age-matched healthy controls (Sulzer et al., 2018). However, it is still unclear whether neuromelanin-sensitive MRI can help to distinguish early stage PD from age-matched healthy controls as the iron concentration within the SN could influence the neuromelanin-generated T1-contrast (Nakamura and Sugaya, 2014). Further, progression and subtype of disease have unclear correlation with neuromelanin, and analysis of neuromelanin involves only SN despite PD being a whole-brain disease.
Diffusion weighted MRI (dMRI) has largely focused on the organization of white matter (WM) fiber tracts in PD (Atkinson-Clement et al., 2017). dMRI encodes information about the orientation and magnitude of the movement of the water molecules within the fiber tracts of the brain, which can be exploited to investigate fiber tract pathology. Four scalar diffusion measures are obtained at every voxel after fitting tensors in dMRI images: fractional anisotropy (FA), axial diffusivity (AD), and radial diffusivity (RD) provide information regarding axonal and myelin organization, while mean diffusivity (MD) conveys information regarding overall diffusivity patterns in the cortex, subcortex, and WM(Mori and Tournier, 2014). Indeed, diffusivity measures obtained using a single-tensor DTI model have been proposed as markers of structural damage in various diseases (Acosta-Cabronero et al., 2012; Aung et al., 2013; Mishra et al., 2017), including PD (Cochrane and Ebmeier, 2013).
DTI measures of SN in PD has been most commonly compared to healthy controls (HC) and reported using both ROI and whole-brain approaches albeit with conflicting results. For instance, previous ROI-based studies have shown an increase (Lenfeldt et al., 2015), decrease (Vaillancourt et al., 2009), and no change (Hirata et al., 2017; Schwarz et al., 2013) in FA of the SN in PD. Although FA is the most commonly reported measure to show differences in SN between PD and HC, differences in other diffusion measures have also been reported for various WM tracts involving the SN(Cochrane and Ebmeier, 2013). Previous meta-analytic approaches have shown decreased FA and non-altered MD (Cochrane and Ebmeier, 2013; Schwarz et al., 2013) of SN in PD. The sample sizes of these meta-analyses were relatively small, and had significant heterogeneity in the studies analyzed, complicating their interpretation. Since PD affects regions beyond SN, whole-brain approaches comparing diffusivity patterns between HC and PD have also been reported (Kamagata et al., 2014; Karagulle Kendi et al., 2008; Zhang et al., 2011). A recent study (Wen et al., 2016) using a tract-based spatial statistics (TBSS) (Smith et al., 2006) approach reported greater FA and reduced MD and RD in callosal, projections and association fibers in early PD. These findings are contradictory, however, to a recent meta-analytic approach (Atkinson-Clement et al., 2017) in PD with a heterogeneous disease duration that reported reduced FA and increased MD within the SN, corpus callosum, and cingulate and temporal cortices, along with an inverse change within the corticospinal tract (CST) of PD. The cause of such disparate findings is not clear, and importantly no previous study has investigated the influence of registration algorithms and the effect of smoothing in evaluating differences in DTI-derived measures between HC and early PD. This, in turn, could significantly impact the findings reported and possibly account for their variable nature.
Hence in this study, we evaluated: (a) the impact of two widely used skeletonwise techniques, Diffusion-Tensor toolkit (DTI-TK) (Zhang et al., 2006) and TBSS(Smith et al., 2006), to understand the impact of the registration algorithms on the DTI-derived measures, as well as investigate their effects on understanding disease duration and severity in early PD; (b) the effect of smoothing in detecting differences in MD between early PD and HC using conventional VBM-based approaches in light of data in previous studies (Cabeen et al., 2017; Jones et al., 2005) showing that different smoothing parameters could influence the outcome of VBM analysis; (c) the impact of the ROI-based approach to understand PD symptomatology from the DTI-derived measures as it has been shown to be most sensitive (Langley et al., 2016); and (d) which among the skeletonwise, VBM, or ROI-based analytical techniques can detect correlation with the clinical measures such as Movement Disorder Society-Sponsored Unified Parkinson's Disease Rating Scale Part III scores (MDS-UPDRS-III), disease duration in PD patients, and Dopamine Transporter Score (DaT) score in early stage PD participants. Both univariate and multivariate statistical techniques were investigated to understand the impact of these varying statistical approaches to the data as well.
We utilized the diffusion dataset of early PD and HC from the Parkinson Progression Markers Initiative (PPMI) (“The Parkinson Progression Marker Initiative (PPMI).,” 2011) database for our study to test these techniques in a well-characterized and standardized PD population in an effort to establish a homogeneous baseline of DTI metrics for the field.
2. Materials and methods
2.1. Participants
All data utilized in the current study were obtained from the PPMI database. The PPMI study was approved by the Institutional Review Board of all the participating sites and written informed consent was obtained from all the participants by the site investigators.
dMRI datasets acquired between 2011 and 2015 were downloaded in 2017 through a standard application process from the PPMI website (http://www.ppmi-info.org). The inclusion criteria for all PD participants in the study were: (a) all diagnostic criteria for PD should be met; (b) the participant should be diagnosed within two years before the initial visit; and (c) at baseline, the participant's Hoehn and Yahr score should be ≤2. In addition, all HC enrolled in the study should be free of any significant neurological deficits. A comprehensive baseline clinical evaluation of cognitive, behavioural, and motor assessment was performed for every participant by the site investigators. Motor severity score and global assessment of cognition were calculated for each participant using the Movement Disorder Society sponsored-Unified Parkinson's Disease Related Scale Part III scores (MDS-UPDRS)-III and Montreal Cognitive Assessment (MoCA) respectively. Only the demographic and clinical variables that were closest in time to the dMRI scan were utilized in this study.
2.2. Image acquisition
The cardiac gated dMRI datasets used in the current study were acquired across 10 different scanning sites on 3T TIM Siemens scanners with a 12-channel head coil. In addition, Dopamine Transporter (DaT) imaging was performed for all participants using Single Photon Emission Computerized Tomography (SPECT). The details of the scanning parameters are available at http://www.ppmi-info.org.
Briefly, a 2D echo-planar diffusion sequence with the following parameters was used at each site for every participant to acquire a near-isotropic dMRI: number of diffusion encoding directions = 64, b-value = 1000 s/mm2, number of non-diffusion (b0) image = 1, matrix size = 112 × 112, slices = 72, in-plane resolution = 1.98 × 1.98 mm2; slice thickness = 2 mm, flip angle = 90o, TR = 900 ms, and TE = 88 ms.
SPECT data for all participants were acquired 4 ± 0.5 hours after injection of 5 millicuries (mCi) of DaTscanTM. The imaging was performed with a 20% symmetric photopeak windows-centered on 159 KeV and 122 KeV with the following parameters: matrix size = 128 × 128; number of projections = 120 or 90, step size = 3o or 4o. The uptake was calculated in bilateral caudate and putamen and was shared for all participants by the site investigators. Briefly, the following procedure was used to compute the uptake in bilateral caudate and putamen: after spatial normalization of SPECT images to a standard template, striatal binding ratio was computed in bilateral caudate and putamen by counting the pixel density and was normalized to the pixel density in the occipital regions for every HC and PD participant.
Only, the “first” available cardiac gated dMRI scan for each participant was used in this study in an effort to emphasize early stage newly diagnosed PD. These criteria yielded the dMRI scans of a total of 152 PD and 72 HC that were included for further preprocessing and quality control.
2.3. Data preprocessing and quality control
Before preprocessing, each participant's dMRI scans were visually inspected for signal dropout or artifacts. After visual inspection, the dMRI scans were preprocessed using FMRIB software library version 5.0.9 (FSL, http://www.fmrib.ox.ac.uk/fsl/). Eddy current distortion correction was performed for each scan of every participant by affine registering all diffusion-weighted imaging (DWI) volumes to the b0 image using eddy_correct. Each participant's movement in x, y, and z-direction in every volume was computed based on the output of eddy_correct. Single tensor was linearly fitted for every voxel inside the brain using dtifit and FA, MD, AD, and RD were estimated in each voxel for every participant. WM mask with a fractional intensity threshold of 0.2 was computed using bet before tensor-fitting to ensure the tensors were estimated only inside the brain voxels.
An estimate of total translational motion during the dMRI scan was calculated using the output file of eddy_correct, an important standardization step in a motorically symptomatic disease such as PD. Briefly, the estimated translation motion in x, y, and z directions between each volume was summed across all the volumes by taking the root mean square motion of the three directions for each participant. Participants with a mean + 1*standard deviation of diffusion motion during the scan greater than a voxel movement in the slice encoding direction were identified and removed from further analysis. Rotation motion was not included in our quality control as rotations and translations are highly correlated during the scanning (Rae et al., 2012). This rigorous quality control process reduced included dMRI scans to a total of 81 PD and 44 HC that were then used in the study.
2.4. Tract-based spatial statistics (TBSS)
FSL's TBSS (Smith et al., 2006) tool (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TBSS/UserGuide) was used to perform skeletonwise statistics on DTI-derived metrics such as FA, AD, RD, and MD in HC and PD. The procedure is outlined at https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TBSS/UserGuide. Briefly, each subject's individual FA map was first nonlinearly registered to FMRIB58_FA template thereby transforming each subject's FA map to a standard space with an isotropic resolution of 1mm3. Next, the standard space FA maps were registered to the MNI152 space for the convenience of display and reporting. Third, the mean FA skeleton representing the centre of the WM tracts common to all subjects was created and thresholded at FA>0.2. Fourth, each participant's FA map in the MNI152 space was projected onto the mean FA skeleton. A similar procedure was conducted for MD, AD, and RD maps by using the registration parameters of the previous step and projecting each participant's diffusivity map onto the same mean FA skeleton. Statistical comparisons were conducted on the skeletonized projection DTI-derived voxelwise maps in the MNI152 space.
2.5. Tensor-based registration (DTI-TK)
Since DTI-TK has been shown to be more accurate than TBSS(Bach et al., 2014), DTI-TK pipeline was used on the same dMRI scans as above to specifically evaluate the acccuracy of FA-based registration against registering with the full tensor images that incorporate local fiber orientations (Zhang et al., 2006). The steps are outlined at http://dti-tk.sourceforge.net/pmwiki/pmwiki.php?n=Documentation.HomePage. Briefly, an initial population-specific template was bootstrapped with the tensor inputs of all participants. Second, the initial population-specific template was refined with affine and deformable alignment to improve the quality of alignment by removing differences in size and shape of local structures. Third, the improved population-specific template was then resampled to isotropic 1mm3 resolution. Fourth, each participant's tensor map was registered to the isotropic population-specific template using diffeomorphic alignments. The scalar diffusivity maps from the template and the registered tensor maps were then obtained for each participant using singular-value decomposition. Of note, once the co-registration of tensor maps of every subject was finished using DTI-TK, the same algorithm for TBSS was used to generate the DTI-derived metrics at every dMRI voxel for all participants following the guidelines outlined in DTI-TK's user manual (http://dti-tk.sourceforge.net/pmwiki/pmwiki.php?n=Documentation.TBSS). A mean FA map representing the centre of the WM tracts common to all subjects was created from the population-specific template in MNI152 space using the TBSS tools, and the skeleton was thresholded at FA>0.2. All the diffusivity maps were then projected onto this mean FA skeleton map using the procedures outlined in TBSS, for statistical comparisons.
2.6. Voxel-based morphometry (VBM) analysis
MD maps, at various smoothing levels from 0mm to 20mm, registered using full tensor information (DTI-TK) in MNI152 space for each participant were then further utilized to investigate the commonly used VBM-based approach in comparing MD changes between PD and HC. In addition, the VBM-based approach was also used to examine the correlation of the DTI-derived metrics in PD with DaT score, disease duration, and disease severity. Similar to Jones et al. (2005), the MD map of every participant was smoothed from 0mm to 20mm using a Gaussian filter with a kernel size = smoothing size/2.3548 (http://mathworld.wolfram.com/GaussianFunction.html) to investigate the effects of smoothing on the VBM-based approach.
2.7. Region of interest (ROI) analysis
Anterior Thalamic Radiation (ATR: left and right), CST (left and right), Cingulum (CGC: left and right), Cingulum-Hippocampal (CGH: left and right), Inferior Frontal-Occipital Fasciculus (IFO: left and right), Inferior Longitudinal Fasciculus (ILF: left and right), Superior Longitudinal Fasciculus (SLF: left and right), Superior Longitudinal Fasciculus-Temporal (SLFt: left and right), Uncinate Fasciculus (UNC: left and right), and Forceps Major and Minor from Johns Hopkins University (JHU) (Hua et al., 2008) probabilistic white matter tract atlas in MNI152 space were then used to derive a comprehensive mask for extracting FA, MD, AD, and RD in each tract for each subject.
In addition, MD values in the 90 cortical and subcortical regions of the Anatomical Atlas Labeling (AAL) template (Tzourio-Mazoyer et al., 2002), and six bilateral subcortical regions of ATAG template (Keuken et al., 2014) were also extracted for each participant. Both AAL and ATAG templates were in MNI152 space.
To validate previous studies including SN(Lenfeldt et al., 2015; Vaillancourt et al., 2009), we also extracted FA, MD, AD, and RD in SN using its probabilistic mask (Murty et al., 2014) in MNI152 space. Of note, we only utilized those voxels in this probabilistic SN mask where there was at least 90% probability of the mask being classified as SN.
2.8. Statistical analysis
Chi-square (χ2) test was used to test the significance of categorical demographic variables as well as head motion between groups. Kruskal-Wallis test was used to check the statistical significance of continuous demographic and clinical variables. Significance was established at p < 0.05 and the values were reported as mean ± standard deviation for each variable.
Since nonparametric statistics have no dependency on the normal distribution of either independent or dependent variables, nonparametric statistical analyses of skeletonwise, VBM-based, and ROI-based DTI-derived metrics were conducted using the permutation analysis of linear models (PALM) toolbox in FSL (Winkler et al., 2014) to evaluate the difference in DTI-derived metrics between HC and PD, along with testing the correlation of DTI-derived metrics with disease duration, MDS-UPDRS-III, and DaT scores in PD. Both two-sample univariate T-Test, and multivariate Hotelling T2 (Hotelling, 1931) statistics were conducted with PALM. Each participant's average motion during the dMRI scan and total brain volume were used as covariates. Since disease duration might be collinear with age, and is an important determinate of DTI signal, age was also used as a covariate in the statistical model. The statistical tests were repeated with and without each participant's scanning site as covariates, in order to evaluate effects of the variance induced by the scanning site on DTI-derived metrics which may be important for multi-site cohort analysis. Threshold-free cluster enhancement (TFCE) (Smith and Nichols, 2009) with the same design matrix was employed for every statistical test with 1000 random permutations.
Of note, all statistical comparisons were corrected for multiple comparisons in PALM. Importantly, all analytical techniques utilized in this study such as skeletonwise, VBM or ROI-based, were independently controlled for multiple comparisons within themselves. Also, skeletonwise statistical results generated from TBSS were controlled independently from skeletonwise statistical results generated from DTI-TK. Correction of multiple comparisons was performed for each analytical technique and each hypothesis. Significance was established at pcorr<0.05. Cohen's d which is a measure of effect-sizes was also computed and reported where applicable.
3. Results
3.1. Demographics and clinical variables
Table 1 outlines the descriptive statistics as mean ± standard deviation and p-values wherever applicable for the demographics, clinical scores, and head motion in PD and HC.
Table 1.
Participant demographics. Results of pairwise statistical comparisons are shown as p-values. Of note, Dopamine Transporter (DaT) score of caudate and putamen in both hemispheres was integrated for each subject. *Significance was established at p < 0.05. NS: Non-significant; NA: Not-applicable; means are reported ±SD.
| Demographics | Healthy Controls (HC) (N = 44) |
Parkinson's Disease (PD) patients (N = 81) |
HC vs PD |
|---|---|---|---|
| Gender | |||
| Female: | 15 | 29 | NS(p = 0.85) |
| Male: | 29 | 52 | |
| Handedness | |||
| Left: | 5 | 7 | NS(p = 0.1) |
| Right: | 34 | 72 | |
| Mixed: | 5 | 2 | |
| Age (years) | 61 ± 10.79 | 61.35 ± 9.93 | NS(p = 1) |
| Years of education | 15.86 ± 3.17 | 15.44 ± 3.03 | NS(p = 0.46) |
| Disease duration (months) | NA | 11.46 ± 13.85 | NA |
| Movement Disorder Society sponsored-Unified Parkinson's Disease Related Scale Part III score (MDS-UPDRS-III) | NA | 18.72 ± 8.13 | NA |
| Montreal Cognitive Assessment (MoCA) Score | 28.16 ± 1.12 | 27.4 ± 2.14 | NS(p = 0.12) |
| Hoehn and Yahr Score (1/2) | NA | 36/45 | NA |
| Dominant affected side | |||
| Left: | NA | 31 | NA |
| Right: | NA | 49 | |
| Mixed: | NA | 1 | |
| Diffusion motion (mm) | 1.25 ± 0.43 | 1.31 ± 0.4 | NS(p = 0.47) |
| Site | |||
| Site 1: | 3 | 12 | NS(p = 0.3) |
| Site 2: | 5 | 7 | |
| Site 3: | 2 | 7 | |
| Site 4: | 6 | 10 | |
| Site 5: | 3 | 5 | |
| Site 6: | 6 | 12 | |
| Site 7: | 1 | 1 | |
| Site 8: | 5 | 13 | |
| Site 9: | 9 | 14 | |
| Site 10: | 4 | 0 | |
| Dopamine Transporter (DaT) score* | 9.55 ± 1.67 | 5.08 ± 1.41 | p < 0.001 |
Means of the participants demographics such as gender, age, years of education, and handedness were not statistically significant (p > 0.05) between the groups. Similarly, there was no statistically significant difference (p = 0.12) in global cognition assessed by MoCA between the groups. PD patients were very early in the disease: mean disease duration was 11.46 ± 13.85 months, disease severity assessed by modified MDS-UPDRS-III score was 18.72 ± 8.13, and 36 and 45 PD were identified to have Hoehn and Yahr score as 1 and 2 respectively. There was a statistically significant difference (p < 0.001) in the cumulative DaT score between the HC and PD, as expected.
The mean motion in HC and PD during the dMRI scan was 1.25 ± 0.43mm and 1.31 ± 0.4mm respectively and was not found to be statistically significant (p = 0.47).
3.2. Are DTI metrics in PD different than in HC?
None of the analytical techniques, namely skeletonwise, VBM or ROI-based found any differences between HC and early stage PD in this cohort. The results were consistent across various smoothing levels employed to test differences in MD between the groups. Further, we found no difference in any DTI-derived metrics between HC and early stage PD with either univariate or multivariate statistical techniques and the findings were not influenced whether scanning site was added as a covariate in the statistical model.
3.3. Is any DTI metric correlated with disease duration in early PD?
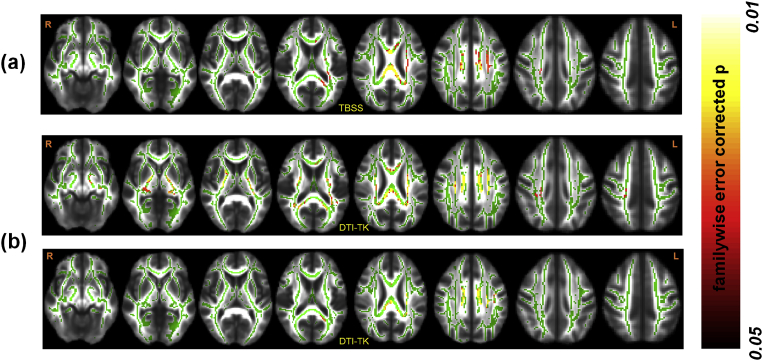
Both skeletonwise techniques, namely TBSS and DTI-TK, revealed a statistically significant negative correlation of FA with disease duration (Fig. 1). TBSS showed that WM tracts in the left hemisphere encompassing left IFO, left CST, left ILF, bilateral ATR, left SLF, along with corpus callosum and bilateral CGC exhibited a significant negative correlation (Mean pcorr: 0.03 ± 0.01, Mean d: 0.44 ± 0.10). This lateralized correlation with disease duration was, however, lost with DTI-TK and WM tracts encompassing corpus callosum, bilateral IFO, bilateral CST, bilateral ILF, bilateral ATR, bilateral SLF, and bilateral CGC exhibited a significant negative correlation (Mean pcorr: 0.02 ± 0.01, Mean d: 0.38 ± 0.11). In addition, RD derived from DTI-TK showed a significant positive correlation (Mean pcorr: 0.04 ± 0.004, Mean d: 0.45 ± 0.1) in WM tracts encompassing left IFO, left CST, left ILF, left ATR, left SLF, left CGC, and corpus callosum with disease duration. These significant correlations were only revealed when scanning site was added as a covariate in the statistical model. Multivariate statistics on skeletonwise DTI metrics revealed no correlation with disease duration with either TBSS or DTI-TK.
Fig. 1.
Skeletonwise results of WM organization in PD. The location of the cluster showing a significantly (pcorr<0.05) negative relationship between FA and disease duration using both TBSS (a) and DTI-TK (b). The top and bottom panel of (b) shows the location of the cluster showing a significantly (pcorr<0.05) negative and positive relationship between FA (top panel) and RD (bottom panel), and disease duration using DTI-TK. R and L represent the right and left hemispheres respectively. Color bar represents the range of p-values in the overlaid cluster.
No correlation of MD with disease duration was found regardless of smoothing and whether scanning site was used as a covariate.
No association of any DTI-derived metric in WM tracts or MD in GM regions were found with ROI-based processing technique. These results were consistent regardless of whether multivariate statistics were employed with DTI-derived metrics in WM tracts and whether scanning site was added as a covariate in the statistical model.
3.4. Is any DTI metric correlated with MDS-UPDRS-III in early PD?
No correlation of MDS-UPDRS-III with any DTI-derived metric was found in our cohort of early stage PD. These results were consistent regardless of analytical technique employed, namely skeletonwise (TBSS and DTI-TK), VBM, and ROI-based, smoothing employed in VBM, multivariate or univariate statistics used, or whether scanning site was added as a covariate in the statistical model.
3.5. Is any DTI metric correlated with cumulative DaT score?
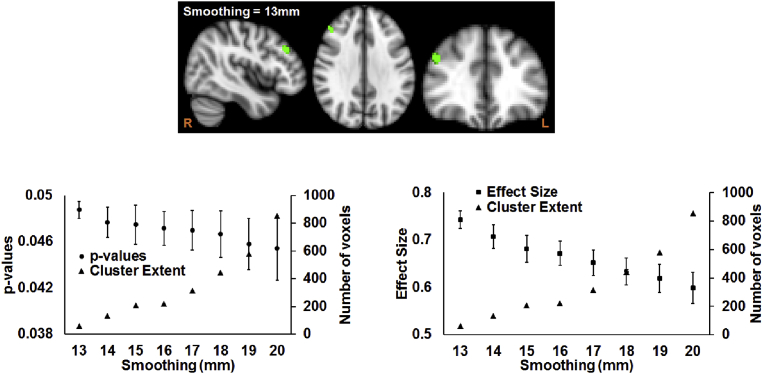
Only MD in the right middle frontal gyrus showed a significantly negative correlation with cumulative DaT score in HC (Fig. 2). This correlation was only revealed when smoothing level was at least 13mm, and scanning site was not added as a covariate in the statistical model. The extent of cluster increased from 60 voxels to 853 voxels when smoothing was increased from 13mm to 20mm. The increase in the cluster size was accompanied with a decrease in Cohen's d, 0.74 ± 0.02 to 0.6 ± 0.03 albeit with not noteworthy increase in pcorr: 0.05 ± 0.0001 to 0.05 ± 0.002, as the smoothing was increased from 13mm to 20mm (bottom panels of Fig. 2).
Fig. 2.
VBM-based results of the effect of smoothing before statistical analysis. (a) Top panel: Location of the cluster, involving right middle frontal gyrus, where MD in HC was significantly (pcorr<0.05) correlated with DaT score. Bottom panel: Scatterplot of the extent of the cluster and p-values as a function of spatial smoothing (left panel), along with scatterplot of the extent of the cluster and effect size as a function of spatial smoothing (right panel) is shown. R and L represent the right and left hemispheres respectively.
Neither skeletonwise (both TBSS and DTI-TK) nor ROI-based processing techniques revealed correlation of any DTI-derived metric with DaT score in either groups. The results were consistent regardless of whether univariate or multivariate statistics was used and whether scanning site was added as a covariate in the statistical model.
Tables 2 and 3 summarizes these results in the tabular format.
Table 2.
Summary of findings. Regions that were found to be significantly different between groups or correlated with clinical variables are listed for each analysis technique to compare DTI-derived measurements in early stage PD. NA: Not-applicable.
| Hypotheses tested | Skeletonwise |
Voxelwise | ROI-based | ||
|---|---|---|---|---|---|
| TBSS | DTI-TK | ||||
| Are DTI-derived measures in PD different than HC? | Findings | No regions found to be significantly different. | No regions found to be significantly different. | No regions found to be significantly different. | No regions found to be significantly different. |
| Effect of Smoothing | NA | NA | No effect of smoothing | NA | |
| Effect of Scanning Site | No effect of scanning site | No effect of scanning site | No effect of scanning site | No effect of scanning site | |
| Univariate or Multivariate? | Both | Both | NA | Both | |
| Are DTI-derived measures correlated with clinical scores? | Findings | Negative correlation with disease duration and FA in the WM tracts of left IFO, left CST, left ILF, bilateral ATR, left SLF, bilateral CGC, and corpus callosum | Negative correlation with disease duration and FA in the WM tracts of corpus callosum, bilateral IFO, bilateral CST, bilateral ILF, bilateral ATR, bilateral SLF, and bilateral CGC. Positive correlation with disease duration and RD in the WM tracts of left IFO, left CST, left ILF, left ATR, left SLF, left CGC, and corpus callosum |
MD of right middle frontal gyrus negatively associated with DaT score in HC | No regions found to be significantly different. |
| Effect of Smoothing | NA | NA | Smoothing>=13mm>=5mm | NA | |
| Effect of Scanning Site | Only when scanning site was used as a covariate | Only when scanning site was used as a covariate | Only when scanning site was not used as a covariate | No effect of scanning site | |
| Univariate or Multivariate? | Univariate | Univariate | NA | NA | |
Table 3.
Consistency of findings across different analytical techniques. Hypotheses were tested for four different analytic technique, namely TBSS skeletonwise, DTI-TK skeletonwise, voxelwise, and ROI-based under two different conditions, (a) when scanning site was used as a nuisance regressor and (b) when scanning site was not used as a nuisance regressor. False-positive rate was controlled for every hypothesis within every DTI-derived metric using non-parametric statistics, and independently controlled for whether scanning site was used a nuisance regressor. ✓ is shown for the columns when the observed conclusion is in accordance with the hypothesis being tested, otherwise they are shown as ✕. For example: Only DTI-TK based skeletonwise analytical technique showed a negative correlation between RD and disease duration for PD participants but only when scanning site was used as a covariate in the statistical model. This finding is indicated by “Only when scanning site was used as a covariate” in the Scanning site used as a covariate column, ✕ in TBSS column within Skeletonwise column, ✓ in DTI-TK within Skeletonwise column, NA for voxelwise (as no voxelwise comparison was conducted for RD), and ✕ for ROI-based technique. NA: Not-applicable.
| Hypothesis Tested | DTI-derived metric | Findings | Scanning site used as a covariate in the statistical model | Analytic technique used |
|||
|---|---|---|---|---|---|---|---|
| Skeletonwise |
Voxelwise | ROI-based | |||||
| TBSS | DTI-TK | ||||||
| HC vs PD | FA | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ |
| MD | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | ✓ | ✓ | |
| AD | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| RD | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| Multivariate | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| Disease duration | FA | Negative correlation | Only when scanning site was used as a covariate | ✓ | ✓ | NA | ✕ |
| MD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | ✓ | ✓ | |
| AD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| RD | Positive correlation | Only when scanning site was used as a covariate | ✕ | ✓ | NA | ✕ | |
| Multivariate | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| MDS-UPDRS-III | FA | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ |
| MD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | ✓ | ✓ | |
| AD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| RD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| Multivariate | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| Cumulative DaT Score | FA | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ |
| MD | Negative correlation in HC (only when smoothing >=13mm) | Only when scanning site was not used as a covariate | ✕ | ✕ | ✓ | ✕ | |
| AD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| RD | No correlation | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
| Multivariate | No difference | Irrespective of whether scanning site was used as a covariate | ✓ | ✓ | NA | ✓ | |
4. Discussion
While carefully controlling for important confounds with stringent standardized criteria in a well-characterized and publicly available multi-site dMRI database of early PD, our study suggests: (i) either skeletonwise approach utilizing different registration techniques such as DTI-TK or TBSS could be utilized (Cabeen et al., 2017) to understand the association of disease duration and DTI-derived metrics, however DTI-TK was found to be more sensitive to disease progression (Fig. 1); (ii) although the VBM-based approach revealed a significant negative correlation of MD with cumulative DaT score in HC, it should be reported with caution as the amount of spatial smoothing could bias the conclusion (Fig. 2); and (iii) care should be taken with ROI-based techniques as no differences in any DTI-derived metrics were found between HC and PD. Furthermore, the findings of our study were sensitive to imaging site but were not sensitive to the choice of univariate nor multivariate statistical analysis (Tables 2 and 3). Overall, our findings suggest that the conclusions for the hypothesis being tested are strongly dependent upon the choice of analytic techniques as different analytical techniques can lead to different conclusions (Tables 2 and 3). In addition, our findings also suggest that there does not appear to be a single brain region (e.g. SN) that could be chosen a priori using scalar DTI-derived metrics that could explain both disease duration and severity in early PD.
There have been conflicting studies with FA of SN as a diagnostic imaging biomarker (Lehericy et al., 2017; Lenfeldt et al., 2015; Menke et al., 2010; Schwarz et al., 2013). Increased MD has also been reported in SN of PD (Eidelberg, 2011; Schwarz et al., 2013). However, the present study does not indicate that any of the DTI-derived scalar metrics of SN or several other subcortical nuclei such as caudate and putamen are significantly different between HC and early PD.
Previous studies (Cabeen et al., 2017; Jones et al., 2005) have shown that results from the conventional VBM-based approach may be affected by spatial smoothing employed before statistical analysis. Similarly, our study also found a smoothing-related correlation of MD and cumulative DaT score in the middle-frontal gyrus of HC using the conventional VBM-based approach: although no statistical difference was observed in MD between HC and PD. We observed a significant negative correlation of MD in HC only when smoothing was greater than or equal to 13mm. This smoothing-related correlation of MD with cumulative DaT score was only revealed when scanning site was not used as a covariate in the statistical model. Overall, this finding warrants further investigation with a longitudinal dataset to further understand the meaning of microstructural pathologic variability across patients including in relationship to motor and non-motor symptoms. Indeed, further understanding of this finding may lead to important and novel insight into the clinically heterogeneous nature of PD. This finding also suggests that data harmonization (Fortin et al., 2017) is necessary while comparing DTI-derived metrics across different scanning sites.
Correlation of DTI-derived metrics with disease progression has been previously reported (Pozorski et al., 2018), but that study was a single-site, not controlled for medication status, and not controlled for early-stage disease. To the best of our understanding, the negative association of disease duration reported here with FA in WM tracts implicated in early PD has not been previously reported. Importantly, the present study revealed preference for DTI-TK based registration technique before statistical analysis. Further, DTI-TK demonstrated a significantly positive correlation of disease duration with left lateralized RD in the same regions as FA suggesting that tensor-based registration may be more accurate than FA-based registration as has been previously hypothesized (Bach et al., 2014). Neither ROI-based nor VBM-based approaches found any association with disease duration in this well-characterized cohort of early PD patients. Our findings thus suggest that DTI-derived FA measurement in the WM skeleton could be used as a potential imaging biomarker to evaluate PD. Of note, extra-axonal diffusion (as measured by RD) spanning WM tracts might also lend support to the recent findings of the sensitivity of “free-water” to monitor (Burciu et al., 2017) and classify typical and atypical Parkinsonism (Planetta et al., 2016), and hence warrants further investigation into the understanding the role of “free-water” in early stage PD beyond SN.
There are several limitations to this study. The distortion correction was only performed using the affine registration of the diffusion-weighted volumes to the non-diffusion-weighted b0 map. Our findings need to be validated with advanced distortion correction techniques to investigate the potential bias induced by off-resonance effects. Importantly, our study did reveal a trend of higher FA in HC in the same tracts where FA was significantly correlated with disease duration in PD participants (70% of the voxels, mean uncorrected p-value: 0.21 ± 0.14; range of uncorrected p–values: 0.0005–0.5). However, the effect size to observe this change was low (Cohen's d: 0.19 ± 0.12). A similar trend was observed with RD in PD (RD in PD > RD in HC) in the same tracts where RD was significantly correlated with disease duration in PD participants (64% of the voxels, mean uncorrected p-value: 0.24 ± 0.14; range of uncorrected p–values: 0.0004–0.5). However, the effect size to observe this change was also low (d: 0.17 ± 0.13). These findings suggest that our cross-sectional analysis was limited by sample size, and longitudinal analysis may reveal steeper changes in FA, MD, and RD in PD than HC over time. However, no longitudinal analysis was conducted as this was beyond the scope of this baseline early PD study. Potential bias introduced by not rotating the gradient vectors after eddy_correct operation (Leemans and Jones, 2009) should be investigated to understand its influence on the overall findings of the study. We did not correct for “free-water” contamination before fitting the tensors (Burciu et al., 2017), as we have used MD as a surrogate of “free-water” (Mishra et al., 2015). Future validating studies should estimate the influence of different analytic techniques on DTI-derived metrics after correcting for “free-water” contamination. Future studies should also evaluate correlations between tract-specific DTI-derived (Mishra et al., 2015) measures and disease duration and severity in PD. Our study found that site influenced the findings when its effects are regressed out as a linear cofactor. However, future studies could evaluate whether harmonization techniques (Fortin et al., 2017) could prove beneficial in improving our understanding of the differences in scalar DTI-derived metrics in early PD as compared to HC, and regarding association with disease severity or duration. Future studies could also evaluate the effect of tissue-specific smoothing (Lee et al., 2009) to understand the bias introduced by spatial smoothing approaches used in our study.
5. Conclusion
The present study found that scalar DTI-derived measures such as FA, MD, and RD should be used as imaging biomarkers but with caution in early stage PD as the conclusions derived are influenced by the choice of the analytic technique used. Furthermore, our study revealed that DTI-TK can detect WM organizational changes evaluated through DTI-derived metrics in early PD, including earlier as compared to TBSS. Longitudinal studies investigating tractography, harmonization, and the role of “free-water” contamination in the DTI-derived metrics are warranted based on our findings. We hope that this study will help standardize the utilization of DTI-derived measures in PD in an effort to improve comparability across studies and time and to minimize variability in reported results as a result of variation in analytic techniques.
Declarations
Author contribution statement
Virendra R. Mishra: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Karthik R. Sreenivasan, Xiaowei Zhuang, Zhengshi Yang, Dietmar Cordes: Contributed reagents, materials, analysis tools or data.
Ryan R. Walsh: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 5P20GM109025, and private grant funds from The Elaine P. Wynn and Family Foundation, The Sam and Peggy Grossman Family Foundation, The Samuel P. Mandell Foundation, The Peter and Angela Dal Pezzo funds, and The Young Scientist Award.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
All the authors had unrestricted access to the data in this study and had final decision to submit for publication. We sincerely thank all the participants of PPMI and Mr. Christopher Bird who helped to download the data from PPMI website to our local server for analysis.
Data used in the preparation of this article were obtained from the Parkinson's Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org. PPMI, a public-private partnership, is funded by the Michael J. Fox Foundation for Parkinson's Research. Other funding partners include a consortium of industry players, non-profit organizations and private individuals: AbbVie, Avid Radiopharmaceuticals, Biogen Idec, Bristol-Meyers Squibb, Covance, GE Healthcare, Genentech, GlaxoSmithKline, Eli Lilly and Company, Lundbeck, Merck, Meso Scale Diagnostics, Pfizer Inc., Piramal Imaging, Roche CNS group, Servier, UCB and Golub Capital.
Contributor Information
Virendra R. Mishra, Email: mishrav@ccf.org.
Ryan R. Walsh, Email: ryan.walsh@barrowneuro.org.
References
- Aarsland D., Creese B., Politis M., Chaudhuri K.R., Ffytche D.H., Weintraub D., Ballard C. Cognitive decline in Parkinson disease. Nat. Rev. Neurol. 2017;13:217–231. doi: 10.1038/nrneurol.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Cabronero J., Alley S., Williams G.B., Pengas G., Nestor P.J. Diffusion tensor metrics as biomarkers in alzheimer's disease. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander G.E. Biology of Parkinson’s disease: pathogenesis and pathophysiology of a multisystem neurodegenerative disorder. Dialogues Clin. Neurosci. 2004;6:259–280. doi: 10.31887/DCNS.2004.6.3/galexander. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson-Clement C., Pinto S., Eusebio A., Coulon O. Diffusion tensor imaging in Parkinson’s disease: review and meta-analysis. NeuroImage Clin. 2017;16:98–110. doi: 10.1016/j.nicl.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung W.Y., Mar S., Benzinger T.L.S. Diffusion tensor MRI as a biomarker in axonal and myelin damage. Imaging Med. 2013;5:427–440. doi: 10.2217/iim.13.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M., Laun F.B., Leemans A., Tax C.M.W., Biessels G.J., Stieltjes B., Maier-Hein K.H. Methodological considerations on tract-based spatial statistics (TBSS) Neuroimage. 2014;100:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Borghammer P., Ostergaard K., Cumming P., Gjedde A., Rodell A., Hall N., Chakravarty M.M. A deformation-based morphometry study of patients with early-stage Parkinson’s disease. Eur. J. Neurol. 2010;17:314–320. doi: 10.1111/j.1468-1331.2009.02807.x. [DOI] [PubMed] [Google Scholar]
- Brooks D.J. Imaging approaches to Parkinson disease. J. Nucl. Med. 2010;51:596–609. doi: 10.2967/jnumed.108.059998. [DOI] [PubMed] [Google Scholar]
- Burciu R.G., Ofori E., Archer D.B., Wu S.S., Pasternak O., McFarland N.R., Okun M.S., Vaillancourt D.E. Progression marker of Parkinson’s disease: a 4-year multi-site imaging study. Brain. 2017;140:2183–2192. doi: 10.1093/brain/awx146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen R.P., Bastin M.E., Laidlaw D.H. A comparative evaluation of voxel-based spatial mapping in diffusion tensor imaging. Neuroimage. 2017;146:100–112. doi: 10.1016/j.neuroimage.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane C.J., Ebmeier K.P. Diffusion tensor imaging in parkinsonian syndromes: a systematic review and meta-analysis. Neurology. 2013;80:857–864. doi: 10.1212/WNL.0b013e318284070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D., Zhuang X., Kaleem M., Sreenivasan K., Yang Z., Mishra V., Banks S.J., Bluett B., Cummings J.L. Advances in functional magnetic resonance imaging data analysis methods using Empirical Mode Decomposition to investigate temporal changes in early Parkinson’s disease. Alzheimer's Dement. Transl. Res. Clin. Interv. 2018 doi: 10.1016/j.trci.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez-Cirarda M., Strafella A.P., Kim J., Peña J., Ojeda N., Cabrera-Zubizarreta A., Ibarretxe-Bilbao N. Dynamic functional connectivity in Parkinson’s disease patients with mild cognitive impairment and normal cognition. NeuroImage Clin. 2018;17:847–855. doi: 10.1016/j.nicl.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidelberg D. Oxford University Press; New York, NY: 2011. Imaging in Parkinson’s Disease. [Google Scholar]
- Fioravanti V., Benuzzi F., Codeluppi L., Contardi S., Cavallieri F., Nichelli P., Valzania F. MRI correlates of Parkinson’s disease progression: a voxel based morphometry study. Parkinsons. Dis. 2015;2015:378032. doi: 10.1155/2015/378032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P., Parker D., Tunc B., Watanabe T., Elliott M.A., Ruparel K., Roalf D.R., Satterthwaite T.D., Gur R.C., Gur R.E., Schultz R.T., Verma R., Shinohara R.T. Harmonization of multi-site diffusion tensor imaging data. Neuroimage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M., Spillantini M.G., Del Tredici K., Braak H. 100 years of Lewy pathology. Nat. Rev. Neurol. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- Hawkes C.H., Del Tredici K., Braak H. A timeline for Parkinson’s disease. Park. Relat. Disord. 2010;16:79–84. doi: 10.1016/j.parkreldis.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Hirata F.C.C., Sato J.R., Vieira G., Lucato L.T., Leite C.C., Bor-Seng-Shu E., Pastorello B.F., Otaduy M.C.G., Chaim K.T., Campanholo K.R., Novaes N.P., Melo L.M., Gonçalves M.R., do Nascimento F.B.P., Teixeira M.J., Barbosa E.R., Amaro E., Cardoso E.F. Substantia nigra fractional anisotropy is not a diagnostic biomarker of Parkinson’s disease: a diagnostic performance study and meta-analysis. Eur. Radiol. 2017;27:2640–2648. doi: 10.1007/s00330-016-4611-0. [DOI] [PubMed] [Google Scholar]
- Holroyd S., Wooten G.F. Preliminary FMRI evidence of visual system dysfunction in Parkinson’s disease patients with visual hallucinations. J. Neuropsychiatry Clin. Neurosci. 2006;18:402–404. doi: 10.1176/jnp.2006.18.3.402. [DOI] [PubMed] [Google Scholar]
- Hotelling H. The generalization of student’s ratio. Ann. Math. Stat. 1931;2:360–378. [Google Scholar]
- Hua K., Zhang J., Wakana S., Jiang H., Li X., Reich D.S., Calabresi P.A., Pekar J.J., van Zijl P.C.M., Mori S. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage. 2008;39:336–347. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.K., Symms M.R., Cercignani M., Howard R.J. The effect of filter size on VBM analyses of DT-MRI data. Neuroimage. 2005;26:546–554. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Kaji Y., Hirata K. Apathy and anhedonia in Parkinson’s disease. ISRN Neurol. 2011;2011:219427. doi: 10.5402/2011/219427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata K., Tomiyama H., Hatano T., Motoi Y., Abe O., Shimoji K., Kamiya K., Suzuki M., Hori M., Yoshida M., Hattori N., Aoki S. A preliminary diffusional kurtosis imaging study of Parkinson disease: comparison with conventional diffusion tensor imaging. Neuroradiology. 2014;56:251–258. doi: 10.1007/s00234-014-1327-1. [DOI] [PubMed] [Google Scholar]
- Karagulle Kendi A.T., Lehericy S., Luciana M., Ugurbil K., Tuite P. Altered diffusion in the frontal lobe in Parkinson disease. AJNR. Am. J. Neuroradiol. 2008;29:501–505. doi: 10.3174/ajnr.A0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuken M.C., Bazin P.-L., Crown L., Hootsmans J., Laufer A., Muller-Axt C., Sier R., van der Putten E.J., Schafer A., Turner R., Forstmann B.U. Quantifying inter-individual anatomical variability in the subcortex using 7 T structural MRI. Neuroimage. 2014;94:40–46. doi: 10.1016/j.neuroimage.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Krajcovicova L., Mikl M., Marecek R., Rektorova I. The default mode network integrity in patients with Parkinson’s disease is levodopa equivalent dose-dependent. J. Neural Transm. 2012;119:443–454. doi: 10.1007/s00702-011-0723-5. [DOI] [PubMed] [Google Scholar]
- Kudlicka A., Clare L., Hindle J.V. Executive functions in Parkinson’s disease: systematic review and meta-analysis. Mov. Disord. 2011;26:2305–2315. doi: 10.1002/mds.23868. [DOI] [PubMed] [Google Scholar]
- Lang A.E., Mikulis D. A new sensitive imaging biomarker for Parkinson disease? Neurology. 2009;72:1374–1375. doi: 10.1212/01.wnl.0000343512.36654.41. [DOI] [PubMed] [Google Scholar]
- Langley J., Huddleston D.E., Merritt M., Chen X., McMurray R., Silver M., Factor S.A., Hu X. Diffusion tensor imaging of the substantia nigra in Parkinson’s disease revisited. Hum. Brain Mapp. 2016;37:2547–2556. doi: 10.1002/hbm.23192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Chung M.K., Lazar M., DuBray M.B., Kim J., Bigler E.D., Lainhart J.E., Alexander A.L. A study of diffusion tensor imaging by tissue-specific, smoothing-compensated voxel-based analysis. Neuroimage. 2009;44:870–883. doi: 10.1016/j.neuroimage.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans A., Jones D.K. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn. Reson. Med. 2009;61:1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- Lehericy S., Vaillancourt D.E., Seppi K., Monchi O., Rektorova I., Antonini A., McKeown M.J., Masellis M., Berg D., Rowe J.B., Lewis S.J.G., Williams-Gray C.H., Tessitore A., Siebner H.R. The role of high-field magnetic resonance imaging in parkinsonian disorders: pushing the boundaries forward. Mov. Disord. 2017;32:510–525. doi: 10.1002/mds.26968. [DOI] [PubMed] [Google Scholar]
- Lenfeldt N., Larsson A., Nyberg L., Birgander R., Forsgren L. Fractional anisotropy in the substantia nigra in Parkinson’s disease: a complex picture. Eur. J. Neurol. 2015;22:1408–1414. doi: 10.1111/ene.12760. [DOI] [PubMed] [Google Scholar]
- Magrinelli F., Picelli A., Tocco P., Federico A., Roncari L., Smania N., Zanette G., Tamburin S. Pathophysiology of motor dysfunction in Parkinson’s disease as the rationale for drug treatment and rehabilitation. Parkinsons. Dis. 2016;2016:9832839. doi: 10.1155/2016/9832839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L. Depression and Parkinson’s disease: current knowledge. Curr. Neurol. Neurosci. Rep. 2013;13:409. doi: 10.1007/s11910-013-0409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles J., Massano J. Cognitive impairment and dementia in Parkinson’s disease: clinical features, diagnosis, and management. Front. Neurol. 2012 doi: 10.3389/fneur.2012.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke R.A., Jbabdi S., Miller K.L., Matthews P.M., Zarei M. Connectivity-based segmentation of the substantia nigra in human and its implications in Parkinson’s disease. Neuroimage. 2010;52:1175–1180. doi: 10.1016/j.neuroimage.2010.05.086. [DOI] [PubMed] [Google Scholar]
- Meszlényi R.J., Hermann P., Buza K., Gál V., Vidnyánszky Z. Resting state fMRI functional connectivity analysis using dynamic time warping. Front. Neurosci. 2017;11:75. doi: 10.3389/fnins.2017.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V., Guo X., Delgado M.R., Huang H. Towards tract-specific fractional anisotropy (TSFA) at crossing-fiber regions with clinical diffusion MRI. Magn. Reson. Med. 2015;74:1768–1779. doi: 10.1002/mrm.25548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V.R., Zhuang X., Sreenivasan K.R., Banks S.J., Yang Z., Bernick C., Cordes D. Multimodal MR imaging signatures of cognitive impairment in active professional fighters. Radiology. 2017;162403 doi: 10.1148/radiol.2017162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Tournier J.-D. second ed. Academic Press; Oxford, UK: 2014. Introduction to Diffusion Tensor Imaging and Higher Order Models. [Google Scholar]
- Murty V.P., Shermohammed M., Smith D.V., Carter R.M., Huettel S.A., Adcock R.A. Resting state networks distinguish human ventral tegmental area from substantia nigra. Neuroimage. 2014;100:580–589. doi: 10.1016/j.neuroimage.2014.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Sugaya K. Neuromelanin-sensitive magnetic resonance imaging: a promising technique for depicting tissue characteristics containing neuromelanin. Neural Regen. Res. 2014;9:759–760. doi: 10.4103/1673-5374.131583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planetta P.J., Ofori E., Pasternak O., Burciu R.G., Shukla P., DeSimone J.C., Okun M.S., McFarland N.R., Vaillancourt D.E. Free-water imaging in Parkinson’s disease and atypical parkinsonism. Brain. 2016;139:495–508. doi: 10.1093/brain/awv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis M. Neuroimaging in Parkinson disease: from research setting to clinical practice. Nat. Rev. Neurol. 2014;10:708–722. doi: 10.1038/nrneurol.2014.205. [DOI] [PubMed] [Google Scholar]
- Pozorski V., Oh J.M., Adluru N., Merluzzi A.P., Theisen F., Okonkwo O., Barzgari A., Krislov S., Sojkova J., Bendlin B.B., Johnson S.C., Alexander A.L., Gallagher C.L. Longitudinal white matter microstructural change in Parkinson’s disease. Hum. Brain Mapp. 2018;39:4150–4161. doi: 10.1002/hbm.24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PPMI The Parkinson progression marker initiative (PPMI) Prog. Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodoehl J., Burciu R.G., Vaillancourt D.E. Resting state functional magnetic resonance imaging in Parkinson’s disease. Curr. Neurol. Neurosci. Rep. 2014;14:448. doi: 10.1007/s11910-014-0448-6. [DOI] [PubMed] [Google Scholar]
- Rae C.L., Correia M.M., Altena E., Hughes L.E., Barker R.A., Rowe J.B. White matter pathology in Parkinson’s disease: the effect of imaging protocol differences and relevance to executive function. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolinski M., Griffanti L., Szewczyk-Krolikowski K., Menke R.A.L., Wilcock G.K., Filippini N., Zamboni G., Hu M.T.M., Mackay C.E. Aberrant functional connectivity within the basal ganglia of patients with Parkinson’s disease. NeuroImage. Clin. 2015;8:126–132. doi: 10.1016/j.nicl.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S.T., Abaei M., Gontu V., Morgan P.S., Bajaj N., Auer D.P. Diffusion tensor imaging of nigral degeneration in Parkinson’s disease: a region-of-interest and voxel-based study at 3 T and systematic review with meta-analysis. NeuroImage. Clin. 2013;3:481–488. doi: 10.1016/j.nicl.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Nichols T.E. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Johansen-Berg H., Rueckert D., Nichols T.E., Mackay C.E., Watkins K.E., Ciccarelli O., Cader M.Z., Matthews P.M., Behrens T.E.J. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Sulzer D., Cassidy C., Horga G., Kang U.J., Fahn S., Casella L., Pezzoli G., Langley J., Hu X.P., Zucca F.A., Isaias I.U., Zecca L. Neuromelanin detection by magnetic resonance imaging (MRI) and its promise as a biomarker for Parkinson’s disease. npj Park. Dis. 2018;4:11. doi: 10.1038/s41531-018-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessitore A., Esposito F., Vitale C., Santangelo G., Amboni M., Russo A., Corbo D., Cirillo G., Barone P., Tedeschi G. Default-mode network connectivity in cognitively unimpaired patients with Parkinson disease. Neurology. 2012;79:2226–2232. doi: 10.1212/WNL.0b013e31827689d6. [DOI] [PubMed] [Google Scholar]
- Tessitore A., Giordano A., De Micco R., Russo A., Tedeschi G. Sensorimotor connectivity in Parkinson’s disease: the role of functional neuroimaging. Front. Neurol. 2014;5:180. doi: 10.3389/fneur.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vaillancourt D.E., Spraker M.B., Prodoehl J., Abraham I., Corcos D.M., Zhou X.J., Comella C.L., Little D.M. High-resolution diffusion tensor imaging in the substantia nigra of de novo Parkinson disease. Neurology. 2009;72:1378–1384. doi: 10.1212/01.wnl.0000340982.01727.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh R.R. Functional imaging markers as outcome measures in clinical trials for Parkinson’s disease. In: Espay A.J., Fernandez H.H., Fox S.H., Galvez-Jimenez N., editors. Parkinson’s Disease: Current & Future Therapeutics & Clinical Trials. Cambridge University Press; 2016. [Google Scholar]
- Watson G.S., Leverenz J.B. Profile of cognitive impairment in Parkinson disease. Brain Pathol. 2010;20:640–645. doi: 10.1111/j.1750-3639.2010.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen M.-C., Heng H.S.E., Ng S.Y.E., Tan L.C.S., Chan L.L., Tan E.K. White matter microstructural characteristics in newly diagnosed Parkinson’s disease: an unbiased whole-brain study. Sci. Rep. 2016;6:35601. doi: 10.1038/srep35601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A.M., Ridgway G.R., Webster M.A., Smith S.M., Nichols T.E. Permutation inference for the general linear model. Neuroimage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnall A.J., Rochester L., Burn D.J. Mild cognitive impairment in Parkinson’s disease. Age Ageing. 2013;42:567–576. doi: 10.1093/ageing/aft085. [DOI] [PubMed] [Google Scholar]
- Zeighami Y., Ulla M., Iturria-Medina Y., Dadar M., Zhang Y., Larcher K.M.-H., Fonov V., Evans A.C., Collins D.L., Dagher A. Network structure of brain atrophy in de novo Parkinson’s disease. Elife. 2015;4 doi: 10.7554/eLife.08440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Yushkevich P.A., Alexander D.C., Gee J.C. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Med. Image Anal. 2006;10:764–785. doi: 10.1016/j.media.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang K., Yu C., Zhang Y., Wu X., Zhu C., Chan P., Li K. Voxel-based analysis of diffusion tensor indices in the brain in patients with Parkinson’s disease. Eur. J. Radiol. 2011;77:269–273. doi: 10.1016/j.ejrad.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Zhuang X., Walsh R.R., Sreenivasan K.R., Yang Z., Mishra V.R., Cordes D. Exploring the dynamics of resting-state networks in Parkinson’s Disease using Co-Activation Pattern. Neuroimage. 2018 doi: 10.1016/j.neuroimage.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]