Abstract
Introduction
Campylobacter jejuni is one of the most frequently reported causes of foodborne bacterial enteric disease worldwide. The main source of these microorganisms is contaminated food, especially of poultry origin. There are several molecular methods for differentiation of Campylobacter isolates at the subgenus level, and one of these is porA-typing based on the sequencing of the major outer-membrane protein (MOMP) encoding gene. The aim of the study was to test the molecular relationship of C. jejuni strains isolated at different points along the poultry food chain and assess the population structure of the isolates.
Material and Methods
A total of 451 C. jejuni were used in the study, and a DNA fragment of 630 bp of the MOMP encoding gene was amplified and sequenced.
Results
One hundred and ten sequence types were identified, with 69 (62.7%) unique to the isolates' origin and 30 not present in the database. The most prevalent nucleotide variant 1 was detected in 37 (8.2%) strains. These isolates were identified in all poultry sources tested, especially in faeces (15 isolates) but also in poultry carcasses and meat (11 isolates in each).
Conclusion
The porA typing method was highly discriminative for C. jejuni of poultry origin since the Simpson's diversity index (D) achieved a value of 0.876, indicating considerable diversity in the bacterial population tested. The method may be further used for epidemiological investigation purposes.
Keywords: poultry, Campylobacter jejuni, porA typing, genetic variation
Introduction
Campylobacter jejuni is one of the most frequently reported causes of foodborne bacterial enteric disease worldwide (12, 17, 18). According to a recent European zoonotic report, a total of 246,307 laboratory-confirmed Campylobacter infections were found in the European Union in 2016, most of them caused by C. jejuni (7). In that year in Poland, only 773 people suffering from this disease were noted, but such a low number of infections may partly be due to underreporting (7). Campylobacters are a part of the natural microflora of animals and birds, and the main transmission route of these microorganisms to humans is ingestion of contaminated food, especially of poultry origin (9, 17, 18, 22). It has been proven that poultry carcasses are most often contaminated during the slaughter process (9, 12, 17).
There are several molecular methods for differentiation of Campylobacter isolates at the subgenus level (13, 14–16). Among them, there is porA-typing, which is based on the sequencing of the major outer-membrane protein-(MOMP) encoding gene (6). It has been detected that the Campylobacter porA gene consists of seven highly variable regions interspersed among conserved sequences and shows potential for the differentiation of isolates from different sources (1, 6). It has also been shown that the porA-typing method is complementary to multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and whole genome sequencing (11).
In the present investigation, typing of C. jejuni isolated from the poultry food chain, i.e. faeces, carcasses, and meat was performed using porA sequencing. The aim of this study was to test the molecular relationship between the isolates and to subtype them to assess the population structure of poultry C. jejuni recovered in Poland.
Material and Methods
Bacterial isolates. A total of 451 C. jejuni isolates were used in the study. The isolates recovered from chicken caeca (n = 151) were obtained using a procedure described previously and then confirmed by PCR (20). The swab samples from broiler carcasses (n = 150) were collected directly after immersion chilling (0 to 4°C) but before further processing. They came from the neck skin and the skin surface under the wings. C. jejuni were isolated from the swab samples and PCR-confirmed as described (19, 20). The Campylobacter isolates from chicken meat (n = 150) were recovered using the ISO 10272-1 standard, and C. jejuni isolates were identified by PCR according to the procedure described for the broiler carcasses (20). The isolates of broiler carcasses were obtained from all 16 Polish geographical regions (provinces), whereas C. jejuni from poultry meat were isolated from samples randomly purchased in the Lublin Province. Detailed information referring to the isolation of all C. jejuni used in the study has been presented previously (21).
Sequencing of the porA gene. A DNA fragment of 630 bp of the MOMP encoding gene used for porA typing was amplified and sequenced with primers MOMP-1 (5′-GATGGTTTAACTCTAGCTGC-3′) or MOMP-3 (5′-GATGGTTTAGTWGGMACAGG-3′) and MOMP-2 (5′-TGAGAAGTTAAGTTTTGGA GAG-3′) as described by Dingle et al. (6), and the reaction conditions were set as follows: initial denaturation at 95°C for 15 min, 35 cycles at 94°C for 30 s, at 50°C for 30 s, and at 72°C for 90 s, and a final extension at 72°C for 5 min.
The PCR products were purified and commercially sequenced by Genomed (Poland) using the BigDye Terminator v. 3.1 kit (Applied Biosystems, USA). The sequencing products were separated in a 3730xl DNA Analyser capillary sequencer (Applied Biosystems), and the DNA sequences were then imported and checked for quality using BioNumerics v. 7.6 software (Applied Maths, Sint-Martens-Latem, Belgium). After that, for designation of sequence numbers, the pubMLST database (http://pubmlst.org/campylobacter) was used. New sequences were submitted to this database for confirmation, and the sequence numbers were received.
Statistical analysis. The discriminatory power of the porA sequencing method was assessed using Simpson’s diversity index (D) with 95% confidence intervals (CI), applying a tool available at www.comparingpartitions.info. An index greater than 0.90 was considered desirable if the typing results were to be interpreted with confidence (10).
Results
C. jejuni porA typing. A total of 110 sequence types were identified among 451 C. jejuni isolates tested, with the most prevalent nucleotide being variant 1, which was detected among 37 (8.2%) isolates (Table 1). This sequence allele type was identified in isolates recovered from all groups of samples. Among 151 isolates of faecal origin, 60 different porA types were detected (Table 1). The most numerous sequence variants were 1 (15 isolates; 9.9%), 296, and 2250 (9 isolates of each; 6.0% each). The remaining 118 isolates were classified into 57 sequence types, occurring in 1 to 8 isolates. Twenty-one nucleotide sequences unique to this source were identified; each of them had one isolate. Additionally, 12 new porA variants with 14 isolates were detected, i.e. 741, 1529 (two isolates), 1827, 2249 (two isolates), 2306, 2307, 2308, 2309, 2310, 2311, 2312, and 2313.
Table 1.
Prevalence of porA sequence types in C. jejuni tested
| Source of isolates | porA sequence types and number of isolates: | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 522 | 6 | 60 | 296 | 7 | 92 | 1,546 | 97 | 164 | 2,106 | 2,250 | Other (number. of different porA sequences)* | ||
| Poultry | faeces | 15 | 8 | 3 | 7 | 9 | 4 | 7 | 8 | 5 | 6 | 0 | 9 | 68 (48) |
| carcasses | 11 | 9 | 12 | 10 | 9 | 7 | 4 | 4 | 6 | 4 | 0 | 0 | 67 (45) | |
| meat | 11 | 12 | 10 | 5 | 3 | 5 | 5 | 3 | 1 | 2 | 12 | 2 | 78 (38) | |
| Total | 37 | 29 | 25 | 22 | 21 | 16 | 16 | 15 | 12 | 12 | 12 | 11 | 213 (98) | |
“Other” includes a total of 97 different porA sequence types, each revealed in one to ten isolates
Isolates recovered from poultry carcasses were classified into 57 porA types, and among them those belonging to variants 6 (12 isolates; 8.0%), 1 (11 isolates; 7.3%), and 60 (10 isolates; 6.7%) predominated. A total of 18 sequences unique to this source were identified from 19 isolates. Eight novel sequence variants, with one isolate in each, were also described (2261, 2301, 2312, 2315, 2316, 2317, 2318, and 2319).
Among C. jejuni isolated from poultry meat, 50 different molecular variants were detected, with the most common sequence types 522 and 2106 each occurring in 12 (8.0%) isolates. Fifteen porA types unique to this source were found, yielded by 20 isolates. Furthermore, 10 new variants were identified, with 1 isolate in each, i.e. 2304, 2305, 2320, 2321, 2322, 2323, 2324, 2325, 2326, and 2327.
All new porA sequence types were submitted to the Campylobacter public database (http://pubmlst.org/campylobacter).
The distribution by isolate source of the most numerous porA sequence types identified among all 451 C. jejuni isolates tested (over 3% of the total number of isolates) is shown in Fig. 1. Isolates with the most common molecular variant, type 1, (37 isolates) were detected in all poultry sources tested, especially in chicken faeces (15 isolates) but also in poultry carcasses and meat (11 isolates in each). A similar relation was observed among isolates with the 522 type (n = 29) where they were identified along the whole poultry food chain. Isolates with the third most numerous porA sequence, variant 6, (n = 25) were mainly extracted from poultry carcasses and meat (12 and 10 isolates, respectively) and only rarely from chicken faeces (only 3 isolates). C. jejuni isolates with the remaining sequence variants are shown in Fig. 1.
Fig. 1.
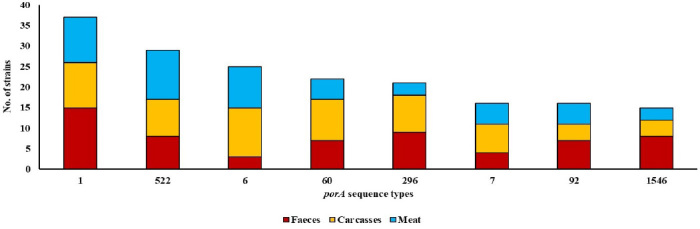
Number of C. jejuni isolates detected by porA sequence type. Only data for the eight most common sequence types detected in more than 3% of the total number of isolates are shown
The prevalence of the most common porA sequence types among C. jejuni isolated from different parts of the poultry food chain is shown in Fig. 2. Among C. jejuni from poultry faeces, isolates with the 1, 296, and 2250 variants were predominant, representing a total of 33 isolates (Fig. 2A). The type 1 sequence was also often identified in isolates from carcasses and meat (22 isolates), although from these sources C. jejuni with the porA sequences 6 and 522 were the most common, respectively, representing 12 isolates of each (Figs 2B and 2C). Isolates with the sequence type 2106 were commonly detected among strains of poultry meat (12 isolates), but they were not identified in faeces or carcasses. Furthermore, one of the most common faeces-isolated variants, 2250 (9 isolates), was rarely identified (2 isolates) or not identified at all among C. jejuni recovered from poultry meat and carcasses, respectively.
Fig. 2.
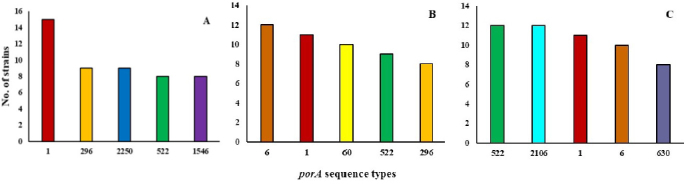
Distribution of the most common porA sequence types among C. jejuni isolated from different poultry food chain sources. Y axis – number of isolates; X axis – porA sequence numbers. A – faeces; B – carcasses; C – meat. The graphs show the sequence types representing more than 5% of the isolates in each group
Population structure of C. jejuni isolates. Overall, a total of 110 porA sequence types were identified, including 69 (62.7%) unique to one origin site of isolates (Table 2). Among them, 30 sequences detected in 32 isolates were not present in the database. The most common porA sequence types found in all 451 poultry C. jejuni isolates tested were 1 and 522, which included 37 (8.2%) and 29 (6.4%) isolates, respectively. Sequence type 1 was the most prevalent among isolates from poultry faeces (15 isolates; 9.9%), whereas the isolates with sequence variant 522 predominated in C. jejuni of meat origin (12 isolates; 8.0%). The remaining numbers of isolates belonging to particular molecular variants are shown in Table 1. Several porA sequence types unique to one source of C. jejuni were identified, i.e. 27 sequences in the isolates from faeces and 21 in isolates from carcasses and meat, respectively (Table 2). On the other hand, a total of 41 different sequences were present among isolates recovered from more than one source along the poultry food chain.
Table 2.
Distribution of C. jejuni porA molecular subtypes according to the source of isolation
| C. jejuni poultry origin (number of isolates) | Total (n = 451) | |||
|---|---|---|---|---|
| Faeces (n = 151) | Carcasses (n = 150) | Meat (n = 150) | ||
| Number sequences of porA | 60 | 57 | 50 | 110 |
| Most prevalent sequence | 1 (15) | 6 (12) | 522 (12); 2106 (12) | 1 (37) |
| Number of new sequences | 12 (14) | 8 (8) | 10 (10) | 30 (32) |
| Number unique to of this sequences origin | 27 (32) | 21 (25) | 21 (48) | 69 (105) |
| Percentage unique to the of sequences origin | 45.0 | 36.8 | 42.0 | 62.7 |
| Number common of to sequences other origins | 33 (119) | 36 (125) | 29 (102) | 41 (346) |
Discriminatory power of porA typing. Overall, the porA typing method was highly discriminative for all C. jejuni used in the study since the Simpson’s diversity index (D) achieved the value 0.876, indicating a considerable diversity in the whole bacterial population tested (Table 3). Taking into account the number of the isolates and sequences, the method was equally discriminative for C. jejuni isolated from all poultry food chain samples (D values by source were 0.967 for meat, 0.970 for carcass, and 0.979 for faecal isolates, i.e. the bacteria from each source showed a similar genetic diversity).
Table 3.
Molecular diversity of C. jejuni isolates tested based on Simpson's diversity index (D)
| Number of: | Origin of isolates: | |||
|---|---|---|---|---|
| Poultry: | Total | |||
| faeces | carcasses | meat | ||
| Isolates | 151 | 150 | 150 | 451 |
| porA sequence types | 60 | 57 | 50 | 110 |
| Diversity index D | 0.970 | 0.979 | 0.967 | 0.876 |
| (95% CI)* | (0.960–0.980) | (0.971–0.988) | (0.958–0.975) | (0.850–0.903) |
CI, confidence intervals with 95% confidence level
Discussion
The results of the porA typing performed in the present study revealed that C. jejuni isolated at three points along the poultry food chain were characterised by high molecular diversity. A total of 110 different sequence types among 451 isolates were identified. Several of them were common to the isolates recovered from all three sources (e.g. sequence types 1, 522, 6, and 60), whereas the others were specific to the isolates of particular origins, e.g. sequence types 1978 from faeces, 61 from carcasses, and 2106 identified only in poultry meat. This finding may suggest that C. jejuni with certain porA sequences may be predominant in the poultry population and in very many instances are able to contaminate chicken carcasses and subsequently poultry meat (15, 16). Furthermore, the frequent identification of several molecular variants of C. jejuni (i.e. with the sequence types 1, 522, 6, 60, and 296) may be due to better adaptation of such genotypes to poultry carcasses than the other types which were only detected occasionally (15). The similarities in the distribution of the same sequence types among C. jejuni isolated from poultry faeces, carcasses, and meat also suggest that the presence of these bacteria in chicken meat may be a result of cross-contamination during the slaughter process.
There are only a few studies related to the molecular characteristics of C. jejuni by the porA typing method. Jay-Russell et al. (11) investigated isolates originating from humans, milk, and the environment, and found great diversity in them, which was then confirmed by MLST. Molecular subtyping of C. coli isolated from diarrhoeal patients and food-producing animals in China revealed that among 113 isolates 52 different porA sequence types were present, mainly 1427 and 687 (11 isolates of each), followed by 1428 (8 isolates) and 915 (7 isolates) (23). None of these sequences were identified in the present study, but a different Campylobacter species was investigated. Another analysis of 583 C. jejuni (n = 540) and C. coli (n = 43) isolates of human origin performed in the United Kingdom showed that they were classified into 63 distinct porA subtypes, with the predominant sequence being type 1, which was also the most common variant in the present investigation (3). These results may suggest that the sequence type 1 is, at least to some degree, persistent in C. jejuni isolates of different origins. It has been previously shown that some genotypes are better adapted than others to certain host environments, including poultry (5). For example, earlier studies demonstrated that the porA sequence types 1 and 41 (not identified in the current analysis) were closely associated with humans and poultry (2, 8, 24).
The results of the current study demonstrated that the typing method based on the sequencing of the C. jejuni porA gene is a reliable molecular tool for rapid differentiation of isolates from the poultry food chain, and in a broader perspective for epidemiological purposes. It was previously shown that the porA-based sequencing approach has sufficient genetic diversity and stability for use as a molecular epidemiological tool (3). The method can also be used for source-tracking genetically related bacterial isolates such as C. jejuni (11). Such molecular subtyping of isolates from different origins provides information about the source attributes and subsequently data for human infection control (4, 13). In the present study, the porA analysis indicated that the majority of the C. jejuni isolates recovered from the poultry food chain shared identical genotypes (similar Simpson's diversity indices), which may suggest that they originated from the same source, i.e. from poultry intestinal contents.
In conclusion, the current investigation highlights the porA sequencing method for molecular differentiation of C. jejuni isolated at different points along the poultry food chain. Using this approach, it was possible to show that isolates of the same species had identical genotypic fingerprints, which may suggest that they originated from the same source. This typing method may be further used for epidemiological investigations.
Acknowledgments
The authors thank Edyta Denis and Katarzyna Półtorak for their technical assistance in the laboratory analyses. Monika Kurpas is also acknowledged with gratitude for calculation of the D values.
Footnotes
Conflict of Interest
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: The study was financially supported by the National Science Centre, Poland, on the basis of Decision UMO-2014/15/B/NZ7/00874.
Animal Rights Statement: None required.
References
- 1.Ahmed M.U., Dunn L., Valcanis M., Hogg G., Ivanova E.P.. Double-locus sequence typing using porA and peb1A for epidemiological studies of Campylobacter jejuni. Foodborne Path Dis. 2014;11:194–199. doi: 10.1089/fpd.2013.1634. [DOI] [PubMed] [Google Scholar]
- 2.Clark C.G., Beeston A., Bryden L., Wang G., Barton C., Cuff W., Gilmour M.W., Ng L.K.. Phylogenetic relationships of Campylobacter jejuni based on porA sequences. Can J Microbiol. 2007;53:27–38. doi: 10.1139/w06-099. [DOI] [PubMed] [Google Scholar]
- 3.Cody A.J., Maiden M.J.C., Dingle K.E.. Genetic diversity and stability of the porA allele as a genetic marker in human Campylobacter infection. Microbiology. 2009;155:4145–4154. doi: 10.1099/mic.0.031047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colles F.M., Maiden M.C.J.. Campylobacter sequence typing databases: applications and future prospects. Microbiol Soc. 2012;158:2695–2709. doi: 10.1099/mic.0.062000-0. [DOI] [PubMed] [Google Scholar]
- 5.Dingle K.E., Colles F.M., Ure R., Wagenaar J., Duim B., Bolton F.J., Fox A.J., Wareing D.R.A., Maiden M.C.J.. Molecular characterisation of Campylobacter jejuni clones: a rational basis for epidemiological investigations. Emerg Infect Dis. 2002;8:949–955. doi: 10.3201/eid0809.02-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle K.E., McCarthy N.D., Cody A.J., Peto T.E.A., Maiden M.C.J.. Extended sequence typing of Campylobacter spp., United Kingdom. Emerg Infect Dis. 2008;14:1620–1622. doi: 10.3201/eid1410.071109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control): The European Union summary report on trends and sources of zoonoses, zoonotic agents, and food-borne outbreaks in 2016. EFSA J 2017. 15:5077. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S., Sahin O., Zhang Q.. Infection-induced antibodies against the major outer membrane protein of Campylobacter jejuni mainly recognize conformational epitopes. FEMS Microbiol Lett. 2007;272:137–143. doi: 10.1111/j.1574-6968.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- 9.Humphrey T., O’Brien S., Madsen M.. Campylobacters as zoonotic pathogens: a food production perspective. Intern J Food Microbiol. 2007;117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Hunter P.R., Gaston M.A.. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jay-Russell M.T., Mandrell M.E., Yuan J., Bates A., Manalac R., Mohle-Boetani M., Kimura A., Lingard J., Miller W.G.. Using major outer membrane protein typing as an epidemiological tool to investigate outbreaks caused by milk-borne Campylobacter jejuni isolates in California. J Clin Microbiol. 2013;51:195–201. doi: 10.1128/JCM.01845-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaakoush N.O., Castaño-Rodriguez N., Mitchell H.M., Man S.M.. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque S., Fournier E., Carrier N., Frost E., Arbeit R.D., Michaud S.. Campylobacteriosis in urban versus rural areas: A case-case study integrated with molecular typing to validate risk factors and to attribute sources of infection. PLoS ONE. 2013;8:e83731. doi: 10.1371/journal.pone.0083731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müllner P., Spencer S.E.F., Wilson D.J., Jones G., Noble A.D., Midwinter A.C., Collins-Emerson J.M., Carter P., Hathaway S., French N.P.. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Gen Evol. 2009;9:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Ravel A., Hurst M., Petrica N., David J., Mutschall S.K., Pintar K., Taboada E.N., Pollari F.. Source attribution of human campylobacteriosis at the point of exposure by combining comparative exposure assessment and subtype comparison based on comparative genomic fingerprinting. PLoS ONE. 2017;12:e0183790. doi: 10.1371/journal.pone.0183790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheppard S.K., Dallas J.F., Strachan N.J.C., MacRae M., McCarthy N.D., Wilson D.J., Gormley F.J., Falush D., Ogden I.D., Maiden M.C.J., Forbes K.J.. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis. 2009;48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skarp C.P.A., Hänninen M.L., Rautelin H.I.K.. Campylobacteriosis: the role of poultry meat. Clin Microbiol Infect. 2016;22:103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Tresse O., Alvarez-Ordóñez A., Connerton I.F.. Editorial: about the foodborne pathogen Campylobacter. Front Microbiol. 2017;8:1908. doi: 10.3389/fmicb.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieczorek K., Denis E., Lynch O., Osek J.. Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in Polish slaughterhouses. Food Microbiol. 2013;34:130–136. doi: 10.1016/j.fm.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Wieczorek K., Kania I., Osek J.. Prevalence and antimicrobial resistance of Campylobacter spp. isolated from poultry carcasses in Poland. J Food Prot. 2013;76:1451–1455. doi: 10.4315/0362-028X.JFP-13-035. [DOI] [PubMed] [Google Scholar]
- 21.Wieczorek K., Wołkowicz T., Osek J.. Antimicrobial resistance and virulence-associated traits of Campylobacter jejuni isolated from poultry food chain and humans with diarrhea. Front Microbiol. 2018;9:1508. doi: 10.3389/fmicb.2018.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wysok B., Wojtacka J., Wiszniewska-Łaszczych A., Szteyn J., Gomółka-Pawlicka M.. Prevalence and antimicrobial resistance of Campylobacter isolates from poultry offals. Med Weter. 2017;73:561–566. [Google Scholar]
- 23.Zhang M., Liu X., Xu X., Gu Y., Tao X., Yang X., Yan G., Zhang J.. Molecular subtyping and antimicrobial susceptibilities of Campylobacter coli isolates from diarrheal patients and food-producing animals in China. Foodborne Path Dis. 2014;11:610–619. doi: 10.1089/fpd.2013.1721. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q., Meitzler J.C., Huang S., Morishita T.. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect Immun. 2000;68:5679–5689. doi: 10.1128/iai.68.10.5679-5689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


