Abstract
Reunion Island is currently experiencing an epidemic caused by Dengue virus type-2 (DENV-2) resulting in over 6,763 cases from austral summer 2017 to winter 2018. Phylogenetic analyses on two non-imported cases of dengue infection from Reunion Island highlight a regional circulation of DENV-2 Cosmopolitan lineage 1 virus on both Reunion Island and the Seychelles.
Keyword: Virology
1. Introduction
Reunion Island is a French overseas department located in the Southwestern Indian Ocean (SWIO) with a population of about 865 000 inhabitants [1]. Since the beginning of 2018, Reunion Island is under the threat of a Dengue type-2 outbreak with more than 6,763 cases diagnosed at the end of 2018 [2]. Although health authorities monitor the surveillance of this epidemic, the genomic data remain limited. In this report, we investigated the molecular characteristics of circulating DENV-2 lineage and the likely relationships that may exist in terms of regional circulation in the neighboring islands of the SWIO, notably with the Seychelles [3].
2. Background
Two autochthonous cases of Dengue virus type-2 infection on Reunion Island. In March–May 2018, two men in their 40–50s living in Saint-Gilles (south) and Saint-Denis (north) cities from Reunion Island showed clinical symptoms compatible with Dengue virus infection. They experienced fever, myalgia, headache, asthenia with moderate Thrombocytopenia, and both have fully recovered one week later. No complications were recorded. Serum samples were collected two or three days after the onset of fever and laboratory tests conducted on early blood samples indicated clear positive RT-qPCR for DENV-2. Since these two patients had not recently travelled to areas endemic for dengue disease, their illnesses were classified as autochthonous cases of DENV-2 infection from Reunion Island.
3. Materials & methods
Total nucleic acids were extracted from the serum samples of two autochthonous cases of dengue fever using the Total RNA Extraction kit (QIAGEN®, Courtabœuf, France), according to the manufacturer's recommendations. The synthesis of cDNA from RNA extracted was carried out using 200 U of ProtoScript II (New England BioLabs®inc., Ips., USA) with random primers according to the manufacturer's recommendations. The resulting cDNA was submitted to a nested-PCR amplification using GoTaq® G2 Hot Start Polymerase (Promega, Mad., USA). To that purpose, we designed degenerated primers targeting a fragment (∼935 bp without primers) of the E-gene encoding the dengue E protein. The first round amplification reaction was performed with primers DENV2-F1 (5'-GTGGTAACACCTCATTCAGGG-3') and DENV2-R1 (5'-CCCACTGCCACATTTCARTTC-3') generating an approximately 1100-bp fragment, whereas the second round used the primers DENV2-F2 (5'-GACACAGGAAAACATGGCAAGG-3') and DENV2-R2 (5'-CTCACAACRCAACCACTATC-3') targeting an approximately 970-bp fragment. The 25-μl reaction mixture contained 2 μl cDNA sample for the first round, and 1 μl of the resulted PCR mixture was used for the second round with 0.1 μM of each primer and 5 U GoTaq polymerase, according to the manufacturer's recommendations. PCR amplification was performed on a PCR System 2700 Thermocycler (ABI Applied BiosystemsTM) as follows: 94 °C for 15 s; 3 cycles of 95 °C for 5 s, 62 °C for 30 s and 72 °C for 30 s; 3 cycles of 95 °C for 5 s, 55 °C for 30 s, 72 °C for 30 s; and 28 cycles of 95 °C for 5 s, 49 °C for 30s, 72 °C for 1 min. Amplified products were visualized in 2% agarose gels stained with GelRed (Biotium Inc., USA) and fragments of expected sizes were excised from the gel, and the both strands were Sanger sequenced (Genoscreen, Lille, France). The quality of sequences was assessed using the Geneious® Pro software package [4], then trimmed and aligned with a set of 110 corresponding E-gene encoding dengue E protein sequences retrieved from GenBank, all being representative of the different sub-genotypes reported for the DENV-2 [5]. The accession numbers for the two sequences from Reunion Island generated in this study are MK570306 and MK570307. Four sequences originated from sera of Israeli travelers who were infected on April 2017 in the Seychelles where a significant dengue outbreak had been occurring since the end of 2015 [6] were also included. These four sequences were kindly provided by Dr. Yaniv Lustig from the Central Virology laboratory in Israel. A GTR + Gamma + I nucleotide substitution model was determined to best fit the data in ModelTest [7] and was used for the phylogenetic analyses. To perform the sub-genotyping, the Bayesian inference computation was used by including two DENV-1 E-gene sequences as outgroup, and employing the MrBayes v3.2.6 [8], with two parallel runs of four Metropolis-coupled chains were initiated for 6M Markov chain Monte Carlo (MCMC) generations, sampling every 1000 generations and discarding the first 25% as burn-in before computing a consensus tree, visualized by FigTree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
4. Results
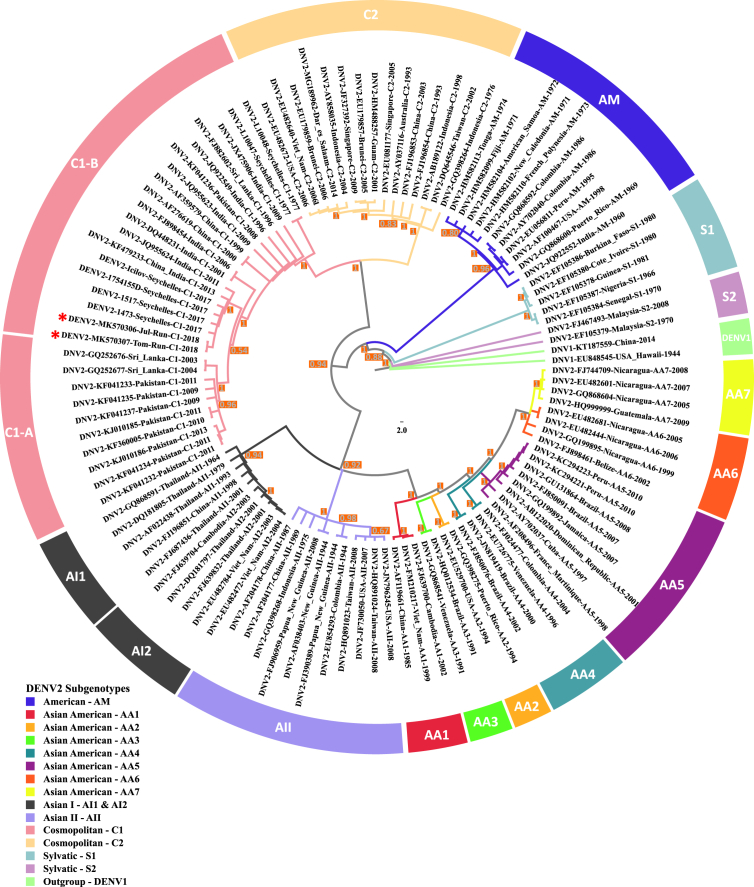
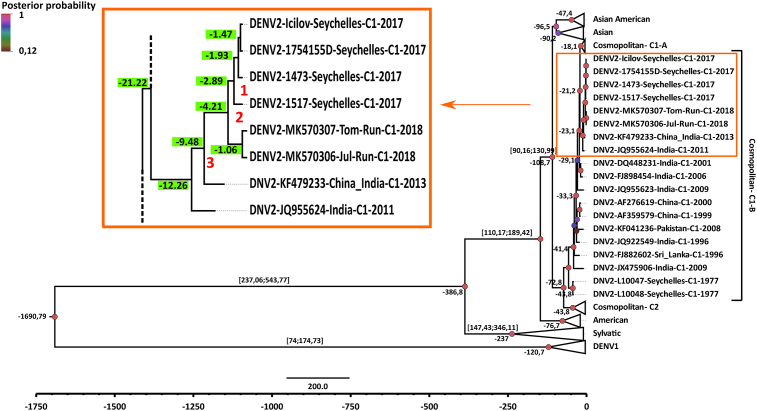
Similar DENV-2 sub-genotype from the Cosmopolitan 1 lineage co-circulates on Reunion Island and the Seychelles. The two sequences from Reunion patients were very closely related to those from the four Israeli travelers, sharing the highest similarity both at level of nucleotides (99.67–99.56%) (Table 1) and amino acids (99.67–99.68%) [9]. Moreover, the Reunionese sequences clustered together within the Cosmopolitan genotype of DENV-2, in accordance with what has been previously reported for strains circulating in the Seychelles [6]. In addition, we showed that the two sequences from Reunion Island fit to lineage I of the Cosmopolitan genotype and more precisely to a C1-B sub-lineage [10] containing sequences from China and India (Fig. 1). The relative geographic proximity between Reunion Island and the Seychelles, as well as the numerous exchanges between their respective populations, allow us to consider that the dengue epidemics currently occurring in the two countries are probably linked, therefore suggesting a co-circulation at a regional level of the cosmopolitan DENV-2 lineage I (Fig. 1). To explore the evolutionary rates and time to the most recent common ancestors (tMRCA), we reconstructed phylogenetic history with the same sequences set by Bayesian MCMC analysis implemented in the Beast package v1.8.3 [11], using an uncorrelated relaxed clock, frequencies empirically based on tips collection dates, and a Bayesian skyline population coalescent model (10 piece-wise constant groups). The analysis was run for an MCMC chain length of 200 million iterations and the maximum clade credibility (MCC) tree was annotated with TreeAnnotator v1.8.3, then visualized with FigTree v1.4.2. The uncertainty in parameter estimates was expressed as 95% highest probability density (HPD). An ESS value > 250 was chosen as a convergence on the posterior distribution. Although taking into account the time inference, the MCC tree topology (Fig. 2) remains globally similar to that of Fig. 1.
Table 1.
Estimates of evolutionary divergence between sequences.
| Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DENV2_IS1473_Seychelles_C1_2017 | 0,0000 | 0,0000 | 0,0000 | 0,0018 | 0,0023 | 0,0029 | 0,0031 | 0,0062 | 0,0062 | 0,0079 | |
| 2 | DENV2_IS1517_Seychelles_C1_2017 | 0,0000 | 0,0000 | 0,0000 | 0,0018 | 0,0023 | 0,0029 | 0,0031 | 0,0062 | 0,0062 | 0,0079 | |
| 3 | DENV2_IS1754155D_Seychelles_C1_2017 | 0,0000 | 0,0000 | 0,0000 | 0,0018 | 0,0023 | 0,0029 | 0,0031 | 0,0062 | 0,0062 | 0,0079 | |
| 4 | DENV2_ISIcilov_Seychelles_C1_2017 | 0,0000 | 0,0000 | 0,0000 | 0,0018 | 0,0023 | 0,0029 | 0,0031 | 0,0062 | 0,0062 | 0,0079 | |
| 5 | DENV2_MK570306_Jul_Run_C1_2018 | 0,0033 | 0,0033 | 0,0033 | 0,0033 | 0,0019 | 0,0033 | 0,0035 | 0,0062 | 0,0062 | 0,0080 | |
| 6 | DENV2_MK570307_Tom_Run_C1_2018 | 0,0044 | 0,0044 | 0,0044 | 0,0044 | 0,0033 | 0,0037 | 0,0037 | 0,0061 | 0,0061 | 0,0079 | |
| 7 | DNV2_JQ955624_India_C1_2011 | 0,0089 | 0,0089 | 0,0089 | 0,0089 | 0,0122 | 0,0133 | 0,0032 | 0,0062 | 0,0063 | 0,0077 | |
| 8 | DNV2_KF479233_China_India_C1_2013 | 0,0089 | 0,0089 | 0,0089 | 0,0089 | 0,0122 | 0,0133 | 0,0111 | 0,0062 | 0,0063 | 0,0079 | |
| 9 | DNV2_L10048_Seychelles_C1_1977 | 0,0388 | 0,0388 | 0,0388 | 0,0388 | 0,0421 | 0,0410 | 0,0399 | 0,0388 | 0,0019 | 0,0070 | |
| 10 | DNV2_L10047_Seychelles_C1_1977 | 0,0410 | 0,0410 | 0,0410 | 0,0410 | 0,0443 | 0,0432 | 0,0421 | 0,0410 | 0,0033 | 0,0071 | |
| 11 | DNV2_MG189962_Dar_es_Salaam_C2_2014 | 0,0642 | 0,0642 | 0,0642 | 0,0642 | 0,0676 | 0,0664 | 0,0642 | 0,0664 | 0,0554 | 0,0576 | |
The number of base differences per site between sequences are shown. Standard error estimate(s) are shown above the diagonal. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.05). The analysis involved 11 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd. All positions containing gaps and missing data were eliminated. There were a total of 903 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [9].
Fig. 1.
Phylogenetic analysis of the envelope E-gene sequence of DENV-2 from Reunion Island, March–May 2018. Bayesian inference computation was performed with a set of 110 corresponding sequences retrieved from GenBank, all being representative of the different sub-genotypes reported for the DENV-2. Two DENV-1 E-gene sequences were included as outgroup. A red star indicates the topological location in the tree for the sequences from Reunion Island. Posterior probabilities are depicted in an orange square at the node level. The branches of the tree are colored according their genotyping origin.
Fig. 2.
Phylodynamic history of the DENV-2 sequences to estimate the introduction dates for the regional circulation – Maximum clade credibility tree of Dengue-2 E-gene sequences. The phylogenetic relationships and temporal evolutionary history have been estimated based on molecular clock analysis. Branch lengths are temporally scaled, and the x-axis presents a time scale (years before present - y.b.p). In the left panel zoom of the main tree, date estimates at the node level are depicted in a green square given in year by past (y.b.p.). The red squares numbered 1, 2 and 3 represent, respectively, the tMRCA of the putative introduction of DENV-2 on the Seychelles, on the Seychelles-Reunion Island, and at the regional level.
5. Discussion
The islands from the SWIO have witnessed several dengue epidemics over the past years. However, data are scattered and when available they mainly referred to post- or inter-epidemic sero-epidemiological studies [12, 13, 14], but seem to express a complex or contrasted epidemiological situation in this multiple insular area. The last reported outbreak that largely affected the SWIO region goes back to 1976–1977 on the Seychelles and subsequently to 1977–1978 on Reunion Island, involving DENV-2 [12, 13]. This outbreak had also very likely affected the Comoros at the same period [15, 16]. A dengue epidemic due to DENV-1 also hit the Comoros and Mayotte in 1993 [15, 17], and later on Madagascar in 2006 [14]. Since 2005, there have been sporadic cases in Reunion Island, mainly imported from neighboring islands including Madagascar, Mayotte (DENV-2, DENV-3) or occasionally from the Caribbean islands (DENV-1) and Thailand (DENV-1, DENV-3). In 2006, the health authorities alerted the inhabitants of Reunion Island about a local and autochthonous circulation of DENV-1, and to a lesser extent, of DENV-2 and DENV-3 [18]. A co-circulation at low-level transmission of DENV-1 and DENV-2 was also observed in 2017. Usually, such low circulation ends when the austral winter arrives, coinciding with the decrease of the vector populations of mosquitoes. Surprisingly, dengue circulation continued throughout this season. On March 2018, Reunion Island was officially declared under dengue epidemic crisis by the health authorities. Positive biologic diagnosis of DENV-2 infection was made by specific RT-qPCR at the National Reference Centre of Arboviruses of Reunion Island. Up to 5,700 cases were reported from November 2017 to July 2018 [19]. Besides, since 2015 the Seychelles have also experienced dengue epidemics mostly due to DENV-1 and DENV-2 [20]. By analyzing the two samples available from consenting adult volunteers, we provided evidence that the DENV-2 circulating on Reunion Island belonged to Cosmopolitan lineage. Access to more samples is required for further epidemiological study. Nevertheless, we undoubtedly established that both the Reunion Island and the Seychelles ongoing epidemics of DENV-2 involved strains that clustered together, and originated probably from India or China. Despite the low number of sequences available for the SWIO, the putative introduction dates of DENV-2 are respectively for the Seychelles and Reunion Island, end of 2015 [tMRCA 2.89 y.b.p., 95% HPD = 1-1 y.b.p.] and end of 2016 [tMRCA 1.06 y.b.p., 95% HPD = 0.03–2.5 y.b.p.], consistently with the recording of the first reported cases for these two geographic locations. Yet, the beginning of this cross-circulation between the SWIO islands was estimated around the end of 2013 [tMRCA = 4.21 y.b.p., 95% HPD = 2.09–6.62 y.b.p.], showing that the DENV-2 probably circulated silently without being detected almost two years before (Fig. 2, red square numbered 2 in the panel). Interestingly, the SWIO lineage including the Indian origin strain has a common ancestor whose tMRCA estimate indicated a circulation date for the whole SWIO area in 2008 (Fig. 2, red square numbered 3 in the panel), that is to say more than 9 years ago. This suggests an endemic circulation at low-level transmission in SWIO neighboring islands. This observation also confirms and reinforces previous sero-epidemiological studies on Mayotte and on the Comoros, which similarly showed that there must have been DENV fluxes between some islands, which occurred quite unnoticed by the relevant authorities [15, 16].
6. Conclusion
This work highlights the circulation of a DENV-2 Cosmopolitan sub-lineage 1 during the ongoing dengue epidemics on Reunion Island. Moreover, we provide evidence that the epidemic strains clustered with sequences detected in the Seychelles, suggesting a co-circulation in the SWIO consecutive to a former introduction from Asia. Future research should include more samples and whole genome sequencing to better characterize the circulating strains on wider genomic scale.
Declarations
Author contribution statement
Hervé Pascalis: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Jonathan Turpin, Gilles Gadea, Célestine Atyame Nten: Analyzed and interpreted the data; Wrote the paper.
Marjolaine Roche, Pascale Krejbich, Philippe Després: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Patrick Mavingui: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the PIMIT funds provided by the French institutions INSERM, CNRS and IRD, and the European Regional Development Funds ERDF PO INTERREG, VECTOBIOMES programme, number RE0009962.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GenBank under the accession numbers MK570306 and MK570307.
References
- 1.PopulationData.net . 2019. Reunion Island, Country and Population Data.https://en.populationdata.net/countries/reunion/ [Google Scholar]
- 2.Santé_Publique_France . Le point Epidémio. CIRE Océan Indien. French National Public Health Agency; 2019. Situation épidémique de la dengue à La Réunion au 8 janvier 2018, N°1.http://invs.santepubliquefrance.fr/fr/content/download/152755/557301/version/106/file/pe_dengue_reunion_080119.pdf [Google Scholar]
- 3.Tortosa P., Pascalis H., Guernier V., Cardinale E., Le Corre M., Goodman S.M., Dellagi K. Deciphering arboviral emergence within insular ecosystems. Infect. Genet. Evol. 2012;12:1333–1339. doi: 10.1016/j.meegid.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waman V.P., Kolekar P., Ramtirthkar M.R., Kale M.M., Kulkarni-Kale U. Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes. PeerJ. 2016;4:e2326. doi: 10.7717/peerj.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lustig Y., Wolf D., Halutz O., Schwartz E. An outbreak of dengue virus (DENV) type 2 cosmopolitan genotype in Israeli travellers returning from the Seychelles, April 2017. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.26.30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali A., Ali I. The complete genome phylogeny of geographically distinct dengue virus serotype 2 isolates (1944–2013) supports further groupings within the cosmopolitan genotype. PLoS One. 2015;10:e0138900. doi: 10.1371/journal.pone.0138900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metselaar D., Grainger C.R., Oei K.G., Reynolds D.G., Pudney M., Leake C.J., Tukei P.M., D'Offay R.M., Simpson D.I. An outbreak of type 2 dengue fever in the Seychelles, probably transmitted by Aedes albopictus (Skuse) Bull. World Health Organ. 1980;58:937–943. PMCID: PMC2395999. [PMC free article] [PubMed] [Google Scholar]
- 13.Zeller H.G. Dengue, arbovirus and migrations in the Indian Ocean. Bull. Soc. Pathol. Exot. 1998;91:56–60. PMID: 9559164. [PubMed] [Google Scholar]
- 14.Ratsitorahina M., Harisoa J., Ratovonjato J., Biacabe S., Reynes J.M., Zeller H., Raoelina Y., Talarmin A., Richard V., Louis Soares J. Outbreak of dengue and Chikungunya fevers, Toamasina, Madagascar, 2006. Emerg. Infect. Dis. 2008;14:1135–1137. doi: 10.3201/eid1407.071521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sissoko D., Ezzedine K., Giry C., Moendandze A., Lernout T., D'Ortenzio E., Pettinelli F., Malvy D. Seroepidemiology of dengue virus in Mayotte, Indian ocean, 2006. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellagi K., Salez N., Maquart M., Larrieu S., Yssouf A., Silai R., Leparc-Goffart I., Tortosa P., de Lamballerie X. Serological evidence of contrasted exposure to arboviral infections between islands of the union of Comoros (Indian Ocean) PLoS Negl. Trop. Dis. 2016;10:e0004840. doi: 10.1371/journal.pntd.0004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisier P., Morvan J.M., Laventure S., Charrier N., Martin E., Ouledi A., Roux J. Dengue 1 epidemic in the Grand Comoro Island (Federal Islamic Republic of the Comores) March-May 1993. Ann. Soc. Belg. Med. Trop. 1994;74:217–229. PMID: 7840689. [PubMed] [Google Scholar]
- 18.D'Ortenzio E., Balleydier E., Baville M., Filleul L., Renault P. Dengue fever in the Reunion Island and in South Western islands of the Indian Ocean. Med. Mal. Infect. 2011;41:475–479. doi: 10.1016/j.medmal.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 19.Santé_Publique_France . Le Point Epidémio. CIRE Océan Indien. French National Public Health Agency; 2018. Situation épidémiologique de la Dengue à La Réunion au 17 juillet 2018, N°56.http://invs.santepubliquefrance.fr/content/download/148411/540180/version/89/file/pe_dengue_reunion_030718.pdf [Google Scholar]
- 20.Sandin A. 8 June 2016. Emergency Plan of Action (EPoA) Seychelles: Dengue Outbreak, MDRSC004.https://reliefweb.int/sites/reliefweb.int/files/resources/MDRSC004fnr.pdf [Google Scholar]